To identify novel susceptibility loci for psoriasis, we undertook a genome-wide association study of 594,224 SNPs in 2,622 psoriatic individuals and 5,667 controls. We discovered associations at eight previously unreported regions of the genome. Seven harboured genes with recognised immune functions (IL28RA, REL, IFIH1, ERAP1, TRAF3IP2, NFKBIA and TYK2). These findings were replicated in 9,079 European samples with six loci generating a P < 5 × 10−8 and two resulting in P < 5×10−7 in the combined dataset. We also report compelling evidence for an interaction between the HLA-C and ERAP1 loci (Pcombined = 6.95×10−6). ERAP1 plays an important role in MHC class I peptide processing. ERAP1 disease variants only influenced psoriasis susceptibility in individuals carrying the HLA-C risk allele, a pattern also observed at the T cell regulator ZAP70 (Pcombined < 5×10−7). Our findings implicate pathways that integrate epidermal barrier dysfunction with innate and adaptive immune dysregulation in psoriasis pathogenesis.
Psoriasis is a common, chronic, skin disease with a complex genetic and environmental etiology, characterised by epidermal hyper-proliferation, vascular remodelling and inflammation1. Prior linkage and association studies have unambiguously identified a disease susceptibility locus of large effect (termed PSORS1), spanning ~ 300kb of the Major Histocompatibility Complex (MHC) class I region on chromosome 6p212. Large-scale resequencing has established HLA-C as the most likely PSORS1 candidate gene3.
Genome Wide Association Studies (GWAS) and analysis of candidate genomic regions have captured eight non-MHC loci which highlight three pathways of considerable biological relevance to psoriasis pathogenesis4-9. These are NF-κB regulated signalling, mechanisms of T cell (particularly Th17) differentiation, and perturbation of the epidermal barrier. To identify additional risk variants we performed a GWAS as a component of the Wellcome Trust Case Control Consortium 2 (WTCCC2), powered by the inclusion of a substantially larger number of discovery samples, compared to previous psoriasis GWAS. We undertook replication of interesting findings in multiple cohorts drawn from across Europe.
Subjects for the GWAS discovery set were recruited from the UK and Ireland and were of self-reported European ancestry (see Supplementary Table 1). Cases were genotyped on the Illumina Human660W-Quad and controls on Illumina custom Human1.2M-Duo (see Supplementary Information), with primary analysis performed on the overlapping set of SNPs. We performed stringent data quality control (see Methods), resulting in a GWAS data set comprising 2,178 individuals with psoriasis and 5,175 controls genotyped at 535,475 SNPs. Principal Components Analysis (PCA) of study data showed the first principal component acted to stratify individuals by population origin (Supplementary Figure 1B). We performed single SNP analysis using score tests under a logistic regression model which assumed multiplicative effects, including the first principal component as a covariate, and accounted for uncertainty in genotype calls as implemented in SNPTEST. After removal of known and replicated psoriasis association loci, the overdispersion factor10 of association test statistics was λGC = 1.045 (see Supplementary Figure 1C).
In Table 1 we show evidence of association in our GWAS data at all other previously reported psoriasis disease risk SNPs. Odds ratios (OR) outside the HLA region, range from 1.12 to 1.49 highlighting substantial power to detect susceptibility loci of small effect. As expected, SNPs within the chromosome 6p21 region showed strong evidence for association with psoriasis (top SNP rs10484554: P = 4 × 10−214, OR = 4.66, 95%CI: 4.23-5.13), which extended across a broad region surrounding the Class I MHC that includes the established risk locus at HLA-C. Comparison of disease models showed a strong departure from the multiplicative model (P = 1.95×10−08). Odds ratios for the heterozygote and homozygote are 5.41 (95%CI: 4.81-6.08) and 10.61 (95%CI: 7.86-14.33) respectively. While not exact, this is comparable to a dominant model, in keeping with previous observations11.
Table 1.
GWAS signals from loci previously reported to be associated with psoriasis.
| Chr | SNP | Positiona | Gene/ Locus |
P value | OR (95% CI) | Reference |
|---|---|---|---|---|---|---|
| 1p31 | rs11209026 | 67478546 | IL23R | 7.13×10−07 | 1.49 (1.27-1.74) | [5] |
| 1q21 | rs4112788 | 150817900 | LCE3D | 3.32×10−10 | 1.29 (1.19-1.40) | [9] |
| 5q31 | rs20541 | 132023863 | IL13 | 2.32×10−02 | 1.12 (1.02-1.24) | [8] |
| 5q33 | rs1024995b | 150456197 | TNIP1 | 3.92×10−05 | 1.27 (1.14-1.44) | [8] |
| 5q33 | rs3213094 | 158683347 | IL12B | 4.93×10−11 | 1.39 (1.26-1.53) | [8] |
| 6p21 | rs10484554c | 31382534 | HLA-C | 4.06×10−214 | 4.66 (4.23-5.13) | [8] |
| 6q23 | rs610604 | 138241110 | TNFAIP3 | 6.54×10−07 | 1.22 (1.13-1.32) | [8] |
| 12q13 | rs2066808 | 55024240 | IL23A | 2.49×10−07 | 1.49 (1.28-1.73) | [8] |
| 20q13 | rs2235617d | 47988384 | ZNF313 | 1.65×10−06 | 1.20 (1.11-1.30) | [4] |
SNPs reported in previous studies with robust GWAS and replication evidence. Odds ratios given for the previously reported risk allele.
