Abstract
Increased transcription of ribosomal RNA genes (rDNA) by RNA Polymerase I is a common feature of human cancer, but whether it is required for the malignant phenotype remains unclear. We show that rDNA transcription can be therapeutically targeted with the small molecule CX-5461 to selectively kill B-lymphoma cells in vivo while maintaining a viable wild-type B cell population. The therapeutic effect is a consequence of nucleolar disruption and activation of p53-dependent apoptotic signaling. Human leukemia and lymphoma cell lines also show high sensitivity to inhibition of rDNA transcription that is dependent on p53 mutational status. These results identify selective inhibition of rDNA transcription as a therapeutic strategy for the cancer specific activation of p53 and treatment of hematologic malignancies.
Introduction
Pol I-dependent transcription of the 47S rDNA dynamically responds to growth signaling and cellular stresses to establish the abundance of the ribosomal RNAs and directly regulates cellular protein translational capacity and thus proliferative growth rate (Jorgensen et al., 2002; Larminie et al., 1998). rDNA transcription takes place in specialized subnuclear domains termed nucleoli that are formed around actively transcribed rDNA repeats in early G1 before being disassembled in mitosis when rDNA transcription is halted. Strikingly, elevated rDNA transcription by Pol I is a stalwart feature of cancer (Barna et al., 2008; Ruggero and Pandolfi, 2003; White, 2005), and enlarged nucleoli, a consequence of hyper-activated rDNA transcription, have been used by pathologists since the late 19th century as a marker of aggressive tumors (Derenzini et al., 2009).
In addition to rRNA and factors associated with ribosome biogenesis, nucleoli are enriched with a large number of other proteins, many of which have no direct function in the synthesis of ribosomes. In many cases, regulated sequestration of these proteins in the nucleoli controls their cellular activity. Because of this, the nucleolus has the potential to control a broad range of cellular functions in addition to rDNA transcription. In particular, nucleolar localization regulates the function of key oncogenes and tumor suppressors such as ARF and MDM2, both of which are critical for the regulation of p53 (TP53) (Boisvert et al., 2007). Thus, Pol I-dependent transcription and nucleolar integrity are pivotal determinants for numerous processes required for the excessive proliferation of cancer cells.
Surprisingly, despite its constant activation in cancer and potential to control critical determinants of malignant transformation, the importance of accelerated Pol I transcription and nucleolar integrity for cancer and their potential as therapeutic targets, remains undefined (Ruggero and Pandolfi, 2003; White, 2005). From a clinical perspective, a key question is whether targeted inhibition of rDNA transcription, generally considered a “housekeeping process” universally required for cell growth and proliferation, can show selectivity for killing malignant cells over normal cells. Furthermore, it is critical to understand the mechanism(s) by which such selectivity might be achieved and identify the tumor types that may therapeutically respond.
MYC is a potent oncogene whose dysregulated expression plays a significant role in human cancer development. MYC also plays a fundamental role in the biogenesis of ribosomes through transcriptional upregulation of 47S rRNA, and transcription of a select group of factors involved in rRNA processing, rRNA transport, and ribosome assembly. Due to the function that MYC plays in regulating Pol I activity and ribosome biogenesis (Arabi et al., 2005; Dai and Lu, 2008; Dang et al., 2006; Grandori et al., 2005; Grewal et al., 2005; Poortinga et al., 2004, 2011; Shiue et al., 2009; van Riggelen et al., 2010), models of MYC driven oncogenesis provide an ideal setting to explore the dependencies between Pol I transcription, ribosome biogenesis, and cancer.
In this study, we used both genetic approaches and a small molecule selective inhibitor of Pol I transcription (CX-5461) (Drygin et al., 2011) to investigate the dependence of tumors on Pol I activity in a murine model of spontaneous lymphoma driven by MYC (Eμ-Myc) (Adams et al., 1985). In addition, we explored the therapeutic potential of inhibiting Pol I transcription in various human hematologic malignancies.
Results
rRNA Transcription and Ribosome Biogenesis Are Accelerated in Eμ-Myc Lymphoma Cells
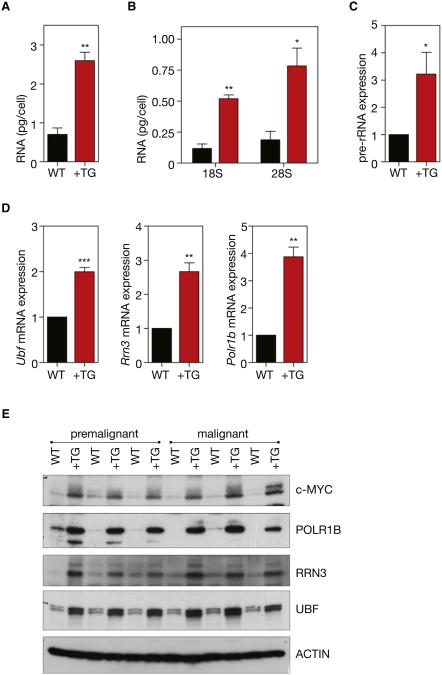
To investigate the role of Pol I transcription in malignancy, we employed a murine model of spontaneous lymphoma (Eμ-Myc) in which MYC is overexpressed in lymphocytes of the B lineage. This genetic model reproduces many of the clinical aspects of Burkitt's lymphoma in humans, including reciprocal chromosomal translocations that juxtapose the c-myc proto-oncogene on chromosome 8 to the immunoglobulin heavy chain locus on chromosome 14, or the κ or λ light chain locus on chromosomes 2 or 22 (Adams et al., 1985; Klein, 1993). As expected from MYC's well-defined role in promoting growth, premalignant B220+ splenic B cells from 4- to 6-week-old Eμ-Myc mice had increased cell volume, increased protein content, and highly accelerated proliferation rates compared to B cells from wild-type littermates (Figure S1 available online) (Iritani and Eisenman, 1999). Despite their much faster cell doubling time, the Eμ-Myc B cells also exhibited higher amounts of both total RNA and ribosomal RNA (rRNA) per cell (Figures 1A and 1B), suggesting that Pol I transcription and cell growth were highly accelerated. Consistent with these findings, B cells from Eμ-Myc mice exhibited robust increases in the level of transcription of rDNA by Pol I, as determined by measuring the abundance of pre-rRNA, which is rapidly processed (half-life of 10–30 min) to mature rRNA (half-life of days to weeks) and can therefore be used as indirect measure of rRNA synthesis rate (White, 2005) (Figure 1C).
Figure 1. Regulation of Pol I Transcription in Transgenic Eμ-MycB Cells.
(A) Premalignant B220+ splenic B cells were isolated from premalignant (4- to6-week-old) Eμ-Myc mice (+TG) and their respective wild-type litter-mates (WT), lysed, and total RNA content per cell determined (p < 0.01, n = 3).
(B) Total 18S (p < 0.01, n = 3) and 28S (p < 0.05, n =3)rRNA per cell, as determined using an Agilent Bioanalyzer.
(C) Pre-rRNA expression determined by quantitative reverse transcription real-time (qRT) PCR of the internally transcribed spacer 1 (ITS1) (+742 to +874) of the 47S pre-rRNA (p < 0.05; n = 5).
(D) Relative mRNA expression of Pol I transcription factors Ubf, Rrn3, and Polr1b, determined by qRT-PCR (Ubf, p < 0.001; Rrn3, p < 0.01; Polr1b, p < 0.01; n = 5).
(E) Western blot analysis of ACTIN, c-MYC and Pol I transcription factors, UBF, RRN3 and POL1RB in premalignant and malignant (tumor bearing) B220+ B cells isolated from +TG mice and their respective WT littermates. Student's two-tailed t test for all comparisons. Error bars represent SEM.
See also Figure S1.
MYC regulates transcription of rDNA through direct activation of Pol I (Arabi et al., 2005; Grandori et al., 2005; Shiue et al., 2009) and by transcriptionally increasing the level of the Pol I subunits and Pol I-specific transcription factors (Grewal et al., 2005; Poortinga et al., 2004, 2011). Consistent with this, both the mRNA and protein abundance of key Pol I specific components including UBF (UBTF), a factor involved in pre-initiation complex formation and rDNA chromatin remodeling (Sanij et al., 2008), RRN3 (also termed TIF-1A), an essential Pol I initiation factor (Yuan et al., 2005), and POLR1B, the second largest subunit of Pol I (Hannan et al., 1998), were all highly elevated in premalignant Eμ-Myc transgenic B cells compared to normal B cells isolated from wild-type littermates (Figures 1D and 1E). Further-more, this increased expression of Pol I-specific components was perpetuated in malignant B cells isolated from tumor bearing Eμ-Myc mice, indicating the continued elevation of this pathway with the development of lymphoma (Figure 1E). This demonstrates that the increased levels of the Pol I factors and rDNA transcription occur during cellular transformation and tumor initiation and are not simply a consequence of transformation.
