Abstract
One fundamental issue regarding stem cells for regenerative medicine is the maintenance of stem cell stemness. The purpose of the study was to test whether small molecules can enhance stem cell properties of mesenchymal stem cells (MSCs) derived from human dental pulp (hDPSCs), which have potential for multiple clinical applications. We identified the effects of small molecules (Pluripotin (SC1), 6-bromoindirubin-3-oxime and rapamycin) on the maintenance of hDPSC properties in vitro and the mechanisms involved in exerting the effects. Primary cultures of hDPSCs were exposed to optimal concentrations of these small molecules. Treated hDPSCs were analyzed for their proliferation, the expression levels of pluripotent and MSC markers, differentiation capacities, and intracellular signaling activations. We found that small molecule treatments decreased cell proliferation and increased the expression of STRO-1, NANOG, OCT4, and SOX2, while diminishing cell differentiation into odonto/osteogenic, adipogenic, and neurogenic lineages in vitro. These effects involved Ras-GAP-, ERK1/2-, and mTOR-signaling pathways, which may preserve the cell self-renewal capacity, while suppressing differentiation. We conclude that small molecules appear to enhance the immature state of hDPSCs in culture, which may be used as a strategy for adult stem cell maintenance and extend their capacity for regenerative applications.
Introduction
Human dental pulp stem cells (hDPSCs) are the first type of dental mesenchymal stem cells (MSCs) isolated and characterized from dental tissues [1]. Since then, significant progress on the understanding of DPSCs and especially the utilization of DPSCs for regenerative purposes has been made. Our group and others have shown that DPSCs may be used to regenerate pulp and dentin in emptied root canal space in both small and large animal models [2,3]. DPSCs also demonstrate many other clinical potentials, including facilitating cardiac angiogenesis and differentiating into neurogenic cells or inducing neural stem cells to differentiate into neural cells for repairing the nervous system [4–7]. Adult stem cells have shown tremendous promise in regenerative and therapeutic medicine evidenced by an increasing number of clinical trials that have been undertaken [8–15]. Besides their obvious potential for tissue regeneration, subpopulations of MSCs have shown a capacity to regulate immune reactions. One prominent feature is their immunosuppressive function, which has been utilized to treat various immunological disorders, including acute graft-vs-host disease and systemic lupus erythematosus clinically [16,17]. The list of MSC versatility appears to keep expanding. Recently, it was shown that bone marrow-derived MSCs (BMMSCs) are capable of effectively suppressing injury-induced hyperalgesia in a rat model [18].
Despite such compelling properties for adult stem cells, the inevitable drawback of these MSCs, including DPSCs, is their limited tissue source and life span in cultures [19]. Each isolation of these cells from tissues is a major undertaking involving acquiring the tissue, transportation to the laboratory, processing the tissue, isolation of the cells, waiting for the initial phase of cell growth, passaging, and characterizing the cells until ready for clinical use. The higher the amount of cells needed for the medical application, the more the amount of tissue has to be taken from the hosts/donors. For this reason alone, researchers have long sought for other sources of stem cells that are longer lasting in cultures, that is, a greater population doubling, and more potent in lineage differentiation. The alternative is embryonic stem cells (ESCs) or more recently, the induced pluripotent stem cells (iPSCs) that are pluripotent and essentially immortal in cultures [20,21]. Unfortunately, the more potent the stem cells are, the more safety concerns they carry with them. For this reason, so far there has been only one preliminary clinical trial reported using ESCs as a cell source [22].
Seeking a way to maintain the stemness of adult stem cells in cultures has been considered an important approach to increase the utility of adult stem cells. Growth factors such as the basic fibroblast growth factor (bFGF) have been tested and shown to be promising [23–26]. However, recombinant protein factors are always difficult to produce and expensive. Small molecules that can be more easily synthesized have played an important role in affecting cell behaviors for various purposes. It has been very difficult to maintain ESCs, especially human ESCs (hESCs) in their undifferentiated state in cultures. Small molecules have been searched to treat ESCs to prevent them from differentiation. A small molecule 6-bromoindirubin-3-oxime (BIO) that activates the Wnt pathway by inhibiting glycogen synthase kinase-3 (GSK-3) has been shown to maintain hESC self-renewal and their expression of pluripotency-associated genes [27]. BIO also increases proliferation of hMSCs [28]. Another small molecule, pluripotin (SC1), has also been identified and shown to enhance ESC self renewal, while inhibiting differentiation [29,30]. The immunosuppressant drug rapamycin produced by streptomyces hygroscopicus has been tested on a number of adult stem cells for its various biological effects, including promoting stemness and preventing differentiation [31,32]. Therefore, in the present study, we aimed to use small molecules/chemical compounds BIO, pluripotin (SC1), and rapamycin to treat DPSCs and determine their effects on the stem cell properties of DPSCs. We show herein that small molecules are capable of affecting DPSC properties by increasing the expression of genes associated with stemness, decreasing their differentiation tendency, and by acting through multiple cellular signaling pathways, including Ras-GAP, ERK1, and mTOR.
Materials and Methods
Sample collection
Normal human impacted third molars (n=12) were collected from healthy patients (eight donors aged 16–24 years) in the Oral Surgery Clinics at the Boston University. Freshly extracted teeth were stored in a cell culture medium and transported to the laboratory for processing. The sample collection process was approved by the Boston University Medical Institutional Review Board (#H-28882).
Cell culture
Isolation of hDPSCs followed a protocol described previously [1,2]. In brief, pulp tissue was removed from the teeth, minced, and digested in a solution of 3 mg/mL collagenase type I and 4 mg/mL dispase (Sigma-Aldrich) for 30–60 min at 37°C. The digested mixtures were passed through a 70-μm cell strainer (Falcon; BD Labware) to obtain single-cell suspensions. Cells were seeded into six-well plates and cultured in the standard medium consisting of the α-Minimum Essential Medium (α-MEM; GIBCO/Invitrogen), supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products, Inc.), 2 mM L-glutamine, 100 μM L-ascorbic acid-2-phospate, 100 units/mL penicillin-G, 100 μg/mL streptomycin, and 0.25 μg/mL fungizone (Gemini Bio-Products, Inc.) and maintained in 5% CO2 at 37°C. Colony formation units of fibroblastic cells (CFU-F) were normally observed within 1–2 weeks after cell seeding and were passaged from one into three wells when they reached ∼70% confluence. Heterogeneous populations of DPSCs were frozen and stored in liquid nitrogen at passages (p) 0–2. Cells were thawed and expanded for experimentation at p3. Cells isolated from each tooth were grown, maintained, and frozen down separately. Cells used for each independent experiment was from one tooth and different donors.
hESCs H9 (obtained from WiCell Research Institute) were grown and maintained on mouse embryonic fibroblasts (feeder cells) with the hESC medium (80% DMEM/F12, 20% knockout serum replacement, 1× nonessential amino acid, 1 mM L-glutamine, and 0.1 μM β-mercaptoethanol) containing 4 ng/mL of the bFGF.
Small molecular treatment and cell viability study
BIO (6-bromoindirubin-3′-oxime; EMD Biosciences), pluripotin (SC1) (Stemgent), or rapamycin (Sigma) each was dissolved in dimethyl sulfoxide (DMSO) at 100, 300, 600, or 900 nM to test for their cell toxicity compared to the control group with DMSO only. For the cell viability study, DPSCs were seeded in 12-well plates at 2–6×104 cells/well and cultured in the absence or presence of small molecules added to the standard hDPSC medium. Cells were harvested at every 2–3 days and subjected to Trypan Blue cell counting in a hemocytometer under the microscope. Nonviable cells were stained and excluded in the counting. Optimal concentrations of small molecules were determined by choosing the concentration with the highest viable cells.