NCBI human genome build 36 coordinates.
r2 = 0.48 (0.49) with previously published SNP rs177283388.
At the HLA locus the results are presented for our top SNP
r2 = 1 with previously published SNP rs4953374. All r2 calculated from 58C and where different, the r2 from HapMap CEU is in brackets.
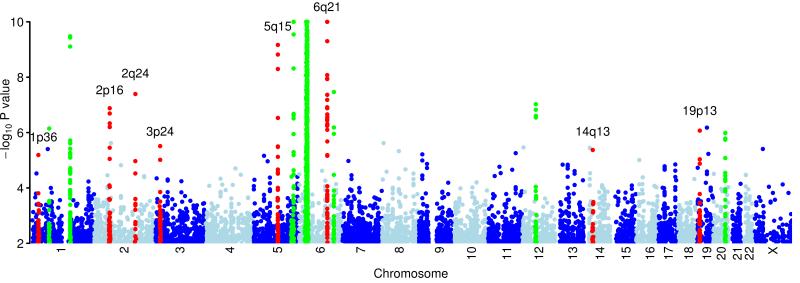
We identified 16 previously unreported loci for further investigation (Figure 1), including all regions with a P < 10−7 and further loci which yield a P ≤10−4 that contain strong biological candidate genes1. Seven independent replication datasets of psoriasis cases and controls were drawn from across Europe (10,060 individuals pre-QC). The most strongly associated SNP, or a surrogate, for each of the 16 previously unreported loci was genotyped using the Sequenom iPlex platform; see Table 2 and Supplementary Tables 2 and 3.
Figure 1.
Plot of genome wide association results.
Genome wide association results from 523,067 SNPs on chromosomes 1-22 and 12,408 SNPs on the X chromosome using the additive model in SNPTEST. The −log10 (P) values are thresholded at 10−10. Regions in red are described in table 2. Regions which have been shown previously to be associated with psoriasis and which replicate in this study are highlighted in green, as described in table 1.
Table 2.
New loci identified in this study
| RAFc | Discovery sample | Replication sample | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | rsID | Positiona | Gene of interest (#)b |
Risk allele |
cases | controls | P scan | OR (95% CI) | P repl | OR (95% CI) | P comb |
| 1p36 | rs4649203 | 24392507 | IL28RA (2) | A | 0.77 | 0.73 | 6.46×10−06 | 1.22 (1.12-1.32) | 1.36×10−03 | 1.13 (1.05-1.22) | 6.89×10−08 |
| 2p16 | rs702873 | 60935046 | REL (5) | G | 0.62 | 0.56 | 1.32×10−07 | 1.23 (1.14-1.32) | 1.41×10−03 | 1.12 (1.04-1.20) | 3.59×10−09 |
| 2q24 | rs17716942 | 162968937 | IFIH1 (5) | A | 0.90 | 0.86 | 4.05×10−08 | 1.38 (1.23-1.54) | 3.82×10−07 | 1.29 (1.17-1.43) | 1.06×10−13 |
| 3p24 | rs6809854d | 18759427 | None | G | 0.23 | 0.19 | 3.09×10−06 | 1.24 (1.14-1.36) | 4.92×10−03 | 1.14 (1.04-1.26) | 1.12×10−07 |
| 5q15 | rs27524 | 96127700 | ERAP1 (2) | A | 0.40 | 0.36 | 6.81×10−10 | 1.27 (1.18-1.38) | 7.96×10−04 | 1.13 (1.05-1.22) | 2.56×10−11 |
| 6q21 | rs240993e | 111780407 | TRAF3IP2 (4) | A | 0.30 | 0.25 | 8.71×10−13 | 1.36 (1.25-1.48) | 3.37×10−09 | 1.25 (1.16-1.34) | 5.29×10−20 |
| rs458017ef | 111802784 | G | 0.09 | 0.06 | 8.11×10−11 | 1.62 (1.40-1.88) | 9.59×10−08 | 1.37 (1.22-1.54) | 2.16×10−16 | ||
| 14q13 | rs8016947 | 34902417 | NFKBIA (1) | C | 0.61 | 0.57 | 4.26×10−06 | 1.19 (1.11-1.29) | 7.89×10−07 | 1.19 (1.11-1.27) | 1.52×10−11 |
| 19p13 | rs12720356g | 10330975 | TYK2 (9) | A | 0.93 | 0.90 | 9.25×10−06 | 1.34 (1.18-1.52) | 8.82×10−07 | 1.40 (1.23-1.61) | 4.04×10−11 |
| rs280519g | 10333933 | A | 0.51 | 0.47 | 8.51×10−07 | 1.20 (1.12-1.30) | 5.93×10−04 | 1.13 (1.05-1.21) | 4.42×10−09 | ||
Evidence for association at loci reported in this study
NCBI human genome build 36 coordinates
The number in brackets shows the number of genes between recombination hotspots
Risk Allele Frequency
This SNP failed Sequenom genotyping so replication genotyping for this locus is at rs7428395 r2=0.97 calculated from 58C (HapMap CEU r2=1)
Separate signals within the same locus, where the marginal P values and OR are reported. For further discussion and details of joint models, see text.