Accelerated rDNA Transcription Is Required for Survival Eμ-Myc Lymphoma Cells
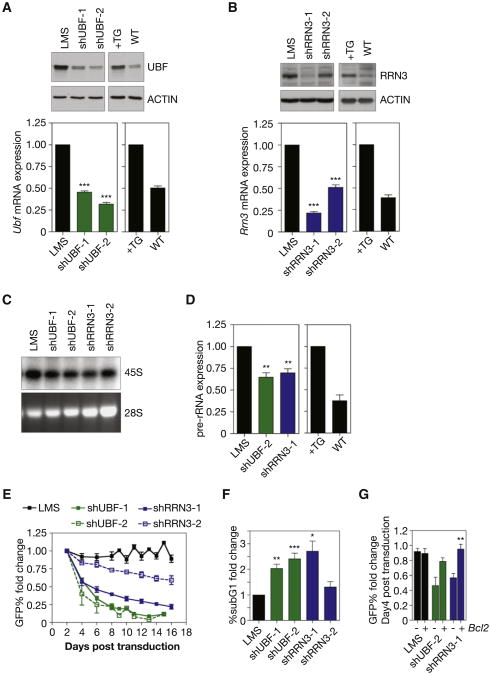
We used RNAi-mediated knockdown of UBF and RRN3 to examine the extent to which the increased rate of Pol I transcription exhibited by the Eμ-Myc B cells was necessary for their proliferative capacity and survival. Eμ-Myc lymphoma cells isolated from tumor bearing mice that grow in culture and are wild-type for p53 were retrovirally transduced with microRNA-based short hairpins (shRNA-mirs) targeting Ubf (shUBF) or Rrn3 (shRRN3) (Sanij et al., 2008). Coexpression of GFP allowed for FACS-based sorting of transduced cells. Transduction efficiency was titered to reduce Ubf and Rrn3 mRNA and protein abundance in the lymphoma cells to near those found in normal B cells (Figures 2A and 2B). Individual knockdown of UBF or RRN3 resulted in a reduction in Pol I transcription by 35%–40%, but notably, not below levels observed in normal B cells (Figures 2C and 2D). To gauge the effect of normalizing Pol I transcription on the proliferation and survival of Eμ-Myc lymphoma cells we performed an in vitro GFP competition assay in which mock transduced Eμ-Myc lymphoma cells were mixed with equal numbers of Eμ-Myc lymphoma cells transduced with either LMS vector alone (adapted from Dickins et al., 2005), (Eμ-Myc/LMS), LMS-shUBF (Eμ-Myc/LMS-shUBF), or LMS-shRRN3 (Eμ-Myc/LMS-shRRN3) and maintenance of the transduced population was followed by FACS. Eμ-Myc/LMS-shUBF and Eμ-Myc/LMS-shRRN3 cells exhibited a profound disadvantage with regard to proliferation and/or survival (Figure 2E). Likewise, expression of shUBF and shRRN3 caused an increased sub-G1 population (Figure 2F) and a 2–3-fold increase in markers of apoptosis including loss of mitochondrial membrane potential (decrease in tetramethylrhodamineethyl [TMRE] retention; Figure S2A) and increased AnnexinV/PI staining (Figure S2B). This suggested that the Eμ-Myc lymphoma cells were eliminated by apoptosis following normalization of Pol I transcription levels. Consistent with the survival disadvantage resulting from an induction of apoptosis, enforced expression of the anti-apoptotic protein BCL-2 (Figure S2C) significantly reduced the disadvantage of the Eμ-Myc/LMS-shUBF and Eμ-Myc/LMS-shRRN3 cells (Figure 2G). Moreover, a long-term competition assay with Eμ-Myc/LMS-shRRN3 cells expressing BCL2 demonstrated that their proliferative capacity was similar to control cells (Figure S2D). These results reveal that a ∼35% reduction in Pol I transcription in Eμ-Myc lymphoma cells results in induction of apoptosis that is not linked to a reduction in proliferation rate. Together these data demonstrate that the process of malignant transformation and cancer cell survival is considerably more dependent upon maintenance of elevated levels of Pol I activity than previously appreciated.
Figure 2. Reducing Pol I Transcription Rates in Eμ-MycLymphoma Cells Using shRNA-mirs Against UBF or RRN3 Strongly Activates Programmed Cell Death.
(A) Western blot analysis of UBF in cultured in Eμ-Myc lymphoma cells transduced with one of two independent shRNA-mirs targeting UBF (shUBF-1 and -2) or empty vector (LMS), and in B220+ splenic B cells isolated from 4- to 6-week-old Eμ-Myc mice (+TG) and their respective wild-type littermates (WT). Relative Ubf mRNA expression determined by qRT-PCR (LMS versus shUBF-1, p < 0.001; LMS versus shUBF-2, p < 0.001; n = 3).
(B) Western blot analysis of RRN3 as for UBF in (a) utilizing two independent shRNA-mirs targeting RRN3 (shRRN3-1 and -2). Relative Rrn3 mRNA expression determined by qRT-PCR (LMS versus shRRN3-1, p < 0.001; LMS versus shRRN3-2, p < 0.001; n = 4).
(C) 47S pre-rRNA synthesis determined by [32P] orthophosphate labeling in cells transduced with shRNA-mirs targeting UBF or RRN3 as compared to LMS alone (representative of n = 2).
(D) Pre-rRNA expression determined by qRT-PCR of the ITS1 in cells transduced with select shRNA-mirs and B cells described in (a) (LMS versus shUBF-2, p < 0.01; LMS versus shRRN3-1, p < 0.01; n = 4).
(E) GFP competition assay of GFP+, transduced Eμ-Myc lymphoma cells (LMS, shUBF, shRRN3) cocultured with GFP–, mock transduced Eμ-Myc lymphoma cells. Data expressed as fold change in percentage of GFP+ cells compared to Day 2 posttransduction (n = 3).
(F) Percentage of apoptotic cells determined by subG1 DNA content analysis expressed as fold change over LMS (6.3% ± 0.61 SEM) (LMS versus shUBF-1, p < 0.01; LMS versus shUBF-2, p < 0.001; n = 3) (LMS versus shRRN3-1, p < 0.05; n = 6). See also Figures S2A and S2B.
(G) GFP competition assay in Eμ-Myc lymphoma cells with BCL2 overexpression, expressed as fold change in percentage of GFP+ cells on Day 4 compared to Day 2 posttransduction (shRRN3-1 p < 0.01; n = 3). One-way ANOVA with Tukey's multiple comparison posttest (a,b,d,f). Student's two-tailed t test (g). Error bars represent SEM. See also Figures S2C and S2D.
CX-5461, A Selective Inhibitor of Pol I Transcription Induces p53-Dependent Apoptotic Cell Deathof Eμ-Myc Lymphoma Cells
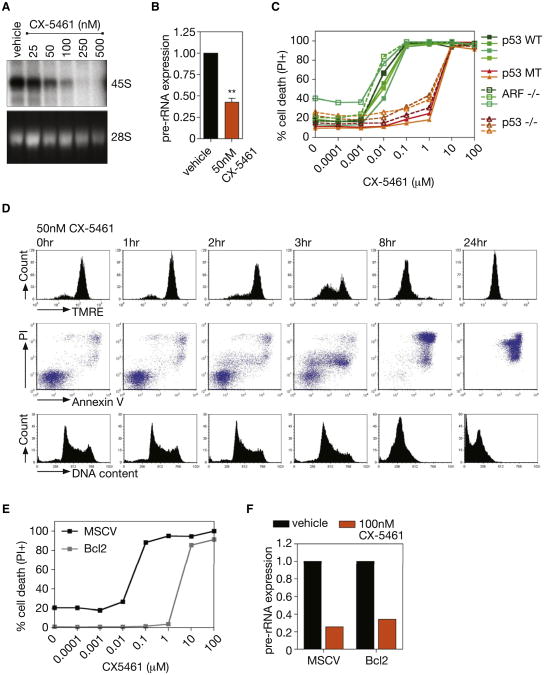
The profound sensitivityofEμ-Myc lymphoma cells to reductions in Pol I transcription led us to question whether this could be exploited therapeutically to treat B cell lymphoma. To test this approach, we utilized a highly selective agent (CX-5461) that inhibits Pol I-driven transcription in the low nanomolar range by preventing the association of the Pol I-specific transcription initiation selectivity factor SL-1 with the rDNA promoter, exhibiting >200-fold selectivity relative to the inhibition of Pol II-driven transcription (Drygin et al., 2011). Eμ-Myc lymphoma cells that had been isolated from tumor-bearing mice and cultured in vitro were exquisitely sensitive to CX-5461 with an IC50 of 27.3 nM ± 8.1 nM for Pol I transcription after 1 hr (Figures 3A, 3B, S3A, and S3B) and IC50 of 5.4 nM ±2.1 nM for cell death after 16 hr (Figure 3C). Under these conditions, transcription of the Pol II-dependent gene, β2m, was not affected (Figure S3C), attesting to the selectively of CX-5461 for Pol I.
