Abstract
Ethanol consumption and poor oral hygiene are risk factors for oral and oesophageal cancers. Although oral streptococci have been found to produce excessive acetaldehyde from ethanol, little is known about the mechanism by which this carcinogen is produced. By screening 52 strains of diverse oral streptococcal species, we identified Streptococcus gordonii V2016 that produced the most acetaldehyde from ethanol. We then constructed gene deletion mutants in this strain and analysed them for alcohol and acetaldehyde dehydrogenases by zymograms. The results showed that S. gordonii V2016 expressed three primary alcohol dehydrogenases, AdhA, AdhB and AdhE, which all oxidize ethanol to acetaldehyde, but their preferred substrates were 1-propanol, 1-butanol and ethanol, respectively. Two additional dehydrogenases, S-AdhA and TdhA, were identified with specificities to the secondary alcohol 2-propanol and threonine, respectively, but not to ethanol. S. gordonii V2016 did not show a detectable acetaldehyde dehydrogenase even though its adhE gene encodes a putative bifunctional acetaldehyde/alcohol dehydrogenase. Mutants with adhE deletion showed greater tolerance to ethanol in comparison with the wild-type and mutant with adhA or adhB deletion, indicating that AdhE is the major alcohol dehydrogenase in S. gordonii. Analysis of 19 additional strains of S. gordonii, S. mitis, S. oralis, S. salivarius and S. sanguinis showed expressions of up to three alcohol dehydrogenases, but none showed detectable acetaldehyde dehydrogenase, except one strain that showed a novel ALDH. Therefore, expression of multiple alcohol dehydrogenases but no functional acetaldehyde dehydrogenase may contribute to excessive production of acetaldehyde from ethanol by certain oral streptococci.
Introduction
Ethanol consumption has been recognized as a risk factor for several types of cancer, including the cancers of the head and neck, liver, colorectum and female breast (Bagnardi et al., 2001). The highest cancer risk is seen for the upper aerodigestive tract, including the oral cavity, throat, voice-box and oesophagus. Recently, poor oral hygiene and tooth loss have been associated with an increased risk of oesophageal cancer (Abnet et al., 2008), suggesting a role for oral micro-organisms in carcinogenesis. Although the oral fungus Candida albicans can produce acetaldehyde directly from glucose through the pyruvate-bypass pathway (Marttila et al., 2013), most oral bacteria, including Streptococcus, do not have pyruvate decarboxylase, the enzyme required for this pathway. Therefore, excessive production of acetaldehyde from ethanol by oral bacteria (Homann, 2001; Kurkivuori et al., 2007) and from ethanol and/or sugar by oral fungi (Uittamo et al., 2009) may contribute to an increased risk of oral–oesophageal cancer.
Alcohol dehydrogenase (ADH) catalyses the conversion of ethanol to acetaldehyde, which can be further converted to acetic acid by acetaldehyde dehydrogenase (ALDH). Therefore, if a bacterium has both active ADH and ALDH, it can metabolize ethanol fully to the harmless acetic acid. However, if a bacterium has active ADH without ALDH, excessive acetaldehyde can be produced from ethanol. Although acetaldehyde production by oral micro-organisms has been reported (Kurkivuori et al., 2007; Meurman & Uittamo, 2008), microbial enzymes involved have not been extensively studied.
Acetaldehyde is a carcinogen in animal models (Woutersen et al., 1986) and causes chromosomal damage, including sister-chromatid exchanges and chromosomal aberrations (Obe & Anderson, 1987). It reacts with 2′-deoxyguanosine to form N2-ethyl-2′-deoxyguanosine (N2-EtdG) to form DNA adducts in animal models of ethanol exposure and in white blood cells of human alcoholics (Vaca et al., 1995). Additionally, acetaldehyde inhibits DNA repair enzymes (Espina et al., 1988). Recently, acetaldehyde has been named as a class I carcinogen for humans by the International Agency for Research on Cancer of WHO (Secretan et al., 2009).
The human body carries about 1014 bacteria in the oral–digestive tract (Dewhirst et al., 2010; Turnbaugh et al., 2007). The metabolic activities performed by these bacteria resemble those of an organ (O'Hara & Shanahan, 2006). Like host cells, many bacteria in the oral cavity and gut can produce acetaldehyde from ingested ethanol (Kurkivuori et al., 2007; Meurman & Uittamo, 2008; Salaspuro, 2003; Väkeväinen et al., 2000, 2001). Even for humans with active ALDH2, nearly all acetaldehyde accumulated in the saliva is of microbial origin (Väkeväinen et al., 2001). Therefore, it is important to study the mechanisms by which oral bacteria produce acetaldehyde from ethanol. In this study, we specifically analysed the enzymes involved in ethanol metabolism in Streptococcus gordonii V2016.
Methods
Bacterial strains, growth conditions and plasmids.
Two groups of oral streptococcal strains were analysed in this study. The first group, obtained from Dr Mark Herzberg of the University of Minnesota, included 14 laboratory strains: Streptococcus sanguinis ATCC 10556, S7, Blackburn, 1239b, 133-79, V2020, V2053, V2054 and V2650 (SK36), and Streptococcus gordonii V685, 488, CHI, V288 and V2016. The second group included 38 clinical strains isolated from the saliva of 12 healthy volunteers. Their species were identified by 16S rRNA gene sequence to be Streptococcus gordonii, S. mitis, S. oralis, S. salivarius and S. sanguinis. The clinical study was approved by the Institutional Review Board of the University of Illinois at Chicago.
The streptococcal strains were grown in Todd–Hewitt (TH) broth or TH broth supplemented with 0.2 % yeast extract (THY) at 37 °C without agitation in a candle jar. For transformation, heat-inactivated horse serum (56 °C for 30 min) was added into TH broth at 5 % (THS). An overnight culture of S. gordonii strain in THS was diluted 1 : 40 into fresh THS. After 2 h of incubation at 37 °C, DNA was added and the bacterial cells were incubated for 1 h and then plated onto TH agar supplemented with appropriate antibiotics (kanamycin, 250 µg ml−1; erythromycin, 10 µg ml−1; or tetracycline, 15 µg ml−1). The plates were incubated at 37 °C for 24 h in a candle jar for selection of transformants. All chemicals and reagents unless otherwise indicated were purchased from Sigma-Aldrich.
Plasmids either as cloning vector or as donors of antibiotic resistance markers included pSF151 (kanamycin resistance, 3.5 kb; Tao, 1998), pAK488 (plasmid carrying the erythromycin resistance cassette from pVA891, 2.1 kb) and pAK560 (plasmid carrying the tetracycline resistance cassette from pVA981, 3.5 kb).
Acetic acid and acetaldehyde production from ethanol.
To detect acetic acid production from ethanol by oral Streptococcus purple broth was used. Each bacterial strain was grown in 5 ml of THY broth overnight at 37 °C. Next, the bacterial cells were harvested by centrifugation and washed in purple broth three times and resuspended in 1 ml of purple broth containing 1 % ethanol. The culture was incubated at 37 °C for 24 h. The change of colour from purple to yellow indicates the production of acetic acid from ethanol.
