Abstract
A group of enzymes, mostly hydrolases or certain transferases, utilize one or a few side-chain carboxyl groups of Asp and/or Glu as part of the catalytic machinery at their active sites. This review follows mainly the trail of studies performed by the author and his colleagues on the structure and function of such enzymes, starting from ribonuclease T1, then extending to three major types of carboxyl peptidases including aspartic peptidases, glutamic peptidases and serine-carboxyl peptidases.
Keywords: ribonuclease (RNase) T1, pepsin, nepenthesin, aspergilloglutamic peptidase, physarolisins I and II
Introduction
The author and his colleagues have been working for over fifty years mainly on the structure and function of proteins and enzymes, especially certain groups of enzymes such as ribonucleases (RNases), peptidases and glutathione S-transferases. The studies on peptidases involved numbers of extra- and intracellular peptidases. In this review, the author follows mainly the trail of his studies on four groups of typical enzymes with a catalytic carboxyl group or groups. They include: Section I - RNase T1 and its homologs, Section II - aspartic peptidases: pepsin, nepenthesin and their homologs, Section III - glutamic peptidases: aspergilloglutamic peptidase and its homologs and Section IV - serine-carboxyl peptidases: physarolisins I and II and their homologs. These enzymes were shown to commonly possess a catalytic carboxyl group or groups at their active sites, which may function as general acid/base or nucleophilic catalysts. Although serine-carboxyl peptidases are classified as a member of serine peptidases, they are tentatively included in this review since they also possess catalytic carboxyl groups.
I. RNase T1 and its homologs
RNase T1 is an extracellular enzyme found by Sato and Egami in 1957 in Taka-Diastase, a commercial enzyme mixture from Aspergillus oryzae, and was shown to hydrolyze specifically the 3′-phosphodiester bond of guanylic acid in RNA unlike the well-studied bovine pancreatic RNase A which is specific for the 3′-cytidylic and 3′-uridylic acids. For this specificity, RNase T1 drew special attention as a specific hydrolyzing agent in RNA structure studies as well as an interesting target in enzyme structure-function studies.1–5) In 1958, I started structure and function studies of this enzyme under the guidance of Prof. Fujio Egami in his laboratory at the Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo.
(i). Purification and characterization.
Previously the enzyme had never been purified to homogeneity; therefore the first step was purification of the enzyme from “Taka-Diastase,” a crude enzyme mixture obtained from the culture medium of Aspergillus oryzae. Fortunately, the timely introduction of DEAE-cellulose chromatography made it possible to purify the enzyme to homogeneity for the first time. Thus the purified enzyme could be submitted to molecular characterization.6–9) It was shown to be composed of a single polypeptide chain of about 100 amino acid residues cross-linked by two disulfide bonds. Preliminary structure-function studies indicated that the enzyme has a His residue or residues essential for activity10) and that the two disulfide bonds can be reduced and then reoxidized with restoration of activity.11) These results were similar to those reported for RNase A, suggesting a similar structure-function relationship between the two enzymes. However, the amino groups were shown to be unimportant for the activity of RNase T1 unlike in RNase A.12)
(ii). Primary structure determination.
To extend the structure-function studies further, determination of the complete primary structure was thought to be prerequisite. Thus, I decided to determine the amino acid sequence of the enzyme. In 1955, F. Sanger’s group first succeeded in the determination of the amino acid sequence of bovine insulin (51 residues). After that, various proteins of molecular weight over 10,000 became the major targets of sequencing. Among these sequence studies, that of RNase A (124 residues) was most advanced and its amino acid sequence was determined in 196013) with some revisions in 1962–1963.
I started large scale preparation of RNase T1 in 1960 and about 3 g of the purified enzyme was obtained from over ten kilograms of Taka-Diastase powder. Then I initiated the sequencing work in the Prof. Egami’s laboratory and completed it in the laboratory of Prof. Toshio Ando at the same department in 1965 including the location of the disulfide bonds.14–19) Thus RNase T1 was shown to be a 104-residue single chain protein cross-linked by two disulfide bonds (Fig. 1a). The amino acid sequence was totally different from that of RNase A. Meanwhile the amino acid sequence of hen egg-white lysozyme (129 residues) was reported in 1963.20,21)
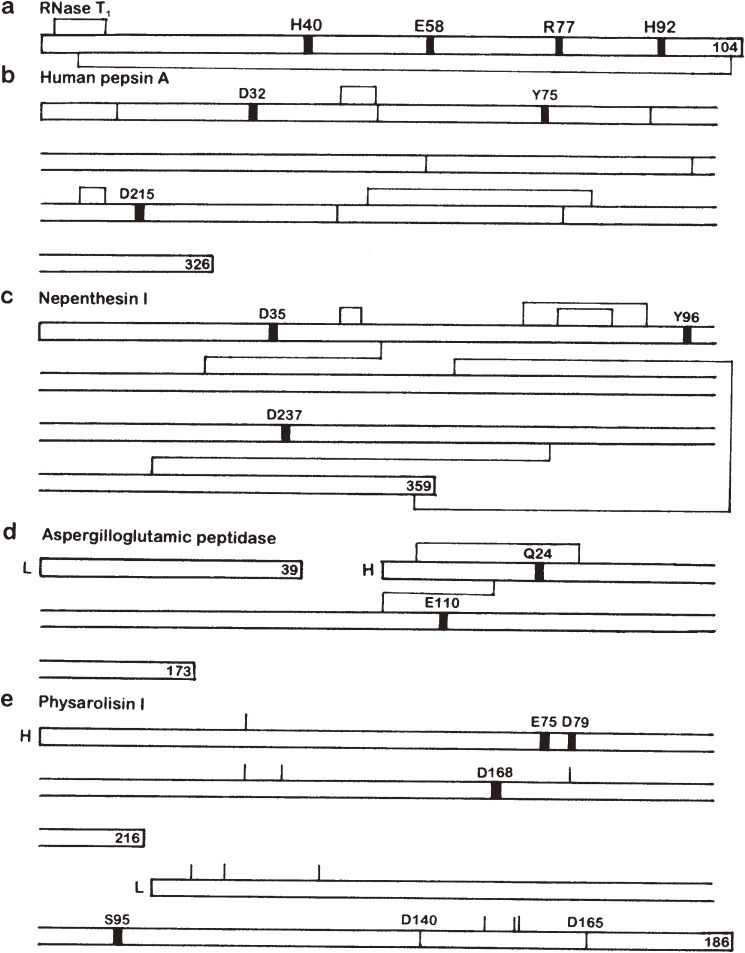
Figure 1.
Gross primary structues and location of the active site residues of some enzymes possessing a catalytic carboxyl group(s). Thick vertical lines in the sequence denote the location of active site residues. Disulfide bonds are shown above and/or below the sequence. L and H stand for L and H chains, respectively. The number at the end of each polypeptide chain indicates the total number of residues. (a) RNase T1. (b) Human pepsin A. The positions corresponding to the intron-exon junctions in the gene are shown with thin vertical lines in the sequence. (c) Nepenthesin I. The disulfide bond pairings were deduced from computer modeling. (d) Aspergilloglutamic peptidase. (e) Physarolisin I. The location of Ca2+-binding Asp residues are also shown with thin vertical lines in the sequence. As the disulfide bond pairings have not been determined, the location of cysteine residues are shown with thin vertical lines above the sequence.
(iii). Discovery of the active site Glu residue.
From 1965 to 1968, I had an opportunity to continue the structure-function studies on RNase T1 as a Research Associate in the laboratory of Profs. Stanford Moore and William H. Stein at the Rockefeller University, New York. For this study, I brought with me a few hundred mg of RNase T1 which had been left after the sequence study. In the Moore-Stein laboratory it had already been shown by A. M. Crestfield et al. that RNase A has two active site His residues, His12 and His119, which are specifically carboxymethylated by reaction with iodoacetate at pH 5.5 with concomitant inactivation of the enzyme.22) While I was in Tokyo, we also showed that RNase T1 was inactivated by reaction with iodoacetate at pH 5.5 like RNase A, but were unable to identify the residue or residues modified. Thus my major work in New York was to identify the critical residue(s) modified by iodoacetate in RNase T1. Iodoacetate had been known to react most rapidly with Cys residues, but might also react potentially with His, Lys and Met residues under certain conditions. The carboxymethyl (CM) derivatives of these residues are all acid-stable; therefore the modified residues can be identified and quantitated by amino acid analysis after acid hydrolysis of the modified proteins. However, no change in amino acid composition, including His residues, of RNase T1 was detected by amino acid analysis after the iodoacetate treatment. Thus the results obtained with RNase T1 were unexpectedly different from those obtained with RNase A.