Figure 3. CX-5461 Induces Rapid, p53-Dependent, Apoptotic Cell Death of Eμ-Myc Lymphoma Cells.
(A) 47S pre-rRNA synthesis in cultured Eμ-Myc lymphoma cells treated with increasing nM concentrations of CX-5461, determined by [32P] orthophosphate labeling (representative of n = 3). See also Figure S3A.
(B) Pre-rRNA expression determined by qRT-PCR in cells treated with 50 nm CX-5461 as compared to vehicle alone (p < 0.01; n = 3). See also Figures S3B and S3C.
(C) CX-5461 cell death curve of cultured Eμ-Myc lymphoma cell lines(including wild-type[WT], mutant [MT], and homozygous deleted [−/−]for indicated genes) 16 hr posttreatment, quantified by percentage of cells incorporating PI (representative of n = 3). See also Figures S3D and S3E.
(D) Induction of apoptosis in cultured Eμ-Myc lymphoma cells treated with 50 nM CX-5461, shown by loss of TMRE staining; increased Annexin V/PI costaining; increased cells with subG1 DNA content. See also Figures S3F-S3H.
(E) CX-5461 cell death curve of a p53 WT Eμ-Myc lymphoma cell line overexpressing BCL2 at 16 hr posttreatment, quantified by percentage of cells incorporating PI (representative of n = 2).
(F) Pre-rRNA expression in an Eμ-Myc lymphoma cell line overexpressing BCL2 in comparison to vector alone (MSCV), and treated with 100 nM CX-5461 as compared to vehicle as determined by qRT-PCR (n = 1). Student's two-tailed t test for all comparisons. Error bars represent SEM.
Loss or mutations of p53 or ARF are frequent lesions in Eμ-Myc driven lymphomagenesis (Eischen et al., 1999), so we tested a panel of Eμ-Myc lymphomas with variable p53 and ARF status for sensitivity to CX-5461. We observed that the ability of CX-5461 to kill lymphoma cells at low nM concentrations (IC50 < 10 nM) was dependent on intact p53 pathway but independent of ARF, which is frequently associated with oncogenic stress-dependent activation of p53. Cells, mutant or null for p53 exhibited a 180-fold decreased sensitivity to CX-5461 (Figure 3C). Importantly this was not due to elevated basal rates of Pol I transcription as the Eμ-Myc clones on the different genetic backgrounds exhibited similar Pol I transcription rates prior to treatment (Figure S3D). Moreover, consistent with our previous study demonstrating that CX-5461 is nongenotoxic, Eμ-Myc lymphoma cells null for ATM, amediator of DNA damage response, exhibited a comparable level of sensitivity to CX-5461 as Eμ-Myc lymphoma cells wild-type for ATM and p53 (Figure S3E).
Cell death in p53 wild-type lymphoma cells in response to CX-5461 was rapid and concomitant with induction of apoptotic markers, including progressive loss of mitochondrial membrane potential, increased AnnexinV/PI staining, and an increased percentage of cells with subG1 DNA content (Figures 3D and S3F–S3H). Enforced expression of BCL-2 in p53 wild-type Eμ-Myc cells reduced the cell death sensitivity to CX-5461 by over 130-fold despite exhibiting comparative inhibition of Pol I transcription (Figures 3E and 3F). Thus, repression of Pol I transcription in Eμ-Myc lymphoma cells by a small molecule inhibitor of Pol I induces rapid p53-dependent cell death by apoptosis.
Inhibition of Pol I Transcription by CX-5461 Induces p53-Mediated Death of Lymphoma Cells via Activation of the Rp-MDM2-p53 Nucleolar Surveillance Pathway
As Pol I transcription rates ultimately determine ribosome levels, it was possible that the apoptotic death of the Eμ-Myc lymphoma cells in response to suppression of Pol I transcription was an indirect response to ribosome insufficiency and thus defects in translation. Indeed, a recent study demonstrated that aberrant translation downstream of MYC hyperactivation is required for MYC's oncogenic potential (Barna et al., 2008). Alternatively, acute perturbations related to many aspects of ribosome biogenesis have been shown to contribute to a “nucleolar stress response,” also called the nucleolar surveillance pathway, which leads to rapid activation of p53, in an ARF-independent manner, and induction of apoptosis well before ribosome insufficiency and translation defects can occur (Boisvert et al., 2007; Boulon et al., 2010; Deisenroth and Zhang, 2010; Lohrum et al., 2003; Macias et al., 2010; Zhang et al., 2003). We examined these mechanisms and found that induction of apoptotic signaling was an immediate response to repression of Pol I transcription and not driven by ribosome insufficiency.
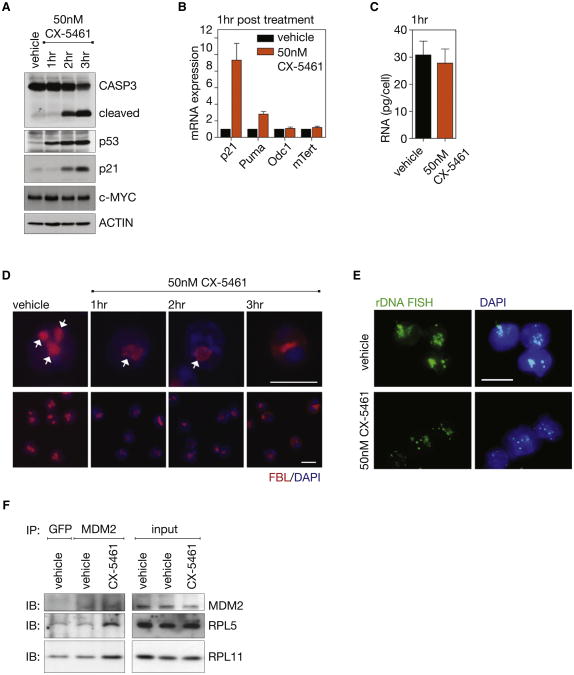
Treatment of Eμ-Myc lymphomas with CX-5461 for as little as 1 hr increased p53 levels followed by transactivation of p53 target genes, p21(Cdkn1a) and Puma(Bbc3), and cleavage and activation of CASPASE 3 (CASP3) at 2 hr (Figures 4A, 4B, and S4A), despite the fact that total RNA levels, which reflect ribosome abundance, were not decreased at these time points (Figure 4C). Notably, neither levels of MYC protein or expression of the MYC transcriptional targets ornithine decarboxylase 1(Odc1) and mTert were affected, demonstrating that the cell death was not due to alterations in MYC activity (Figures 4A and 4B). These findings are consistent with the induction of apoptosis being a direct result of disruption of Pol I transcription, and activation of nucleolar stress, rather than an indirect effect of ribosome insufficiency. To further examine this mechanism we monitored nucleolar integrity using a prototypical nucleolar marker protein fibrillarin (FBL), nucleophosmin (B23), and nucleolin (NCL), which demonstrate both nucleolar and nuclear localization, and by fluorescence in situ hybridization (FISH) for the 47S rDNA repeats. Within 1 hr of treatment with 50 nM CX-5461, the multiple nucleoli normally found in Eμ-Myc lymphoma cells had fused to form a single nucleolus per cell that exhibited reduced FBL staining (Figure 4D). Further analysis revealed that the altered FBL staining was accompanied by relocalization of a proportion of B23 and NCL to the periphery of the single nucleoli (Figure S4B). This correlated with condensation of the rDNA within the nucleoli to punctate foci, which is characteristic of a reduced Pol I transcription rate (Figures 4E and S4C) (McStay and Grummt, 2008). Activation of p53 by the Rp-MDM2-p53 nucleolar surveillance pathway has been attributed in part to the release of ribosomal proteins (RP), in particular RPL5 and RPL11, from the nucleolus and their binding to MDM2, a p53 E3 ubiquitin ligase. This results in the release of p53 from MDM2 and the subsequent accumulation of p53 (Deisenroth and Zhang, 2010). To test if CX-5461 acts, in part through the nucleolar surveillance pathway to activate p53, we immunoprecipitated MDM2 from Eμ-Myc lymphoma cells following CX-5461 treatment and examined the levels of RPL5 and RPL11 that coimmunoprecipitated with MDM2. Because MDM2 is a transcriptional target of p53, it was important to avoid possible confounding effects of increased levels of immunoprecipitated MDM2. A time course following treatment of cells with CX-5461 demonstrated that MDM2 expression increased dramatically beyond the 1 hr time point (Figure S4D); therefore we performed the coimmunoprecipitation experiments after 1hr treatment, prior to changes in MDM2 levels. Consistent with a nucleolar surveillance mechanism, coimmunoprecipitation experiments demonstrated an increase in the amount of RPL5 and RPL11 associated with MDM2 in p53 wild-type Eμ-Myc lymphoma cells in response to 1 hr treatment with CX-5461 relative to vehicle-treated cells (Figure 4F). Together with the data shown in Figures 1, 2, and 3, these findings provide independent lines of evidence for an absolute dependence of Eμ-Myc lymphoma cells on high rates of Pol I transcription for their survival, whereby perturbations in rRNA synthesis result in immediate induction of a nucleolar stress response leading to p53-dependent apoptotic cell death.