Purple broth based (PBB)-Schiff’s agar was used for detecting acetaldehyde production from ethanol by oral Streptococcus. Schiff’s reagent made with a mixture of pararosaniline and sodium bisulfite has been widely used to detect acetaldehyde (Lillie, 1977). A previously described protocol (Conway et al., 1987) was modified. Briefly, 8 ml of pararosaniline (2.5 mg ml−1 100 % ethanol) and 100 mg of sodium bisulfite were added to 400 ml batches of precooled (45 °C) PBB agar medium containing 1 % peptone, 0.5 % sodium chloride, 0.1 % beef extract and 1.5 % agar. Plates were freshly made for each assay.
The bacteria were grown overnight at 37 °C in THS broth supplemented with 1 % ethanol to induce ADH expression in a candle jar. The cells were harvested by centrifugation, washed three times and resuspended in sterile saline. Bacterial cell suspension was dropped onto the PBB-Schiff’s agar. The incubation was carried out in the dark at 37 °C for 24–48 h. Red colour developed in and around the bacterial growth would indicate positive acetaldehyde production from ethanol.
Construction of adh mutants in S. gordonii V2016.
Standard recombinant DNA techniques were employed (Sambrook et al., 1989). Multiple pairs of oligonucleotides (Integrated DNA Technologies) used in this study are shown in Table 1. Chromosomal DNA was prepared by the glass bead method (Ranhand, 1974). PCR products were purified using the QIAquick PCR Purification kit (Qiagen). DNA restriction enzymes were used under the conditions specified by the manufacturer (New England BioLabs).
Table 1. Oligonucleotides used in this study.
| Oligonucleotide (5′→3′) | Sequence |
| Construction of ΔadhA (SGO_0565) | |
| adhA-F1 | AAGTTTGAGGAACCTTGATGAT |
| adhA-R1 | CGTAGGATCCCTTCGTGACCAAGGAT |
| adhA-R2b | ATGAGACTTTGGCATGAGGCC |
| Construction of ΔadhB (SGO_1774) | |
| adhB-F1b | GCCTTTATTTCCGACGACCGCG |
| adhB-R1 | AGCTGGATCCAACCGCTCCGTCACCA |
| adhB-F2 | GCATCTCGAGCAGCCTCCGTCACGACTT |
| adhB-R2 | TATCAGCGGCCGGTGCCTTGA |
| Construction of ΔadhE (SGO_0113) | |
| adhE-F0 | TAAGCGAAAGTGTTTACAAA |
| adhE-R1 | GCACCGAAGCAGAATTCTTC |
| adhE-F1 | GATCGGATCCACCCATCTGCTCAAGAA |
| adhE-R3 | CTGTCTTAGCTGGACGTGTAC |
| Construction of SGO_0273 insertion mutant | |
| Sgo-273-F2 | GGAACAATGATCAGTGACCCTGG |
| Sgo-273-R2 | GTACTAGCGTTCCAATAGCTGTGC |
| Construction of SGO_0440 insertion mutant | |
| Sgo-440-F1 | AGTTGGCGATCGGGTAACAG |
| Sgo-440-R1 | GAACAGCAGCCAAAGCTTGC |
| Construction of SGO_0841 insertion mutant | |
| Sgo-841-F1 | CTGAGTTAGCTGCAGTTCCT |
| Sgo-841-R1 | TCTTGATTGACTTGAGCTGAATG |
The V2016 adhA deletion mutant was obtained by transforming the wild-type V2016 strain with a 5.5 kb linear DNA construct containing two DNA ends flanking the adhA gene and a 3.5 kb tetracycline resistance cassette. To obtain the cassette, plasmid pAK560 originally derived from pVA981 (Lindler & Macrina, 1986) was digested with BamHI and SalI. The left-end DNA was obtained by PCR with primers adhA-F1 and adhA-R1 to generate a DNA fragment of 1.06 kb, which was digested with BamHI. The right-end DNA was obtained by PCR with primers adhA-F1 and adhA-R2b to generate a 2.6 kb DNA, which was cut with SalI and subjected to agarose gel electrophoresis. The DNA fragment of 1.1 kb was isolated and purified. After ligation with the tetracycline resistance cassette at 16 °C for 16 h with T4 DNA ligase, the ligated DNA was purified with a QIAquick PCR Purification kit and served as the template to perform a long PCR with the EmeraldAmp Max enzyme (Takara Bio). The resulting 5.5 kb PCR product was used to transform S. gordonii V2016. The ΔadhA mutant was selected on TH agar containing tetracycline at 15 µg ml−1 and confirmed by PCR with primers adAh-F1 and adhA-R2b (Lau et al., 2002).
Likewise, the V2016 ΔadhB mutant was obtained by transforming the wild-type V2016 with a linear DNA construct (4.4 kb) containing two DNA ends flanking the adhB gene and a 2.1 kb erythromycin resistance cassette. To obtain the cassette, plasmid pAK488 originally derived from pVA891 (Macrina et al., 1983) was digested with BamHI and XhoI. The left end 1.6 kb DNA fragment was obtained by PCR with primers adhB-F1b and adhB-R1 and was digested with BamHI. The right-end DNA (1.15 kb) obtained with primers adhB-F2 and adhB-R2 was digested with XhoI. The two DNA fragments were ligated to the erythromycin resistance cassette. The ligated DNA was amplified with a long PCR with the EmeraldAmp Max enzyme. The PCR product (4.4 kb) was used to transform S. gordonii V2016. The ΔadhB mutant was selected on TH agar containing erythromycin at 10 µg ml−1 and was confirmed by PCR with primers adhB-F1 and adhB-R2.
The V2016 ΔadhE mutant was obtained by transforming the wild-type V2016 with a linear DNA construct containing two DNA ends flanking the adhE gene and a 3.5 kb kanamycin resistance cassette. To obtain the cassette, plasmid pSF151 (Tao, 1998) was digested with EcoRI and PstI. The left-end DNA (1.0 kb) obtained by PCR with primers adhE-F0 and adhE-R1 was digested with EcoRI. The right-end DNA was obtained with primers adhE-F1 and adhE-R3 to generate a 1.9 kb DNA, which was cut with PstI and subjected to agarose gel electrophoresis. The DNA fragment of 1.1 kb was isolated from the agarose gel and purified. The two DNA ends were ligated to the kanamycin resistance cassette. The ligated DNA was amplified with a long PCR with the EmeraldAmp Max enzyme. The PCR product was about 5.6 kb and was used to transform S. gordonii V2016. The ΔadhE mutant was selected on TH agar containing kanamycin at 250 µg ml−1 and was confirmed by PCR with primers adhE-F0 and adhE-R3.
After the three Δadh mutants were constructed, chromosomal DNAs were isolated from each mutant and used to transform other mutants to create three double mutants, adhAB, adhAE, adhBE and a triple mutant, adhABE.
Analysis of ADH and ALDH activities.