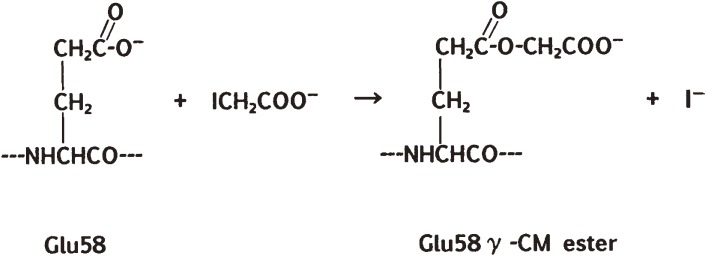
To identify the reaction site, I performed the carboxymethylation experiment using 14C-labeled iodoacetate, and found that the reaction occurred in 1:1 stoichiometry with the enzyme and that the 14C-CM group introduced into the protein was liberated as one equivalent of 14C-glycolic acid upon acid hydrolysis. These results suggested that the reaction might have occurred with a carboxyl group or groups in the enzyme. This supposition was consistent with the fact that the introduced 14C-CM group was fairly labile to weakly alkaline conditions and to treatment with hydroxylamine. Based on these results, I intended to isolate a proteolytic peptide fragment containing the 14C-CM group and identify the modified residue in it. This task was rather difficult due to spontaneous liberation of the labeled CM group during the isolation procedures. Finally, however, I was able to isolate a 14C-labeled short peptide, derived from residues 57 to 62: Tyr-Glu-Trp-Pro-Ile-Leu, from a peptide mixture obtained by limited Nagarse (subtilisin) digestion. Hydrolysis of the peptide with aminopeptidase M yielded most of the expected amino acids (but no Glu) plus a new component, which was proved to be the γ-CM ester of Glu (Fig. 2). These experiments established the presence of an unusually reactive γ-carboxyl group at Glu58 in the active site of RNase T1. The iodoacetate reaction was inhibited by substrate analogs 2′- or 3′-guanylic acid, by phosphate or citrate ions, and by Zn2+ or Cu2+, which are all inhibitors of the enzyme. These results strongly indicated that Glu58 is part of the active site of the enzyme. Moreover, the reaction was also inhibited by 8M urea, and thus required the native three-dimensional (3D) structure of the enzyme.
Figure 2.
The reaction of iodoacetate with the γ-carboxyl group of Glu58 in RNase T1.
Thus in 1967, we reported these results, i.e., the first identification by chemical modification of a specific carboxyl group (Glu58) in the active site of an enzyme.23) Meanwhile, in 1965 it was suggested first by X-ray crystallography that in hen egg-white lysozyme, Glu35 and Asp52 were the active site residues.24)
(iv). Identification of the active site Arg and His residues.
RNase T1 is an acidic protein with only 5 residues of basic amino acids: 1 Arg, 1 Lys and 3 His, per molecule of protein. This is in contrast to RNase A, which is a basic protein with 4 Arg, 10 Lys and 4 His per molecule. Since RNases act on RNA and polyribonucleotides having negatively charged phosphate groups, some basic residue is thought to interact with the phosphate group. In RNase A, Lys41 was shown to interact with the phosphate group in the substrate by X-ray crystallography.25) In our earlier study on RNase T1, one to two His residues, but not the sole Lys (Lys41), was shown to be important for the activity, but no information was available for the role of the single Arg (Arg77) due to the lack of a suitable Arg-specific chemical modification reagent that can be used under mild conditions. To investigate the role of Arg77, I decided to explore a new reagent specific for Arg residues in proteins. Among many dicarbonyl compounds examined using RNase A (as a model protein) and amino acid mixtures, phenylglyoxal (PGO) was eventually found to be most useful as an Arg-specific modification reagent that can be used under neutral to mildly alkaline conditions.26–28) Two PGO molecules were shown to react with one Arg residue. The reaction product of PGO with Arg was not stable under acid hydrolysis conditions but did not regenerate free Arg. Therefore the extent of modification could be measured as a loss of Arg after acid hydrolysis followed by amino acid analysis. After return to Tokyo in 1968, I investigated the reaction of PGO with RNase T1, and showed that it indeed inactivates the enzyme by specific reaction with Arg77 at the active site.29) This is the first time that a specific Arg residue was shown by chemical modification to reside in the active site of an enzyme. Since then PGO has found wide use as an Arg-specific reagent for many enzymes and proteins. Later, similar results were also obtained by modification of Arg77 with ninhydrin (i.e., 1,2,3-indantrione monohydrate).30)
Then my attention was turned to elucidation of the role of His residues in the enzyme. Through studies using rose bengal-catalyzed photooxidation and reaction with iodoacetamide, His40 and His92 were deduced to be involved in the active site.31–35) Interestingly iodoacetamide, but not iodoacetate, reacts with these His residues at pH 8.0, but not at pH 5.5. Later the pKa values of the three His residues were determined by NMR spectroscopy to be 7.26, 7.92 and 7.80 for His27, His40 and His92, respectively.36)
(v). Analysis of interaction with substrate analogs.
In order to elucidate the mechanism of interaction of RNase T1 with its substrates, the binding affinities of various nucleotides and nucleosides and their derivatives were analyzed using the gel-filtration method of Hummel and Dryer. The results indicated that the N1 position, 2-amino group, 6-oxo group, 7-amino group and phosphate groups (especially at the 3′ position) are involved in the interaction with the enzyme.37) However, the groups in the enzyme which are involved in the specific binding with the guanine portion remained undetermined.
(vi). 3D structure analysis.
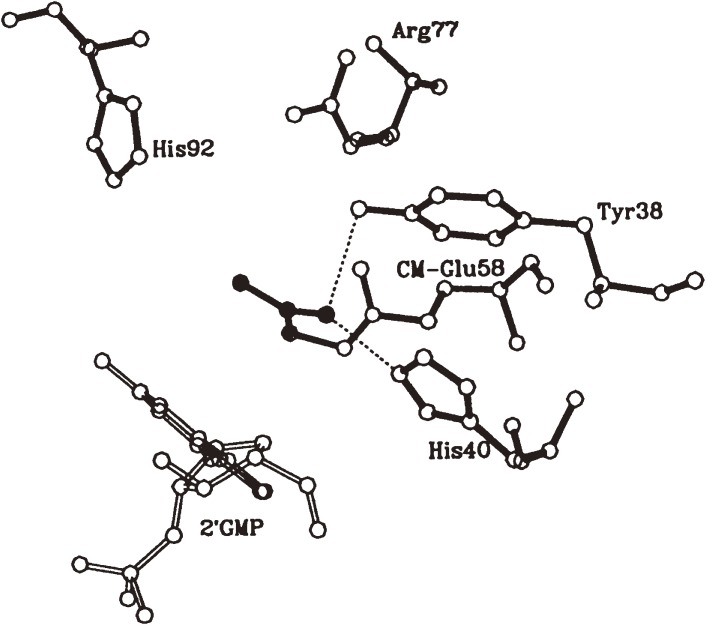
In 1982, Heinemann and Saenger first reported the crystal structure of RNase T1 complexed with 2′-GMP, in which Glu58, His40, His92 and Arg77 were shown to reside in close proximity with each other in the active site38) as previously predicted mainly from chemical modification studies. The crystal structure was also consistent with the results obtained previously using gel filtration regarding the interaction of guanine base with the enzyme. In addition, it was revealed that the region from Tyr42 to Tyr45 interacts with the guanine base in the substrate through multiple hydrogen bonds and that Tyr45 is stacked with the guanine base.
Later we elucidated the crystal structure of CM-RNase T1·2′-GMP complex (Fig. 3a and 4)39) and the solution structure of RNase T1,40) the overall structures of which were essentially identical with the RNase T1·2′-GMP strucure.38) A structural comparison of CM-RNase T1·2′-GMP with intact RNase T1·2′-GMP revealed that the CM group is present in the catalytic site and prevents the phosphate of nucleotide from coming into the phosphate binding site. This is thought to be the major cause of inactivation of the enzyme by carboxymethylation. No structural difference was found in the specific guanine binding site between CM-RNase T1·2′-GMP and RNase T1·2′-GMP. This explains my earlier observation37) that CM-RNase T1 possessed almost the same binding ability toward guanosine as intact RNase T1, whereas the binding ability toward 2′ or 3′-guanylic acid was considerably lowered by carboxymethylation of Glu58. These results are partially consistent with those obtained by molecular dynamics simulation and energy minimization calculation.41) The increased thermal stability of the enzyme by carboxymethylation was also explained by hydrogen bonding interaction of the CM group with Tyr38 and His40 and electrostatic interaction of the CM group with Arg77 and His92. On the other hand, the specific binding of the carboxyl group of iodoacetate to Tyr38 and His40 was indicated to serve as orienting the reagent in the carboxymethylation reaction of the enzyme.
Figure 3.