Figure 4. CX-5461 Activates p53 via the Nucleolar Stress Response in Eμ-MycLymphoma Cells.
(A) Western blot analysis over 3 hr of cleavage of CASPASE 3 (CASP3), total p53, p21, and c-MYC in a p53 wt wild-type Eμ-Myc lymphoma cell line treated with 50 nM CX-5461 in culture. See also Figure S4A.
(B) Relative p21, Puma, Odc1, and mTert mRNA expression determined by qRT-PCR (n = 3).
(C) Total RNA per cell (p > 0.05; n = 3) in cells from (a) at 1 hr of CX-5461 treatment.
(D) Nucleolar disruption shown in cells from (a) by fibrillarin (FBL) immunofluorescence with DAPI counterstain, nucleoli indicated by white arrows. See also Figure S4B.
(E) rDNA fluorescent in situ hybridization (FISH) with DAPI counterstain 2 hr post CX-5461 treatment. See also Figures S4C and S4D.
(F) Coimmunoprecipitation of ribosomal proteins L5 (RPL5) andL11(RPL11) with MDM2 (as comparedto GFP control)in vehicle-treated Eμ-Myc lymphoma cells overexpressing BCL2 cells versus cells treated with 500 nM CX-5461, as shown by western immunoblot (IB) analysis. Images taken at 60× magnification. Scale bar represents 10 μm. Student's two-tailed t test for all comparisons. Error bars represent SEM. See also Figure S4D.
Inhibition of Pol I Transcription by CX-5461 Selectively Induces p53-Mediated Cell Death of Lymphoma Cells In Vivo while Sparing Normal B Cells
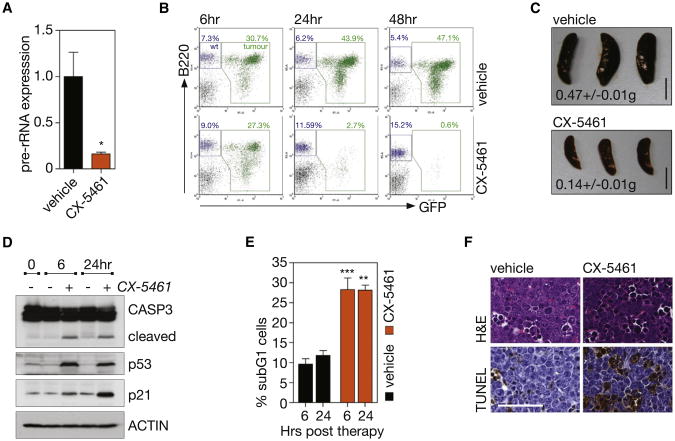
We next examined whether inhibition of Pol I transcription and thus activation of the Rp-MDM2-p53 nucleolar stress pathway could be used to selectively kill malignant B cells in vivo. C57BL/6 mice with established disease from transplanted Eμ-Myc lymphoma that is wild-type for p53 (Clone 4242) were treated with a single oral dose of CX-5461 (50 mg/kg) or vehicle. The Eμ-Myc tumor cells infiltrating the lymph nodes showed marked sensitivity to CX-5461, with cells exhibiting an 84% repression in Pol I transcription at 1 hr posttreatment (Figure 5A), also confirmed by RNA chromogenic in situ hybridization (CISH) for 47S pre-rRNA levels in the spleen (Figures S5A and S5B). Moreover, CX-5461 induced a rapid reduction in tumor burden in the lymph nodes (3.1% GFP-tagged malignant cells ± 0.20 for CX-5461-treated versus 34% GFP-tagged malignant cells ± 5.5 for vehicle-treated mice at 24 hr post therapy, p < 0.01) (Figures 5B and S5C) and a concomitant reduction of spleen size to within the normal range (0.14 g ± 0.01 for CX-5461-treated versus 0.47 g ± 0.01 for vehicle-treated mice at 24 hr posttherapy, p < 0.001) (Figure 5C). Consistent with the in vitro studies, reduction in malignant B cell numbers in vivo in response to CX-5461 therapy was preceded by the rapid activation of p53 and p21 and markers of apoptosis within 6 hr of drug administration (Figures 5D–5F and S5D).
Figure 5. Therapeutic Administration of CX-5461 Selectively Kills Transplanted p53 Wild-type Eμ-MycLymphoma Cells In Vivo.
(A) Pre-rRNA expression in tumor bearing lymph nodes of mice transplanted with p53 wild-type Eμ-Myc lymphoma cells (clone 4242) 1 hr posttreatment with 50 mg/kg CX-5461, determined by qRT-PCR (p < 0.05; n = 4). See also Figures S5A and S5B.
(B) FACS analysis of tumor burden in lymph node; stained with antibody against pan B cell marker B220, tumor cells GFP+ (green) (representative of n = 3). See also Figure S5C.
(C) Mean spleen weight ± SEM at 24 hr post therapy (p < 0.001; n = 3). Scale bar represents 1 cm.
(D) Western blot analysis at 6 and 24 hr of cleavage of CASP3, total p53, and p21 in the lymph node.
(E) Quantitation of apoptotic cells in lymph node, determined by subG1 DNA content analysis (vehicle versus CX-5461, p < 0.001 at 6 hr, p < 0.01 at 24 hr; n = 3). See also Figure S5D.
(F) Hematoxylin and eosin (H&E) and TUNEL stained lymph node sections at 6 hr post therapy; images taken at 40× magnification. Scale bar represents 50 μm. Student's two-tailed t test (a). One-way ANOVA with Tukey's multiple comparison posttest (c,e). Error bars represent SEM.