Flow cytometry
The mouse anti-human antibodies used for flow cytometry analysis were the following: Alexa Fluor-conjugated NANOG, OCT4, SOX2, and STRO-1; phycoerythrin (PE)-conjugated CD34; fluorescein isothiocyanate (FITC)-conjugated CD45, CD73, CD90, and CD105. Nonimmune, isotype-matched and -conjugated mouse IgG1 and IgM (Invitrogen) antibodies were used as negative controls.
For direct staining of cell surface antigens, cell aliquots of DPSCs (2–5×105 cells) were washed, resuspended in a buffer [0.1% FBS in phosphate-buffered saline (PBS)], and incubated with conjugated PE, FITC, or Alexa Fluor antibodies according to the manufacturer's recommendations. For intracellular antigens, single-cell suspensions were first fixed in 4% paraformaldehyde in PBS for 10 min, washed with the buffer, permeabilized in 0.1% TritonX100 for 10 min, washed with the buffer again, and incubated with the antibodies for 1 h at 4°C in the dark, and then washed twice and re-suspended in 0.1% FBS/PBS for analysis on a flow cytometer (FACSCalibur; BD Biosciences) using the CellQuest ProTM software (BD Biosciences). These heterogeneous populations of DPSCs were tested positive for STRO-1, CD73, CD90, and CD105, and negative for CD34 and CD45, typical of mesenchymal cell type. For detecting the protein expression of pluripotency-associated genes NANOG, OCT4, and SOX2, a hESC line H9 was used as a positive control (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd).
Cell proliferation and cell cycle analysis
For cell proliferation studies, hDPSCs (p3) were seeded at densities of 0.3–0.5×105/well of 12-well plates and cultured in the absence or presence of small molecules. Cells were then harvested and counted at different time points. For cell cycle analysis, cells were seeded at a density of 0.5–3×105/10-cm dish and cultured for 5 days. The medium was replaced every 2–3 days in the presence or absence of chemicals. At subconfluence, DPSCs were harvested into single-cell suspensions, collected by centrifugation, and washed twice with PBS. The cells were suspended in 0.5 mL PBS, fixed by adding 4.5 mL ice-cold 70% ethanol dropwise with gentle vortex, and incubated at 4°C overnight. After further washing, cell suspensions were stained with the propidium iodide staining solution (100 μg/mL RNAse A, 40 μg/mL propidium iodide; Sigma) at 37°C for 30 min, filtered, and subjected to a flow cytometry system (BD Biosciences LSRII) for cell cycle analysis. Cell cycle distributions were analyzed in the form of percentage of cells in the G1, S, and G2 phases.
Multiple lineage differentiation
Odonto-/Osteogenic differentiation
Cells were seeded into 12- or 24-well plates, grown to ∼70% confluence, and incubated in a differentiation medium containing 10 nM dexamethasone, 10 mM β-glycerophosphate, 50 μg/mL ascorbate phosphate, 10 nM 1, 25 dihydroxyvitamin D3, and 10% FBS for 4 weeks. This differentiation medium has been used for osteogenic studies for osteoblast differentiation. DPSCs mainly undergo odontogenic differentiation; therefore, we designated the term the odonto-/osteogenic differentiation medium. Cultures were then fixed in 60% isopropanol, and mineralization of the extracellular matrix stained with 1% Alizarin Red S (ARS) [2]. For quantitative analysis, the culture wells were washed three times with dH2O, fixed with 70% ice-cold ethanol, and the ARS stain was dissolved in the cetylpyridinium chloride (CPC) buffer (10% CPC (w/v) in 10 mM Sodium Phosphate Buffer, Sigma) for 1 h. Three aliquots in 200 μL of ARS/CPC extract from each well were then transferred to a 96-well reading plate and quantified by absorbance measurement at 550 nm by a spectrophotometer (Bio-Rad). In parallel experiments, cells were harvested at the end of differentiation induction for analysis of odonto/osteogenic gene expression by real time polymerase chain reaction (qPCR).
Adipogenic differentiation
Cells were seeded into 12- or 24-well plates, grown to subconfluence, and incubated in an adipogenic medium containing 1 μM dexamethasone, 1 μg/mL insulin, 0.5 mM 3-isobutyl-1-methylxantine (IBMX), and 10% FBS for 8 weeks. Cells were then fixed in 10% formalin for 60 min, washed with 70% ethanol, and lipid droplets were stained with 2% (w/v) Oil Red O reagent for 5 min and washed with water [2]. For quantitative analysis of the staining, 60% isopropanol was added to each well to extract Oil Red O for 10 min. The supernatant was collected and transferred into a 96-well plate for spectrophotometry at 540 nm. Parallel experiments were performed and cells were harvested at the end of differentiation induction for analysis of adipogenic gene expression by qPCR.
Neurogenic differentiation
Neurogenic differentiation was initiated by adding 10 ng/mL of the bFGF (BD Biosciences) to the medium of subconfluent DPSCs for at least 24 h, followed by adding the neural induction medium consisting of the αMEM with a 10 ng/mL bFGF, 10 μM forskolin, 25 mM KCl, 2 mM valproic acid, and 5 μg/mL insulin (the latter four items all from Sigma) [33].
After 5 weeks, cells were harvested and analyzed by qPCR for the expression of the neural cell markers.
Reverse transcription polymerase chain reaction (RT-PCR) and real time polymerase chain reaction (qPCR)
Total cellular RNA was isolated using an RNeasy mini kit (Qiagen) with DNase I (Invitrogen) to remove the genomic DNA contaminant. The extracted RNA (1 μg) was reverse transcribed to generate the first-strand cDNA using Superscript II (Invitrogen). The produced cDNA was used as a template for each reverse transcription polymerase chain reaction (RT-PCR) and real time polymerase chain reaction (qPCR). cDNA, gene-specific primers (200 nM final concentration) (designed according to published cDNA Genbank sequences shown in Table 1), and 45 μL of Platinum Blue SuperMix containing 1 U Taq DNA Polymerase (Invitrogen) in a 50 μL final volume underwent PCR processes with the following program in the Thermocycler machine (AB Applied Biosystems): Initial denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 53°C–58°C (depending on the primers used) for 30 s, 72°C for 60 s, and a final extension of 72°C for 5 min. The resulting RT-PCR products were run on a 1.5% agarose gel with ethidium bromide, and gel images were captured and quantified with BIO-RAD Image Lab 3.0 system.
Table 1.