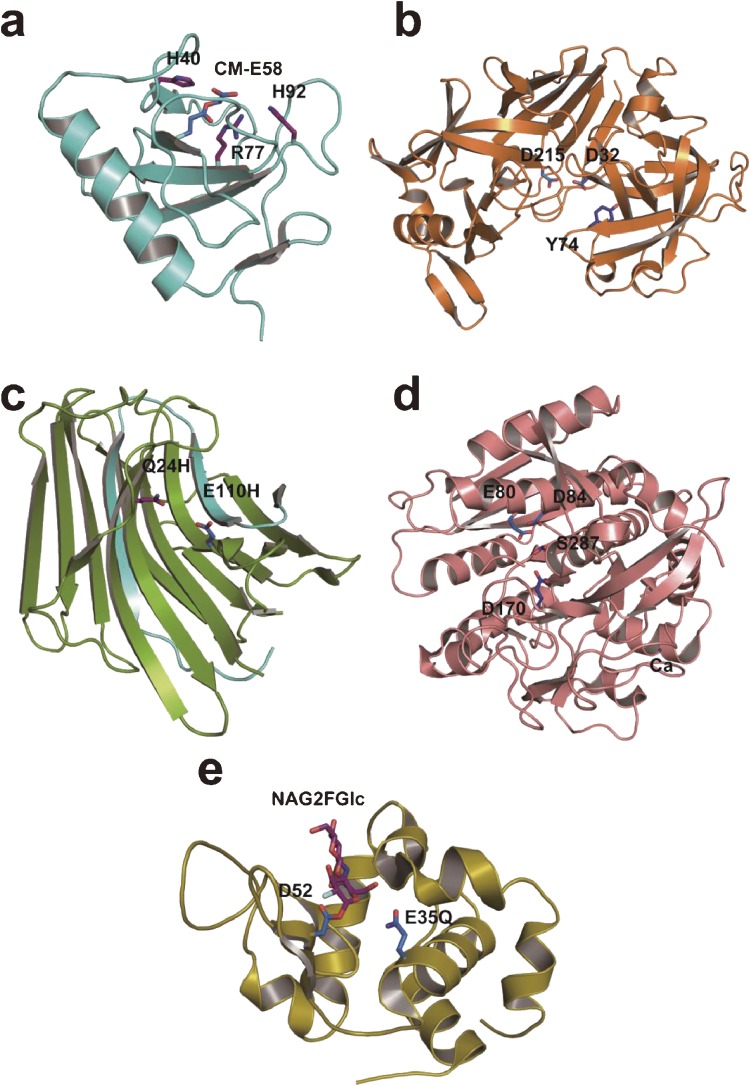
3D structures of some enzymes with a catalytic carboxyl group(s). Active-site residues are shown in stick representation. (a) RNase T1 carboxymethylated at Glu58 (PDB ID: 1DET). (b) Porcine pepsin A (PDB ID: 4PEP). (c) Aspergilloglutamic peptidase (PDB ID: 1Y43). (d) Sedolisin (PDB ID: 1GA4). (e) Hen egg-white lysozyme covalently bound with a substrate analog (PDB ID: 1H6M). NAG2FGlc, 2-acetamido-2-deoxy-β-d-glucopyranosyl-(1→4)-2-deoxy-2-fluoro-β-d-glucopyranosyl group.
Figure 4.
Active site geometry of CM-RNase T1-2′-GMP complex. The CM group is shown in black.
(vii). Proposed catalytic mechanisms.
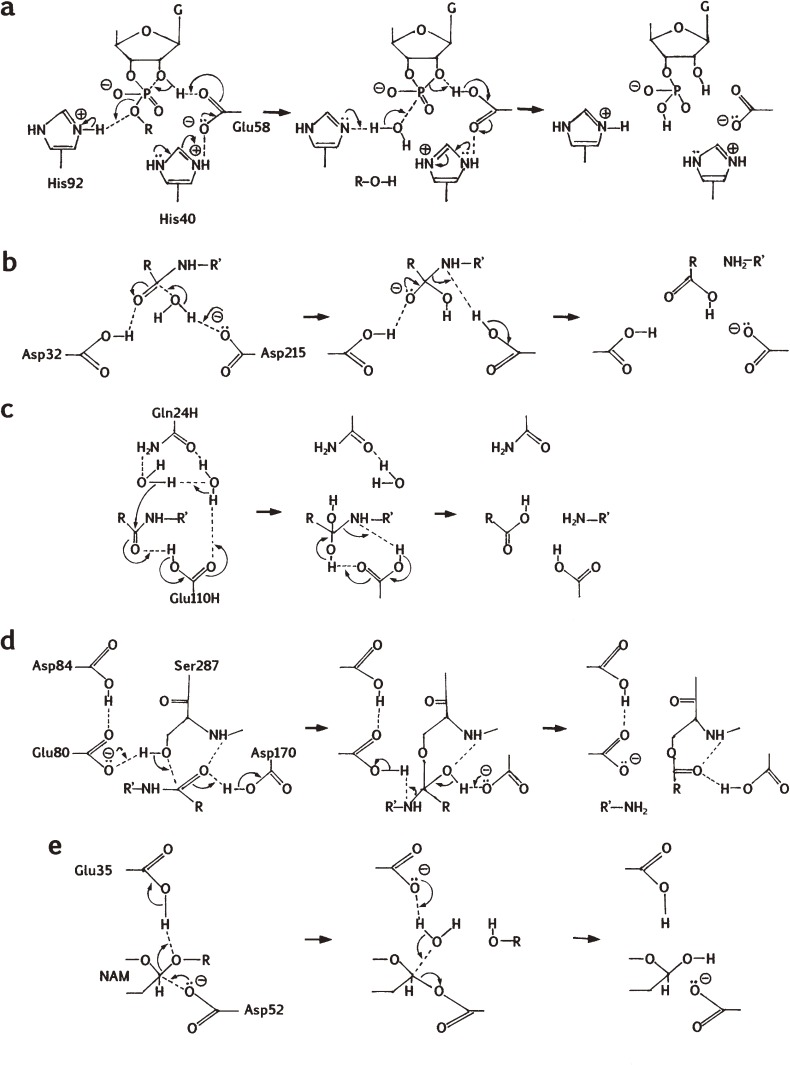
Prior to the 3D structure determination, I proposed in 1970 a catalytic mechanism as based mainly on the results obtained by chemical modification studies as shown in Fig. 5a.31) In this mechanism Glu58 and His (His40 or His92) are implicated as a general base and a general acid, respectively, in the first stage of catalysis (i.e., transphosphorylation), and vice versa in the second stage (i.e., hydrolysis). The mechanism was later shown to be consistent with the results of 3D structural analysis.38)
Figure 5.
Proposed catalytic mechanisms of some enzymes with a catalytic carboxyl group(s). (a) RNase T1. The mechanism is based on refs. 31 and 44. The first step is intramolecular transphosphorylation, and the second step is hydrolysis. G, guanine. (b) Porcine pepsin A. The mechanism is based on ref. 96. (c) Aspergilloglutamic peptidase. The mechanism is based on ref. 186, and includes two water molecules found in the crystal structure, but the possibility that the reaction proceeds without the second water molecule cannot be excluded. (d) Sedolisin. The mechanism is based on ref. 174. Only the acylation process is shown. (e) Hen egg-white lysozyme. The mechanism is based on ref. 205. NAM, N-acetylmuramic acid.
Meanwhile Nishikawa et al. investigated the roles of the active-site residues by site-directed mutagenesis.42,43) They found among others that the His40Ala and His92Ala mutants are inactive, but that the Glu58Ala mutant is partially active. Based on those results, they proposed a modified catalytic mechanism similar to that of RNase A, in which His40 and His92 are the major catalytic residues where the ionized form of Glu58 binds to and assists the role of His40.43) On the other hand, Steyaert and his colleagues performed more extensive site-directed mutagenesis, kinetics and X-ray studies.44–47) They found among others that the His40Lys mutant is active, and eventually concluded as follows. In the native enzyme, Glu58 and His92 are the major catalytic residues as originally proposed, and His40 (protonated form) assists the role of Glu58 (ionized form) as a general base in the first stage of catalysis (Fig. 5a). On the other hand, in the Glu58Ala mutant, His40 partially substitutes the role of Glu58. The Glu58/His92 mechanism is consistent with the pKa values of the activity profile obtained by kinetic studies4,44) and with the result of our modeling studies.48) Anyway Glu58 and His40 are very closely located with each other in the active site and are both involved in catalysis (Fig. 5a). In this connection it should be noted that His40 (RNase T1 numbering) is missing in the prokaryotic RNases, RNases St, Ba and Bi, which are distantly related homologs of RNase T1.49) Anyway, the catalytic roles of Glu and His residues in RNase T1 continue to draw interest in further investigation.
(viii). Physico-chemical analyses of the conformation and its changes.
Thermal denaturation of CM-RNase T1 was analyzed by one dimensional NMR,50) which showed that the whole molecule was denatured apparently simultaneously and that the melting temperature was by 9℃ elevated by carboxymethylation. The marked elevaton of the melting temperature indicated that the enzyme molecule was considerably stabilized by introduction of one CM group into Glu58. This stabilization can be explained by hydrogen bonding and electrostatic interactions of the CM group with nearby residues as described already based on the crystal structure of CM-RNase T1.39)
On the other hand, thermal denaturation of RNase T1 was also analyzed by two-dimensional NMR.51) The results indicated that the thermal denaturation was apparently a two-state transition, but that microscopically it proceeded as a continuous but stepwise process where the α-helix region lost the structure first, followed by the β-sheet region involving the active site. This is to our knowledge the first case where a denaturation process of a protein was analyzed by two-dimensional NMR to clarify such a microscopic change.
(ix). Comparative studies on RNase T1 homologs.
Later, we sequenced two RNase T1 homologs, RNase U1 from Ustilago spherogena52) and RNase N1 from Neurospora crassa,53) which were shown to have the four active site residues (His40, Glu58, Arg77 and His92 in RNase T1) in common. While sequencing RNase U1 and N1, I noticed that there is one region in the RNase T1 sequence that needs revision. Thus after the revision, the sequence of residues 71–73: Pro-Gly-Ser, was corrected to Gly-Ser-Pro.54) Further, one of the Glu residues in RNase U1 was shown to be specifically carboxymethylated by iodoacetate like Glu58 in RNase T1, and His and Arg residues were also shown to be in the active site by chemical modifications.55,56) Those active-site amino acid residues were also shown to be conserved in several other RNase T1 homologs.4,57)
II. Pepsin, nepenthesin and their homologs
Pepsinogens are the zymogens of pepsins, the major gastric acid peptidases involved in food protein digestion under acidic conditions.58) When I was working at the Rockefeller University, structure-function studies of porcine pepsinogen A and pepsin A were in progress there in the laboratory of S. Moore and W.H. Stein and also in that of G.E. Perlmann. Porcine pepsin A was partially sequenced and suggested to have a critical carboxyl group at the active site that was specifically modified by diazoacetyl-d,l-norleucine methyl ester (DAN) in the presence of cupric ions.59) Therefore, porcine pepsin A and RNase T1 appeared to be common in that both share an essential carboxyl group at their active sites. These results led me to have deeper interest in structure-function studies of pepsins and related acid peptidases. At that time, acid peptidases (later called aspartic peptidases) were less studied than serine and cysteine peptidases regarding structure and function relationships.