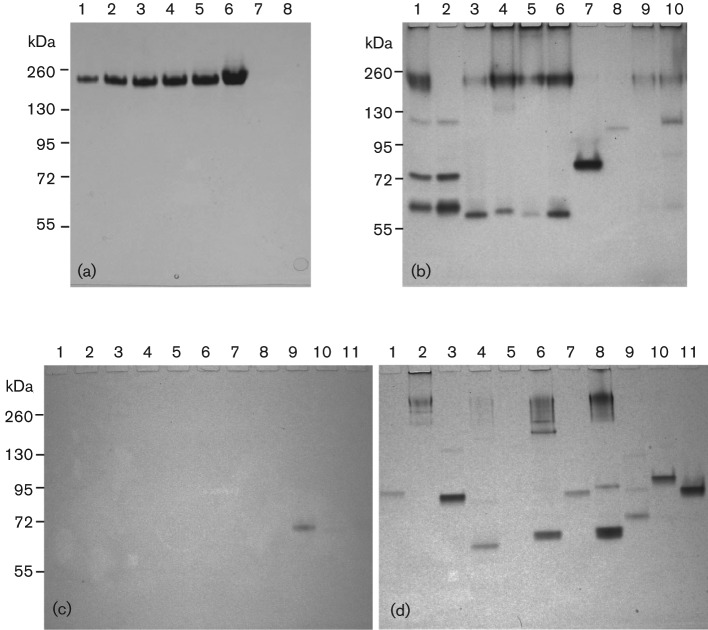
ADH and ALDH were determined by a specific enzyme activity gel assay (zymogram) improved from several methods described previously (Gabriel, 1971; Grell et al., 1968; Muto et al., 2000). Nitro blue tetrazolium (NBT) in the presence of phenazine methosulfate (PMS) reacts with NADP produced by dehydrogenases to produce an insoluble blue-purple formazan. This NBT-PMS reaction can be used to visualize ADH and ALDH in polyacrylamide gels. Bacteria were grown overnight in 15 ml THY broth supplemented with appropriate testing substrate (1 % ethanol, 1 % methanol, 0.2 % 1-propanol, 0.2 % 2-propanol, 0.2 % 1-butanol, 0.2 % tertiary-butanol or 0.5 % threonine) to induce the expression of each substrate-metabolizing enzyme. The metallic cofactors Fe2+ and Zn2+ required by these enzymes were provided by addition of 0.01 % FeCl2 (w/v) and 0.01 % ZnSO4 (w/v). The bacterial cells were harvested, washed and resuspended in PBS (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4; pH 7.4). The cells were treated with lysozyme for 1 h at 37 °C before being disrupted by glass beads with a TurboMix beater for 5 min. Aliquots of the crude bacterial lysates were electrophoresed on 10 % Criterion TGX pre-cast gels (Bio-Rad) in the tris-glycine buffer. The gel was briefly washed in PBS after electrophoresis. The ADH bands were visualized by incubating the gel for 1 h at 37 °C in the dark in 100 mM sodium phosphate buffer (pH 8.8) containing 0.4 mg ml−1 NAD, 0.008 mg ml−1 PMS, and 0.2 mg ml−1 NBT. To improve the detection sensitivity, ethanol, 1-butanol, tertiary-butanol, 1-propanol, 2-propanol and methanol were added at 1.5 M (Membrillo-Hernandez et al., 2000; Muto et al., 2000). l-threonine was added at 10 mM in 100 mM glycine-KCl/KOH buffer (pH 10.4) (Ueatrongchit & Asano, 2011). For determining the ALDH activity, the gel was incubated in 100 mM potassium phosphate buffer pH 7.4 with the same ingredients as above and containing 100 mM acetaldehyde (Membrillo-Hernandez et al., 2000). Yeast ADH and ALDH (Sigma-Aldrich) were used as positive controls. In addition to S. gordonii V2016, three other S. gordonii strains (V288, CHI, 110-3) and five S. sanguinis laboratory strains (133-79, S7, Blackburn, SK36, ATCC 10556) and 11 oral Streptococcus isolates, including four strains producing only acetaldehyde, two strains producing both acetic acid and acetaldehyde and five strains producing only acetic acid from ethanol (see legend to Fig. 4 for strain names), were also analysed for both ADH and ALDH activities.
Fig. 4.
(a) ALDH zymogram: 1–6, Saccharomyces cerevisiae ALDH controls: 1, 0.1 U; 2, 0.25 U; 3, 0.5 U; 4, 0.75 U; 5, 1 U; 6, 3 U; 7, blank; 8, S. gordonii V2016wt. (b) ADH zymogram: 1–3, S. gordonii V2016wt, V2016ΔadhE and V288; 4 and 5, S. sanguinis S7 and Blackburn; 6 and 7, S. gordonii CHI and 110-3; 8–10, S. sanguinis 133-79, SK36 and ATCC 10556. (ALDH zymogram of these strains was negative; data not shown). (c) ALDH zymogram: 1–4 (produced only acetaldehyde from ethanol), S. salivarius 101-1; S. sanguinis 104-5; S. salivarius 109-2, and S. sanguinis 109-3; 5 and 6 (produced both acetaldehyde and acetic acid from ethanol), S. oralis 108 and S. mitis 110-5; 7–11 (produced only acetic acid from ethanol), S. salivarius 101-7, S. mitis 104-4, S. salivarius 107-2, 110-1 and 110-4. (d) ADH zymogram of the same 11 strains displayed in (c). Note: only S. salivarius 107-2 displayed an ALDH band, which is different from AdhE. S. oralis 108 did not show detectable ADH.
Growth study.
The V2016 wild-type and seven mutants, ΔadhA, ΔadhB, ΔadhE, ΔadhAB, ΔadhAE, ΔadhBE and ΔadhABE, were analysed for growth in THY and THY supplemented with 1 % ethanol or 1 % acetaldehyde. Each strain was grown in three tubes of 5 ml THY broth overnight with serial diluted inoculations. On the second morning, the culture at mid-exponential phase was transferred with a 1 : 100 dilution to the three different testing media and incubated in a 37 °C water bath. The optical density at 600 nm was measured every 30 min with a Genesys 20 Spectrophotometer. To better present the bacterial growth data, optical density readings as a function of time in the exponential growth phase were converted to doubling time.
Results
Isolation of strains producing acetaldehyde from ethanol
Bacterial metabolism of ethanol involves two steps. The first step is conversion of ethanol to acetaldehyde by ADH. The second step is conversion of acetaldehyde to acetic acid by ALDH. If a bacterium has both ADH and ALDH, it can convert ethanol to acetic acid, which reduces pH and can be detected by colour change in the purple broth. However, if a bacterium has only ADH but no ALDH, it can only produce acetaldehyde from ethanol without further converting it to acetic acid.
