Abstract
Abundant evidence suggests a central role for the amyloid-β (Aβ) peptide in Alzheimer’s disease (AD) pathogenesis. Production and clearance of different Aβ isoforms have been established as targets of proposed disease-modifying therapeutic treatments of AD. However, previous studies used multiple sequential purification steps to isolate the isoforms individually and quantitate them based on a common mid-domain peptide. We created a method to simultaneously purify Aβ isoforms and quantitate them by the specific C-terminal peptides in order to investigate Aβ isoform physiology in the central nervous system. By using standards generated from in vitro metabolic labeling, the relative quantitation of four peptides representing total amount of Aβ (Aβ-Tot), Aβ38, Aβ40 and Aβ42 were achieved, both in cell culture and human CSF. Standard curves for each isoform demonstrated good sensitivity with very low limits of detection and high accuracy. Because the assay does not require antibody development for each Aβ isoform peptide, significant improvements in the throughput and accuracy of isoform quantitation were achieved.
Keywords: Alzheimer’s disease, Amyloid beta isoforms, Cerebrospinal fluid, Mass Spectrometry, Relative quantitation
Introduction
Alzheimer disease (AD)1 is associated with an excess amount of amyloid-beta (Aβ) in the brain. Therefore, metabolic studies involving the kinetic rates of Aβ are an attractive clinical approach to understand the pathophysiology of AD and apply it to clinical trials of disease-modifying therapeutic treatments. The metabolic rates of isoforms of different proteins have been implicated in several human diseases [1]; in AD, several Aβ isoforms support a causal role, for example Aβ42 [2, 3, 4, 5], Aβ43 [6] and modified forms including pyrolization[2, 7], oxidation, isomerization [8], and partial proteolytic degradation. Metabolic turnover of Aβ has clear implications under the amyloid hypothesis, which states that AD is caused by an imbalance between Aβ production and clearance, resulting in increased amounts of Aβ in various forms such as monomer, oligomer, insoluble fibrils, and plaques in the Central Nervous System (CNS) [1]. Dominantly inherited AD is caused from mutations in amyloid precursor protein (APP), Presenilin 1 or Presenilin 2 genes. These mutations are thought to cause AD at an early age due to the over production of Aβ, or an increase in Aβ42 to Aβ40 ratio [9]. It has been reported that abundance Aβ15, Aβ17 and other shorter N-terminal variants in the cerebrospinal fluid (CSF) of AD patients compared to normal patients indicates the progression from normal to AD [10] even more so when APP is glycosylated at Tyr-10 of the Aβ domain, leading to increasing γ-secretase activity at these locations [11].
Aβ is a cleavage product of APP, through sequential proteolytic processing by β- and γ-secretases [12]. The γ-secretase, which cuts at the C-terminal end of the Aβ peptide, cleaves within the transmembrane region of APP to generate a number of Aβ isoforms of 36–43 amino acid residues in length. Three major forms of Aβ detected in the CSF are Aβ40, Aβ38, and Aβ42 [13, 14] with other minor abundance forms 15, 16, 17, 34, 37, and 39 amino acids in length [15, 16, 17]. Although Aβ40 is produced at higher levels, Aβ42 is more hydrophobic and prone to form aggregates. Importantly, Aβ42 is more neurotoxic and seems to be of particular importance for plaque formation [2, 18], while Aβ40 appears to be the most abundant peptide deposited in cerebral vessels [19]. It has been shown that the relative concentration of Aβ42, in addition to the total Aβ concentration are critical factors in the rate of amyloidogenesis [20]. The length of the C-terminal seems to be critical for the oligomerization and neurotoxicity [6] of the Aβ peptide, thus longer peptide variants seeding Aβ polymerization in vivo.
Previously, we studied the metabolism of Aβ40 and Aβ42 isoforms independently, by serial immunoprecipitation (IP) with different antibodies directed to Aβ42 and Aβ40 [21, 22, 23]. We found that AD was associated with slowed clearance of both Aβ42 and Aβ40 [23], indicating that Aβ clearance mechanisms may be critically important in the development of AD [24]. However, each sample required separate IP, enzymatic digestion and injection for liquid chromatography (LC) and mass spectrometry (MS) analysis of each isoform, thus duplicating the amount of lab work for each isoform studied. A recent study quantitated the Aβ isoforms in human CSF by analyzing the intact peptides [25]. However, the sizes of the intact Aβ peptides were too large and not suitable for the stable isotope labeling kinetics (SILK) quantitation used in our study. Another study quantitated the Aβ isoforms in rat tissues [26] using tryptic digests. Aβ tryptic digests, however, generate isoform specific peptides that do not ionize efficiently due to the formation of multiple charge states and oxidized forms during MS. To overcome these issues, we developed a more efficient and specific quantitative isoform characterization and kinetics method to accelerate SILK studies. This method utilized one antibody which bound to a common epitope in order to purify all isoforms from the same sample at the same time. We then cleaved the peptides with a metalloendopeptidase [27] to produce a C-terminal peptide unique to each of the Aβ isoforms. We adopted triple quadrupole mass spectrometry, operated in the selected reaction monitoring (SRM) mode, for targeted quantitation of multiple Aβ isoform peptides due to its high specificity and its ability to monitor many product ions of a large number of peptides.