(i). Active site studies on pepsin and related acid (aspartic) peptidases.
In the early 1970’s, the DAN-reactive active site residues of porcine pepsin A and some related acid peptidases were identified as an Asp residue at the specific sites of the enzymes (later shown to be Asp215 in porcine pepsin A) by using DAN and similar diazo reagents.60) In addition another active site Asp residue (later shown to be Asp32) was identified in porcine pepsin A by using 1,2-epoxy-3-(p-nitrophenoxy)propane (EPNP).61) Following these studies, we showed that porcine cathepsin D, bovine rennin (later called chymosin) and acid peptidases from insectivorous plants and several fungi were also inhibited by DAN, EPNP and pepstatin, indicating a wider range of distribution of these peptidases,62–68) and identified the DAN-reactive Asp residue in bovine chymosin.69) Later, we also identified chemically the two active site Asp residues in Rhizopus chinensis acid peptidase.70) More recently we synthesized a series of peptide derivatives containing EPNP moiety and showed that these affinity labeling reagents inactivate porcine pepsin A much more rapidly than EPNP.71)
Through these studies, it became clear that porcine pepsin A and related acid peptidases have two active site Asp residues in common, and hence that they should be called ‘aspartic peptidases’. The occurrence of these residues as well as Tyr75 (porcine pepsin A numbering) at the active site has since been confirmed in all ordinary aspartic peptidases including nepenthesin and its homologs. A catalytic mechanism involving these residues was first proposed by James et al.72)
(ii). Purification, characterization and primary structure determination.
From 1974 to 1984 I worked in a newly established laboratory of the Department of Biochemistry at Primate Research Institute, Kyoto University, Inuyama, Aichi. Pepsinogen/pepsin studies at the protein level were performed there mostly in collaboration with Dr. Takashi Kageyama. We purified pepsinogens A and C from primates and several other mammalian species, characterized them and determined their primary structures.73–84) The sequence data were used to construct phylogenic trees and investigate their phylogenic relationships. We also determined the primary structures of the signal peptides of prepepsinogens from human and other animals.85,86)
The primary structure of the prosequence (i.e., activation peptide) of porcine pepsin A was determined at the protein level by Ong and Perlmann87) and by Pedersen and Foltmann88) and that of the porcine pepsin A moiety by Tang et al.,89) thus leading to the complete primary structure of porcine pepsinogen A. This was the first primary structure determination of an aspartic peptidase. Later we deduced the primary structure of porcine prepepsinogen A based on the cDNA sequence, and revised the above structure derived from protein analysis.90) On the other hand, we determined the complete primary structures of both monkey pepsinogens A and C.83,84) Thus the primary structure of pepsinogen C (progastricsin) was determined by us for the first time.
When analyzing cathepsin D in monkey tissues, we found, purified and characterized pepsinogens C in lung91–93) and prostate.94) Pepsinogen C was found for the first time in lung, but its physiological role has not yet been clarified. On the other hand, we purified cathepsin D-like enzyme (so-called ‘slow-moving protease’) to homogeneity from human stomach,95) which was later shown to be identical with cathepsin E. Thus we purified cathepsin E for the first time.
(iii). 3D structures and proposed catalytic mechanism of aspartic peptidases.
So far the 3D structures of many aspartic peptidases have been determined by X-ray crystallography. In each case the overall 3D structure is bilobal with two domains separated by a deep cleft where the two catalytic Asp residues are located to form the catalytic site. As an example, the 3D structure of porcine pepsin A is shown in Fig. 3b. Each enzyme is rich in β-sheet, and a Tyr residue (corresponding to Tyr75 in porcine pepsin) is present nearby. Although the 3D structures of nepenthesin and its homologs have not yet been solved, they should be similar to those of pepsin-type aspartic peptidases. A proposed catalytic mechanism96) is shown in Fig. 5b. One of the active-site Asp residues (Asp215 in porcine pepsin) is ionized and the other (Asp32 in porcine pepsin) is protonated. The water oxygen is involved in nucleophilic attack to the carbonyl carbon of the peptide bond to form an intermediate with a tetrahedral carbon at the substrate carbonyl position. Hydrogen donation from Asp215 to the substrate nitrogen results in the breaking of the peptide bond.
(iv). Studies on the activation mechanism of pepsinogen: discovery of one-step activation.
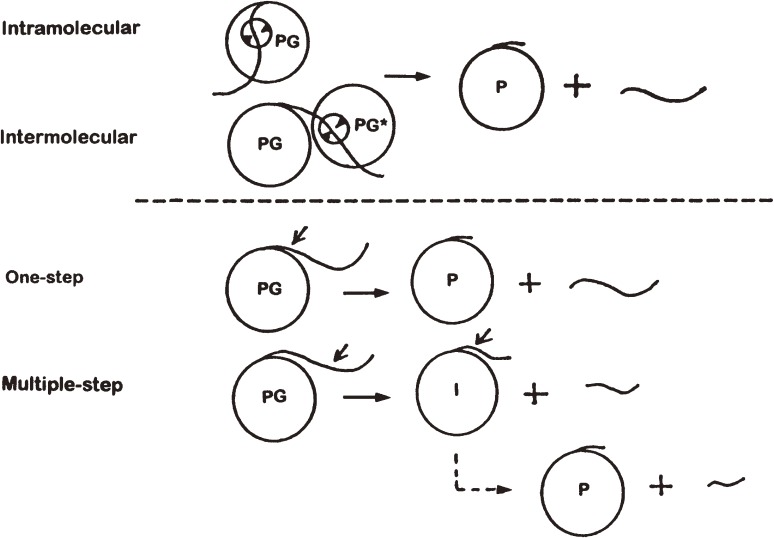
Figure 6 shows schematically the activation pathways of pepsinogen to pepsin. Kinetic studies indicated that the first step of activation of porcine pepsinogen A to pepsin A at pH below 3 takes place by autocatalytic cleavage of the propeptide through intramolecular reaction.96) However, the first cleavage site was not definitely established. In porcine pepsinogen A, it was assumed that the first cleavage occurs in the middle of the propeptide segment with the final cleavage at the boundary between the propeptide and the pepsin moiety. Thus the activation may proceed as a two-step process or a multiple step process in which an additional cleavage(s) occurs stepwise after the first cleavage point. On the other hand, the single, complete-length propeptide was never isolated before from the activation mixture; thus one-step activation had never been proved. To shed light on this issue, we examined the course of activation of porcine and monkey pepsinogens A under restricted conditions (at 4℃) to slow down the rate of activation. As a result, we could obtain a single, intact propeptide in the early phase of activation of both pepsinogens A, demonstrating for the first time the occurrence of one-step activation.97–101) Later we compared the activation processes of several other animal pepsinogens.102) These studies indicated that the one-step and multiple-step activations may take place more or less simultaneously; the ratio of the two processes depends on the differences in the amino acid sequence of the propeptide followed by the N-terminal few residues of the pepsin moiety and the peptide bond specificity of the pepsin moiety. It still remains to be clarified, however, whether the one-step activation occurs by an intramolecular reaction or intermolecular reaction.
Figure 6.
Pathways of activation of pepsinogen to pepsin. PG, pepsinogen; PG*, pepsinogen, intermediate or pepsin; P, pepsin; and I, activation intermediate. Two-step activation is shown as an example for multiple-step activation.
(v). Studies on the gene structures and function of pepsinogens: the first determination of the structure of an aspartic peptidase gene.
In 1983 we succeeded to determine the structure of human pepsinogen A gene using the Maxam-Gilbert method and showed that the gene was composed of nine exons separated by eight introns.103,104) Thus human pepsinogen A was one of several proteins whose gene sequences were elucidated in the early period of gene sequencing. It was the first gene structure elucidated for aspartic peptidases. This allowed us to deduce the complete primary structure of human prepepsinogen A (total 403 residues), composed of a 15-residue signal sequence, a 62-residue prosequence and a 326-residue enzyme. Figure 1b shows the gross primary structure of human pepsin A thus deduced, including the exon-intron junction points. In 1984 I returned to the Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, and continued studies on aspartic peptidases. The studies included the structure and function analyses of human pepsinogen A and C genes105–108) and rat pepsinogen C cDNA109) and gene110) and cloning of frog pepsinogen C cDNA111) and tuna pepsinogens A and C cDNAs.112)
We also investigated the relationship between the extent of methylation/demethylation and tissue-specific expression of rat and human pepsinogen genes.113–118) The results indicated that the low level of pepsinogen gene methylation is closely correlated with its tissue- and cell-specific expression and that the extent and pattern of pepsinogen gene methylation is markedly different between normal and embryonic or neoplastic tissues. These results show that the tissue-specific expression of pepsinogen is regulated by methylation and demethylation of its gene.
(vi). Clinical application: serum pepsinogen test.