Primers Used for Reverse Transcription Polymerase Chain Reaction and Real Time Polymerase Chain Reaction Analysis
| Gene function | Gene | Primer (5′— 3′) sense Antisense | Product size (bp) |
|---|---|---|---|
| Pluripotency associated | NANOG | TAATAACCTTGGCTGCCGTCTCTG | 150 |
| GCCTCCCAATCCCAAACAATACGA | |||
| OCT-4 | CAGTGCCCGAAACCCACAC | 161 | |
| GGAGACCCAGCAGCCTCAAA | |||
| SOX-2 | ACACCAATCCCATCCACACT | 224 | |
| GCAAACTTCCTGCAAAGCTC | |||
| Senescence-associated | P16 | CCCAACGCACCGAATAGTTAC | 135 |
| CACGGGTCGGGTGAGAGT | |||
| Odonto/Osteogenic markers | ALP | CCACGTCTTCACATTTGGTG | 196 |
| AGACTGCGCCTGGTAGTTGT | |||
| BSP | AAAGTGAGAACGGGGAACCT | 161 | |
| GATGCAAAGCCAGAATGGAT | |||
| CBFA1 | TTTGCACTGGGTCATGTGTT | 156 | |
| TGGCTGCATTGAAAAGACTG | |||
| OCN | GGCAGCGAGGTAGTGAAGAG | 230 | |
| CTGGAGAGGAGCAGAACTGG | |||
| Adipogenic markers | LPL | AGTGGCCAAATAGCACATCC | 186 |
| CCGAAAGATCCAGAATTCCA | |||
| PPARγ2 | TCCATGCTGTTATGGGTGAA | 193 | |
| TCAAAGGAGTGGGAGTGGTC | |||
| Neurogenic markers | βIII tubulin | CAGATGTTCGATGCCAAG | 164 |
| GGGATCCACTCCACGAAGTA | |||
| Nestin | AACAGCGACGGAGGTCTCTA | 220 | |
| TTCTCTTGTCCCGCAGACTT | |||
| NFM | CGACCTCAGCAGCTACCAGGACAC | 200 | |
| CAGTGATGCTTCCTGCAATGTGCT | |||
| Housekeeping | GAPDH | CAAGGCTGAGAACGGGAAGC | 194 |
| AGGGGGCAGAGATGATGACC |
For qPCR, the same cDNA was used with SYBR Green PCR Master Mix (AB Applied Biosystems). The PCR was performed using 7500 Real-Time PCR System (AB Applied Biosystems) with the following thermal cycling conditions: 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, 53°C–58°C (depending on the primers used) for 30 s, 60°C for 1 min, cycled to step 3 for 40 cycles. PCR were performed using the human-specific sense and anti-sense primers (Table 1). A relative quantitative analysis method was performed to quantify the relative gene expression compared to the level of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Western blot
hDPSCs were harvested and lysed in an ice-cold Triton X-100/β-octylglucoside buffer (10 mM imidazole, 100 mM NaCl, 1 mM MgCl2, 5 mM Na2EDTA, 1% Triton X-100, and 0.87 mg/mL β-octylglucoside) containing 0.5 μM phenylmethylsulfonyl fluoride and 1% protease inhibitor. Lysates were centrifuged at 12,000 rpm for 20 min. The supernatant was collected and protein concentrations were determined using a BCA (bicinchoninic acid) protein assay (Pierce). The Laemmli sample buffer (Bio-Rad) was added to the lysates, which were then heated to 95°C for 5 min. Typically, 15 μg of the total cellular protein was used for comparison of expression levels of chemically treated DPSCs and nontreated controls. Whole cell lysates were fractionated on a 7.5% SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, blocked with 5% nonfat dry milk, and incubated with primary antibodies to selected proteins. The following rabbit anti-human primary antibodies were used: anti-P70S6 kinase, anti-phosphorylated (p)-P70S6 kinase, anti-ERK1/2, anti p-ERK1/2, anti-mTOR, anti p-mTOR (Cell Signaling Technology), and anti-RAS-GAP (Abcam). Membranes were reprobed with anti-GAPDH antibodies (Chemicon) for control of equal loading. Protein-specific detection was carried out with horseradish peroxidase-labeled anti-rabbit secondary antibodies (GE Healthcare) and Immun-Star HRP Chemiluminescent Kit (Bio-Rad). The luminescent protein bands were detected by x-ray films.
Data analysis
All experiments were performed at least in triplicate. All values are expressed as mean±SD. Statistical analyses of the data were performed using the Student's t-test when only two groups were compared. One-way analysis of variance (ANOVA) was used followed by Tukey's honestly significant difference (HSD) when comparing three groups or more. Values of P less than 0.05 were considered statistically significant.
Results
Effects of small molecules on cell viability and proliferation
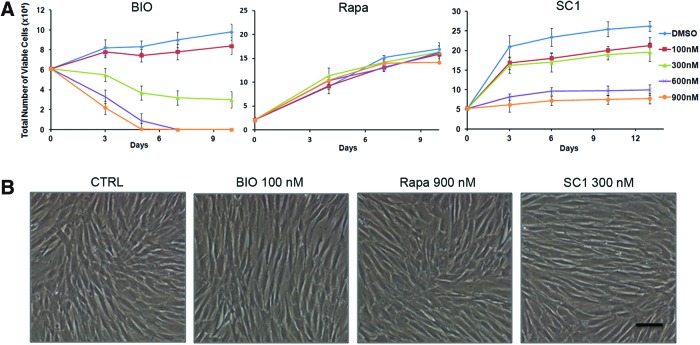
We first determined the optimal doses for each small molecule that did not affect the viability of DPSCs. The graphs in Fig. 1A show that BIO at 100 nM and SC1 at 300 nM had a minimal effect on cell viability, while at higher concentrations, the cell viability dramatically decreased. Rapamycin did not affect cell viability even at the highest concentration tested (900 nM). There were no or minimal observable morphological changes of the cells observed in the treated cells (Fig. 1B). With the presence of these small molecules under the selected nontoxic concentrations, cells maintained viability after expansion in cultures to >10 passages.
FIG. 1.
Cell viability analysis after treatment with small molecules. Dental pulp stem cells (DPSCs) were seeded in 12-well plates and treated with BIO, Rapa (rapamycin), or SC1 of various concentrations. At different time points, cells were harvested and the viable cells were counted. (A) Cell viability counts over time. (B) Cell images after 10 days in cultures at indicated concentrations. Scale bar: 80 μm. Only dimethyl sulfoxide (DMSO) was present in control cells. Data represent three independent experiments each performed in triplicate. Color images available online at www.liebertpub.com/scd
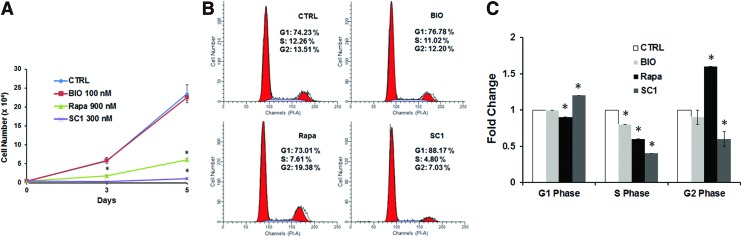
Cell proliferation was tested comparing treated and nontreated controls. There was a significant decrease after treatment with rapamycin and SC1 (P<0.05), but not BIO (P>0.05) (Fig. 2A). Cell cycle analysis using propidium iodide to label DNA showed that lower percentage of S phase, while a higher percentage of G2 phase was observed in cells treated with rapamycin compared to control cells (P<0.05). For the SC1-treated group, there was a higher percentage of cells at the G1 phase, while less at S and G2 phases (P<0.05). BIO treatment showed no obvious change of the cell numbers in the cell cycle interphases compared to the nontreated controls (Fig. 2B, C).
FIG. 2.