This SNP failed Sequenom genotyping so replication genotyping for this locus is at rs465969 r2=0.73 calculated from 58C (HapMap CEU r2=0.82).
Separate signals within the same locus, where the marginal P values and OR are reported. For further discussion and details of joint models, see text.
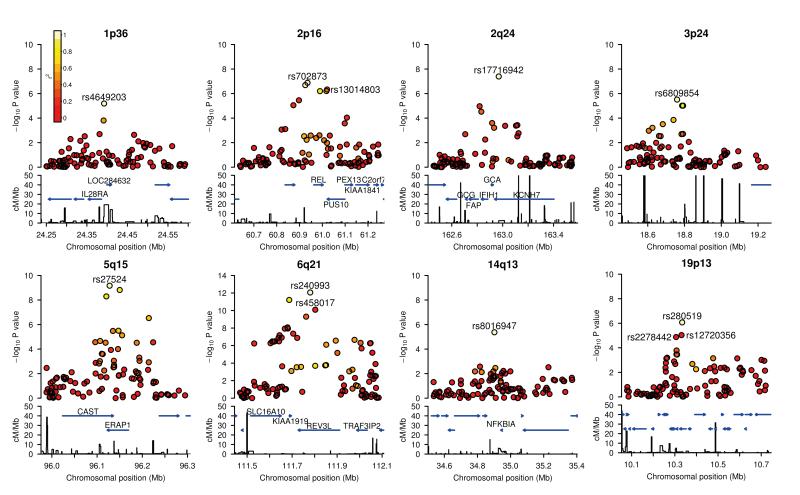
Eight regions showed strong evidence of association and independent replication (analysed using fixed-effects methods), six with P < 5 × 10−8 and two with P < 5 × 10−7 in the combined GWAS and replication dataset, with OR estimates in the replication sample ranging from 1.12 (chromosome 2p16) to 1.40 (chromosome 19p13). See Table 2, and Figure 2 for the regional signal plots. Closer analysis (see Supplementary note) of the original signal at the 2p16, 6q21, and 19p13 loci revealed multiple statistically independent SNP associations. Two of these were tested and confirmed in our replication study, see Table 2. Full details are given in Supplementary Table 4. Where there are signals at two SNPs within a locus it is difficult from our data to rule out the possibility that the pattern of association is generated by a single untyped SNP. Regardless of this, we note that under a multiplicative model, the effect of carrying an additional copy of each of the risk alleles at 2p16, 6q21, and 19p13 is 1.40 (95%CI: 1.25-1.56), 1.66 (95%CI: 1.44-1.92), and 1.52 (95%CI: 1.31-1.75) respectively. One important consequence of the potential allelic heterogeneity, is that the overall genetic effect at the locus is larger than that associated with the single SNP with the lowest P value.
Figure 2.
Regional association plots.
The -log10 (P) values for SNPs at eight new loci are shown on the upper part of each plot. SNPs are coloured based on their r2 with the labeled hit SNP which has the smallest P value in the region. r2 is calculated from the 58C control data. Where more than one SNP is labeled, there is evidence for multiple signals at the locus (see text). The bottom section of each plot shows the fine scale recombination rates estimated from HapMap individuals and genes are marked by horizontal blue lines. Genes within the recombination region of the hit SNP are labeled except for 19p13, where the genes are: GLP-1, FDX1L, RAVER1, ICAM3, TKY2, CDC37, PDE4A, KEAP1, S1PR5.