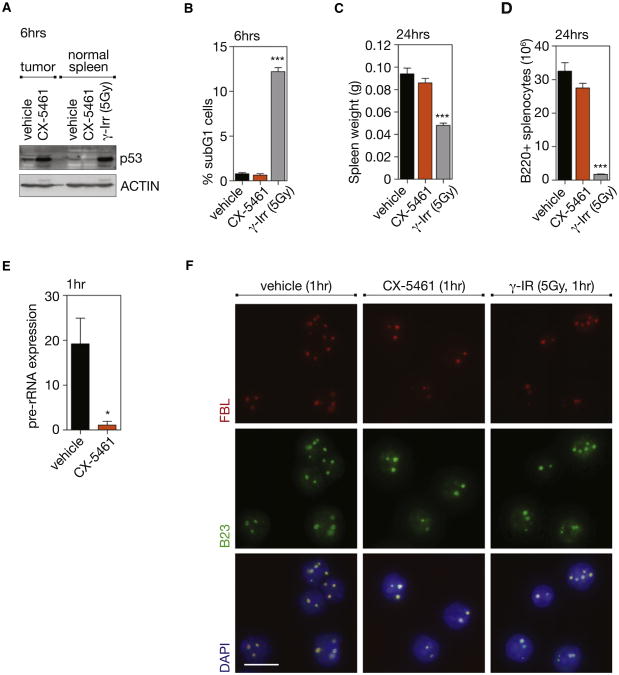
Although CX-5461 activated p53 and induced apoptosis among malignant cells, it did not trigger these responses in the normal spleen cells of wild-type nontumor bearing mice (Figures 6A and 6B) and did not affect either spleen size or B cell numbers (Figures 6C and 6D). The lack of a cytotoxic effect in normal spleens cells was not due to lack of inhibition of Pol I transcription as CISH demonstrated robust reductions in 47S pre-rRNA levels in the spleens of wild-type nontumor bearing mice as observed in tumor bearing mice (Figures 6E and S6A) Further-more, the nucleolar integrity of normal bone marrow cells from mice treated with CX-5461 was maintained, as determined by FBL and B23 immunofluorescence (Figure 6F). By comparison, exposure of nontumor bearing mice to 5Gy of γ-irradiation, aclinically appropriate dose for hematologic malignancies, resulted in marked elevation of p53 levels, apoptosis, reduced spleen weight, and B cell numbers (Figures 6A–6D). We also examined the response of B220+ B cells from the spleens and bone marrow of 4- to 6-week-old premalignant Eμ-Myc mice. These cells also demonstrated profound sensitivity to CX-5461 as characterized by p53 activation and markers of apoptotic cell death (increased CASP3 cleavage and sub-G1 cells) whereas the normal B cells from age-matched littermate mice failed to show p53 activation or increased cell death (Figures S6B–S6E). Together these data demonstrate the therapeutic response to inhibition of Pol I transcription by CX-5461 in vivo is highly selective for premalignant and malignant cells, which differentiates CX-5461 from genotoxic therapies such as γ-irradiation.
Figure 6. Therapeutic Administration of CX-5461, in Contrast to γ-Irradiation, Does Not Kill Normal B Cells In Vivo.
(A) Western blot analysis of total p53 in tumor-bearing lymph nodes (tumor) (Figure 5) or spleen from wild-type (WT) mice (normal spleens) treated for 6 hr with vehicle, 40 mg/kg CX-5461, or 5 Gy γ-irradiation (γ-Irr).
(B) Quantitation of apoptotic cells in spleens of WT mice treated with vehicle, 40mg/kg CX-5461or5 Gyγ-Irr; determinedby subG1 DNA content analysis (vehicle versus γ-Irr, p < 0.001; n = 5).
(C) Spleen weight of WT mice treated for 24 hr with vehicle, 40 mg/kg CX-5461 or 5 Gy γ-Irr (vehicle versus γ-Irr, p < 0.001; n = 5).
(D) Number of B220+ cells in WT spleen at 24 hr post therapy, as determined by FACS and comparative cell counts on whole organ suspensions (vehicle versus γ-Irr, p < 0.001; n = 5).
(E) Pre-rRNA expression in spleen from WT mice at 1 hr posttreatment with vehicle or 40 mg/kg CX-5461, as determined by CISH using an anti-sense probe to the ITS1 (p < 0.05; n = 3 spleens, mean of six fields of comparative regions per spleen; %threshold area). See also Figure S6A.
(F) FBL and B23 immunofluorescence with DAPI counterstain on bone marrow from WT mice 1 hr posttreatment with vehicle, 40 mg/kg CX-5461, or 5 Gy γ-Irr; images taken at 60× magnification. Scale bar represents10 μm. One-way ANOVA with Tukey's multiple comparison posttest (b–d). Student's two-tailed t test (e). Error bars represent SEM. See also Figure S6.
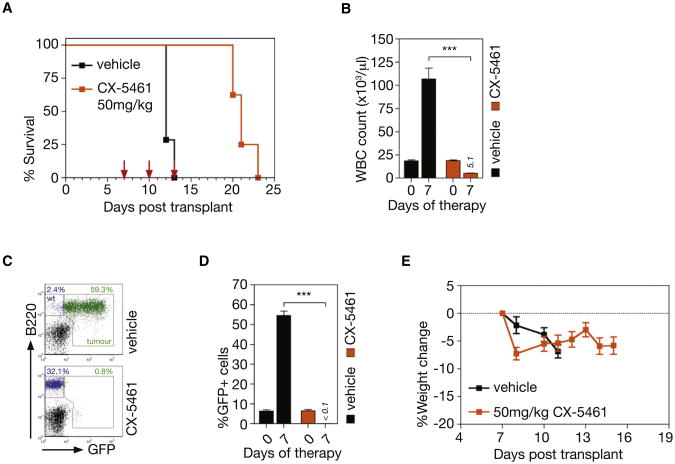
To further demonstrate the therapeutic effect of CX-5461 in this model, mice with established transplanted p53 wild-type Eμ-Myc lymphoma (clone 107) were treated with three doses of CX-5461 at 50 mg/kg once every 3 days. This regimen significantly prolonged the survival of the tumor-bearing mice (Figure 7A, p < 0.0001) and restored the white blood cell count to within the normal range (Figure 7B). Indeed, the day following the last dose of CX-5461, there were few identifiable GFP-tagged tumor cells in the peripheral blood (Figures 7C and 7D). Notably, there was preservation and restoration of the recipient-derived nonmalignant normal B cell population, again indicating selective eradication of malignant disease (Figure 7C, bottom panel). Moreover, more prolonged dosing with CX-5461 also showed minimal effects on normal B cells (data not shown). CX-5461 had no discernible adverse effect on the health of the mice and the measured body weights showed minimal deviation below that at the commencement of therapy and in comparison to vehicle-treated mice (Figure 7E). To assess the ability of prolonged dosing of CX-5461 to extend survival, mice transplanted with p53 wild-type Eμ-Myc tumors (clone 4242) were maintained on continuous repeat dosing of CX-5461 at 40 mg/kg once every 3 days (Figure S7A). This treatment regimen reproducibly increased the average survival compared to the three repeat dose regimen with the mice going through a stage of apparent disease remission (no detectable tumor cells in peripheral blood at day 7/8) before eventually relapsing (Figures S7B– S7E). CX-5461 was also evaluated in a murine genetic model of acute myeloid leukemia (AML1/ETO9a+Nras) that reproduces many of the clinical aspects of leukemias in human patients that carry AML1/ETO fusion genes (Zuber et al., 2009). As with therapy in the Eμ-Myc model, transplanted AML1/ETO9a+Nras leukemic cells showed marked sensitivity to CX-5461 in vivo, undergoing p53-dependent apoptotic cell death within 6 hr of treatment of a single 40 mg/kg dose of CX-5461 (Figures S7F– S7I). This apoptotic response was comparable to 100 mg/kg cytarabine, a cytotoxic drug frequently used in treatment of patients with AML.
Figure 7. Therapeutic Administration of CX-5461 Increases Survival from Transplanted p53 Wild-type Eμ-MycLymphoma.
(A) Kaplan-Meier curves showing increased survival of mice transplanted with p53 wild-type Eμ-Myc lymphoma (clone 107) treated with three doses of CX-5461 at 50 mg/kg in comparison to vehicle (dosing day indicated by arrows) (p < 0.0001; vehicle, n = 7; drug, n = 8).
(B) White blood cell (WBC) count at 0 and 7 days poststart of therapy (Day7 vehicle versus Day7 CX5461 p < 0.001; vehicle, n = 7; drug, n = 8).
(C) FACS analysis of tumor burden as measured by GFP+ B cells (B220+) in peripheral blood at 7 days posttherapy.
(D) Quantitation of FACS analysis (c) of tumor burden in the peripheral blood at 7 days poststart of therapy (p < 0.0001; vehicle, n = 7; drug, n = 8).
(E) Percent change in body weight of mice on therapy (vehicle, n = 7; drug, n = 8). Logrank test (a). One-way ANOVA with Tukey's multiple comparison posttest (b,d). Error bars represent SEM. See also Figure S7.