Cell proliferation and cell cycle analysis of DPSCs treated with small molecules. (A) Cell proliferation analysis. Total cell counts at days 0, 3, and 5 after small molecule treatments. (B) Representative data of flow cytometry analysis of cell cycle. After 5 days of treatment, cells were analyzed and the percentage of cells in different cell cycle interphases G1, S, and G2 are indicated. (C) Quantitative comparison of the levels of different cell cycle phases. CTRL: control without small molecule treatments (only DMSO); BIO at 100 nM, SC1 at 300 nM, and Rapa (rapamycin) at 900 nM in DMSO. DPSCs at P3 were used. *Significantly different between treated and CTRL (ANOVA; P<0.05). Data from one representative experiment performed in triplicate. Color images available online at www.liebertpub.com/scd
Increased STRO-1 and pluripotent gene expression following small molecule treatment
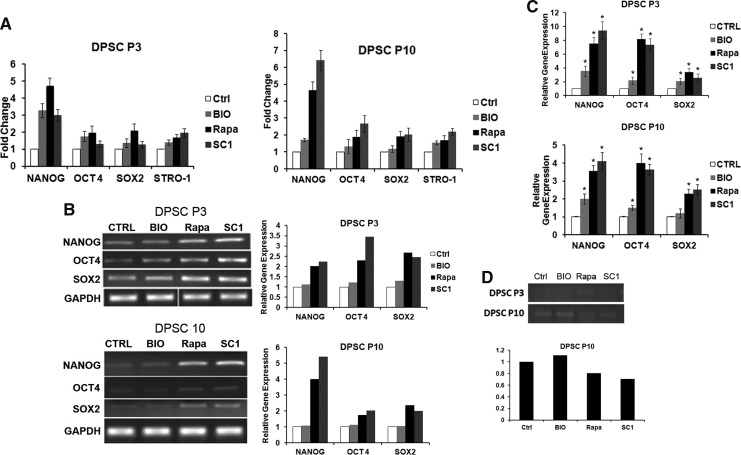
STRO-1 is associated with the immaturity of MSCs as its expression decreases when the cell passage number increases. NANOG, OCT4, and SOX2 are known to be associated with pluripotency, and their expression has been used to indicate stem cell immaturity [20]. We therefore measured the expression of these pluripotent genes in DPSCs after being treated by the selected small molecules. Both protein and mRNA expression levels were examined. We first tested their expression after treatment in cultures for 3, 5, or 7 days. We found that day 5 revealed the most difference between the treated and nontreated group under this experimental setting, and therefore, more extensive comparison was performed for the day 5 time point. The results in Fig. 3A show that protein levels of STRO-1 and pluripotent genes NANOG, OCT4, and SOX2 all had some levels of increase in DPSCs treated by these small molecules. STRO-1 was mildly increased in response to the three small molecule treatments. For pluripotent gene expression at p3, protein levels of NANOG had the most increase by SC1, BIO, and rapamycin treatments, whereas OCT4 and SOX2 only mildly responded to BIO and rapamycin. At a higher passage (p10), NANOG was more drastically responsive, while OCT4 only mildly responsive to SC1 and rapamycin treatment (flow cytometry data presented in Supplementary Fig. S1). The effects of these small molecules appeared to continue after their withdrawal as shown in Supplementary Fig. S2 that DPSCs expressed increased pluripotency-associated genes after continuous culturing and passaged compared to the nontreated controls.
FIG. 3.
Expression of stem cell-associated and senescence-associated genes after small molecule treatment. (A) Flow cytometry analysis of STRO-1 and pluripotency-associated gene protein products after 5 days of small molecule treatments. Expression levels of STRO-1 and pluripotent genes after small molecule treatments of DPSCs at passages 3 and 10. (B) RT-PCR gel images of amplified target genes and the band intensities were quantitated as shown in bar charts on the right. (C) qPCR analysis showing relative levels of gene expression with or without small molecule treatments for 5 days. * Significantly different between treated and CTRL (ANOVA; P<0.05). (D) RT-PCR analysis of p16Ink4A expression after 5 days of small molecule treatment of DPSCs at passages 3 and 10. RT-PCR gel images of amplified target genes are shown in the top panel and the band intensities of DPSCs at P10 were quantitated as shown in the bottom graph. Data represent two independent experiments each performed in triplicate. RT-PCR, reverse transcription-polymerase chain reaction; qPCR, real time polymerase chain reaction.
More quantifiable analysis to see the increase of gene expression was verified at the mRNA levels examined by RT-PCR and especially by qPCR. As shown in Fig. 3B and C, the profiles of the increased pluripotent mRNA expression are similar to those of protein levels. The consistent part of the qPCR data and RT-PCR data is that SC1 and Rapa, not BIO, considerably increased all three pluripotent gene expressions (Fig. 3C).
Decreased expression of p16 after small molecule treatment
p16Ink4A or the cyclin-dependent kinase inhibitor 2A, (CDKN2A) is a tumor suppressor protein that plays an important role in regulating the cell cycle. Elevated p16 is associated with cell senescence [34]; therefore, we tested whether small molecule treatment of DPSCs would decrease the expression of p16. Our data presented in Fig. 3D show that p16 levels at passage 3 were too low after PCR amplification for any meaningful analysis. p16 expression in DPSCs was increased at passage 10 and treatment with SC1 and Rapa, not BIO, decreased the p16 expression.
Effects of small molecule treatment on DPSC differentiation capacities
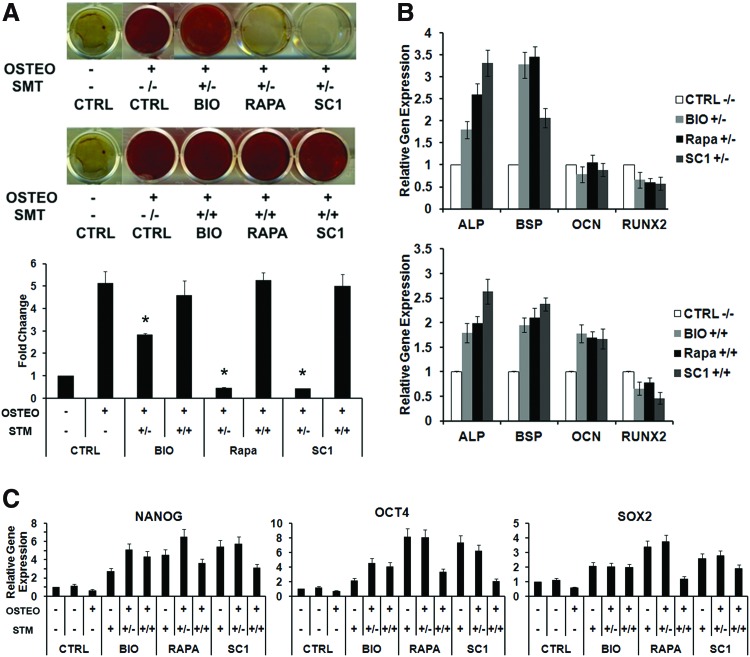
To determine whether DPSC differentiation is affected after the treatment of small molecules, we examined multiple differentiation pathways. Under odonto/osteogenic induction, cells exhibited stronger odonto/osteogenicity with the presence of small molecules. Interestingly, if the cells were pretreated with the small molecules for 5 days first and followed by odonto/osteogenic induction without the continual presence of the small molecules, the level of odonto/osteogenicity is lower than those with a continual presence of the small molecules (Fig. 4A). This phenomenon was drastic when cells were pretreated with SC1 and rapamycin first, and then without their presence during the odonto/osteogenic induction.
FIG. 4.