Methods
Preparation of H4 APP695ΔNL Cell Media Standards
Immortalized H4 APP695ΔNL neuroglioma cells expressing human Aβ were used to prepare 13C6-leucine-labeled Aβ standards in vitro. Cells were grown until near confluency in DMEM supplemented with 10% dialyzed, fetal bovine serum. The media was changed to serum-free DMEM/Ham’s F-12, lacking leucine and supplemented with 105 mg/L leucine (or 0–20% 13C6-leucine, 98% 13C6, Cambridge Isotope Laboratories, Andover, MA). Labeled media (containing 13C6-leucine) was serially diluted (0, 1.25%, 2.5%, 5%, 10% and 20%) with unlabeled media to generate six sets for use in the production of standards for use to generate calibration curves. Labeled media from cell culture containing secreted proteins was collected after 3 days of labeling, replacing it with fresh serum-free media with 0–20% 13C6-leucine each time. Labeled media was filtered, pooled, aliquoted, then stored at −80°C until use.
In vivo Labeling
The labeling was done as described previously [21, 22, 23]. All human studies were approved by the Washington University Human Studies Committee and the General Clinical Research Center (GCRC) Advisory Board. Informed consent was obtained from all participants and they were screened to be in good general health and without neurologic disease. Briefly, the participants were admitted to the Washington University Medical Center and a 13C6-labeled leucine solution was infused through an IV for 9 hours. Six mL of CSF was obtained through a lumbar catheter every hour for 36 hours. All 37 sampled CSF samples were processed as described below under “Isolation of Aβ and digestion” sub-section, followed by digestion with 2.5 ng of metalloendopeptidase (Lys-N), then analysis by LC/SRM. The percent labeled Aβ at each hour time point was determined for the Aβ isoforms.
Isolation of Aβ and digestion
HJ5.1, an anti-human Aβ monoclonal antibody directed against amino acids 13–28, was coupled to CNBr-activated sepharose beads per the manufacturer’s instructions (GE Healthcare). The IP mixture comprised of 1 mL of cell culture media or 800 μl CSF, 12.5 μL 100x protease inhibitor cocktail (Roche), 20μl of a solution containing U:15N- labeled Aβ38 (1.5 ng), Aβ40 (10 ng) and Aβ42 (1 ng) as external standards (rPeptide, Bogart, GA), 110 μl of 5M guanidine hydrochloride, and 30 μL of HJ5.1-beads slurry. The mixtures were rotated for 2 hours at room temperature. The beads were then centrifuged at 4,000x G and the supernatant was removed. The beads were washed with 1 mL 0.5 M guanidine hydrochloride solution followed by two washes with 1 mL of 25 mM ammonium bicarbonate solution, with centrifugation and aspirating the supernatant away each time. The beads in the final wash were aspirated to dryness and then neat formic acid was added to elute Aβ from the antibody-bead complex. The formic acid supernatant was transferred to a new polypropylene tube and dried in a vacuum dryer for about 30 to 60 minutes. The dried Aβ complex was re-suspended in 25 mM ammonium bicarbonate solution and ready for protease digestion. A suspension of 2.5 ng Metalloendopeptidase (Lys-N) (Associates of Cape Cod Inc., MA) in 25 mM ammonium bicarbonate was added (estimated 1:10 Lys-N to protein ratio), and incubated for 16 hours at 37 °C. The digest was dried and reconstituted first by adding 2 μl of neat formic acid (98%) to the dried spot and vortexing to mix. This was followed by adding 20% dimethyl sulfoxide (DMSO), for a final resuspension solution concentration of 10% formic acid and 18% DMSO. The resuspended digest was centrifuged at 20,000x G for 15 minutes and then transferred to autosampler vials for running on the LC/SRM system.