With purified human pepsinogens A and C (clinically called pepsinogens I and II, respectively) at hand, we explored for the clinical use of them in collaboration with K. Miki and M. Ichinose at the Faculty of Medicine, The University of Tokyo. We prepared specific antisera for pepsinogens I and II, established their radioimmunoassay methods, and analyzed the serum pepsinogens I and II contents of many human serum samples from normal and clinical patients.119–121) Unexpectedly, it turned out that the ratio of pepsinogens I and II in the serum can be utilized for screening of extensive chronic gastritis, which may be a potential cause of gastric cancer.122,123) Thus the serum pepsinogen test has found wide use as a primary screening method in mass examination for potential gastric cancer.124) Recently the method has become more refined as combined with a Helicobacter pylori test and this improved method is called the ABC method.125)
(vii). Comparative studies on pepsin-related aspartic peptidases.
We extended our structure-function studies on pepsinogens and pepsins to those on other homologous aspartic peptidases, including human pepsinogens and pepsins126,127) and non-mammalian pepsinogens and pepsins,128–133) cathepsin D,92,134–138) cathepsin E,139–149) filarial parasite aspartic peptidases (from Brugia malayi),150) HIV-1 protease,151,152) Drosophyla copia protease,153) rhizopuspepsin (from Rhizopus chinensis),154,155) and aspergillopepsin I (from Aspergillus niger).156,157)
(viii). Nepenthesin and related aspartic peptidases: discovery of a new type of aspartic peptidases.
Carnivorous plants secrete various proteases in the digestive fluids to digest prey proteins for nutrition. Previously, however, no such protease was completely purified and characterized mainly due to the difficulty to obtain enough amounts of samples. To overcome this problem, we collected a large amount of the digestive fluid of Nepenthes distillatoria in the mountain area of Sri Lanka where it grows abundantly. As a result, we could for the first time purify to homogeneity the digestive enzyme ‘nepenthesin’ and characterize them.158–161) A few mg each of the purified enzymes, nepenthesin I and II, were obtained from 30 l of the digestive fluid. The enzymes were then shown to have the following interesting properties.
First, they had unusually high temperature and pH stabilities as compared with ordinary pepsin-type aspartic peptidases. They were stable for several weeks at 50℃ in a wide range of pH from 2 to 10. To our knowledge, such an unusual stability of proteinases, especially of nepenthesin I, was never reported previously. These enzymes, especially nepenthesin I, are thought to be so designed that they are capable of remaining active in a wide range of pH at relatively high temperatures to work for a long time in the pitcher fluid in a tropical habitat. This stability was thought to be a result of evolutionary adaptation to their specific environments. Unexpectedly, however, they were later found to be less stable to certain denaturing agents such as urea and guanidine hydrochloride than pepsin A.162) It seems to be not so important for nepenthesins to have higher stability toward such denaturing agents which do not exist in nature. As far as we know, there is scarcely any protein known with such a dual stability, and therefore nepenthesin may be useful as an interesting target in studying the mechanism of protein denaturation.
Secondly, the complete primary structures of nepenthesins I and II (437 and 438 residues, respectively, as preproenzymes) were deduced from cDNA sequencing. The prepro-nepenthesin I was composed of a 24-residue putative signal sequence, a 54-residue putative propeptide, and a 359-residue mature enzyme, and prepro-nepenthesin II was composed of a 24-residue putative signal sequence, a 55-residue putative propeptide and a 359-residue mature enzyme. The gross structure of nepenthesin I is shown in Fig. 1c. The results indicated that both nepenthesins had essentially the same active site motif but twice as many disulfide bonds (i.e., 6 disulfide bonds/molecule) as hitherto-known pepsin-type aspartic peptidases (cf. Figs. 1b and 1c). These disulfide bonds should contribute greatly to the pH/temperature stability of these enzymes. In addition, nepenthesin I contained carbohydrate, which should also contribute to the stability.
Thirdly, these enzymes had low homology in amino acid sequence with ordinary aspartic peptidases, and did not contain the plant-specific insertion sequence characteristic of plant vacuole-derived aspartic peptidases. Instead, they contained unique insertion sequences characteristic of nepenthesin, the role of which remains to be elucidated. We also investigated the occurrence of nepenthesin-type aspartic endopeptidases in the digestive fluids of other carnivorous plants and partially characterized them.163) The enzymes from Dionaea muscipula (Vinus’s fly-trap), Drosera (sundew) sp. and Cephalotus follicularis were named “dionaeasin”, “droserasin” and “cephalotusin”, respectively.164)
In addition, database search for homologous protein genes revealed that many genes orthologous to the nepenthesin-type enzyme genes are encoded in the genome of plants such as Arabidopsis thaliana and Oryza sativa. In the case of Arabidopsis proteins, nearly 60 genes are encoded and many of them were shown to be expressed at the mRNA level in various tissues including leaves, stems, seeds, and pods, suggesting ubiquitous occurrence and multiple functions of the corresponding proteases in the tissues of A. thaliana.165) They are thought to be intracellular aspartic peptidases but their physiological roles are largely unknown. I propose the names “arabidopsin” and “oryzapsin” for these groups of enzymes from A. thaliana and O. sativa, respectively.
The phylogenic tree based on the primary structures of pepsin-type and nepenthesin-type aspartic peptidases revealed that these two types of aspartic peptidases are clearly separated from each other as distinct groups. As a result, the major aspartic peptidase groups (MEROPS family A1) are now classified into two large groups, pepsin type (subfamily A1A) and nepenthesin type (subfamily A1B). It should be noted that in addition to family A1, several other minor aspartic peptidase families are known to date including Copia transposon peptidase (Drosophila melanogaster)153) (family A11/subfamily A11A).
III. Aspergilloglutamic peptidase and its homologs
Since 1976 when we found that an acid endopeptidase from Aspergillus niger var. macrosporus (previously called ‘Proctase A’, then A. niger acid proteinase A or aspergillopepsin II) was insensitive to pepsin-type proteinase inhibitors, such as DAN, EPNP and pepstatin A, it continued to attract our attention and was long been thought to be a kind of non-pepsin type (or pepstatin-insensitive) aspartic peptidase.166–169) Recently it was shown not to be an aspartic peptidase but a glutamic peptidase which has a catalytic Glu residue at the active site and thus renamed aspergilloglutamic peptidase.170,171) Meanwhile, extensive structure-function studies were also performed by Oda and his collaborators with its homolog, scytalidopepsin B, later renamed scytalidoglutamic peptidase.172–174) In this review, I describe mainly our results obtained with aspergilloglutamic peptidase.
(i). Primary structure determination.
The primary structure of aspergilloglutamic peptidase (total 212 residues) was determined at the protein level.175) The enzyme was composed of a 39-residue light chain and a 173-residue heavy chain bound non-covalently with each other (Fig. 1d). No homology was found with ordinary pepsin-type enzymes. On the other hand, the primary structure of the precursor form (282 residues) was deduced by sequencing the enzyme gene.176) Thus the preproenzyme was shown to be composed of an 18-residue putative signal peptide, a 41-residue propeptide, the light chain, an 11-residue intervening peptide and the heavy chain linked in this order.
It was shown later that the propeptide portion was inhibitory to the enzyme. Therefore we synthesized a number of the propeptide fragments and tested their inhibitory action toward the enzyme.177) As a result, the specific sequence of a few residues in the central part of the propeptide was shown to contribute most effectively to the inhibitory activity. These peptide regions were thought to bind specifically to the active site of the enzyme, thus exerting greater inhibition. They were also found to protect significantly the enzyme from thermal inactivation. Thus the specific binding of these peptides to the enzyme was also thought to increase its thermal stability.
(ii). Discovery of the active site Glu residue.
To identify the active site residues, we performed site-directed mutagenesis studies on all Asp and Glu residues in the enzyme. The results indicated that Glu110 in the heavy chain is of critical importance for the activity and presumably is one of the catalytic residues.170,178) This residue was the only acidic residue the mutation of which resulted in complete loss of activity. We therefore suggested that the enzyme may be called a ‘glutamic peptidase,’ the occurrence of which had not been known before. In addition, however, Asp14 in the heavy chain was suggested to be important for the activity. The role of this residue, however, remained unknown until it was solved by X-ray crystallographic studies. In the previous study,170) Gln24 in the heavy chain was also mutated to Glu with complete loss of activity; however, we misinterpreted the result as due to structural instability of the mutant.170) Later studies of site-directed mutagenesis were performed on all the hydrophilic amino acid residues other than acidic residues, and Gln24 in the heavy chain was definitely confirmed to be the second critically important residue,179) which was consistent with the results of X-ray crystallographic analysis.
(iii). 3D structure analysis.
To shed further light on the structure-function relationships, we attempted to elucidate the 3D structure of the enzyme by X-ray crystallography. We obtained three kinds of the enzyme crystals (types I, II and III) under different conditions.180,181) Type I contained one molecule, and types II and III each contained two molecules of the monomeric enzyme per asymmetric unit. The type I enzyme was solved to a resolution of 1.4 Å (Fig. 3c).182) It had a β-sandwich structure composed of two domains of multiple anti-parallel β-sheets, and Glu110 and Gln24 in the heavy chain were suggested to be the catalytic residues. On the other hand, Asp14 in the heavy chain, suggested to be important by site-directed mutagenesis, was found to reside somewhat apart from the active site and was thought to be involved in the construction of the 3D structure near the active site.