Effects of small molecules on DPSC odonto/osteogenesis. (A) DPSCs were pretreated or nonpretreated with small molecules for 5 days before differentiation induction for 4 weeks. (−/−): no pretreatment and no small molecules presence during odonto/osteogenic induction; (+/−): with pretreatment, but no small molecules presence during odonto/osteogenic induction; (+/+): with pretreatment and with small molecules presence during odonto/osteogenic induction; CTRL (control); OSTEO (odonto/osteo induction); SMT (small molecule treatment). Cell cultures were stained with Alizarin Red and representative data are shown in the top panel and the quantitative Alizarin Red measurements are shown in the bottom graph. *Significantly different between treated, odonto/osteogenic induced, and CTRL (ANOVA; P<0.05). (B) qPCR analysis of odonto/osteogenic gene expression of DPSCs under odonto/osteogenic induction treated by small molecules. (C) qPCR analysis of pluripotent gene expression by DPSCs treated with small molecules and underwent odonto/osteogenesis. Data represent two independent experiments each performed in quintuplicate. Color images available online at www.liebertpub.com/scd
We next determined the effects of small molecule treatment on odonto/osteogenic marker expression. As shown in Fig. 4B, if cells were pretreated with small molecules, and then the small molecules removed during the odonto/osteogenic induction, the expression of odonto/osteogenic gene markers ALP and BSP was enhanced, whereas RUNX2 and OCN expression decreased. When the cells were pretreated with small molecules, which were also present during odonto/osteogenic induction, the ALP and BSP expression was also enhanced as well as OCN except RUNX2.
In examining the pluripotency-associated gene expression during the odonto/osteogenic differentiation, we found that the pluripotent gene expression is reduced when the odonto/osteogenic differentiation is enhanced, while their expression is increased when the differentiation is impeded, particularly in the cases of SC1 and rapamycin treatments (Fig. 4C).
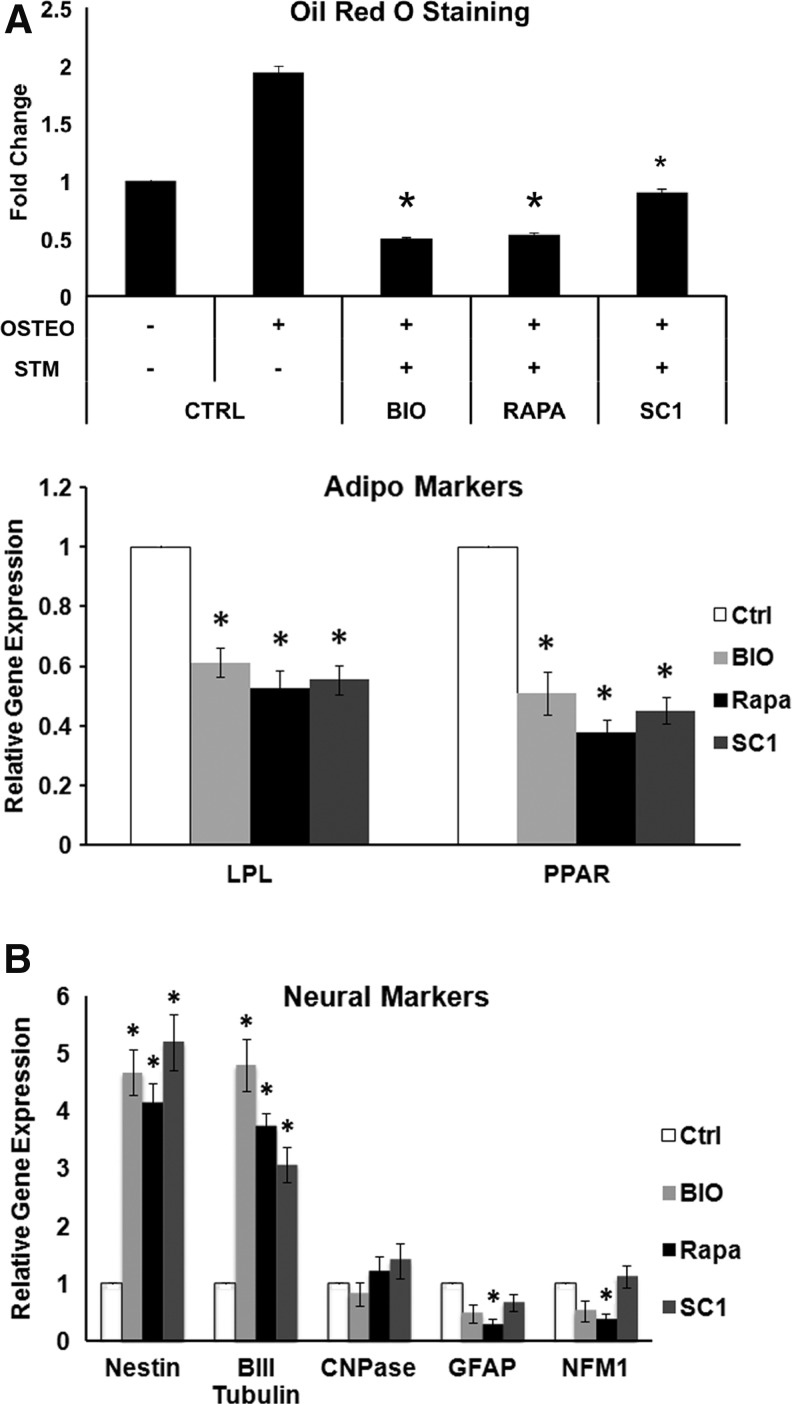
Pretreatment of cells with small molecules and their continuous presence during the adipogenic and neurogenic induction media resulted in cell death, therefore, the effects of this treatment protocol on the two differentiation pathways could not be observed. However, pretreatment of cells with small molecules and then without their presence during the adipogenic and neurogenic induction, we were able to observe the effects without cell death. In the adipogenic group, cell cultures showed decreased Oil Red staining compared to the nontreated controls with lower gene expression of adipogenic markers LPL and PPARγ2 (Fig. 5A). In neurogenic experiments, the expression of neural markers nestin and β-III tubulin was increased, whereas the expression of CNPase, GFAP, and NFM did not change or decrease compared to the nontreated controls (Fig. 5B).
FIG. 5.
Effects of small molecules on DPSC adipogenesis and neurogenesis. DPSCs were pretreated or nontreated with small molecules for 5 days before differentiation induction; 8 weeks for adipogenic induction and 5 weeks for neurogenic induction. (A) Top graph: quantitative Oil Red O stain measurement; bottom graph: qPCR analysis of adipogenic gene expression by DPSCs pretreated with small molecules for 5 days. *Significantly different between treated, adipogenic induced, and CTRL (ANOVA; P<0.05). (B) qPCR analysis of neurogenic gene expression after a 5-week neurogenic induction of DPSCs pretreated with small molecules for 5 days. *Significantly different between treated and CTRL (ANOVA; P<0.05). Data represent two independent experiments each performed in quintuplicate.
Effects of SC1 and rapamycin on signal transduction pathways
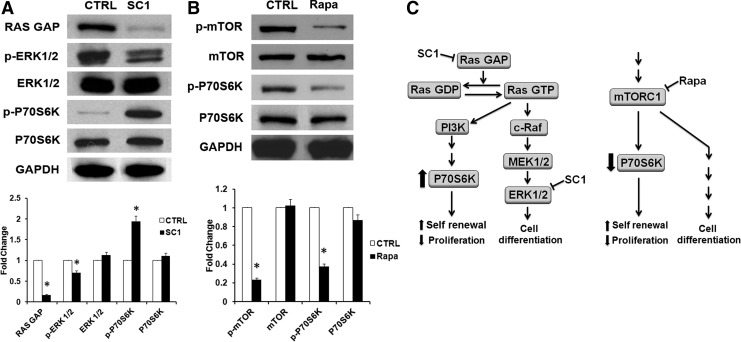
SC1 was first shown to allow propagation of murine ES cells in an undifferentiated, pluripotent state in the absence of feeder cells, serum, and leukemia inhibitory factor (LIF). SC1 functions through dual inhibition of Ras-GAP and ERK1 [29]. Our western blot analysis of treated DPSCs shown in Fig. 6A revealed that SC1 activated Ras by inhibition of Ras-GAP function, which led to the increased phosphorylation of P70S6K (p-P70S6K), a mitogen-stimulated serine/threonine kinase regulated by the PI3K pathway. SC1 also inhibited the phosphorylation of ERK1 (44 kDa).