Identification of SRM transitions and nanoLC tandem MS
Separation of peptides was done using an Eksigent NanoLC-2D-Ultra (Eksigent Technologies, Dublin, CA) operated in a 1D mode at a flow rate of 500 nL/min. The peptides were detected in SRM mode on TSQ Vantage triple quadrupole MS (ThermoFisher Scientific, San Jose, CA) equipped with a nanospray source and a column heater (Phoenix S&T, Chester, PA). Samples were kept at 4°C in an autosampler, and a 5 μL aliquot was injected each time onto a nano column packed in-house (150-μm × 15 cm) with a 3 μm Zorbax SB300 C18 column packing material (Agilent Technologies, Santa Clara, CA). Solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid in acetonitrile. The gradient was 20% B to 65% B in 15 min followed by 65% B to 95% B in 5 min, then to 20% B in 5 min and re-equilibration for another 5 min. The TSQ Vantage was operated in positive ion mode using a spray voltage of 1.2 kV, and capillary temperatures of about 300°C. Synthetic Aβ42 was digested with Lys-N and infused into the TSQ Vantage for tuning and optimization on the C-terminal peptide (KGAIIGLMVGGVVIA). The peak widths for Q1 and Q3 were both set at 2.0 Da (FWHM) and a collision pressure in Q2 of 2.0 mTorr. Based on the results, the transition ions of other Aβ isoform specific peptides were predicted in silico, using the Pinpoint Software (ThermoScientific). A list of SRM precursor and product ions (Supplementary Table 1) were created and imported into the instrument control software for quantitating the peptides. The various SRM transitions were validated using Aβ standards produced in media by cell culture. Aβ was immunopurified from the media, Lys-N digested, and data on Aβ28–X fragments were acquired on the TSQ Vantage MS. The SRM profiles were quantitated and analyzed using the Xcalibur software.
Results
Sample Preparation
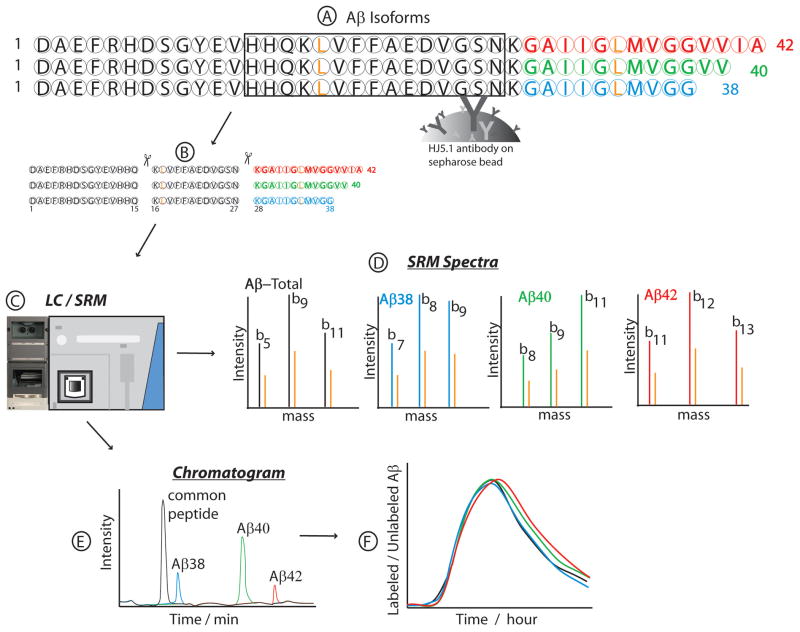
The HJ5.1 antibody-bead conjugate has been extensively studied and the IP efficiency for Aβ isoform complex was found to be nearly 100%. This efficiency was monitored by measuring the SRM peak areas detected of two known synthetic standards. The first of the standards was U:15N-labeled Aβ external standards that was spiked into the sample before the IP process to correctly identify the retention times of the various Aβ isoforms that were measured from the matrix of cell culture media and CSF proteins. They also were used to check IP efficiency and Aβ recovery after sample drying steps. It was also combined with a second external standard, a 13C6-leucine labeled C-terminal Lys-N digest of Aβ42 (KGAIIGLMVGGVVIA) which was spiked into sample just before and after digestion to check proteolytic cleavage efficiency. ELISA quantitation methods were also used to monitor the binding efficiency of the antibody during the IP process, but no significant amounts of Aβ were detected in the post-IP supernatant. Typical yields were 97% for antibody recovery of Aβ and 99% after eluting the Aβ with formic acid. This showed that our method recovered almost all the Aβ isoforms, and there was little to no acid hydrolysis during the time (approximately 30 min) that samples were in formic acid solution. Our resuspension techniques also recovered almost 100% of Aβ after samples were dried. The amount of protease used was also optimized and digestion efficiency was typically 99% for Aβ. The general overview of the procedure was illustrated in Figure 1, where HJ5.1 antibody was illustrated to be capturing all Aβ isoforms in step (A), followed by Lys-N digestion in step (B) at the N-terminal end of Lysyl-residues. Step C showed the diagram for LC and MS that were used to generate SRM spectra in (D), in the order of eluted peptides in the chromatogram shown in step (E). Protease cleavage of Aβ by Lys-N generated peptides which were unique to the Aβ isoforms, plus another peptide from the domain common to all the isoforms. Specifically, cleavage of Aβ by Lys-N was at the N-terminal sites of lysyl amino acid residues at positions 16 and 28. Three fragments were produced, but only two are of interest in leucine labeling kinetic studies, as those two peptides have one leucine each in the peptide sequence. Of the two leucine-containing peptides, one at the C-terminal is unique to each C-terminal Aβ isoform. The chromatographic separation of the Aβ digest was monitored for peptides representing the amount of Aβ-total (Aβ16-27, KLVFFAEDVGSN), Aβ38 (Aβ28-38, KGAIIGLMVGG),Aβ40 (Aβ28-40, KGAIIGLMVGGVV) and Aβ42 (Aβ28-42, KGAIIGLMVGGVVIA). The SRM peaks for labeled and unlabeled peptides were calculated over time, usually 36 hours, and those ratios are plotted against the time points of CSF sampling to generate SILK profile illustrated in Figure 1(F).