While most of the novel disease-associated intervals (as defined by recombination hotspots) in Table 2 span multiple transcripts, all but one contain at least one immune-related gene. These genes are strong biological candidates, and it is striking that they collectively cluster into a limited number of pathways with recognised roles in innate and adaptive immunity. For instance, ERAP1 in the chromosome 5q15 associated region encodes an amino-peptidase regulating the quality of peptides bound to MHC class I molecules, such as HLA-C12. The SNP rs30187 within this interval is also significantly associated with ankylosing spondylitis13. Given the established link between psoriasis and the development of spondyloarthropathy14, the finding of an association at the ERAP1 locus in both conditions is noteworthy, particularly as the coding SNP rs30187 also generated a very significant P value in our GWAS (P = 1.5 × 10−9, OR= 1.27, 95% CI: 1.13-1.31). Conditional analyses in our association data do not distinguish significantly between our top SNP rs27524, and the coding variant rs30187.
The chromosome 6q21 region spans four known genes (Figure 2). Of these, TRAF3IP2 encodes Act1, an adaptor protein essential for IL17-dependent NF-κB activation and Th17-mediated inflammatory responses15. Remarkably, genes participating in NF-κB and IL17-mediated signalling are also found in three further disease susceptibility regions identified by this study. NFKBIA, the only gene lying within the 14q13 associated region, encodes a key inhibitory protein that inactivates NF-κB dimers in the absence of inflammatory stimuli16. Likewise, REL, which maps to the associated region on chromosome 2p16, encodes one of four subunits found in NF-κB dimers and has recently been implicated in risk for rheumatoid arthritis16,17. Finally, the interval on chromosome 19p13 includes TYK2, which encodes a kinase promoting IL17 transcription via STAT3 phosphorylation18. Common and low frequency variants of TYK2 confers risk in multiple autoimmune diseases, namely Type I diabetes and Systemic lupus erythematosus19.
It is noteworthy that TYK2 also regulates type I and type III interferon (IFN) signaling20, suggesting that innate IFN-activating anti-microbial response pathways may contribute to initiation or maintenance of disease progression. This would be consistent with our observation of disease-associated variants in the proximity of IL28RA, which encodes a type III IFN receptor subunit21. Pathogenic involvement of IFN signaling pathways is also supported by the association signal detected on chromosome 2q24. In this case, the associated region encompasses the IFIH1, FAP and GCA genes together with part of KCNH7 as shown in Figure 2. Of these, IFIH1 encodes an innate receptor triggering type I IFN in response to microbial infection22 and both rare and common IFIH1 variants are associated with type I diabetes23. The SNP rs1990760 (minor allele frequency 0.38) has a strong signal in our sample and type 1 diabetes but the correlation between this SNP and rs17716942 is low (r2 = 0.3). Further resequencing of IFIH1 will be required to determine the properties of rare variation in relation to psoriasis risk.
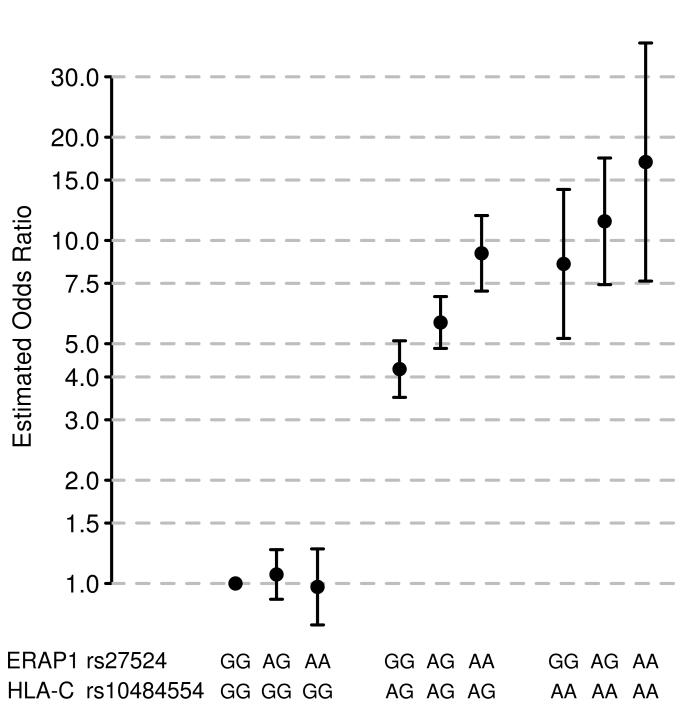
Given the coincidence of many signals within a small number of pathways, we looked for statistical interactions (see methods) between the top SNPs in each of the 17 known and confirmed novel regions in Tables 1 and 2, fitting a dominant model at the HLA-C SNP rs10484554. One pair of SNPs, (rs10484554 (HLA-C) and rs27524 at ERAP1), generated strong evidence for interaction in the discovery, replication and combined data (Pdiscovery = 2.45×10−5, Preplication = 0.027, Pcombined = 6.95×10−6) (see Supplementary Table 5). Figure 3 shows that when stratified by genotypes at the two loci, the ERAP1 SNP only has an effect in individuals carrying at least one copy of the risk allele at rs10484554.