Inhibition of Pol I Transcription by CX-5461 Induces Apoptotic Cell Death of Human Hematological Malignancies with Sensitivity Correlating with p53 Mutational Status
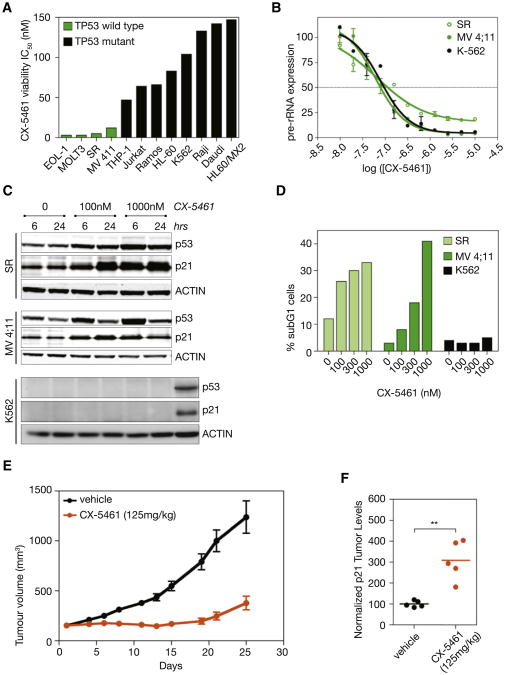
To translate our findings with CX-5461 to human hematologic malignancies, we evaluated CX-5461 against a genetically diverse panel of cells derived from human hematologic cancers. Consistent with the data from the murine models (Figure 3C), viability assays demonstrated that human hematologic cancer cells wild-type for p53 were significantly more sensitive to CX-5461 (median IC50 = 12 nM) than p53 mutant cells (median IC50 = 230 nM) (Figure 8A). Three cell lines, SR (large cell immunoblastic lymphoma), MV 4;11 (biphenotypic B myelomonocytic leukemia), and K562 (chronic myelogenous leukemia) were examined in further detail. Although all three cell lines exhibited similar IC50 s for Pol I transcription in response to CX-5461 (Figure 8B), the cell viability IC50 for the SR and MV 4;11 cell lines (IC50 = 5 nM and 12 nM, respectively) that are wild-type for p53, were 10–20-fold less than for K562 cells (IC50 = 104 nM) that carry a c.406_407insC nonsense mutation in p53 and hence do not express a stable p53 transcript. Consistent with this, CX-5461 induced rapid p53 accumulation and induction of the p53 target p21 (Figure 8C), in addition to nucleolar disruption (Figure S8B) and subsequent induction of apoptosis as determined by induction of a subG1 population (Figure 8D) and Caspase 3/7 activity (after 24 hr) in both the SR and MV 4:11 lines (Figure S8A). In contrast, the p53-deficient K562 cells did not undergo apoptosis in response to CX-5461 (Figures 8D and S8A).
Figure 8. Human Leukemia and Lymphoma Cell Lines Treated with CX-5461.
(A) The result of 96 hr treatment with CX-5461 on viability of human leukemia and lymphoma cell lines with different p53 status (p < 0.0009 comparing TP53 wild-type [WT] IC50 to TP53 mutant IC50).
(B) Effect of CX-5461 on Pol I transcription in human cancer cell lines, as determined by qRT-PCR to the 5′ externally transcribed spacer (5′ ETS) region of the 47S pre-rRNA. Error bars represent SEM.
(C) CX-5461 induces p53 and p21 in p53 WT SR and MV 4;11 cell lines, but not in the p53 null K562 cell line, as determined by western blot analysis. A549 cell lysates used as positive control for p53 and p21 in K562 blots (far right lane).
(D) CX-5461 induces apoptosis in p53 WT SR and MV 4;11 cell lines, but not in the p53 null K562 cell line, as determined by subG1 DNA content analysis. See also Figures S8A and S8B.
(E) Activity of CX-5461 in vivo against MV 4;11 xenograft model as measured by increased tumor volume over days of therapy (n = 10). Error bars represent SEM. See also Figure S8C.
(F) CX-5461 induces p21 in MV 4;11 xenografts, as determined by western blot analysis of p21 normalized to ACTIN using densitometry (p < 0.001; n = 5). Student's two-tailed t test.
To determine whether this apoptotic response of cells derived from human hematologic malignancies following inhibition of Pol I transcription could be translated into a therapeutic index in vivo, MV 4:11 cells were transplanted subcutaneously into nude mice and the effect of CX-5461 on tumor growth examined. Notably, CX-5461 administered once weekly for 3 weeks demonstrated potent antitumor activity (Figure 8E), was associated with increased tumor levels of the p53 transcriptional target gene p21 (Figure 8F), and was well tolerated (Figure S8C).
Together these studies establish that Pol I transcription can be selectively targeted in vivo using a small molecule inhibitor of Pol I to preferentially eradicate hematologic malignant cells by activation of p53, while simultaneously maintaining a normal populations of cells of the same lineage.
Discussion
Herein we have used RNAi to the essential Pol I-specific transcription factors UBF and RRN3 to provide robust genetic evidence that accelerated Pol I transcription is required for survival of malignant cells. Specifically we have show that reductions in rDNA transcription rate by as little as 35% result in apoptotic cell death of Eμ-Myc lymphoma cells rather than slower growing tumor cells as might be predicted. Using a newly described small molecule inhibitor of Pol I transcription, CX-5461, we extended these findings to demonstrate that dysregulation of the synthesis of rRNA plays an essential role in cancer cell biology in vivo, thus providing an important alternative approach for the treatment of malignant diseases. One of the most striking and unanticipated findings of these studies was the demonstration that the apoptotic death induced by reductions in Pol I transcription occurred rapidly as the result of immediate activation of p53 following perturbations of the nucleolus rather than an indirect effect resulting from reduced ribosome accumulation and defective protein translation. This is in contrast to what was previously demonstrated in the Eμ-Myc lymphoma model following ribosomal protein haploinsufficiency (Barna et al., 2008). Indeed when apoptosis was blocked in Rrn3 knockdown Eμ-Myc cells, the resultant 35% reduction in rDNA transcription rate did not result in a proliferative disadvantage of these cells. Thus contrary to the commonly accepted belief, rDNA transcription rates are not always rate limiting for optimal proliferative growth of mammalian cells. Our data demonstrate that MYC's control of Pol I transcription and nucleolar integrity is required for its oncogenic potential, independent of its function in regulating ribosome levels, protein translation, and proliferative growth. These findings also lend strong support for the evolving paradigm of the nucleolus being a key regulator of biology of the cancer cell distinct from its role in determining the abundance of ribosomes (Boisvert et al., 2007).
The rapid disruption of the nucleolus and subsequent induction of p53-mediated apoptotic cell death in response to CX-5461 is consistent with induction of the nucleolar stress pathway (also called the ribosome biogenesis surveillance/sensing pathway) (Boulon et al., 2010; Deisenroth and Zhang, 2010; Fumagalli et al., 2009) that acutely monitors the integrity of ribosome biogenesis pathway and activates p53 when the fidelity of this complex process is dysregulated. This pathway can be initiated by the accumulation of free ribosomal proteinsfollowing perturbation of ribosome biogenesis (Boulon et al., 2010; Deisenroth and Zhang, 2010) or MYC activation (Macias et al., 2010). The release of free ribosomal proteins from the nucleolus has been shown to sequester the p53 ligase MDM2 leading to elevated p53 levels and to directly increase p53 translation, the net result of which is induction of apoptosis (Boulon et al., 2010; Deisenroth and Zhang, 2010). Thus, the ribosome surveillance pathway (ribosomal protein-MDM2-p53) works independently of ARF and functions in parallel and cooperatively with ARF-dependent MYC oncogenic stress pathways to restrain MYC oncogenesis (Macias et al., 2010). Most likely, not only does impaired ribosome biogenesis act as a trigger to induce p53, but ribosome biogenesis is also a homeostatic regulator in normal cells, tightly linking on-going production of newly synthesized ribosomes with p53 levels (Boulon et al., 2010; Deisenroth and Zhang, 2010; Fumagalli et al., 2009). Here we show how this process can be exploited as a therapeutic strategy to selectively kill malignant cells. Moreover, as a nongenotoxic, cancer-specific activator of p53, CX-5461 is ∼300-fold more potent at killing Eμ-Myc lymphoma cells (IC50 for cell death of 5.4 nM) than Nutlin-3A, a selective small-molecule MDM2 antagonist and p53 activator (IC50 for cell death of 1600 nM, results not shown), that is currently in clinical trials (Saha et al., 2010).