A total of 52 oral Streptococcus strains (14 laboratory and 38 clinical strains) were analysed for their capacity for acetic acid and acetaldehyde production from ethanol. There were only two species of laboratory strains, S. gordonii and S. sanguinis, while the 16S rDNA analysis of clinical strains revealed five species, S. gordonii, S. mitis, S. oralis, S. salivarirus and S. sanguinis. Only 17 strains of S. mitis, S. oralis and S. salivarius produced acetic acid from ethanol, while 19 strains of all five Streptococcus species produced acetaldehyde. Some strains produced only acetic acid without detectable acetaldehyde, while others produced only acetaldehyde without detectable acetic acids, and still others produced both or neither from ethanol. Among all the strains tested, S. gordonii V2016, S. oralis 108 and S. mitis 110-5 showed the most abundant production of acetaldehyde. However, S. oralis 108 and S. mitis 110-5 also showed production of acetic acid from ethanol, but S. gordonii V2016 showed only acetaldehyde production from ethanol. Therefore, V2016 was selected for further study of its enzymes involved in acetaldehyde production from ethanol.
Mutant construction in S. gordonii V2016
By in silico analysis of the S. gordonii genome (Vickerman et al., 2007), we have identified three genes: adhA (SGO_0565; 1023 bp), adhB (SGO_1774; 1038 bp), and adhE (acdH, SGO_0113; 2652 bp) that encode putative ADHs. These three genes were subjected to PCR-ligation mutagenesis with different antibiotic resistance markers to achieve allelic exchange (Lau et al., 2002). Each mutant was confirmed genotypically by PCR. Due to insertion of the antibiotic resistance cassette, the size of the PCR DNA fragment became larger in the mutant than that in the wild-type with primers flanking the insertion site (not shown). By combinational DNA transformation, three double gene deletion mutants, ΔadhAB, ΔadhAE and ΔadhBE, and one triple gene deletion mutant, ΔadhABE, were obtained. Additionally, three more genes, SGO_0273 (1005 bp), SGO_0440 (1047 bp) and SGO_0841 (993 bp), which encode putative Zn-binding dehydrogenases, were mutated by insertion duplication with the plasmid pSF151 (Tao, 1998).
Acetaldehyde production from ethanol by S. gordonii adh mutants
The acetaldehyde production by the wild-type and various gene-deletion mutants of V2016 was tested on the PBB-Schiff’s agar plate. A positive reaction to acetaldehyde is indicated by a pinkish red colour, and a negative reaction by white colour. As shown in Fig. 1, the wild-type strain displayed the strongest production, the single gene knockout mutants a slightly reduced production, and double gene knockout mutants a much reduced production; the triple knockout mutant failed to produce acetaldehyde.
Fig. 1.
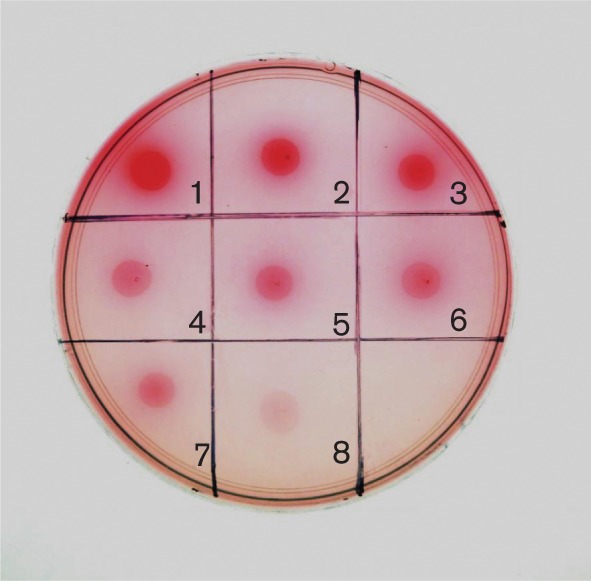
Acetaldehyde production by S. gordonii on PBB-Schiff’s agar. 1, V2016wt; 2, ΔadhA; 3, ΔadhB; 4, ΔadhAB; 5, ΔadhE; 6, ΔadhAE; 7, ΔadhBE; 8, ΔadhABE.
ADH profiles of the S. gordonii V2016 wild-type and adh mutants
The ADH activities of V2016 and its various adh mutants were analysed with the NBT-PMS zymogram method. First, the approximate size of each ADH was estimated by testing each adh gene deletion mutant against the wild-type on an ADH zymogram. As shown in Fig. 2, the wild-type V2016 displays all three functional ADH enzymes (lane 1). The ΔadhA mutant lacks three bands between 55 and 72 kDa (lane 2). The ΔadhB mutant misses a single band near 130 kDa (lane 3). The ΔadhE mutant misses top two bands around 260 kDa (lane 4). Because the actual molecular sizes of these enzymes cannot be determined by the native polyacrylamide gel the sizes and shapes of these enzymes can only be estimated. For example, since three bands are related to AdhA, this enzyme may take three different forms (e.g. monomer, dimer and/or trimer).
Fig. 2.
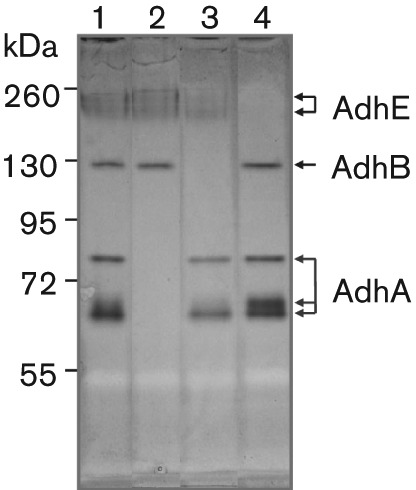
S. gordonii V2016 ADH distribution analysis by zymogram without added Fe2+ and Zn2+: 1, V2016wt; 2, ΔadhA; 3, ΔadhB; 4, ΔadhE. Note: missing band(s) of each Δadh mutant indicates the location(s) of the target ADH.
The DNA sequences of adhA and adhB both show a Zn-binding domain, but only AdhA showed enhanced ADH activity after Zn2+ supplementation and only in the absence of AdhE (Fig. 3a). Supplementing Fe2+ significantly enhanced AdhE activity (Fig. 3a, lanes 1–4) suggesting that the AdhE protein is an Fe-dependent ADH. However, in mutants with adhE inactivation (Fig 3, lanes 5 and 7), the activity of AdhA is enhanced, but the activity of AdhB is not. The AdhB protein apparently has one (Fig. 2) or two conformations (Fig. 3). The second AdhB band showed up only when Zn2+ was added to the growth medium and detection buffer, and when AdhE was present.
Fig. 3.