Figure 3.
Statistical interaction between ERAP1 and HLA-C genotypes.
Odds ratio estimates for ERAP1 genotypes at rs27524 stratified by HLA-C genotypes at rs10484554. Odds ratios were estimated by fitting a logistic regression model which included the first principal component as a covariate and a parameter for each of the possible two-locus genotypes. Note that odds are measured relative to the most protective two-locus genotype (which by definition has an odds ratio of 1). Bars represent 95% confidence intervals calculated from the standard error of the estimates of the genotype parameters in logistic regression model.
Very few convincing examples of interactions between complex disease loci have been reported in humans24-26, perhaps because the power to detect these is limited unless the causal SNPs or very good surrogates have been typed27. The finding that variation at the ERAP1 locus is only associated with disease in individuals carrying the HLA-C risk allele is particularly interesting biologically, because of ERAP1’s role in class I peptide presentation. It is also noteworthy that the odds ratio for rs27524 is 1.43 (1.21-1.69) in the HLA-C positive subgroup, compared to the estimate of 1.27 (1.18-1.38) from the GWAS on the entire dataset (in the replication data these estimates were 1.23 (1.09-1.39) and 1.13 (1.05-1.22) respectively). Were this phenomenon to be more widespread, it would impact on the proportion of broad-sense heritability explained by GWAS findings.
Next, we looked systematically across the genome for other loci which might only have an effect on one or other of the HLA-C risk backgrounds. We divided individuals in our discovery study into two groups, depending on whether they carried zero copies (“HLA-C negative” individuals: 831 cases and 3,985 controls) or one/two copies (“HLA-C positive” individuals: 1,347 cases and 1,190 controls) of the risk allele at rs10484554, the index SNP for the HLA-C locus, and undertook separate GWAS in each group (Supplementary Figure 2 Supplementary Table 6).
Four other loci generated evidence for association in the GWAS of HLA-C positive individuals. One of these, SNP rs17695937, near ZAP70, also displayed a clear signal in the replication data, generating a Pcombined = 2.37×10−7. Several of the other loci also warrant further investigation (Supplementary Figure 2). The identification of risk variation at ZAP70 together with ERAP1 in individuals with HLA-C risk alleles provides further genetic evidence that psoriasis may in part be caused by dysregulation of HLA-restricted CD8 T cells. ZAP70 is a tyrosine kinase that binds to the T cell receptor-associated chain, CD3-zeta (CD247) following the TCR engagement of MHC-peptide complexes, and is critical for setting the response threshold that in turn governs the T cell selection events which purge the T cell compartment of autoreactivity28. Finally, although we followed-up several SNPs from the HLA-negative GWAS, none replicated (Supplementary Table 6).
We undertook additional analyses of the relationship between the known classical HLA risk allele, Cw*0602 and typed SNPs. Based on molecular typing in a subset of 725 of the 58C individuals, the correlation between Cw*0602 genotypes (0, 1, or 2 copies of Cw*0602) and our top SNP, rs10484554, is r2= 0.7. We imputed the HLA-C genotypes from our SNP data29 (see Methods). Association testing with these imputed genotypes showed a stronger signal for the classical allele than for any single SNP yet conditional analyses are not definitive in ruling out either variant as the primary source of the signal (see supplementary data). We reexamined the HLA-C/ERAP1 interaction in an attempt to ascertain whether it was driven by the top SNP (rs10484554) or classical allele at HLA-C, and likewise whether the top SNP (rs27524) or the previously reported coding SNP (rs30187) at ERAP1 was more closely involved. Although evidence for interaction existed for all four pairs of markers (Supplementary Table 7), the dominant model at the HLA Cw*0602 with an interaction at ERAP1 SNP rs27524 best fits the GWAS data. Conditional analyses are not definitive in ruling out either variant at either site as the primary source of the interaction signal.
Collectively, the candidate genes identified in this study are relevant to the question of whether psoriasis is primarily an epidermal or immune dysfunction disorder. The present data provide evidence for an integrated model for the pathogenesis of psoriasis that combines skin barrier function (e.g. variants in LCE3B/3C); innate-immune pathogen sensing focused on NF-κB and IFN signalling (e.g. NFKBIA, REL, TYK2, IFIH1, IL28RA); and adaptive immunity involving CD8 T lymphocytes (ERAP1 and ZAP70) and Th17 cell responses (e.g. TRAF3IP3, TYK2, IL23R). Fine mapping, functional characterisation and meta-analysis in additional psoriasis cohorts can now determine the detailed molecular consequences of genetic variation on these pathways. Finally, this study also provides one of the first compelling examples of an interaction between two GWAS loci, and the success of our separate GWAS in HLA-C positive individuals suggests that similar analyses might be valuable in other auto-immune diseases.