The data herein also demonstrate that Pol I transcription can be therapeutically targeted to selectively activate p53 and kill malignant cells of hematologic origin in vivo while maintaining a viable population of wild-type cells of the same lineage. Indeed, normal B cells isolated from the bone marrow of wild-type mice treated in vivo with CX-5461 at doses sufficient to kill malignant Eμ-Myc B cells, did not exhibit disruption of their nucleoli, activation of p53 or induction of apoptosis despite having a significantly reduced rate of Pol I transcription. In contrast, treatment of wild-type mice with doses of γ-irradiation similar to those used therapeutically for human hematologic malignancies induced robust apoptotic cell death associated with p53 activation in normal B cells. This striking tumor cell selective activity following inhibition of Pol I transcription by CX-5461 is most likely a function of the robust upregulation of ribosome biogenesis we have shown to occur during MYC driven lymphomagenesis sensitizing cells to the induction of the nucleolar stress pathway. The MYC-induced upregulation of rRNA synthesis also requires a stoichiometric elevation in the levels of ribosomal proteins in the nucleolus. Inhibition of rRNA synthesis in cancer cells would result in excessive accumulation of free ribosomal proteins and subsequent activation of the nucleolar stress pathway and p53 as described above. Consistent with this model, treatment of Eμ-Myc lymphoma cells with CX-5461 promoted enhanced association of ribosomal proteins L5 and L11 with MDM2 and subsequent activation of p53. Furthermore, premalignant B cells of 4- to 6-week-old Eμ-Myc transgenic mice also displayed p53-mediated increased sensitivity to CX-5461 providing evidence that MYC overexpression alone, independent of a fully malignant phenotype, may serve to prime the nucleolar stress pathway in cells. Given that a range of other oncogenes including PI3K, AKT, and Ras also play important roles regulating ribosome biogenesis (Chan et al., 2011; Hannan et al., 2003; Ruggero and Pandolfi, 2003; Stefanovsky et al., 2001; White, 2005), our findings indicate that selective activation of the ribosome biogenesis surveillance pathway to activate p53 using small molecule inhibitors of Pol I is likely to be therapeutically useful in the treatment of a wide range of tumors in addition to those under control of MYC. Indeed, we demonstrated that CX-5461 potently induced p53-dependent apoptosis of non-MYC driven tumor cells in vivo using a transgenic model of AML in which the leukemic cells coexpress the AML/ETO9a fusion protein with NRas. However, we note that increased expression of MYC is common in hematologic malignancies even when it is not the primary driving event (for example AML/ETO9a leukemia cells have elevated MYC levels). Thus we cannot discount the possibility that MYC may be a common factor in conferring sensitivity to apoptotic death in hematologic malignancies in response to Pol I inhibition.
We also demonstrated that CX-5461 also exhibits potent antigrowth capacity in a broad range of human hematological tumor cell lines with sensitivity being dependent on the genetic and functional status of p53. Importantly, p53 gene mutations are rare at diagnosis in common cancers of hematopoietic origin (Saha et al., 2010), suggesting that inhibition of Pol I may be a broadly applicable therapeutic approach for treatment of numerous human hematological cancers. In support of this, CX-5461 demonstrated potent antitumor activity in a xenograft model of biphenotypic B myelomonocytic leukemia. Intriguingly, human cell lines from solid tumors do not generally demonstrate p53 dependent apoptotic responses following Pol I transcription inhibition (Drygin et al., 2011) but instead undergo senescence and autophagy. This differential response highlights the dissimilarity in the crosstalk between the nucleolar stress and cell death in different types of cancer. It is possible that hematological malignancies have a unique nucleolar biology that makes them especially susceptible to activation of p53 and apoptotic cell death following acute perturbations of ribosome biogenesis. In support of this notion, mutations in genes encoding ribosomal proteins that cause defects in ribosome biogenesis and activate p53 is emerging as a shared paradigm among bone marrow failure syndromes such as Diamond Blackfan Anemia (DBA) and the acquired 5q syndrome myelodysplastic syndrome (del [5q] MDS) that both exhibit increased cancer susceptibility. Together these data suggest that patients with hematologic malignancies represent a highly sensitive population for the initial trial of CX-5461 in humans.
In summary, our work demonstrates that hematologic tumor cells depend upon hyperactivated rDNA transcription and integrity of Pol I transcription for survival and this dependency is vulnerable to perturbations that induce apoptosis via a process that is independent of ribosome levels or effects on proliferation. Further, we show that a selective inhibitor of Pol I transcription, CX-5461, causes cancer-specific activation of p53 and induction of apoptosis that results in the improvement of outcome in in vivo models of leukemia and lymphoma.
Experimental Procedures
Eμ-Myc Transgenic Mice, B Cell Purification, and Lymphoma Cell Lines
All animal work was performed with approval from the Peter MacCallum Cancer Centre Animal Experimentation Ethics Committee and Cylene Pharmaceuticals' Institutional Animal Care and Use Committee. Eμ-Myc mice were maintained as heterozygote on a C57BL/6 background. Primary B cells were purified from the spleen of transgenic mice and their wild-type littermates with anti-B220 conjugated microbeads (Miltenyi Biotech) using the autoMACS system (Miltenyi Biotech) according to manufacturer's instructions Lymphoma cell lines were generated from lymph nodesof tumor bearing Eμ-Myc/+, Eμ-Myc/p53+/−, Eμ-Myc/Arf+/− mice, and Eμ-Myc/Atm−/−. Cell lines were determined as mutant for p53 via sequencing or assessment of protein molecular weight via western blotting, in addition to exhibiting resistance to etoposide. For in vivo drug studies, 1 × 105 Eμ-Myc lymphoma cells were injected into the tail veins of wild-type C57BL/6 mice. Peripheral blood was sampled by retro-orbital bleeding into 10 mM EDTA. For in vivo drug studies with the mouse model of AML, Scott Lowe generously provided leukemia cells isolated from the spleens of tumor-bearing mice. Leukemia cells (1 × 106), AML1/ETO9a NRas p53 wt, or p53−/−, were injected into the tail vein of irradiated (2×3 Gy) recipient C57BL/6 mice. Leukemia onset/progression was monitored by peripheral blood analysis and single dose of 40 mg/kg CX-5461 was administrated when ∼40%-50% GFP+ cells in the peripheral blood were detectable. Lymphoma cell lines were maintained in Anne Kelso DMEM supplemented with 10% FBS, 100 μM L-asparagine (Sigma), 0.5% b-mercaptoethanol, and penicillin/streptomycin/glutamine (GIBCO). Cell volume was determined using a Beckman Coulter Z series. shRNA-mediated knockdown was achieved via retroviral transduction as described (Schmitt et al., 2000) using the CSHL vector system LMS (Dickins et al., 2005) with the short-hairpin sequences listed in Supplemental Experimental Procedures. Transduced populations were selected by sorting for GFP expressionon a BD FACS Vantage SE. Bcl2 overexpression was achieved by sorting for Cherry expression following retroviral transduction with a MSCV vector expressing Bcl2 cDNA.
Orthophosphate Labeling
Lymphoma cells (1.5 × 106) were cultured in 3 ml media in the presence of 500 μCi of 32P orthophosphate for 20 min. Cells were harvested on ice and RNA extracted using the QIAGEN RNAeasy kit, according to the manufacturer's instructions. RNA (7 μg) was run overnight on a 1.2% MOPS/formaldehyde agarose gel. The gel was dried using a Bio-Rad gel dryer, exposed overnight to a phospho imager screen and scanned using a Typhoon TRI variable mode imager (GE Healthcare). The radioactivity of each band was determined using the Image Quant TL software (GE Healthcare).
Immunofluorescence and FISH
For immunofluorescence, cells were fixed in a 2% paraformaldehyde suspension and spun onto slides. Cells were stained with antibodies listed in Supplemental Experimental Procedures followed by the appropriate secondary, either 594 conjugated goat anti-rabbit IgG (Molecular Probe, A21442) or 488 conjugated goat anti-mouse IgG1 (Jackson ImmunoResearch Laboratories). Stained cells were fixed again with 4% formaldehyde, counterstained with DAPI, and visualized on an Olympus BX-51 microscope, with images captured using the SPOT Advance image acquisition software (Diagnostic Instruments). For FISH, 1 μg of BAC clone (RP23-225M6) was biotin-labeled using a nick translation kit (Roche), according to the manufacturer's instructions. Labeled probe (100 ng) was precipitated in ethanol together with 30 μg salmon sperm DNA and 18 mg COT1 carrier DNA (Invitrogen) and dissolved in 30 ml hybridization buffer (50% formamide, 20% dextran sulfate in 2× SSC). Probes were denatured at 95°C for 5 min then incubated at 37°C for 1 hr. Cells were fixed in paraformaldehyde and spun onto slides as described above. Slides were fixed in methanol/acetic acid (3:1) for 5 min at room temperature and dried then dehydrated through an ethanol series. Slides were incubated in 70% formamide/2× SSC for 10 min at 83°C and then dehydrated through an ethanol series. Slides were hybridized overnight at 37°C in a humidified chamber and then washed in 50% formamide/2× SSC at 42°C for 15 min and 0.1 × SSC at 60°C for 15 min. Streptavidin-Alexa fluor 488 was added for 1 hr at 37°C and slides were then washed in 0.05% Tween-20/4× SSC for 15 min. Slides were mounted in DAPI and microscopy was performed as above. Quantitation was performed using MetaMorph (Molecular Devices, Sunnydale, CA).