FIG. 6.
Western blot analysis of signaling events in DPSCs after treatment with SC1 or rapamycin (Rapa). Cells were treated with 300 nM SC1 (A) or 900 nM Rapa (B) for 5 days and subjected to protein extraction for detection of signaling factors RAS GAP, phosphorylated (p)-ERK1(44)/2(42), and p-P70S6K shown in (A) or p-mTOR and p-p70S6K in (B). GAPDH: glyceraldehyde 3-phosphate dehydrogenase, CTRL: control nontreated. The band intensities were quantitated as shown in the bottom panel. *Significantly different between treated and CTRL (t-test; P<0.05). (C) Schematic summary and hypothesis of the effects of SC1 and Rapa on Ras GAP and mTORC1 signaling pathways. Data represent two independent experiments each performed in triplicate.
Rapamycin is known to inhibit mTORC1 by associating with its intracellular receptor FKBP12 (FK506 binding protein-12). The FKBP12-rapamycin complex binds directly to the FKBP12-rapamycin binding (FRB) domain of mTOR forming FKBP-rapamycin-associated protein (FRAP)/mTOR complex; as such, inhibiting the activity of mTORC1. Our results showed that treatment of DPSCs with rapamycin reduced phosphorylated mTOR (p-mTOR) and P70S6K (p-P70S6K) levels (Fig. 6B). The reduction of p-P70S6K was in opposite to the effect of SC1.
Discussion
Maintaining stem cell properties in vitro has been a challenge in the field of regenerative medicine. Identifying small molecules instead of recombinant proteins or via gene transduction to retain the stemness of cultured stem cells have been a focus of interest in stem cell biology. Along this line of thought, we selected three small molecules known to affect stem cell properties and determine whether they can enhance the stemness of DPSCs. Our findings revealed that these three chemicals affect DPSCs in various ways. BIO has shown to enhance hMSC proliferation and maintain hESCs and mouse ESCs (mESCs) in an undifferentiated state [27,28]. In our present studies, while BIO did not increase DPSC pluripotent gene expression, it appeared to prevent full differentiation of DPSCs into the three lineages tested. SC1 and rapamycin had a more conspicuous effect on DPSC stemness by increasing pluripotent gene expression and prevent cell differentiation.
ESCs have been a valuable tool to study stem cell self-renewal and differentiation because they can grow in cultures and be maintained in an undifferentiated pluripotent state. Most ESCs in the colonies are undergoing self-renewal with a symmetrical division such that, these cells can propagate indefinitely. However, to maintain this state for mESCs, it requires the presence of feeder cells and LIF along with other specific chemicals. To effectively maintain their ESC state, a small molecule SC1 has been tested and found to be effective in maintaining mESC stemness [29]. In our present studies, we showed that SC1 had similar effects on DPSCs to those on mESCs in some aspects. However, to understand self-renewal of MSCs in cultures is more difficult compared with ESCs because the self-renewal MSCs cannot be assessed or observed easily. For MSCs, their self-renewal is a slow process, while their progenitor cells undergo transit rapid proliferation in response to stimulation in vivo. hDPSCs normally proliferate rapidly in culture at low passages (∼p3) with a population doubling time ∼22 h [35]. At the nontoxic concentration, SC1 and rapamycin decreased the proliferation rate, while BIO had little to no effect. Both SC1 and rapamycin altered the percentage of cells in the phases of the cell cycle by locking cells in the G1 and G2 phases, respectively, leading to a lower proliferation rate. This is in contrast to the effect of bFGF treatment of stem cells from apical papilla (SCAP) that the cell proliferation rate was increased by the growth factor [26].
To assess whether the small molecules enhance the stemness of DPSCs, examining the expression levels of pluripotency-associated genes NANOG, OCT4, and SOX2 is an important approach as these genes are downregulated in differentiated cells, while maintained at high levels in ESCs. SC1 and rapamycin, more conspicuously than BIO, increased the levels of these pluripotent genes, while suppressing p16Ink4a, which is a cyclin-dependent kinase inhibitor (CKI) that prevents cells from entering the cell cycle phases and is associated with cell senescence [36]. Therefore, it appears that SC1 and rapamycin help DPSCs maintain their stemness by boosting the pluripotent gene expression as well as inhibiting the cell senescence process. Additionally, STRO-1, which has been associated with the stemness of some types of MSCs such as BMMSCs and DPSCs, was also increased in DPSCs by these three small molecules in various degrees. There is a concern regarding the induction of a more primitive state that may cause changes in the karyotype due to the induction of various oncogenes. We performed standard cytogenetic analysis of DPSCs from three different donors treated with small molecules. Cells examined showed a normal karyotype and the representative data are presented in Supplementary Fig. S3 (Materials & Methods for karyotyping are also in the Supplementary Data). Although the results appeared normal, it should be noted that a low level of mosaicism or a small structural aberration may be undetected due to the limitation of the band level. Standard karyotyping of iPSCs does not detect abnormality as evidenced by numerous reports, including ours [21], while somatic coding nucleotide mutations and aberrant epigenetic profiles have been detected in iPSCs via genome sequencing and DNA methylation analysis [37,38]. Therefore, extensive genome sequencing and epigenetic analysis will be needed to determine the safety of small molecule-treated cells for clinical applications.
The small molecules appeared to prevent cells from differentiating to the end point. It is interesting that DPSC pre-exposure to the small molecules prevent the cells from differentiating, whereas when the osteo/odontogenic stimulus was added simultaneously with the small molecules, the differentiation is enhanced. Further investigation of the mechanism underlying this phenomenon is required. Similarly, small molecules decreased adipogenic gene expression. Neurogenic early markers nestin and β-III tubulin were increased by the three molecules, but not the late markers CNPase, GFAP, and NFM. In regard to cell death observed in experiments when cells were pre-exposed to small molecules followed by continuous exposure of small molecules during the adipogenic and neurogenic induction, the possible explanation is the activation of intracellular pathways that leads to cell apoptosis.
The actions of SC1 and rapamycin were further studied because of their potent upregulation of pluripotent genes and downregulation of p16 as well as their regulation of differentiation. Upon examining the signaling pathways that SC1 and rapamycin interfere, it was confirmed that both molecules act on the intracellular signaling factors in DPSCs as they do on other cell types. SC1 inhibited Ras-GAP leading to the increase of Ras signaling via the PI3 kinase pathway, which promotes DPSC self-renewal. SC1 also inhibited ERK1, which blocked ERK1-dependent cell differentiation (Fig. 6A). These actions of SC1 on DPSCs are the same as on mESCs. SC1 reduces the mESC proliferation rate shown in a previous report [29] and in our present study on DPSCs. Rapamycin has been shown to inhibit osteogenic differentiation of rat bone marrow MSCs [39] and odonto/osteogenic differentiation and mineralization of SHED (stem cells from human exfoliated deciduous teeth) by restricting the synthesis of dentin sialoprotien expression [40]. Rapamycin is also known to inhibit adipogenesis of preadipocytes [41]. For many cell types, including our findings in this present study, rapamycin slows down cell proliferation [31,32,42–45], as it decreases p-P70S6K (Fig. 6B). Phosphorylation of P70S6K and 4EBP by mTORC1 allows protein translation and, thus, facilitates cell growth and proliferation. Lowered p-P70S6K in DPSCs by rapamycin treatment may decrease cell proliferation. In general, reduced cell proliferation and growth by rapamycin treatment is considered to prevent stem cell exhaustion, thereby conserving their stemness. Spermatogonial stem cells isolated from mice treated with rapamycin form more and larger colonies and express higher levels of oxidative stress response genes than those from the untreated group, indicating their stemness and antiaging capacity [46].