Prior to our report on the 3D structure of aspergilloglutamic peptidase, that of scytalidopepsin B, an enzyme homologous to the present enzyme, was reported, which was the first paper on the 3D structure of this group of enzymes.172–174) The structure of the present enzyme was very similar to that of scytalidopepsin B, and Glu110 and Gln24 in the heavy chain of the present enzyme corresponded to Glu190 and Gln107 in the active site of the latter enzyme. Based on the 3D structure, they first proposed a Glu-Gln catalytic dyad. Later the importance of these residues were confirmed by site-directed mutagenesis studies.183) Eventually, scytalidopepsin B and the present enzyme were classified as ‘glutamic peptidases’ (MEROPS family G1), a new class of endopeptidases, and renamed ‘scytalidoglutamic peptidase’ or ‘eqolisin’,172) and ‘aspergilloglutamic peptidase’,171) respectively. It should be noted that another type of glutamic peptidase (i.e., pre-neck appendage protein (bacteriophage phi-29)) has very recently been found and classified as MEROPS family G2, which has a Glu and an Asp residue at the active site.184)
The X-ray analysis of the types II and III of the present enzyme was performed to a resolution of 1.6 Å. The type II enzyme contained two monomeric enzymes bound in a two-fold symmetry. The type III enzyme had a unique structure in which the C-terminal region of the light chain of one of the molecules bound to the active site cleft of the other molecule like a part of a substrate.185) This form mimics the enzyme-activation product complex produced upon autoproteolysis, and provides a structural clue that could help to clarify the activation mechanism. This type of dimeric structure of a peptidase was reported for the first time.
(iv). Proposed catalytic mechanism.
To obtain a clue to the catalytic mechanism, pH dependences of activity toward various oligopeptides were examined.186) The pH dependence of activity toward substance P, which was known to be cleaved specifically at the Phe-Gly bond by the enzyme, was rather broad and the maximum kcat/Km value was obtained at pH 4–5. The kcat/Km value dropped fairly rapidly toward pH 7 and implication of a group of pKa ∼ 6.3 was suggested. On the other hand, in the acidic region, the kcat/Km value dropped very slowly and over 50% of the maximum kcat/Km value was obtained even at pH 1.6. Similar results were obtained with other peptide substrates with some variations. At pH 1.0, the enzyme showed more than 50% of the maximum activity toward most peptides examined, and no significant drop of activity was observed in one case. These results also suggested the implication of a group with pKa of around 6. This group was thought most likely to be the carboxyl group of Glu110 in the heavy chain, although at above pH 6, alkaline denaturation of the enzyme took place simultaneously.
On the other hand, the pH dependence curves of hemoglobin and casein digestion had a maximum at pH 1–2, and suggested the implication of a protonated group of pKa 2–4 and deprotonated group of pKa less than 1. The former group may correspond to Glu110 in the heavy chain. The difference between protein and peptide hydrolysis is not certain, but the decrease in activity at pH below 1 may be due to acid denaturation of the enzyme.
Based on these results, we proposed a catalytic mechanism (Fig. 5c).186) In this mechanism, Glu110 in the heavy chain, as a general acid, gives a proton to the carbonyl oxygen of the cleavage site peptide bond in the initial stage of hydrolysis. Then Glu110 acts as a general base to draw a proton from the neighboring water molecule, and the resulting OH− group attacks nucleophilically the carbonyl carbon atom of the substrate to generate a tetrahedral intermediate. Finally, the tetrahedral intermediate is decomposed to result in peptide bond cleavage through reversal of the electron flow. Preceding our studies, a similar, but slightly different mechanism was proposed for scytalidoglutamic peptidase,172,173) in which the catalytic Glu acts as a general base to draw a proton from water molecule so that the resulting OH− group is able to attack the scissile peptide bond nucleophilically in the initial stage of hydrolysis. In this case, however, the pKa value of the catalytic Glu should possibly be lower than 1. Since both enzymes are homologous to each other, the catalytic mechanism should be the same. Thus further elaborate studies are necessary to draw a definitive conclusion.
(v). Mechanism of proenzyme activation.
To investigate the activation profile of the proenzyme, we incubated recombinant proenzyme (264 residues) at pH 5.25 or 3.5 and 4℃, and the course of activation was followed by SDS-PAGE and the resulting fragments were analyzed by mass spectroscopy.187) As a result, it was shown that the Glu12–Ala13 bond of the prosegment was cleaved first, followed by stepwise cleavages in both the N-terminal region and the intervening sequence region thus producing the final active enzyme via multiple intermediates. The first cleavage may occur either intramolecularly or intermolecularly, but the following cleavages are thought to occur intermolecularly. So far no studies have been performed on the activation mechanism of the proforms of other glutamic peptidases. However, since they do not have an intervening sequence, the course of activation would be simpler and may proceed stepwise from the N-terminus.
(vi). Stability studies.
The present enzyme is known to be rapidly inactivated between pH 6 and 7. This is similar to the alkaline denaturation of pepsin, but more complicated due to the two-chain structure of the enzyme. The native conformation of the enzyme is thought to be stabilized by strong noncovalent interaction between the light chain and the heavy chain. We analyzed the pH-dependent denaturation process of the enzyme using differential scanning calorimetry,188) and small-angle X-ray scattering (SAX), circular dichroism (CD) and gel filtration.189) The midpoint values for the unfolding were found to be significantly different among the methods used, suggesting the existence of an intermediate state during the unfolding. Further analyses of the SAX data showed that the heavy chain just after the dissociation still kept molecular compactness and that it gradually increased its dimensions as the pH was further raised. Noncoincidence of the two phenomena (i.e., chain dissociation and swelling) led to elucidation of a novel intermediate state during unfolding. The results of kinetic studies of the unfolding process by pH-jump methods using time-resolved SAX and CD were consistent with the above results.190)
IV. Physarolisins I and II and their homologs
Serine-carboxyl peptidases are a kind of pepstatin-insensitive acid peptidases and are classified in the MEROPS peptidase data as family S53 enzymes including sedolisin and its homologs, which possess catalytic serine and carboxyl groups at the active sites. They are structurally related to subtilisin. Their structure and function relationships have been extensively studied mainly by Oda and his coworkers,174,191–193) including the 3D structures and catalytic mechanisms, as will be referred to later. In this paper, I describe mainly our results obtained with physarolisins I and II, which are members of serine-carboxyl peptidases, but with unique characteristics.
The slime mold Physarum polycephalum is known to undergo a drastic change in cellular organization through degradation and reorganization of membrane structures. In this process, certain intracellular proteases are thought to play crucial roles. Among various protease activities present in the plasmodia of P. polycephalum, the acid protease activity had been shown to exhibit quantitative changes during spherule formation. We found that this acid protease activity was due to the presence of two kinds of acid peptidases. They were initially called physaropepsins I and II, but later studies showed that they are not pepsin-type peptidases, but were members of serine-carboxyl peptidases. Therefore their names were changed to physarolisins I and II.194) Although these enzymes are members of serine enzymes, I included them in this review as members of apparently the same category for convenience. Physarolisins as well as arabidopsins are the only major intracellular enzymes, in addition to cathepsins D and E, among the three types of peptidases studied, and elucidation of their physiological roles is awaited.
IV (A). Physarolisin I
(i). Purification and characterization.
The enzyme was purified to homogeneity from the plasmodia of P. polycephalum by detergent extraction, acid precipitation and successive chromatographies.195) The enzyme was shown to have a two-chain structure, in which a 31-kDa heavy chain and a 23-kDa light chain were linked by disulfide bonds (Fig. 1e). The heavy chain contained carbohydrate. The enzyme hydrolyzed hemoglobin optimally at pH 1.7, but not hydrolyzed N-acetyl-d,l-phenylalanyl-3,5-diiodo-l-tyrosine, a synthetic peptide substrate for pepsin. Among aspartic peptidase-specific inhibitors, DAN inhibited the enzyme, but EPNP and pepstatin A did not. These results indicated that the enzyme was not a pepsin-type aspartic peptidase. On the other hand, oxidized insulin B chain was hydrolyzed most rapidly at the Gly8–Ser9 bond. However, later studies showed that the P1 and/or P1′ positions of the major cleavage sites in a combinatorial peptide mixture were occupied by bulky hydrophobic or aromatic residues such as Leu, Phe, Tyr etc.196) The present enzyme lacked the tripeptidylpeptidase activity toward Ala-Ala-Phe-4-methylcoumaryl-7-amide, a good substrate for CLN2, and was insensitive to Ala-Ala-Phe-chloromethyl ketone, a strong inhibitor for CLN2, indicating that the present enzyme is not a tripeptiylpeptidase. It had apparently the same cleavage specificity toward the chromogenic substrate Lys-Pro-Ile-Glu-Phe*Phe(NO2)-Arg-Leu (*, cleavage site) used as sedolisin and had a similar kcat value. However, the Km value was roughly ten times higher than that of sedolisin, suggesting a marked difference in substrate specificity.
(ii). Primary structure determination.