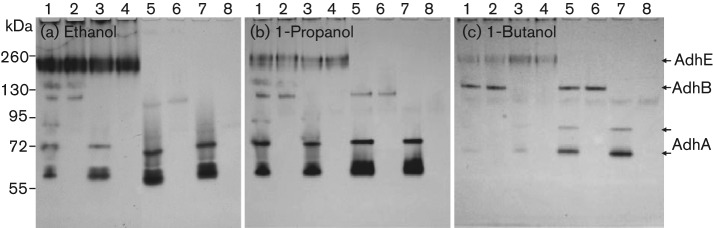
Analysis of substrate preference of S. gordonii V2016 ADH. Fe2+ and Zn2+ were added to the growth medium and zymogram detection solution. 1, V2016wt; 2, ΔadhA; 3, ΔadhB; 4, ΔadhAB; 5, ΔadhE; 6, ΔadhAE; 7, ΔadhBE; 8, ΔadhABE. Note: in the wild-type strain, ethanol is the preferred substrate for AdhE, 1-propanol is the preferred substrate for AdhA and 1-butanol is the preferred substrate for AdhB .
S. gordonii V2016 ADH substrate specificities
In addition to ethanol, we also tested other alcohols, methanol, 1-propanol, 2-propanol, 1-butanol and tertiary-butanol, and the amino acid threonine with the NBT-PMS zymogram. Except for methanol and tertiary-butanol, which showed no activity, all other tested substrates showed varied activities with these three primary ADHs (Figs 3 and 4). The preferred substrates for AdhA, AdhB and AdhE were 1-propanol, 1-butanol and ethanol, respectively. Additionally, two new dehydrogenases for threonine and 2-propanol were observed. An insertion-inactivation study showed that the dehydrogenase encoded by SGO_0440 was specific for threonine. However, none of the mutants had lost the enzyme activity for 2-propanol. The specificities of five dehydrogenases to various tested substrates are listed in Table 2.
Table 2. Substrate specificity of S. gordonii V2016 dehydrogenases.
| Substrate | AdhA | AdhB | AdhE | S-AdhA | TdhA |
| Acetaldehyde | − | − | − | − | − |
| Methanol | − | − | − | − | − |
| Ethanol | +* | + | +++ | − | − |
| 1-Propanol | +++ | + | + | − | − |
| 2-Propanol | + | − | − | +++ | − |
| 1-Butanol | +* | +++ | + | ± | − |
| tert-Butanol | − | − | − | − | − |
| Threonine | + | ± | − | − | ++ |
When AdhE was present, AdhA activity was low, but when AdhE was absent, AdhA activity was high.
S. gordonii V2016 does not have detectable ALDH activity
Because adhE has homology to the ALDH/ADH dual function AdhE in other bacteria (Koo et al., 2005), it is important to test whether S. gordonii V2016 AdhE also has dehydrogenase activity for acetaldehyde. As shown in Fig. 4a, we tested S. gordonii V2016 with the ALDH of Saccharomyces cerevisiae (Sigma) as the positive control. However, under this assay condition, we did not observe ALDH activity from proteins isolated from S. gordonii V2016 (Fig. 4a, lane 8).
ADH/ALDH profiles vary among different strains of oral Streptococcus
As shown in Fig. 4b, among four S. gordonii strains tested, only V2016 showed all three ADHs. S. gordonii V288 and CHI showed AdhA and AdhE, but no AdhB. However, S. gordonii 110-3 showed only one ADH similar to AdhA. Among five S. sanguinis strains tested, S7 and Blackburn showed only AdhA and AdhE. 133-79 showed only a weak AdhB. SK36 showed only a weak AdhE. Although ATCC 10556 showed four bands representing AdhA, B and E, their activities are relatively weak. As shown in Fig 4(c, d), three groups of oral Streptococcus strains, including four strains producing only acetaldehyde, two strains producing both acetic acid and acetaldehyde, and five strains producing only acetic acid from ethanol, were tested for both ALDH and ADH. Although most strains showed one or more ADHs, only one strain, S. salivarius 107-2 showed an ALDH activity band, which is significantly smaller than AdhE.
Effect of adh gene deletions on bacterial growth in medium containing ethanol or acetaldehyde
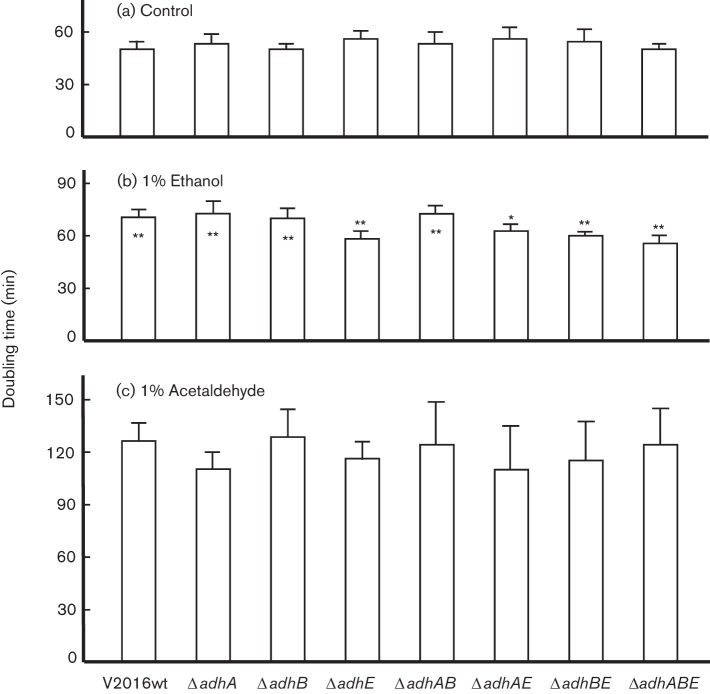
To evaluate if deletions of adh genes could affect bacterial growth in medium containing ethanol or acetaldehyde, we performed a growth study for S. gordonii V2016 and its seven adh mutants. The results are presented as doubling time (Fig. 5). Deletion of any of these three adh genes did not show an apparent difference in the growth doubling times in THY without supplemental ethanol. However, when ethanol was added at 1 %, the growth slowed with significantly longer doubling times for the wild-type and the ΔadhA and/or ΔadhB mutants. The four mutants containing adhE deletion had largely the same doubling times when growing in THY alone and in THY supplemented with 1 % ethanol. In comparison with the wild-type, these four mutants showed significantly shorter doubling times in THY supplemented with 1 % ethanol. All eight strains displayed significantly longer doubling times in THY supplemented with 1 % acetaldehyde.
Fig. 5.
Doubling times of S. gordonii V2016wt and seven Δadh mutants grown in THY or THY containing 1 % ethanol or 1 % acetaldehyde. * Statistical differences by Student’s t-test (*P<0.05; **P<0.01). When * is on top of the data bar, it represents significant difference between the doubling time of this strain and its wild-type growing in the same medium. When * is inside the data bar, it represents significant difference between the same strain growing in THY and THY containing 1 % ethanol. All strains growing in THY containing 1 % acetaldehyde had significantly longer doubling times than when grown in other media. Each data bar represents the mean of five measurements plus standard deviation.