Histology
Harvested tissues were fixed in 10% neutral buffered formalin overnight followed by paraffin wax embedding. Hematoxylin and eosin staining (H&E) was performed on 4 μm sections. TUNEL staining was performed on 4 μm sections using an Apoptag Peroxidase In Situ Apoptosis Detection kit (Chemicon, S7100), according to the manufacturer's instructions. For CISH, 4 μm sections were dewaxed in xylene followed by an ethanol series. Epitope recovery was performed with incubation at 120°C in 10 mM sodium citrate (pH 6.0) followed by the addition of proteinase K(12.5 ng/ml) and finally, fixation in formaldehyde (4%). Hybridization was performed overnight at 37°C with digoxigenin-labeled oligonucleotide rRNA anti-sense probe to the ITS1. Sense probe hybridization was performed under identical conditions on serial sections as a negative control. Sections were incubated with anti-Digoxigeninalkaline phosphatase (Roche, 11093274910) and signal was detected in 100 mM NaCl, 100 mM Tris (pH 9.5) buffer containing 0.18 mg/ml BCIP, 0.34 mg/ml NBT (Roche, 11 681 451 001), and 240 μg/ml levamisole (Sigma). Tissues were counter stained with nuclear fast red stain. Sections were visualized on an Olympus BX-51 and images captured using the SPOT Advance image acquisition software. Quantitation was performed using MetaMorph (Molecular Devices).
MV 4;11Xenograft Model
Female immunocompromised mice CrTac:Ncr-Foxn1nu (6–8 weeks old) were obtained from Taconic Farms (Germantown, NY). Animals were maintained under clean room conditions in sterile filter top cages. Animals received sterile rodent chow and water ad libitum. All procedures were conducted in accordance with the Institute for Laboratory Animal Research Guide: The Care and Use of Laboratory Animals. Xenografts were initiated by subcutaneous injection of 5 × 106 MV4;11 cells into the right hind flank region of each mouse. When tumors reached a designated volume of ∼150 mm3, animals were randomized and divided into Vehicle (50 mM NaH2PO4, pH 4.5) or CX-5461 treatment groups of 10 mice per group. CX-5461 was administered intraperitoneally once a week at 125 mg/kg for the length of 25 days. Tumor volumes and body weights were measured twice weekly. The length and width of the tumor were measured with calipers and the volume calculated using the following formula:tumor volume = (length × width2)/2. Tumor growth inhibition (TGI) was determined on the last day of study according to the formula: TGI (%) = [100 – (VfD – ViD)/(VfV – ViV) × 100], where ViV is the initial mean tumor volume in vehicle-treated group, VfV is the final mean tumor volume in vehicle-treated group, ViD is the initial mean tumor volume in drug-treated group, and VfD is the final mean tumor volume in drug-treated group.
Supplementary Material
Significance.
Morphologic abnormalities of the nucleolus, the site of ribosomal gene (rDNA) transcription by RNA Polymerase I (Pol I), have been recognized as diagnostic for cancer for over a century. Nevertheless, an unresolved question has been whether the accelerated ribosome biogenesis responsible for the nucleolar changes is required for malignancy. Here we provide the definitive evidence, both genetic and pharmacologic, that accelerated rDNA transcription and nucleolar integrity are necessary for oncogenic activity. Further, we show that Pol I transcription can be targeted in vivo to treat lymphoma and leukemia through the nongenotoxic activation of p53-dependent apoptosis, while sparing normal cells. Thus, selective inhibition of Pol transcription, can serve as a therapeutic strategy for the treatment of cancer.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia project grants; NHMRC Research Fellowship to R.D.H.; NHMRC Postgraduate Research Scholarship, GSK Postgraduate Research Scholarship and Leukaemia Foundation Postdoctoral Fellowship to M.J.B.; Cancer Council of Victoria Sir Edward Weary Dunlop Clinical Research Fellowship and NHMRC Research Fellowship to G.A.M.; The John T. Reid Charitable Trusts and Mrs. Margaret Ross AM; and in part by Cylene Pharmaceuticals. We thank Joseph Trapani and James Whisstock for critical evaluation of the manuscript, Brian McStay for anti-sera to RRN3 and Sinisa Volarevic for anti-sera to rpL5. We also thank Rachael Walker, Kym Stanley, Analia Lesmana, Kerry Ardley, Susan Jackson, Jeannette Valentan, Kathryn Kinross, Petranel Ferrao, Ralph Rossi, and Sarah Ellis for technical assistance.
M.J.B., G.P., G.A.M., and R.D.H. were responsible for the overall concept and design of experiments. D.D., K.A., M.H., M.K.S., D.M.R., W.G.R., S.W.L., and R.W.J provided essential materials. M.J.B., G.P., E.S., N.Hein, A.P., C.C., L.C., N.H., C.P., J.B., D.D., G.A.M., and R.D.H. were responsible for the collection and assembly of data. M.J.B., G.P., E.S., N.Hein, C.C., M.W., D.D., K.A., R.W.J., R.B.P., G.A.M., and R.D.H. were involved in data analysis and interpretation. M.J.B., G.P., E.S., C.C., M.W., D.D., K.A., W.G.R., S.W.L., R.W.J., R.B.P., G.A.M., and R.D.H. were involved in the writing of the manuscript and all authors approved the final form. D.D., K.A., N.H., C.P., J.B., M.H., M.K.S., D.M.R., and W.G.R. are affiliated with Cylene Pharmaceuticals in being current or past employees of the company. However, this did not influence the conduct of the research described in this manuscript and had no bearing on the decision to submit this paper for publication.
Footnotes
Supplemental Information: Supplemental Information includes eight figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.ccr.2012.05.019.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, Fahlén S, Hydbring P, Söderberg O, Grummt I, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M, et al. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal. 2011;4:ra56. doi: 10.1126/scisignal.2001754. [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Montanaro L, Treré D. What the nucleolus says to a tumour pathologist. Histopathology. 2009;54:753–762. doi: 10.1111/j.1365-2559.2008.03168.x. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho C, O'Brien S, Proffitt C, Omori M, Haddach M, Schwaebe M, Siddiqui-Jain A, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxyterminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan RD, Hempel WM, Cavanaugh A, Arino T, Dimitrov SI, Moss T, Rothblum L. Affinity purification of mammalian RNA polymerase I. Identification of an associated kinase. J Biol Chem. 1998;273:1257–1267. doi: 10.1074/jbc.273.2.1257. [DOI] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci USA. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Klein G. Multistep evolution of B-cell-derived tumors in humans and rodents. Gene. 1993;135:189–196. doi: 10.1016/0378-1119(93)90065-b. [DOI] [PubMed] [Google Scholar]
- Larminie CG, Alzuherri HM, Cairns CA, McLees A, White RJ. Transcription by RNA polymerases I and III: a potential link between cell growth, protein synthesis and the retinoblastoma protein. J Mol Med. 1998;76:94–103. doi: 10.1007/s001090050196. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindström MS, Zhang Y. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K, Wall M, Brandenburger Y, Palatsides M, Pearson RB, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Wall M, Sanij E, Siwicki K, Ellul J, Brown D, Holloway TP, Hannan RD, McArthur GA. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011;39:3267–3281. doi: 10.1093/nar/gkq1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Saha MN, Micallef J, Qiu L, Chang H. Pharmacological activation of the p53 pathway in haematological malignancies. J Clin Pathol. 2010;63:204–209. doi: 10.1136/jcp.2009.070961. [DOI] [PubMed] [Google Scholar]
- Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Rosenthal CT, Lowe SW. Genetic analysis of chemoresistance in primary murine lymphomas. Nat Med. 2000;6:1029–1035. doi: 10.1038/79542. [DOI] [PubMed] [Google Scholar]
- Shiue CN, Berkson RG, Wright AP. c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene. 2009;28:1833–1842. doi: 10.1038/onc.2009.21. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell. 2001;8:1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Gröne HJ, Schütz G, Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, McCurrach ME, Yang MM, Dolan ME, Kogan SC, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.