Figure 1.
Schematic diagram of the quantitative isoform characterization and kinetics assay, by using stable isotope labeling kinetics (SILK) technique. Aβ peptide isoforms were purified from the same sample source (media or CSF) using common epitope antibody bound to beads (HJ5.1) (A) and digested with Metalloendopeptidase (Lys-N) to generate isoform specific peptides (B). The digest was separated and quantitated by LC/SRM (C), resulting in unique SRM spectra from each peptide (D). The chromatographic elution profile of the various peptides as detected by SRM is as shown in (E). The least hydrophobic peptide common to all isoforms elutes first, followed closely by the peptide for Aβ38, then Aβ40 and Aβ42. Isotopic enrichment of each Aβ isoform was calculated as the ratio of the SRM peak area for labeled peptide to unlabeled peptide. This relative quantitation of chromatographic SRM peak ratios was computed for each time point of CSF collection, to generate a graph of Aβ isoform kinetics. Leucine residues (L) (colored gold) indicate 13C6-Leucine labeling sites in the peptides. The unique C-terminal residues were highlighted in different colors to distinguish one unique isoform peptide from the other. Aβ42 is colored red, Aβ40 green and Aβ38 blue. The spectra were illustrated in colors to match those colors used for the C-terminal peptides they were generated from, with their corresponding gold-colored spectra from 13C6-Leucine labeling.
LC/SRM analysis of Aβ Isoforms
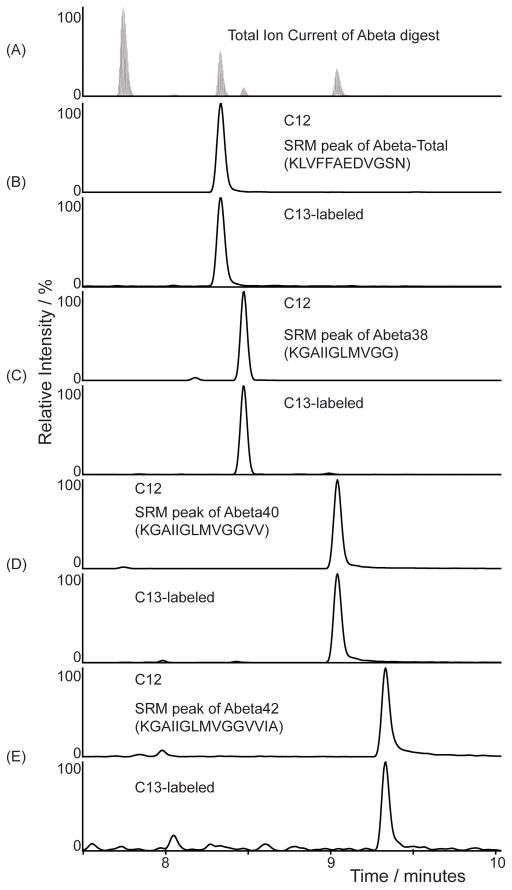
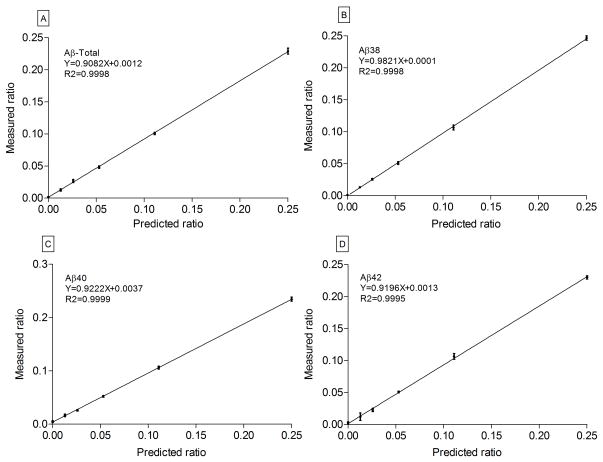
All four peptides eluted and ionized well on LC/MS, except the Aβ42 peptide. Due to its extreme hydrophobic nature, significant ‘tailing’ was noticed on most columns tested. For effective elution, a combination of a 3 micron particle size column and a column heater were employed. This combination significantly reduced tailing of peaks and excessive back pressures. All 4 peptides were separated and detected from the same sample as shown in Figure 2. Both the endogenous 12C form and the stable isotope 13C6-leucine labeled form of the peptides co-eluted as expected. The IP coupled with the LC/SRM method reduced interfering signals to <0.1% as detected in the chromatograms. In figure 2(A), the SRM peak shows the elution profiles of the Aβ peptides that were detected by LC/SRM after Lys-N digestion. All four peptides were well-separated and detected from the same sample as shown in figure 2(A–E), with the mid-domain peptide representing total Aβ eluting first, followed by Aβ38, Aβ40 and Aβ42. Due to the wide dynamic range, the SRM peak for Aβ42 is not visible on the chromatogram. The peaks (figure 2B to E) represent the ion pairs for 12C (top) with its corresponding 13C labeled peptides (bottom) as detected by LC/SRM. The SRM peaks were for (b) Aβ-Total (m/z 663.3 and 666.3) and Aβ isoforms (C) Aβ38 (m/z 508.3 and 511.3), (d) Aβ40 (m/z 607.4 and 610.4) and (e) Aβ42 (m/z 699.4 and 702.4). The full list of transitions used to acquire the SRM data has been reported as supplementary table S1. For the relative quantitation performed here to generate the SILK profile, the endogenous unlabeled 12C peptide was set as the internal standard and the 13C6-labeled peptide was set as the unknown (or target peptide), as required by the Xcalibur data processing software (ThermoScientific). The percent of 13C6 leucine incorporated into Aβ was determined by area ratios of labeled to unlabeled peptide peaks from the SRM