ONLINE METHODS
Samples
Cases
A total of 6,523 samples were assembled for this investigation of which 2,735 were processed for the GWAS discovery set and 3,788 for the replication set. For all samples, psoriasis was defined as psoriasis vulgaris (chronic plaque type psoriasis) the commonest manifestation of the disease. A total of 2,178 DNA samples from unrelated individuals of European ancestry, recruited from five centres in England, Scotland and Ireland, were included in the GWAS discovery set, having passed pre- and post-genotyping control filters (see below). Phenotypic data and blood samples were collected after research ethics approval was received at each site and patients had given written, informed consent. The replication set of DNA was derived from patients with psoriasis recruited in several European countries (Austria, Germany, Italy, Netherlands, Spain, Sweden, and UK) after research ethics approval and written informed consent was given. In total 3,174 DNA samples passed quality control for replication.
Controls
A total of 10,639 control individuals passed the quality control filters (see below). 5,175 individuals from the WTCCC2 common control set were used in the discovery GWAS. This included 2,501 healthy blood donors from the United Kingdom Blood Service (UKBS) collection and 2,674 individuals from the 1958 Birth Cohort (58C) dataset27. 5,464 individuals were used in the replication dataset including 2,717 from the People of the British Isles (PoBI) collection. The remaining controls were taken from six other European populations selected to match the case individuals. These collections have been described elsewhere (Supplementary Table 1).
Details of DNA sample preparation and genotyping are given in the Supplementary Note.
Quality Control (QC)
Full details of QC procedures are given in the Supplementary Note. In brief:
Samples
We used a recently developed Bayesian clustering approach to infer and exclude outlying individuals on the basis of call rate, heterozygosity, relatedness and ancestry30, and for an additional artifact which led to spurious associations, where for some samples SNP intensities in one channel were systematically higher than in the other. We also excluded one of each pair of related individuals, as identified by a hidden Markov model.
SNPs
SNPs were excluded unless the (Fisher) information for the allele frequency was very close to unity, or if minor allele frequency was very low (<0.01%), or for extreme departures from Hardy-Weinberg equilibrium (except in the MHC), or if they showed a strong plate effect. Cluster plots of SNPs showing putative associations were inspected manually (Supplementary Figure 3).
The QC measures excluded 444 case and 492 control individuals, and 9.8% of SNPs. Of the 2,178 case samples which passed quality control, 345 have self-reported Irish ancestry (Supplementary Table 8).
Statistical methods
We performed Principal Component Analysis (PCA) on a subset of 205,842 post-QC SNPs (none from the MHC), selected so as to minimize the contribution from regions of extensive strong linkage disequilibrium and to ensure that only genome-wide effects are detected. Principal component scores were computed for the combined dataset of post-exclusion case and control samples. On inspection, the first principal component acts to differentiate UK from Irish individuals (see Supplementary Figure 1A) suggesting that the major determinant of the variance in genome-wide patterns of diversity between sample individuals was due to differences in ancestry. To guard against possible artifacts due to population structure, we repeated the primary association analysis excluding the Irish cases. Reassuringly, this yielded comparable association signals after allowing for the reduction in sample size (data not shown). As no further principal components driven by genome-wide patterns exhibited significant differences between cases and controls we used only the first principal component to control for population stratification in subsequent analyses.
We used a logistic regression model with case/control status as the response variable, the first principal component as a covariate and the genotype at a particular SNP as the explanatory variable. Genome wide association tests were carried out at each SNP with uncertainty in genotype calls modeled using missing data likelihoods as implemented in SNPTEST. Unless otherwise stated, we assumed that the change in odds of case status due to each copy of the allele was multiplicative. To investigate patterns of association between SNPs, for example to look for interactions or secondary signals, we used the statistical package R31.
It has become standard practice in GWAS to refer to the odds ratio associated with a particular allele or haplotype, which we estimate as eβ, where β is the maximum likelihood estimate of the coefficient describing the effect of each predictor on the response in the assumed model. We note however that, as is true of this study and many others, where the controls are taken at random from the population without reference to disease status, β is actually the log of the relative risk and not the log of the odds ratio32.
Imputation was performed using IMPUTE233, which adopts a two-stage approach using both a haploid reference panel and a diploid reference panel. We used two methods to investigate possible secondary signals within GWAS association regions, standard frequentist conditional analyses and the program GENECLUSTER34 which adopts a Bayesian approach to look for primary and secondary association signals at known and putative SNPs. See Supplementary Note for full details and for additional pre-imputation SNP QC.