Discussion
Ethanol consumption (Bagnardi et al., 2001) and poor dental health (Homann, 2001; Homann et al., 2001) are two major risk factors for cancers of the upper aerodigestive tract including the oral cavity, throat, voice-box and oesophagus. Although the exact mechanism by which ethanol consumption causes cancer is unknown, local production of carcinogenic agents by micro-organisms is suspected (Meurman & Uittamo, 2008). Ethanol itself is not carcinogenic, but it can be oxidized to carcinogenic acetaldehyde in the oral cavity by ADHs of oral micro-organisms (Muto et al., 2000). It has been reported that many species of oral streptococci can produce acetaldehyde from ethanol (Kurkivuori et al., 2007). However, little is known about ethanol metabolic enzymes in these bacteria. To our knowledge, this is the first report on molecular characterization of ethanol metabolic enzymes in an oral streptococcal species.
With acetaldehyde detection agar we isolated an oral strain of Streptococcus that produces a high level of acetaldehyde from ethanol. We chose to analyse a high acetaldehyde-producing strain instead of an average acetaldehyde producer because the former can allow us to study a broader range enzymes involved in the bacterial production of acetaldehyde from ethanol. However, existing zymogram methods were not sensitive enough to detect multiple ADH activities from crude bacterial samples. Therefore, we developed a more sensitive method, the NBT-PMS zymogram, which allowed us to detect multiple ADHs simultaneously on the same gel with crude bacterial lysates. By knocking out three adh genes individually and in various combinations, we found that S. gordonii V2016 has three primary ADHs, AdhA, AdhB and AdhE, which all recognize ethanol as a substrate, but their preferred substrates were 1-propanol, 1-butanol and ethanol, respectively. Due to different substrate preferences, their roles in bacterial ethanol metabolism may vary. Additionally, we have also identified a secondary ADH, S-AdhA, which specifically recognizes the secondary alcohol, 2-propanol, and a dehydrogenase specific for threonine. These two dehydrogenases, however, do not recognize ethanol as their substrate (Figs 2 and 3a), despite the fact that AdhA recognizes both threonine and 2-propanol as its substrate (Fig. 6).
Fig. 6.
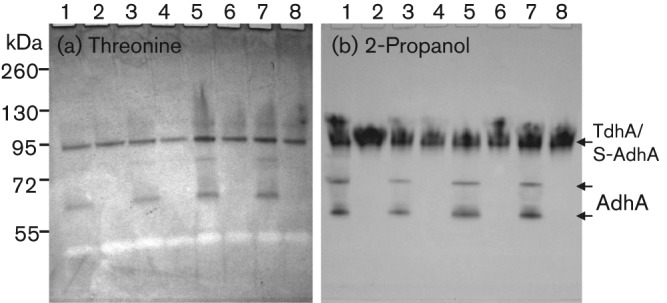
Identification of two novel dehydrogenases in S. gordonii, the threonine dehydrogenase (TdhA) and the secondary alcohol dehydrogenase (S-AdhA) by zymograms. 1, V2016wt; 2, ΔadhA; 3, ΔadhB; 4, ΔadhAB; 5, ΔadhE; 6, ΔadhAE; 7, ΔadhBE; 8, ΔadhABE. Note: AdhA reacted with both threonine and 2-propanol because mutants with ΔadhA did not show these bands.
The insertion-inactivation study confirmed that the dehydrogenase encoded by SGO_0440 was specific for threonine. We therefore named this gene tdhA, encoding the threonine dehydrogenase. However, none of the mutations in the three loci encoding putative dehydrogenases, SGO_0273, SGO_0440, and SGO_0841, had inactivated the enzyme activity for 2-propanol. Therefore, the gene encoding the dehydrogenase specific for the secondary alcohol remains to be determined. The substrate specificity analysis (Table 2) showed that three ADHs of S. gordonii V2016 all recognize a broad range of substrates besides ethanol, but the activities of S-AdhA and TdhA were quite specific to their preferred substrates. It appears to be disadvantageous for a bacterium to have multiple ADHs that all produce the toxic metabolite from ethanol. Having multiple different ADHs may offer the bacterium competitive growth advantage in the environment by making it capable of utilizing multiple different nutrient substrates.
There may be cross-regulation of activity among the three ADHs in S. gordonii V2016. As shown in Fig. 2, when the activity of AdhE was weak due to the lack of its cofactor Fe2+ or missing due to gene deletion, the activities of AdhA and AdhB were relatively strong. However, when Fe2+ and Zn2+ were added to the growth medium, the AdhE activity increased substantially (Fig. 3a, lanes 1–4) but the activities of both AdhA and AdhB were suppressed, possibly by the increased activity of AdhE. However, when the adhE gene was deleted, the activity of AdhA increased (Fig. 3a, lanes 5, 7), but not AdhB, which appeared to be relatively independent from AdhE regulation. A similar scenario was also observed when 1-propanol (Fig. 3b) and 1-butanol were used as substrates (Fig. 3c). These results suggest that AdhE may be the major ADH in S. gordonii. When its activity is upregulated, the activities of other ADHs, especially the AdhA, are reduced.
The significant increase in bacterial doubling time of all eight strains indicates that acetaldehyde is more toxic than ethanol. A similar effect is also reported in a study with yeast (Brendel et al., 2010). Therefore, mutants that lack the enzyme for the production of acetaldehyde can be more tolerant to ethanol than the wild-type (Brown et al., 2011). The growth study (Fig. 5) showed that all four mutants containing ΔadhE when grown in THY containing 1 % ethanol had no significant increase in doubling times comparing with growth in control THY. However, the ΔadhA and/or ΔadhB mutants showed increased doubling times like the wild-type when grown in THY containing 1 % ethanol. This suggests that AdhA and AdhB may be less involved in acetaldehyde production from ethanol than AdhE in S. gordonii V2016.
The S. gordonii genomic data (Vickerman et al., 2007) showed that this bacterium has a gene (acdH, SGO_0113) encoding the putative dual functional ALDH/ADH (AdhE). However, S. gordonii V2016 did not show any detectable ALDH activity with the optimized NBT-PMS zymogram. To make sure that this method is sensitive enough to detect microbial ALDH, we used S. cerevisiae ALDH as a positive control. The zymogram detected ALDH activity as low as 0.1 U. This method has also detected ALDH from another oral Streptococcus strain, S. salivarius 107-2 (Fig. 4c). Therefore, the zymogram method should be reliable and the negative result indicated that S. gordonii V2016, as well as other tested oral Streptococcus strains, did not have any detectable ALDH activity. Because the adhE gene of these oral streptococci is highly homologous to adhE genes in other bacteria (Koo et al., 2005) that encode a bifunctional ALDH/ADH, there might be a mutation(s) in its ALDH domain. This finding, together with findings on oral Neisseria (Muto et al., 2000), indicates that genetic polymorphisms in ALDH in bacteria may exist similar to those seen in humans (Druesne-Pecollo et al., 2009; Hiyama et al., 2007). Because most tested oral streptococcal strains showed multiple ADHs, but no ALDH, the enzyme distribution bias may contribute to their excessive production of acetaldehyde from ethanol.