experiments. From the media standards, calibration curves were generated and shown in figure 3, where relative quantitation of unlabeled to labeled Aβ ratios for (a) Aβ-Total using Aβ16-27, (b) Aβ38 using Aβ28-38, (c) Aβ40 using Aβ28-40 and (d) Aβ42 using Aβ28-42, showed accurate and precise measurements. For each data point, the area under the SRM peaks were calculated for labeled and unlabeled peptides and expressed as a ratio. The measured ratios of 13C6-Leucine labeling were plotted against the predicted ratios used in producing the standards. All four calibration curves, done in quadruplicates, indicated high linear correlations (R2= 0.999) with slopes of at 0.99 least 0.9, (figure 3) with minimal error. The standard deviations ranged from 0.001 to 0.006, indicating the assay was reproducible and robust with a very low limit of quantitation (LOQ) of <1% 13C6-labeling. Aβ concentration in CSF was determined previously by enzyme-linked immunosorbant assays (ELISA) [28]. The amounts of Aβ42 were in the range of 206–685 pg/ml (45 – 152 fmol), which gives an average of about 100 fmol of Aβ recovered in a 20 μl per sample after each IP. Thus, with an injection of 5 μl, 25 fmol were routinely analyzed. From our standard curves in figure 3, it was estimated that this assay has a lower LOQ of at least 1% of 13C6-labeled peptides, equivalent to 250 attomoles of Aβ42.
Figure 2.
The SRM chromatogram of Aβ isoforms detected from media standards labeled to 10% 13C6-Leucine incorporation. The ion chromatograms demonstrate the elution profiles of the C-terminal peptides generated from the various Aβ isoforms from the same CSF sample, and aligned below the total ion count (TIC) plot (A). The peptides were eluted off the LC column based on their hydrophobicity. The first to elute was the peptide common to all isoforms, followed closely by the peptide for Aβ38, then Aβ40 and Aβ42. Peaks of ion pairs of unlabeled and 13C6-Leucine labeled peptides, as detected by SRM were plotted below the TIC plot with their retention times. Among the 4 different peptides monitored were theAβ-Total, common peptide contributed by all major isoforms of Aβ, which eluted first (B). This was followed by the C-terminal peptides for Aβ38 (C), then Aβ40 (D), and Aβ42 (E). Isotopic enrichment of each Aβ isoform was calculated as the ratio of the area of the SRM peak for labeled peptide divided by the area of the unlabeled peptide. Sample labeled to 10% 13C6-Leucine labeling was used as an example to show that the endogenous and 13C6-Leucine labeled pairs co-elute.
Figure 3.
Calibration curve plots of measured vs. predicted ratios of labeled Aβ isoform standards representing Aβ-Total (A), Aβ38 (B), Aβ40 (C) and Aβ42 (D). Labeled media was serially diluted (0, 1.25%, 2.5%, 5%, 10% and 20%) with unlabeled media to generate six sets for use in the production of standards for these curves. Aβ isoforms were immunoprecipitated from the media, Lys-N digested and the unique fragments were analyzed on TSQ Vantage MS. There were four biological replicates for each sample set. Due to very low errors in measurement, the point markers were not shown. Indicated instead were the standard deviations at the point marker locations. Measured ratio of labeled to unlabeled peptides (mean ± standard deviation between replicates), of each set are shown on the Y-axis with the predicted values on the X-axis. The linear regression lines show an accuracy (slope) of at least 0.9 and a precision (R2) of at least 0.999.