We used standard “fixed-effect” meta-analysis techniques to analyse the replication data. Specifically, in each population, we fitted a logistic regression model for case/control status with no covariates at each SNP with a single parameter for the genetic effect (a multiplicative effect on the risk scale, additive on the log-odds scale). We then assumed a fixed underlying genetic effect, common to all populations, for the SNP, which we estimated by combining the effect estimates across populations, weighting by the precision of the estimate within each population. P-values, effect size estimates and confidence individuals for each population, and combined across populations are shown in Supplementary Table 2, see also Supplementary Figure 4. As is typical for GWAS replication studies which type a small number of SNPs, testing for possible substructure within populations is not possible. Our replication results involve averaging across separate results for seven populations. This provides robustness against false positives due to substructure in some populations, and it is encouraging that most or all replication populations typically exhibit evidence for the signal. We confirmed the power of this replication cohort by genotyping SNPs rs4085613 and rs4112788 which tag the known LCE3B/3C psoriasis risk locus6,9, where we observed replication P values of 2.33 × 10−15 and 4.44 × 10−16, respectively.
To look for interactions between associated loci we considered all pairs of index SNPs from Tables 1 and 2 and for each pair we compared two logistic regression models. In the first (null) model, a separate parameter for each SNP specifies the multiplicative increase in the odds of disease with each additional copy of the risk allele for that SNP. We compared this, via a likelihood ratio test, to a model for interaction in which there is a third parameter which modifies the effect associated with carrying risk alleles at both SNPs. There are many different ways to model both the marginal effects at each SNP and their joint effect35, and so many approaches to search for interactions. The procedure we adopted is perhaps the simplest (multiplicative model marginally at each SNP and a single additional parameter for multiplicative interactions). Having identified the interaction between the HLA-C and ERAP1 loci, we examined the nature of the effect, and observed that it takes the simple, and easily interpretable, form in which additional copies of the index SNP at ERAP1 only affect disease risk in HLA-C positive individuals. To test for interactions between SNPs in the replication data we included a categorical covariate indicating population membership to the logistic regression model. We used this framework to compare two models, via a likelihood ratio test, which included or excluded a term for the interaction between the two SNPs.
HLA Imputation
Imputation of classical HLA genotypes was performed using published statistical methodology29 and a reference panel of over 1,500 samples with classical HLA and SNP genotype data from the 1958 Birth Cohort and the International HapMap Project36. When predicting whether an individual carries one or more copies of HLACW*0602, the primary risk allele for psoriasis, we estimate false positive rate to be 1% (no call threshold), and false negative rate < 1%. In the replication study, four SNPs were typed in the HLA (rs10484554, rs3906272, rs12111032 and rs2243868), two of which (rs12111032 and rs2243868) form a near-perfect (false positive rate < 1%) haplotype tag for CW*0602 (alleles A and G on the plus strand respectively). Imputations were performed by phasing the four SNPs with PHASE37 and assuming the AG haplotype to be a perfect tag. From the independent validation study we estimate the false positive rate of this approach to be 9% (no call threshold), the decrease in accuracy resulting from uncertainty in phasing. As a check, we then applied the imputation approach we used in the replication data to the GWAS samples. Comparison of the imputations made using the four SNPs to those made from the full GWAS data show 1.4% of Cw*0602 presence/absence predictions to be discrepant.
Data Availability
A list of the SNPs in the discovery scan exhibiting P < 10−4 are available in Supplementary Table 9. Researchers can gain access to the data by applying to the data access committee (www.ebi.ac.uk/ega/).
Supplementary Material
Acknowledgements
The principal funding for this study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (083948/Z/07/Z). We also thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, A. Wilk, C.R. Stribling and S. Taylor of the Wellcome Trust Sanger Institute’s Sample and Genotyping Facilities for technical assistance. We thank Dan Davison for making available his program “Shellfish” for calculating principal components in large genetic data sets. Case collections were supported by the Netherlands Organization for Health Research and Development (P.L.Z.); the Swedish Medical Research Council, Karolinska Institutet, Karolinska University Hospital, Psoriasis Foundation, AFA, and Welander Finsen Foundation (M.S.); the Association for the Defence of Psoriasis Patients (G.N.); Psoriasis Association and the Cecil King Memorial Foundation (M.J.C.); the Swedish Psoriasis Association (L.S.); the German Research Foundation (Tr 228/5-4 and Re 679/10-4) and The Interdisciplinary Centre for Clinical Research (IZKF B32/A8) of the University of Erlangen-Nuremberg (A.R.); the Spanish Ministry of Science and Innovation (grant SAF 2008-00357) and the “Generalitat de Catalunya” Departments of Health and Universities and Innovation (X.E.); the Genetic Repository in Ireland for Psoriasis and Psoriatic Arthritis (GRIPPsA), the Dublin Centre for Clinical Research (DCCR, funded by the Irish Health Research Board), The Wellcome Trust, and Science Foundation Ireland (R.McM.); National Institute for Health Research, Manchester Biomedical Research Centre (J.W., C.E.M.G., R.B.W., H.S.Y.); Arthritis Research UK (J.W., grant 17552). P.D. was supported in part by a Wolfson–Royal Society Merit Award and A.O. was supported by a PhD studentship from The Generation Trust. We also acknowledge support from the UK Medical Research Council (R.C.T., F.O.N., A.H., JNB grant G0601387), the Wellcome Trust (F.O.N., grant 078173/Z/05/Z) and the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre awards to Guy’s & St. Thomas’ NHS Foundation Trust in partnership with King’s College London (J.K, M.E.W, C.G.M, F.O.N, A.Hayday and J.N.B) and the NIHR award to Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology for a Specialist Biomedical Research Centre for Ophthalmology (A.C.V). We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02, and of the UK National Blood Service controls funded by the Wellcome Trust. We thank W. Bodmer and B. Winney for use of the People of the British Isles DNA collection, which was funded by the Wellcome Trust.