There has been a lack of clarity in defining stem cell stemness in vitro. Cells that have a greater proliferation rate are considered stem cells [47]. Therefore, growth conditions such as adding growth factors or small molecules, or under low oxygen, that increase stem/progenitor cell proliferation in vitro may be considered as enhancing stemness [26,28,48]; whereas agents that decrease cell proliferation such as SC1 and rapamycin are also considered as having the same effect. Although increased p-P70S6K in cells normally increases cell proliferation and growth, SC1-induced p-P70S6K increase slows down cell proliferation. This opposite finding on the p70S6K levels after SC1 and rapamycin also implicates that increased or decreased cell proliferation may not be related to self-renewal as we hypothesized in Fig. 6C. Other signaling events outside of the scheme depicted in the illustration are likely causing the increase of pluripotent gene expression, which may be a more important indicator for self-renewal capacity.
Taken together, small molecules used in our studies appear to enhance the stemness of cultured hDPSCs at different capacities evidenced by the increased expression of the MSC marker STRO-1 and the pluripotent markers NANOG, OCT4, and SOX2, and decreased expression of the senescent gene p16. Along with the reduced tendency to differentiate into the three cell lineages tested, the results suggest that hDPSCs were guided to a more immature state. The increase of pluripotency-associated genes also appears to persist after the withdrawal of the small molecule treatment. The findings in the present study provided a basis from which a protocol may be established to maintain cultured DPSC stemness and subsequently verify their in vivo tissue regeneration capacity.
Supplementary Material
Acknowledgments
This work was supported, in part, by grants from the National Institutes of Health R01 DE019156 (G.T.-J.H.), and by an Endodontic Research Grant from the American Association of Endodontists Foundation (M.A.-H.). The authors wish to thank Sonia Kim for her assistance in the cell viability test in the study. The Cancer Center Core Cytogenetics laboratory is supported by the National Institute of Health, National Cancer Institute, Cancer Center Support Grant P30 CA21765, and ALSAC.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Gronthos S. Mankani M. Brahim J. Robey PG. Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang GT. Yamaza T. Shea LD. Djouad F. Kuhn NZ. Tuan RS. Shi S. Stem/Progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng Part A. 2010;16:605–615. doi: 10.1089/ten.tea.2009.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iohara K. Imabayashi K. Ishizaka R. Watanabe A. Nabekura J. Ito M. Matsushita K. Nakamura H. Nakashima M. Complete pulp regeneration after pulpectomy by transplantation of CD105+ stem cells with stromal cell-derived factor-1. Tissue Eng Part A. 2011;17:1911–1920. doi: 10.1089/ten.TEA.2010.0615. [DOI] [PubMed] [Google Scholar]
- 4.Gandia C. Arminan A. Garcia-Verdugo JM. Lledo E. Ruiz A. Minana MD. Sanchez-Torrijos J. Paya R. Mirabet V, et al. Human dental pulp stem cells improve left ventricular function, induce angiogenesis, and reduce infarct size in rats with acute myocardial infarction. Stem Cells. 2008;26:638–645. doi: 10.1634/stemcells.2007-0484. [DOI] [PubMed] [Google Scholar]
- 5.Wang J. Wang X. Sun Z. Yang H. Shi S. Wang S. Stem cells from human-exfoliated deciduous teeth can differentiate into dopaminergic neuron-like cells. Stem Cells Dev. 2010;19:1375–1383. doi: 10.1089/scd.2009.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang AHC. Snyder BR. Cheng PH. Chan AW. Putative dental pulp-derived stem/stromal cells promote proliferation and differentiation of endogenous neural cells in the hippocampus of mice. Stem Cells. 2008;26:2654–2663. doi: 10.1634/stemcells.2008-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai K. Yamamoto A. Matsubara K. Nakamura S. Naruse M. Yamagata M. Sakamoto K. Tauchi R. Wakao N, et al. Human dental pulp-derived stem cells promote locomotor recovery after complete transection of the rat spinal cord by multiple neuro-regenerative mechanisms. J Clin Invest. 2012;122:80–90. doi: 10.1172/JCI59251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamout B. Hourani R. Salti H. Barada W. El-Hajj T. Al-Kutoubi A. Herlopian A. Baz EK. Mahfouz R, et al. Bone marrow mesenchymal stem cell transplantation in patients with multiple sclerosis: a pilot study. J Neuroimmunol. 2010;227:185–189. doi: 10.1016/j.jneuroim.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Duijvestein M. Vos AC. Roelofs H. Wildenberg ME. Wendrich BB. Verspaget HW. Kooy-Winkelaar EM. Koning F. Zwaginga JJ, et al. Autologous bone marrow-derived mesenchymal stromal cell treatment for refractory luminal Crohn's disease: results of a phase I study. Gut. 2010;59:1662–1669. doi: 10.1136/gut.2010.215152. [DOI] [PubMed] [Google Scholar]
- 10.Bang OY. Lee JS. Lee PH. Lee G. Autologous mesenchymal stem cell transplantation in stroke patients. Ann Neurol. 2005;57:874–882. doi: 10.1002/ana.20501. [DOI] [PubMed] [Google Scholar]
- 11.Lee JS. Hong JM. Moon GJ. Lee PH. Ahn YH. Bang OY. A long-term follow-up study of intravenous autologous mesenchymal stem cell transplantation in patients with ischemic stroke. Stem Cells. 2010;28:1099–1106. doi: 10.1002/stem.430. [DOI] [PubMed] [Google Scholar]
- 12.Horwitz EM. Gordon PL. Koo WK. Marx JC. Neel MD. McNall RY. Muul L. Hofmann T. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci U S A. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen SL. Fang WW. Ye F. Liu YH. Qian J. Shan SJ. Zhang JJ. Chunhua RZ. Liao LM. Lin S. Sun JP. Effect on left ventricular function of intracoronary transplantation of autologous bone marrow mesenchymal stem cell in patients with acute myocardial infarction. Am J Cardiol. 2004;94:92–95. doi: 10.1016/j.amjcard.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 14.Koc ON. Day J. Nieder M. Gerson SL. Lazarus HM. Krivit W. Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH) Bone Marrow Transplant. 2002;30:215–222. doi: 10.1038/sj.bmt.1703650. [DOI] [PubMed] [Google Scholar]
- 15.Quarto R. Mastrogiacomo M. Cancedda R. Kutepov SM. Mukhachev V. Lavroukov A. Kon E. Marcacci M. Repair of large bone defects with the use of autologous bone marrow stromal cells. N Engl J Med. 2001;344:385–386. doi: 10.1056/NEJM200102013440516. [DOI] [PubMed] [Google Scholar]
- 16.Sun L. Akiyama K. Zhang H. Yamaza T. Hou Y. Zhao S. Xu T. Le A. Shi S. Mesenchymal stem cell transplantation reverses multiorgan dysfunction in systemic lupus erythematosus mice and humans. Stem Cells. 2009;27:1421–1432. doi: 10.1002/stem.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Blanc K. Frassoni F. Ball L. Locatelli F. Roelofs H. Lewis I. Lanino E. Sundberg B. Bernardo ME, et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 2008;371:1579–1586. doi: 10.1016/S0140-6736(08)60690-X. [DOI] [PubMed] [Google Scholar]