In vivo Aβ isoform metabolism
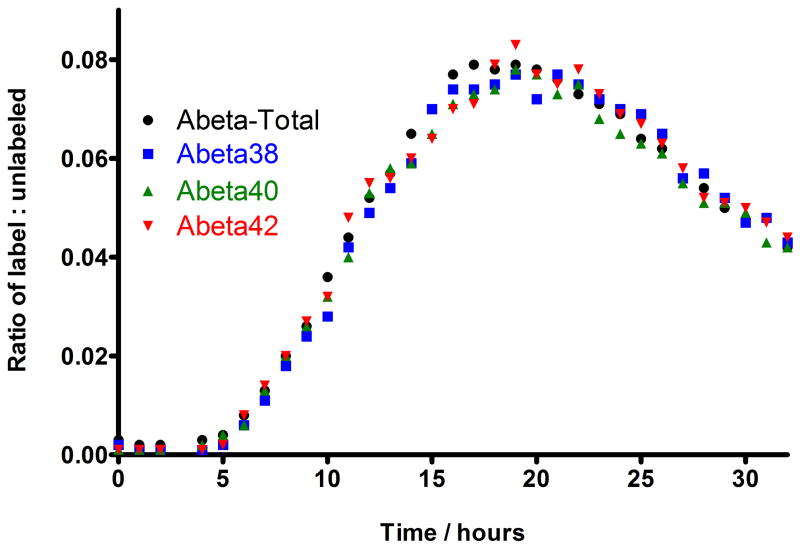
In vivo CSF measurements were done in a healthy human participant. The SILK study (figure 4) demonstrated that the metabolism of the Aβ isoforms Aβ38, Aβ40 andAβ42 could be monitored in a single injection from each sample. The pattern of production and clearance of each Aβ isoform is similar and comparable to prior published reports [21, 22, 23]. In figure 4, the SILK profiles of Aβ38 (blue squares), Aβ40 (green triangles), and Aβ42 (red triangles) are presented for each Aβ isoform. The 13C6-leucine isotopic enrichments of Aβ38, Aβ40, and Aβ42 recovered from CSF were compared from a healthy participant to establish the relationship between Aβ isoform metabolic kinetics. The start time of 13C-label incorporation and the time to reach peak 13C-labeling in each Aβ isoform was measured and found to be the same for all the isoforms. Start time was after 4 hours and peak time point was between 18 to 20 hours. The 13C-labeling from all isoforms also cleared in the same fashion. In fact, the time course of all isoforms cannot be distinguished from each other.
Figure 4.
SILK profiles of Aβ isoforms to demonstrate the relative quantitation of production and clearance of Aβ isoforms in healthy human’s CSF over 36 hours. The isotopic enrichment time courses of Aβ 38 (blue squares), 40 (green triangles) and 42 (red triangles) are presented for each Aβ isoform, in relation to total Aβ (black circles). The kinetics curve shows there was no detectable incorporation of label in the first 4 hours. This was followed by an increase in percent labeled Aβ (production phase) at hours 5 to 15, which plateaued near steady-state levels of labeled leucine (up to about 10% at time points of between 18 to 20 hours), before decreasing (clearance phase) over the last 12 hours of the study (from time point 21 to 32 hours). The steady-state value (10%) was estimated as the average amount of labeled leucine in the CSF measured during labeling. From these curves, the Fractional Synthesis Rate (FSR) and Fractional Clearance Rate (FCR) of each isoform could be calculated as previously described [21]. The kinetic curves of Aβ38, Aβ40, and Aβ42 quantitated from CSF were compared to show the relationship between Aβ isoform metabolic kinetics.
Discussion
The relative quantitation of Aβ isoforms was previously done by immunopurification of each individual isoform by specific antibodies in a serial fashion, which required expensive and lengthy IP development for each isoform. Further, specificity was a concern due to cross-reactivity by nonspecific binding of Aβ isoform species. The assay described herein was faster, more economical, and a more specific way to quantify Aβ isoforms for metabolism studies. Simultaneous studies of isoforms from the same sample, including the ones for which antibodies do not exist, provided more reliable data and avoided issues of the specificity of purifying Aβ isoforms. With this method, we demonstrated the measurement of Aβ38, Aβ40 and Aβ42 production and clearance rates in in vivo experiments, successfully reproduced our previous results [23], and enabled the study of a larger number of participants within a shorter period of time. The separation and quantitation of Aβ isoforms, especially Aβ42, were challenging, due to the extremely high hydrophobicity of the peptides, strong self-aggregation, non-specific binding tendencies and peptide separation by LC. Due to higher than normal peptide hydrophobicity and the previous unavailability of analytical detection tools, this method of analysis was not considered plausible or deemed too difficult to do. For example, the C-terminal fragments generated upon tryptic cleavage of Aβ, i.e. Aβ29-x, were too long and are not well suited for analysis by LC-MS because of their high hydrophobicity [6]. In this study, a combination of harsher solubility techniques, smaller particle size of LC column used, and the use of a column heater enabled the detection and quantitation of these very hydrophobic Aβ peptides. For our LC/SRM, we analyzed Aβ28-x which was generated with Lys-N. This introduced an amino-terminal lysyl-residue into the peptide sequence with better ionization efficiency of the Aβ C-terminal peptides, in contrast to the use of other proteases such as trypsin. To aid the solubility of the peptides, we used higher amounts of formic acid than normal (10%) and 20% dimethyl sulfoxide (DMSO) to re-suspend the peptides. Also, using a column heater at high temperature lowered the viscosity of the mobile phase, and increased the diffusivity of solvents which showed as lower back pressures. The Aβ peptides were stable at the operating column temperature of 75°C. This quick assay can measure the physiology of Aβ isoforms in in vitro cell culture and human body fluids by simultaneously measuring the Aβ isoforms’ in vivo metabolic rates as demonstrated with the SILK profiles. The technique enables the field to demonstrate whether Aβ42, Aβ40 and Aβ38 have similar or different kinetic rates under health or diseased states. We have been able to demonstrate that the kinetics of all Aβ isoforms measured were the same for a healthy participant in our AD study. This assay will accelerate Aβ metabolism research and facilitate a better understanding of the pathophysiology of the brain in AD. This information may lead to improved diagnostic testing and more precise pharmacodynamics of proposed disease-modifying treatments.