Footnotes
URLs, SNPTEST, http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html. WTCCC2, http://www.wtccc.org.uk/ccc2/.
Competing financial interests The authors declare no competing financial interests
References
- 1.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 2.Veal CD, et al. Family-based analysis using a dense single-nucleotide polymorphism-based map defines genetic variation at PSORS1, the major psoriasis-susceptibility locus. Am J Hum Genet. 2002;71:554–64. doi: 10.1086/342289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair RP, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–51. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capon F, et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum Mol Genet. 2008;17:1938–45. doi: 10.1093/hmg/ddn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cargill M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–90. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Cid R, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–5. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollox EJ, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–5. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang XJ, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet. 2009;41:205–10. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 10.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 11.Jenisch S, et al. Linkage analysis of human leukocyte antigen (HLA) markers in familial psoriasis: strong disequilibrium effects provide evidence for a major determinant in the HLA-B/-C region. Am J Hum Genet. 1998;63:191–9. doi: 10.1086/301899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.York IA, et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002;3:1177–84. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 13.Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan MA. Update on spondyloarthropathies. Ann Intern Med. 2002;136:896–907. doi: 10.7326/0003-4819-136-12-200206180-00011. [DOI] [PubMed] [Google Scholar]
- 15.Qian Y, et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8:247–56. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 16.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Gregersen PK, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–3. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oyamada A, et al. Tyrosine kinase 2 plays critical roles in the pathogenic CD4 T cell responses for the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:7539–46. doi: 10.4049/jimmunol.0902740. [DOI] [PubMed] [Google Scholar]
- 19.Wallace C, et al. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2009;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z, et al. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol. 2007;81:7749–58. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 22.Wilkins C, Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 22:41–7. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lincoln MR, et al. Epistasis among HLA-DRB1, HLA-DQA1, and HLA-DQB1 loci determines multiple sclerosis susceptibility. Proc Natl Acad Sci U S A. 2009;106:7542–7. doi: 10.1073/pnas.0812664106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orsmark-Pietras C, et al. Biological and genetic interaction between tenascin C and neuropeptide S receptor 1 in allergic diseases. Hum Mol Genet. 2008;17:1673–82. doi: 10.1093/hmg/ddn058. [DOI] [PubMed] [Google Scholar]
- 26.Smyth DJ, et al. PTPN22 Trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with HLA class II genotypes. Diabetes. 2008;57:1730–7. doi: 10.2337/db07-1131. [DOI] [PubMed] [Google Scholar]
- 27.The Wellcome Trust Case-Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picard C, et al. Hypomorphic mutation of ZAP70 in human results in a late onset immunodeficiency and no autoimmunity. Eur J Immunol. 2009;39:1966–76. doi: 10.1002/eji.200939385. [DOI] [PubMed] [Google Scholar]
- 29.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–4. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2010. [Google Scholar]
- 32.Schouten EG, et al. Risk ratio and rate ratio estimation in case-cohort designs: Hypertension and cardiovascular mortality. Statistics in Medicine. 1993;12:1733–1745. doi: 10.1002/sim.4780121808. [DOI] [PubMed] [Google Scholar]
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Z, Cardin N, The Wellcome Trust Case Control Consortium. Donnelly P, Marchini J. A Bayesian Method for Detecting and Characterizing Allelic Heterogeneity and Boosting Signals in Genome-Wide Association Studies. Statistical Science. 2009;24:430–450. [Google Scholar]
- 35.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–7. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 36.de Bakker PI, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A list of the SNPs in the discovery scan exhibiting P < 10−4 are available in Supplementary Table 9. Researchers can gain access to the data by applying to the data access committee (www.ebi.ac.uk/ega/).