Then we cloned and sequenced the cDNA for this enzyme, and further characterized it.197) As a result, the enzyme was shown to be synthesized as a 575-residue preproform, in which a 173-residue prepropeptide, a 216-residue enzyme heavy chain and a 186-residue enzyme light chain are linked in this order. A gross structure of the enzyme moiety is shown in Fig. 1e. Sequence comparison showed that the enzyme is homologous to serine-carboxyl peptidase family (MEROPS Family S53) enzymes. The sequence identity was 28% with CLN2/tripeptidylpeptidase-1, 17% with sedolisin and 18% with kumamolisin. Strong inhibition by DFP, a serine peptidase inhibitor, of the enzyme confirmed that it is a serine-carboxyl peptidase. The catalytic residues (Glu75, Asp79 and Asp168 in the heavy chain and Ser95 in the light chain) and the Ca2+-binding site residues (Asp140 and Asp165 in the light chain) were well conserved.193) The two-chain structure is not shared with other serine-carboxyl peptidases. The C-terminal region of the heavy chain and the N-terminal region of the light chain in the proenzyme resided in a 23–32 residue insert sequence which was not present in other homologs and this region was indicated by homology modeling to form a loop protruding from the remaining part of the enzyme molecule, which should permit the proteolytic cleavage to generate the two chains. In the course of these studies, we also obtained structural evidence that scytalidopepsin A is also a serine-carboxyl peptidase and proposed a new name ‘scytalidolisin’ for this enzyme.198)
(iii). Identification of a unique serine-carboxyl peptidase with a DAN-reactive site.
The enzyme was inhibited by DAN in the presence of cupric ions. DAN was known to inhibit aspartic peptidases, but not any other enzymes including other serine-carboxyl peptidases. In the present enzyme, the DAN-reactive site was shown to be Asp140 in the light chain (Fig. 1e).197) The Asp residue corresponding to this residue had been shown not to reside in the active site but to be essential for activity by site-directed mutagenesis and to be one of the Ca2+-binding site residues by X-ray crystallography in other homologs.191–193) To our knowledge, this is the first time that DAN was found to specifically inactivate an enzyme other than aspartic peptidases by reacting with a specific Asp residue. Presumably the modification by DAN should destroy the essential Ca2+-binding site, thus resulting in inactivation of the catalytic machinery of the present enzyme. The introduced DAN moiety appeared to be located not in the active site of the enzyme, unlike the case of pepsin, but in the cavity formed between Thr142 and Lys155 in the light chain which might act as an affinity binding site to enhance the reaction. This supposition was consistent with the fact that the other known serine-carboxyl peptidases, which were insensitive to DAN, did not contain both of these (or similar) residues at the corresponding positions.
(iv). 3D structure and proposed catalytic mechanism.
The 3D structures of serine-carboxyl peptidases were determined for some enzymes such as sedolisin and kumamolisin, and the catalytic mechanisms have been proposed.174,191–193) Ser287/Glu80/Asp84 catalytic triad and Asp170 are thought to be involved in catalysis in sedolisin. As an example, the 3D structure and the proposed catalytic mechanism of sedolisin are shown in Fig. 3d and Fig. 5d, respectively. In the proposed mechanism for sedolisin, Glu80 acts as a general base to accept a proton from Ser287 during the nucleophilic attack and then as a general acid to protonate the leaving group during the cleavage of the scissile bond. Asp170 acts as a general acid to protonate the carbonyl oxygen of the P1 residue during the formation of the tetrahedral intermediate and as a general base for the formation of the acyl-enzyme (Fig. 5d). Since physarolisins I and II are their homologs and the residues corresponding to the catalytic residues in sedolisin are all conserved, their 3D structure and the catalytic mechanism should be similar to them.
IV (B). Physarolisin II
(i). Purification and characterization.
We found a physarolisin I homolog encoded in the php gene199) of P. polyephalum. To characterize the enzyme, we expressed in an E. coli expression system the encoded protein in which the N-terminal putative signal peptide (16 residues) was removed from the N-terminus and a His tag added to the C-terminus. The expressed 38 kDa soluble protein (one chain protein of 340 residues) was purified to homogeneity by chromatography on His-Bind®.200) Interestingly, the enzyme activity was maximum at 16–22℃, 50% at 5℃ and 32℃ and 5% at 40℃. When incubated at pH 4.2 and 18℃, the original enzyme disappeared rapidly due to autoproteolysis (half-life, about 5 min). In the initial stage, an intermediate form lacking the N-terminal 6-residue sequence was produced. This sequence may be the propeptide sequence. This change was largely stopped by prior treatment of the enzyme with DFP. On the other hand, the enzyme was more stable at pH 4.2 and 37℃ (half-life, about 6 h), and very stable at pH 7.5. Secondary structure changes analyzed by CD indicated that the enzyme was fully denatured at pH 7.9 and 37℃, that the enzyme recovered the folded structure by lowering pH to 4.2, and that DFP-treatment at pH 4.2 and 37℃ did not change the CD spectrum.
The enzyme was shown to hydrolyze oxidized insulin B chain most rapidly, followed by glucagons, substance P and Lys-Pro-Ile-Glu-Phe*Phe(NO2)-Arg-Leu (*, cleavage site) at pH 4.2 and 18℃. The optimum pH for oxidized insulin B chain was 4.2, which was close to those of other homologs. The major cleavage sites were Ala14-Leu15 and Phe25-Tyr26, which were quite different from those of physarolisin I, whose major cleavage sites were Gly8-Ser9, Leu11-Val12 and Cys(SO3H)19-Gly20. On the other hand, hemoglobin, casein, azocol and Ala-Ala-Phe-4-methylcoumaryl-7-amide, a good substrate for CLN2/tripeptidylpeptidase, were not hydrolyzed at pH 3.6–7.5 and 37℃. The enzyme activity was strongly inhibited by DFP, but not by pepstatin A and Ala-Ala-Phe-CH2Cl, a strong tripeptidylpeptidase inhibitor. The enzyme thus resembled physarolisin I in the effects of inhibitors, but unlike physarolisin I it was not inhibited by DAN.
(ii). Primary structure.
The primary structure of the enzyme had low homology with other serine-carboxyl peptidases including physarolisin I (sequence identity, 16%), but the catalytic residues (a Ser-Glu-Asp catalytic triad and an additional Asp) and the Ca2+-binding site Asp residues were well conserved. Therefore, this enzyme was thought to be a member of serine-carboxyl peptidases. The enzyme is unique in that it has no propeptide corresponding to those in the other known serine-carboxyl peptidases. The latter possess a long N-terminal propeptide of approximately 170–240 residues. During autolysis, the N-terminal six-residue segment appeared to be removed. At present, it is not clear whether this segment acts as the propeptide. The expressed and purified enzyme preparation at pH 7.9 had a denatured form as examined by CD spectroscopy. However, the enzyme was found to be folded very quickly when exposed to pH 4.2 to generate the enzyme activity.
(iii). Identification as a unique cold-adapted enzyme.
As described above, the present enzyme was considerably different from other serine-carboxyl peptidases including physarolisin I. The enzyme was most active at 16–22℃, indicating that it is a kind of cold-adapted enzymes. To our knowledge, it is the first cold-adapted enzyme found among peptidases. P. polycephalum grows optimally in a similar temperature range, and therefore the enzyme appeared to be well adapted to the growing temperature. Interestingly, the stability of the enzyme was also highly dependent on temperature; the enzyme was unstable at 18℃ due to autolysis, but not at 37℃. These results suggested that the activity and amount of the enzyme was strictly regulated by environmental temperature. The rapid autolysis at 18℃ seemed to indicate that the enzyme possesses a fairly flexible conformation at the cold temperature, which should also be favorable for expression of the enzyme activity under such conditions. This assumption was consistent with the fact that the cold-adaptation of an enzyme is generally achieved through such a flexible structure of the enzyme molecule. On the other hand, it is notable that the enzyme was less active but more stable at higher temperatures, such as 37℃, despite the fact that no gross conformational change was observed between the temperatures at 37℃ and 20℃ in the CD spectroscopy. This was in sharp contrast with the fact that other cold-adapted enzymes are usually heat-labile at or above a moderate temperature. In the case of physarolisin II, the temperature-stability profile may be at least partly explained by the preferred autodigestion at around the optimal temperature for activity. To establish physarolisin II as a unique cold-adapted enzyme, further studies will be necessary, including 3D structure study.
The cold adaptation, as well as the rapid autolysis, restricted substrate specificity and unique timing of the gene replication, together suggest that the enzyme might play a very unique role in vivo, possibly regulated by the change of environmental temperature, through processing a specific or a limited kind of substrate(s). Thus, the enzyme stands out in both functional and structural features among the serine-carboxyl peptidase family.
Concluding remarks
Thus far I followed the trait of our studies on those enzymes in which one or two active-site carboxyl groups play a key role or an equally important and leading role with other groups as general acid/base or nucleophilic catalysts. Carboxyl groups are widely utilized as indispensable catalytic residue(s) in various enzymes, notably in hydrolases and certain transferases. Among hydrolases and transferases, they include certain RNases, peptidases, glycosidases etc., and I focused attention to typical ones including RNase T1 and certain peptidases in this review. To our knowledge, only a limited number of enzymes with such catalytic carboxyl group(s) are known so far in other groups of enzymes than hydrolases and transferases, such as triosephosphate isomerase (catalytic Glu),201) glucosamine 6-phosphate isomerase (or deaminase) (catalytic Asp),202) phosphofructokinase (catalytic Asp)203) etc., where the catalytic Glu/Asp residues are also thought to function as general acid/bases.
Among many RNases, RNase T1 and its homologs are, to my knowledge, the only enzymes to be called “carboxyl RNases.” To be more strict, they should be called such as Glu/His/His-RNases. However, they may be also called “histidine RNases” since they have catalytic histidines. On the other hand, RNase A (bovine pancreas) and its homologs should be called His/His-RNases (or “histidine RNases”).