Supplementary Material
Acknowledgments
Funding: The Hope Center for Neurological Disorders at Washington University (R.J.B.), NIA K23 AG030946 (R.J.B.), NINDS RO1-NS065667 (R.J.B.), and NIH-supported UL1 RR024992, TL1 RR024995 and KL2 RR 024994 (ICTS CRU), Washington University Biomedical Mass Spectrometry Research Facility (P30-RR000954, DK020579), and Washington University Nutrition Obesity Research Center (DK056341).
We thank the participants for their time and contributions and Dr. David Holtzman, for the donation of the HJ5.1 antibody. None of the above stated competing interests alter our adherence to all the Analytical Biochemistry’s policies on sharing data and materials.
Footnotes
Abbreviations used: AD, Alzheimer’s disease; Aβ, amyloid beta; CSF, cerebrospinal fluid; IP, immunoprecipitation: ELISA, enzyme-linked immunosorbant assay; LC, liquid chromatography; MS, mass spectrometry; SRM, selected reaction monitoring; DMSO, dimethyl sulfoxide; LLOQ, lower limit of quantitation.
Competing interests: R.J.B. co-founded C2N Diagnostics and is on the scientific advisory board of C2N Diagnostics which did not support this work. Washington University has a pending patent on material presented in this report with Drs. Bateman and Mawuenyega listed as inventors.
Authors Contribution
Conceived and designed experiments: KGM and RJB. Performed experiments: KGM, TK and WS. LC/SRM and data analysis: KGM. Wrote the paper: KGM, TK, RJB. All authors revised the manuscript for important intellectual content and gave final approval of the version to be published.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 3.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 4.Levy-Lahad E, Wijsman EM, Nemens E, Anderson L, Goddard KA, Weber JL, Bird TD, Schellenberg GD. A familial Alzheimer’s disease locus on chromosome 1. Science. 1995;269:970–3. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 5.Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–6. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 6.Welander H, Franberg J, Graff C, Sundstrom E, Winblad B, Tjernberg LO. Abeta43 is more frequent than Abeta40 in amyloid plaque cores from Alzheimer disease brains. J Neurochem. 2009;110:697–706. doi: 10.1111/j.1471-4159.2009.06170.x. [DOI] [PubMed] [Google Scholar]
- 7.Saido TC, Iwatsubo T, Mann DM, Shimada H, Ihara Y, Kawashima S. Dominant and differential deposition of distinct beta-amyloid peptide species, A beta N3(pE), in senile plaques. Neuron. 1995;14:457–66. doi: 10.1016/0896-6273(95)90301-1. [DOI] [PubMed] [Google Scholar]
- 8.Roher AE, Lowenson JD, Clarke S, Wolkow C, Wang R, Cotter RJ, Reardon IM, Zurcher-Neely HA, Heinrikson RL, Ball MJ, et al. Structural alterations in the peptide backbone of beta-amyloid core protein may account for its deposition and stability in Alzheimer’s disease. J Biol Chem. 1993;268:3072–83. [PubMed] [Google Scholar]
- 9.Kumar-Singh S, Theuns J, Van Broeck B, Pirici D, Vennekens K, Corsmit E, Cruts M, Dermaut B, Wang R, Van Broeckhoven C. Mean age-of-onset of familial alzheimer disease caused by presenilin mutations correlates with both increased Abeta42 and decreased Abeta40. Hum Mutat. 2006;27:686–95. doi: 10.1002/humu.20336. [DOI] [PubMed] [Google Scholar]
- 10.Portelius E, Westman-Brinkmalm A, Zetterberg H, Blennow K. Determination of beta-amyloid peptide signatures in cerebrospinal fluid using immunoprecipitation-mass spectrometry. J Proteome Res. 2006;5:1010–6. doi: 10.1021/pr050475v. [DOI] [PubMed] [Google Scholar]
- 11.Halim A, Brinkmalm G, Ruetschi U, Westman-Brinkmalm A, Portelius E, Zetterberg H, Blennow K, Larson G, Nilsson J. Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc Natl Acad Sci U S A. 2011;108:11848–53. doi: 10.1073/pnas.1102664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–41. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 13.Portelius E, Brinkmalm G, Tran AJ, Zetterberg H, Westman-Brinkmalm A, Blennow K. Identification of novel APP/Abeta isoforms in human cerebrospinal fluid. Neurodegener Dis. 2009;6:87–94. doi: 10.1159/000203774. [DOI] [PubMed] [Google Scholar]