The ADH zymograms (Fig. 4b, d) showed great ADH polymorphism among 20 oral Streptococcus strains representing five different species. Because bacterial ADHs show broad substrate preferences and their ADH profiles vary, these bacteria may metabolize ethanol differently. One of the acetic acid producers, S. salivarius 107-2, showed a positive band for ALDH (Fig. 4c). Its size is not within the range of AdhE. It may be a novel ALDH. Because other acetic acid producers did not show ALDH activity bands, these bacteria either have very weak ALDH or use different mechanisms to produce acetic acid from ethanol. In addition to enzymic pathways, ethanol can also be oxidized by non-enzymic free radical pathways to produce acetaldehyde (Reinke et al., 1994; Welch et al., 2002). This might explain why S. oralis 108 showed no detectable ADH activity (Fig. 4d) but still produced excessive acetaldehyde from ethanol.
AdhE is highly conserved and may have multiple functions depending upon different bacterial species. For example, in Leuconostoc, AdhE is a bifunctional ALDH/ADH (Koo et al., 2005). In Escherichia coli (Nnyepi et al., 2007) and Streptococcus bovis (Asanuma et al., 2004), AdhE has three distinct enzymic activities: ADH, acetaldehyde-CoA dehydrogenase and pyruvate formate-lyase deactivase. In Listeria, AdhE is also a major adhesion protein and is located on the cell surface (Jagadeesan et al., 2010). In Thermoanaerobacter mathranii, AdhE is a bifunctional ALDH/ADH responsible for ethanol production (Yao & Mikkelsen, 2010).
In the East Asian population of humans, a rather high percentage (up to 30 %) carry a defective ALDH2, which is caused by a point mutation resulting in a Glu to Lys substitution at the amino acid position 487, and is referred to as ALDH2*487Lys [previous symbol: ALDH2*2 (Lewis & Smith, 2005; Yokoyama et al., 1998)]. In this study, we observed that in most strains of oral Streptococcus tested, the AdhE protein has only ADH and no ALDH activity. This is also true in Neisseria (Muto et al., 2000). This indicates that the adhE gene of these bacteria might have lost its ability to express functional ALDH during the course of evolution. Based on a recent study on bacterial evolution (Martincorena et al., 2012), if a gene is non-essential for bacterial survival, more mutations can be accumulated in comparison with genes that are essential. Because adhE is non-essential, a random mutation in adhE could be allowed and be passed down to the offspring. The questions are how many bacterial species carry such a mutation in their adhE gene and which base substitution(s) may inactivate its ALDH activity.
In summary, we have analysed S. gordonii V2016, a strain that produced abundant acetaldehyde from ethanol. We found that this bacterium displayed three different ADHs that all oxidize ethanol to acetaldehyde, but did not show a detectable ALDH. Analyses of 19 additional strains of S. gordonii, S.mitis, S. oralis, S. salivarius and S. sanguinis all showed similarly varied enzyme profiles of ADHs without detectable ALDH except one strain. Therefore, activities of multiple ADHs but no ALDH in most oral streptococci may contribute to the excessive production of acetaldehyde from ethanol. As a result, these bacteria can contribute to alcohol-associated oral and oesophageal carcinogenesis in the human host.
Acknowledgements
This work was supported by a grant from NIH National Cancer Institute (CA162537). We thank Dr Mark Herzburg for sending us 14 oral Streptococcus laboratory strains.
Abbreviations:
- ADH
alcohol dehydrogenase
- ALDH
acetaldehyde dehydrogenase
- NBT
nitro blue tetrazolium
- PMS
phenazine methosulfate
References
- Abnet C. C., Kamangar F., Islami F., Nasrollahzadeh D., Brennan P., Aghcheli K., Merat S., Pourshams A., Marjani H. A., et al. (2008). Tooth loss and lack of regular oral hygiene are associated with higher risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 17, 3062–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma N., Yoshii T., Hino T. (2004). Molecular characteristics and transcription of the gene encoding a multifunctional alcohol dehydrogenase in relation to the deactivation of pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Arch Microbiol 181, 122–128. [DOI] [PubMed] [Google Scholar]
- Bagnardi V., Blangiardo M., La Vecchia C., Corrao G. (2001). Alcohol consumption and the risk of cancer: a meta-analysis. Alcohol Res Health 25, 263–270. [PMC free article] [PubMed] [Google Scholar]
- Brendel M., Marisco G., Ganda I., Wolter R., Pungartnik C. (2010). DNA repair mutant pso2 of Saccharomyces cerevisiae is sensitive to intracellular acetaldehyde accumulated by disulfiram-mediated inhibition of acetaldehyde dehydrogenase. Genet Mol Res 9, 48–57. [DOI] [PubMed] [Google Scholar]
- Brown S. D., Guss A. M., Karpinets T. V., Parks J. M., Smolin N., Yang S., Land M. L., Klingeman D. M., Bhandiwad A., et al. (2011). Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci U S A 108, 13752–13757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. (1987). Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol 169, 2591–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E., Chen T., Izard J., Paster B. J., Tanner A. C., Yu W. H., Lakshmanan A., Wade W. G. (2010). The human oral microbiome. J Bacteriol 192, 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druesne-Pecollo N., Tehard B., Mallet Y., Gerber M., Norat T., Hercberg S., Latino-Martel P. (2009). Alcohol and genetic polymorphisms: effect on risk of alcohol-related cancer. Lancet Oncol 10, 173–180. [DOI] [PubMed] [Google Scholar]
- Espina N., Lima V., Lieber C. S., Garro A. J. (1988). In vitro and in vivo inhibitory effect of ethanol and acetaldehyde on O6-methylguanine transferase. Carcinogenesis 9, 761–766. [DOI] [PubMed] [Google Scholar]
- Gabriel O. (1971). Locating enzymes on gels. Methods Enzymol 22, 578–604. [Google Scholar]
- Grell E. H., Jacobson K. B., Murphy J. B. (1968). Alterations of genetics material for analysis of alcohol dehydrogenase isozymes of Drosophila melanogaster. Ann N Y Acad Sci 151, 441–455. [DOI] [PubMed] [Google Scholar]
- Hiyama T., Yoshihara M., Tanaka S., Chayama K. (2007). Genetic polymorphisms and esophageal cancer risk. Int J Cancer 121, 1643–1658. [DOI] [PubMed] [Google Scholar]
- Homann N. (2001). Alcohol and upper gastrointestinal tract cancer: the role of local acetaldehyde production. Addict Biol 6, 309–323. [DOI] [PubMed] [Google Scholar]
- Homann N., Tillonen J., Rintamäki H., Salaspuro M., Lindqvist C., Meurman J. H. (2001). Poor dental status increases acetaldehyde production from ethanol in saliva: a possible link to increased oral cancer risk among heavy drinkers. Oral Oncol 37, 153–158. [DOI] [PubMed] [Google Scholar]