- 18.Guo W. Wang H. Zou S. Gu M. Watanabe M. Wei F. Dubner R. Huang GT. Ren K. Bone marrow stromal cells produce long-term pain relief in rat models of persistent pain. Stem Cells. 2011;29:1294–1303. doi: 10.1002/stem.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang GT-J. Gronthos S. Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dental Res. 2009;88:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Yan X. Qin H. Qu C. Tuan RS. Shi S. Huang GT-J. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells Dev. 2010;19:469–480. doi: 10.1089/scd.2009.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz SD. Hubschman J-P. Heilwell G. Franco-Cardenas V. Pan CK. Ostrick RM. Mickunas E. Gay R. Klimanskaya I. Lanza R. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;25(379):713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 23.Solchaga LA. Penick K. Porter JD. Goldberg VM. Caplan AI. Welter JF. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2005;203:398–409. doi: 10.1002/jcp.20238. [DOI] [PubMed] [Google Scholar]
- 24.Kim JH. Lee MC. Seong SC. Park KH. Lee S. Enhanced proliferation and chondrogenic differentiation of human synovium-derived stem cells expanded with basic fibroblast growth factor. Tissue Eng Part A. 2011;17:991–1002. doi: 10.1089/ten.TEA.2010.0277. [DOI] [PubMed] [Google Scholar]
- 25.Zeng YA. Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J. Huang GT. He W. Wang P. Tong Z. Jia Q. Dong L. Niu Z. Ni L. Basic fibroblast growth factor enhances stemness of human stem cells from the apical papilla. J Endod. 2012;38:614–622. doi: 10.1016/j.joen.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato N. Meijer L. Skaltsounis L. Greengard P. Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman MD. Takahata M. Benoit DSW. 6-Bromoindirubin-3′-oxime (BIO) induces proliferation of human mesenchymal stem cells (hMSCs. Bioengineering Conference (NEBEC), 2011 IEEE 37th Annual Northeast; Troy, New York. 2011. pp. 1–2. [Google Scholar]
- 29.Chen S. Do JT. Zhang Q. Yao S. Yan F. Peters EC. Scholer HR. Schultz PG. Ding S. Self-renewal of embryonic stem cells by a small molecule. Proc Natl Acad Sci U S A. 2006;103:17266–17271. doi: 10.1073/pnas.0608156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W. Wei W. Shi C. Zhu J. Ying W. Shen Y. Ye X. Fang L. Duo S, et al. Pluripotin combined with leukemia inhibitory factor greatly promotes the derivation of embryonic stem cell lines from refractory strains. Stem Cells. 2009;27:383–389. doi: 10.1634/stemcells.2008-0974. [DOI] [PubMed] [Google Scholar]
- 31.Hoogduijn MJ. Crop MJ. Korevaar SS. Peeters AM. Eijken M. Maat LP. Balk AH. Weimar W. Baan CC. Susceptibility of human mesenchymal stem cells to tacrolimus, mycophenolic acid, and rapamycin. Transplantation. 2008;86:1283–1291. doi: 10.1097/TP.0b013e31818aa536. [DOI] [PubMed] [Google Scholar]
- 32.Izumi K. Inoki K. Fujimori Y. Marcelo CL. Feinberg SE. Pharmacological retention of oral mucosa progenitor/stem cells. J Dental Res. 2009;88:1113–1118. doi: 10.1177/0022034509350559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebrahimi B. Yaghoobi MM. Kamal-abadi AM. Raoof M. Human dental pulp stem cells express many pluripotency regulators and differentiate into neuronal cells. Neural Regen Res. 2011;6:2666–2672. [Google Scholar]
- 34.Reznikoff CA. Yeager TR. Belair CD. Savelieva E. Puthenveettil JA. Stadler WM. Elevated p16 at senescence and loss of p16 at immortalization in human papillomavirus 16 E6, but not E7, Transformed Human Uroepithelial Cells. Cancer Res. 1996;56:2886–2890. [PubMed] [Google Scholar]
- 35.Huang GTJ. Shagramanova K. Chan SW. Formation of odontoblast-like cells from cultured human dental pulp cells on dentin in vitro. J Endod. 2006;32:1066–1073. doi: 10.1016/j.joen.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Baker DJ. Wijshake T. Tchkonia T. LeBrasseur NK. Childs BG. van de Sluis B. Kirkland JL. van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lister R. Pelizzola M. Kida YS. Hawkins RD. Nery JR. Hon G. Antosiewicz-Bourget J. O'Malley R. Castanon R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gore A. Li Z. Fung HL. Young JE. Agarwal S. Antosiewicz-Bourget J. Canto I. Giorgetti A. Israel MA, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isomoto S. Hattori K. Ohgushi H. Nakajima H. Tanaka Y. Takakura Y. Rapamycin as an inhibitor of osteogenic differentiation in bone marrow-derived mesenchymal stem cells. Journal of Orthopaedic Science. 2007;12:83–88. doi: 10.1007/s00776-006-1079-9. [DOI] [PubMed] [Google Scholar]
- 40.Kim J-K. Baker J. Nor JE. Hill EE. mTor plays an important role in odontoblast differentiation. J Endod. 2011;37:1081–1085. doi: 10.1016/j.joen.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 41.Cho HJ. Park J. Lee HW. Lee YS. Kim JB. Regulation of adipocyte differentiation and insulin action with rapamycin. Biochem Biophys Res Commun. 2004;321:942–948. doi: 10.1016/j.bbrc.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 42.Sun CL. Ham DS. Park HS. Kim JW. Cho JH. Song KH. Son HY. Yoon KH. Rapamycin suppresses the expansion and differentiation of porcine neonatal pancreas cell clusters. Transplantation. 2010;90:717–724. doi: 10.1097/TP.0b013e3181eceaaf. [DOI] [PubMed] [Google Scholar]
- 43.Chen TG. Chen JZ. Wang XX. Effects of rapamycin on number activity and eNOS of endothelial progenitor cells from peripheral blood. Cell Prolif. 2006;39:117–125. doi: 10.1111/j.1365-2184.2006.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H. Feng G. Wu L. Fu S. Liu P. Yang W. Zhang X. The effects of rapamycin on lens epithelial cell proliferation, migration, and matrix formation: an in vitro study. Mol Vis. 2010;16:1646–1653. [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J. Li D. Wang F. Assessing the function of mTOR in human embryonic stem cells. Methods Mol Biol. 2012;821:361–372. doi: 10.1007/978-1-61779-430-8_23. [DOI] [PubMed] [Google Scholar]
- 46.Kofman AE. McGraw MR. Payne CJ. Rapamycin increases oxidative stress response gene expression in adult stem cells. Aging (Albany NY) 2012;4:279–289. doi: 10.18632/aging.100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holstein TW. David CN. Cell cycle length, cell size, and proliferation rate in hydra stem cells. Dev Biol. 1990;142:392–400. doi: 10.1016/0012-1606(90)90360-u. [DOI] [PubMed] [Google Scholar]
- 48.D'Ippolito G. Diabira S. Howard GA. Roos BA. Schiller PC. Low oxygen tension inhibits osteogenic differentiation and enhances stemness of human MIAMI cells. Bone. 2006;39:513–522. doi: 10.1016/j.bone.2006.02.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.