- 14.Portelius E, Zetterberg H, Dean RA, Marcil A, Bourgeois P, Nutu M, Andreasson U, Siemers E, Mawuenyega KG, Sigurdson WC, May PC, Paul SM, Holtzman DM, Blennow K, Bateman RJ. Amyloid-beta1-15/16 as a Marker for gamma-Secretase Inhibition in Alzheimer’s Disease. J Alzheimers Dis. 2012 doi: 10.3233/JAD-2012-120508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–66. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 16.Wiltfang J, Esselmann H, Bibl M, Smirnov A, Otto M, Paul S, Schmidt B, Klafki HW, Maler M, Dyrks T, Bienert M, Beyermann M, Ruther E, Kornhuber J. Highly conserved and disease-specific patterns of carboxyterminally truncated Abeta peptides 1-37/38/39 in addition to 1-40/42 in Alzheimer’s disease and in patients with chronic neuroinflammation. J Neurochem. 2002;81:481–96. doi: 10.1046/j.1471-4159.2002.00818.x. [DOI] [PubMed] [Google Scholar]
- 17.Portelius E, Tran AJ, Andreasson U, Persson R, Brinkmalm G, Zetterberg H, Blennow K, Westman-Brinkmalm A. Characterization of amyloid beta peptides in cerebrospinal fluid by an automated immunoprecipitation procedure followed by mass spectrometry. J Proteome Res. 2007;6:4433–9. doi: 10.1021/pr0703627. [DOI] [PubMed] [Google Scholar]
- 18.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–7. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 19.Gravina SA, Ho L, Eckman CB, Long KE, Otvos L, Jr, Younkin LH, Suzuki N, Younkin SG. Amyloid beta protein (A beta) in Alzheimer’s disease brain. Biochemical and immunocytochemical analysis with antibodies specific for forms ending at A beta 40 or A beta 42(43) J Biol Chem. 1995;270:7013–6. doi: 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- 20.Jan A, Gokce O, Luthi-Carter R, Lashuel HA. The ratio of monomeric to aggregated forms of Abeta40 and Abeta42 is an important determinant of amyloid-beta aggregation, fibrillogenesis, and toxicity. J Biol Chem. 2008;283:28176–89. doi: 10.1074/jbc.M803159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman RJ, Munsell LY, Morris JC, Swarm R, Yarasheski KE, Holtzman DM. Human amyloid-beta synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat Med. 2006;12:856–61. doi: 10.1038/nm1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bateman RJ, Munsell LY, Chen X, Holtzman DM, Yarasheski KE. Stable isotope labeling tandem mass spectrometry (SILT) to quantify protein production and clearance rates. J Am Soc Mass Spectrom. 2007;18:997–1006. doi: 10.1016/j.jasms.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- 25.Lame ME, Chambers EE, Blatnik M. Quantitation of amyloid beta peptides Abeta(1-38), Abeta(1-40), and Abeta(1-42) in human cerebrospinal fluid by ultra-performance liquid chromatography-tandem mass spectrometry. Anal Biochem. 2011;419:133–9. doi: 10.1016/j.ab.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe K, Ishikawa C, Kuwahara H, Sato K, Komuro S, Nakagawa T, Nomura N, Watanabe S, Yabuki M. A new methodology for simultaneous quantification of total-Abeta, Abetax-38, Abetax-40, and Abetax-42 by column-switching LC/MS/MS. Anal Bioanal Chem. 2012;402:2033–42. doi: 10.1007/s00216-011-5648-1. [DOI] [PubMed] [Google Scholar]
- 27.Taouatas N, Drugan MM, Heck AJ, Mohammed S. Straightforward ladder sequencing of peptides using a Lys-N metalloendopeptidase. Nat Methods. 2008;5:405–7. doi: 10.1038/nmeth.1204. [DOI] [PubMed] [Google Scholar]
- 28.Bateman RJ, Wen G, Morris JC, Holtzman DM. Fluctuations of CSF amyloid-beta levels: implications for a diagnostic and therapeutic biomarker. Neurology. 2007;68:666–9. doi: 10.1212/01.wnl.0000256043.50901.e3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.