- Jagadeesan B., Koo O. K., Kim K. P., Burkholder K. M., Mishra K. K., Aroonnual A., Bhunia A. K. (2010). LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria, promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 156, 2782–2795. [DOI] [PubMed] [Google Scholar]
- Koo O. K., Jeong D. W., Lee J. M., Kim M. J., Lee J. H., Chang H. C., Kim J. H., Lee H. J. (2005). Cloning and characterization of the bifunctional alcohol/acetaldehyde dehydrogenase gene (adhE) in Leuconostoc mesenteroides isolated from kimchi. Biotechnol Lett 27, 505–510. [DOI] [PubMed] [Google Scholar]
- Kurkivuori J., Salaspuro V., Kaihovaara P., Kari K., Rautemaa R., Grönroos L., Meurman J. H., Salaspuro M. (2007). Acetaldehyde production from ethanol by oral streptococci. Oral Oncol 43, 181–186. [DOI] [PubMed] [Google Scholar]
- Lau P. C., Sung C. K., Lee J. H., Morrison D. A., Cvitkovitch D. G. (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49, 193–205. [DOI] [PubMed] [Google Scholar]
- Lewis S. J., Smith G. D. (2005). Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev 14, 1967–1971. [DOI] [PubMed] [Google Scholar]
- Lillie R. D. (1977). H. J. Conn's Biological Stains: a Handbook on the Nature and Uses of the Dyes Employed in the Biological Laboratory, 9th edn Baltimore, MD: The Williams and Wilkins Co. [Google Scholar]
- Lindler L. E., Macrina F. L. (1986). Characterization of genetic transformation in Streptococcus mutans by using a novel high-efficiency plasmid marker rescue system. J Bacteriol 166, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Evans R. P., Tobian J. A., Hartley D. L., Clewell D. B., Jones K. R. (1983). Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25, 145–150. [DOI] [PubMed] [Google Scholar]
- Martincorena I., Seshasayee A. S., Luscombe N. M. (2012). Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature 485, 95–98. [DOI] [PubMed] [Google Scholar]
- Marttila E., Bowyer P., Sanglard D., Uittamo J., Kaihovaara P., Salaspuro M., Richardson M., Rautemaa R. (2013). Fermentative 2-carbon metabolism produces carcinogenic levels of acetaldehyde in Candida albicans. Mol Oral Microbiol. 10.1111/omi.12024. [Epub ahead of print] 10.1111/omi.12024 [DOI] [PubMed] [Google Scholar]
- Membrillo-Hernandez J., Echave P., Cabiscol E., Tamarit J., Ros J., Lin E. C. (2000). Evolution of the adhE gene product of Escherichia coli from a functional reductase to a dehydrogenase. Genetic and biochemical studies of the mutant proteins. J Biol Chem 275, 33869–33875. [DOI] [PubMed] [Google Scholar]
- Meurman J. H., Uittamo J. (2008). Oral micro-organisms in the etiology of cancer. Acta Odontol Scand 66, 321–326. [DOI] [PubMed] [Google Scholar]
- Muto M., Hitomi Y., Ohtsu A., Shimada H., Kashiwase Y., Sasaki H., Yoshida S., Esumi H. (2000). Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: implications for carcinogenesis in upper aerodigestive tract. Int J Cancer 88, 342–350. [PubMed] [Google Scholar]
- Nnyepi M. R., Peng Y., Broderick J. B. (2007). Inactivation of E. coli pyruvate formate-lyase: role of AdhE and small molecules. Arch Biochem Biophys 459, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara A. M., Shanahan F. (2006). The gut flora as a forgotten organ. EMBO Rep 7, 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obe G., Anderson D. (1987). International Commission for Protection against Environmental Mutagens and Carcinogens. ICPEMC Working Paper No. 15/1. Genetic effects of ethanol. Mutat Res 186, 177–200. [DOI] [PubMed] [Google Scholar]
- Ranhand J. M. (1974). Simple, inexpensive procedure for the disruption of bacteria. Appl Microbiol 28, 66–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke L. A., Rau J. M., McCay P. B. (1994). Characteristics of an oxidant formed during iron (II) autoxidation. Free Radic Biol Med 16, 485–492. [DOI] [PubMed] [Google Scholar]
- Salaspuro M. P. (2003). Acetaldehyde, microbes, and cancer of the digestive tract. Crit Rev Clin Lab Sci 40, 183–208. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Secretan B., Straif K., Baan R., Grosse Y., El Ghissassi F., Bouvard V., Benbrahim-Tallaa L., Guha N., Freeman C., et al. (2009). A review of human carcinogens–Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol 10, 1033–1034. [DOI] [PubMed] [Google Scholar]
- Tao L. (1998). Streptococcal integration vectors for gene inactivation and cloning. Methods Cell Sci 20, 59–64. [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueatrongchit T., Asano Y. (2011). Highly selective L-threonine 3-dehydrogenase from Cupriavidus necator and its use in determination of L-threonine. Anal Biochem 410, 44–56. [DOI] [PubMed] [Google Scholar]
- Uittamo J., Siikala E., Kaihovaara P., Salaspuro M., Rautemaa R. (2009). Chronic candidosis and oral cancer in APECED-patients: production of carcinogenic acetaldehyde from glucose and ethanol by Candida albicans. Int J Cancer 124, 754–756. [DOI] [PubMed] [Google Scholar]
- Vaca C. E., Fang J. L., Schweda E. K. (1995). Studies of the reaction of acetaldehyde with deoxynucleosides. Chem Biol Interact 98, 51–67. [DOI] [PubMed] [Google Scholar]
- Väkeväinen S., Tillonen J., Agarwal D. P., Srivastava N., Salaspuro M. (2000). High salivary acetaldehyde after a moderate dose of alcohol in ALDH2-deficient subjects: strong evidence for the local carcinogenic action of acetaldehyde. Alcohol Clin Exp Res 24, 873–877. [PubMed] [Google Scholar]
- Väkeväinen S., Tillonen J., Salaspuro M. (2001). 4-Methylpyrazole decreases salivary acetaldehyde levels in aldh2-deficient subjects but not in subjects with normal aldh2. Alcohol Clin Exp Res 25, 829–834. [PubMed] [Google Scholar]
- Vickerman M. M., Iobst S., Jesionowski A. M., Gill S. R. (2007). Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol 189, 7799–7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch K. D., Davis T. Z., Aust S. D. (2002). Iron autoxidation and free radical generation: effects of buffers, ligands, and chelators. Arch Biochem Biophys 397, 360–369. [DOI] [PubMed] [Google Scholar]
- Woutersen R. A., Appelman L. M., Van Garderen-Hoetmer A., Feron V. J. (1986). Inhalation toxicity of acetaldehyde in rats. III. Carcinogenicity study. Toxicology 41, 213–231. [DOI] [PubMed] [Google Scholar]
- Yao S., Mikkelsen M. J. (2010). Identification and overexpression of a bifunctional aldehyde/alcohol dehydrogenase responsible for ethanol production in Thermoanaerobacter mathranii. J Mol Microbiol Biotechnol 19, 123–133. [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Muramatsu T., Ohmori T., Yokoyama T., Okuyama K., Takahashi H., Hasegawa Y., Higuchi S., Maruyama K., et al. (1998). Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis 19, 1383–1387. [DOI] [PubMed] [Google Scholar]