Major carboxyl peptidases we studied included the following three types of enzymes: (1) pepsin- and nepenthesin-type aspartic peptidases, which should be called more strictly as Asp/Asp-peptidases, (2) glutamic peptidases to be called more strictly as Glu/Gln-peptidases, and (3) serine-carboxyl peptidases to be called more strictly such as Ser/Glu/Asp/Asp-peptidases. Among these enzymes, serine-carboxyl peptidases may be classified as both serine peptidases and carboxyl peptidases. However, considering the central catalytic role of the active-site serine residue, it should be reasonable to classify them as serine peptidases. Serine peptidases such as trypsin and chymotrypsin, are known to have a Ser/His/Asp catalytic triad. In these cases, however, the Asp residue was shown to work only as an anionic species to stabilize the transition state.
On the other hand, many glycosidases are known to have an acidic residue dyad at the catalytic sites, which are thought to play key roles in catalysis.204) They are classified into two groups, (1) retaining glycosidases including hen egg-white lysozyme and (2) converting glycosidases. In the former, one of the catalytic acidic residues are thought to work as a general acid/base, while the other as a nucleophilic group. For example, in hen egg-white lysozyme, Glu35 is thought to operate as a general acid/base catalyst whereas Asp52 is thought to be involved in the formation of a glycosyl enzyme intermediate by direct nucleophilic attack at the C1-position of N-acetylmuramic acid moiety205) (Figs. 3e and 5e). The possibility for such a covalent intermediate mechanism might be present in some other groups of enzymes. In some retaining glycosidases, however, the C2 acetoamide group is assumed to be involved in catalysis without the nucleophilic carboxyl group. On the other hand, the converting glycosidases are generally thought to use both carboxyl groups as general acid/bases.
Taken together, I would like to propose for most of these enzymes with a catalytic carboxyl group or groups the generic name “carboxyl enzymes” like those used for other groups of enzymes, such as thiol (cysteine) enzymes, serine enzymes and metalloenzymes. Further studies, however, would be necessary to define the realm of “carboxyl enzymes.”
Acknowledgements
I am especially grateful to Prof. Tamio Yamakawa for recommending me to write this review. I thank all my colleagues who collaborated with me in certain aspects of the present studies. I also thank Dr. Keiko Kubota at the Institute of Molecular and Cellular Biosciences, The University of Tokyo for helping me to prepare Fig. 3.
Abbreviations
- RNase
ribonuclease
- CM
carboxymethyl
- PGO
phenylglyoxal
- 3D
three-dimensional
- DAN
diazoacetyl-d,l-norleucine methyl ester
- EPNP
1,2-epoxy-3-(p-nitrophenoxy)propane
- SAX
small-angle X-ray scattering
- CD
circular dichroism
Profile
Kenji Takahashi was born in 1934 in Nagano and graduated from the Department of Chemistry, Faculty of Science, The University of Tokyo in 1957. In 1962, he received a Ph.D. degree at the Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, and continued his work in the same Department as an Assistant from 1963 to 1973, and as a Lecturer in 1973. In 1965, he determined the complete amino acid sequence of ribonuclease T1, and received a Young Scientist Award from The Japanese Biochemical Society. From 1965 to 1968 he worked as a Research Associate at the Rockefeller University on the active sites of enzymes and identified a glutamic acid at the active site of ribonuclease T1. After return to Japan, he worked as a Professor at the Department of Biochemistry, Primate Research Institute, Kyoto University from 1974 to 1984, mainly on the structure and function of proteolytic enzymes, especially carboxyl peptidases. From 1984 to 1995, he continued his work as a Professor at the Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo (Professor Emeritus in 1995), then from 1995 to 2005 as a Professor at the School of Life Sciences, Tokyo University of Pharmacy and Life Sciences (Professor Emeritus in 2005). Since 2005 to date he has been a Visiting Professor of The Metropolitan University, Tokyo and also associated with The Nippon Dental University.
References
- 1).Egami F., Takahashi K., Uchida T. (1964) Ribonucleases in Taka-Diastase: properties, chemical nature, and applications. Prog. Nucleic Acid Res. Mol. Biol. 3, 59–101 [DOI] [PubMed] [Google Scholar]
- 2).Takahashi, K., Uchida, U. and Egami, F. (1970) Ribonuclease T1—structure and function. Advan. Biophys. (ed. Kotani, M.). Vol. 1, Univ. Tokyo Press, Tokyo, pp. 53–98. [PubMed] [Google Scholar]
- 3).Takahashi, K. (1971) Ribonuclease T1 In Proteins. Structure and Function (eds. Funatsu, M., Hiromi, K., Imahori, K., Murachi, T. and Narita, K). Vol. 1, Kodansha, Tokyo, pp. 285–331. [Google Scholar]
- 4).Takahashi, K. and Moore, S. (1982) Ribonuclease T1 The enzymes (ed. Boyer, P.D.). Vol. XV, Part B, 3rd ed., Academic Press, New York, pp. 435–468. [Google Scholar]
- 5).Kojima M., Tanokura M., Takahashi K. (2000) Structure-function relationship of ribonuclease Tl based on molecular structure. Bioimages 8, 45–55 [Google Scholar]
- 6).Takahashi K. (1961) The structure and function of ribonuclease T1. I. Chromatographic purification and properties of ribonuclease T1. J. Biochem. 49, 1–8 [Google Scholar]
- 7).Takahashi K. (1962) The structure and function of ribonuclease T1. II. Further purification and amino acid composition of ribonuclease T1. J. Biochem. 51, 95–108 [DOI] [PubMed] [Google Scholar]
- 8).Takahashi K. (1962) The structure and function of ribonuclease T1. III. Amino- and carboxyl-terminal sequences of ribonuclease T1. J. Biochem. 52, 72–81 [DOI] [PubMed] [Google Scholar]
- 9).Takahashi K. (1966) The structure and function of ribonuclease T1. VII. Further investigations on amino acid composition and some other properties of ribonuclease T1. J. Biochem. 60, 239–245 [DOI] [PubMed] [Google Scholar]
- 10).Yamagata S., Takahashi K., Egami F. (1962) The structure and function of ribonuclease T1. IV. Photooxidation of ribonuclease T1. J. Biochem. 52, 261–266 [DOI] [PubMed] [Google Scholar]
- 11).Yamagata S., Takahashi K., Egami F. (1962) The structure and function of ribonuclease T1. VI. Reduction of disulfide bonds of ribonuclease T1. J. Biochem. 52, 272–274 [DOI] [PubMed] [Google Scholar]
- 12).Shiobara Y., Takahashi K., Egami F. (1962) The structure and function of ribonuclease T1. V. Deamination of ribonuclease T1. J. Biochem. 52, 267–271 [DOI] [PubMed] [Google Scholar]
- 13).Hirs C.H.W., Moore S., Stein W.H. (1960) The sequence of the amino acid residues in performic acid-oxidized ribonuclease. J. Biol. Chem. 235, 633–647 [PubMed] [Google Scholar]
- 14).Takahashi K. (1965) The amino acid sequence of ribonuclease T1. J. Biol. Chem. 240, 4117–4119 [PubMed] [Google Scholar]
- 15).Takahashi K. (1971) The structure and function of ribonuclease T1. XIII. Isolation and amino acid sequences of tryptic peptides from performic acid-oxidized ribonuclease T1. J. Biochem. 70, 477–495 [DOI] [PubMed] [Google Scholar]
- 16).Takahashi K. (1971) The structure and function of ribonuclease T1. XIV. Isolation and amino acid composition of chymotryptic peptides from performic acid-oxidized and heat-denatured ribonuclease T1. J. Biochem. 70, 603–615 [DOI] [PubMed] [Google Scholar]
- 17).Takahashi K. (1971) The structure and function of ribonuclease T1. XV. Amino acid sequences of chymotryptic peptides from performic acid-oxidized and heat-denatured ribonuclease T1—the complete amino acid sequence of ribonuclease T1. J. Biochem. 70, 617–634 [DOI] [PubMed] [Google Scholar]
- 18).Takahashi K. (1971) The structure and function of ribonuclease T1. XVI. Isolation and amino acid sequences of peptic peptides from heat-denatured ribonuclease T1. J. Biochem. 70, 803–815 [DOI] [PubMed] [Google Scholar]
- 19).Takahashi K. (1971) The structure and function of ribonuclease T1. XVII. Isolation and amino acid sequences of papain and subtilisin peptides from ribonuclease T1—the complete covalent structure of ribonuclease T1. J. Biochem. 70, 945–960 [DOI] [PubMed] [Google Scholar]
- 20).Canfield R.E. (1963) The amino acid sequence of egg white lysozyme. J. Biol. Chem. 238, 2698–2707 [PubMed] [Google Scholar]
- 21).Jollès J., Jauregui-Adell J., Bernier I., Jollès P. (1963) La structure chimique du lysozyme de blanc d'oeuf de poule: étude détaillée. Biochim. Biophys. Acta 78, 668–689 [DOI] [PubMed] [Google Scholar]
- 22).Crestfield A.M., Stein W.H., Moore S. (1963) Alkylation and identification of the histidine residues at the active site of ribonuclease. J. Biol. Chem. 238, 2413–2419 [PubMed] [Google Scholar]
- 23).Takahashi K., Stein W.H., Moore S. (1967) The identification of a glutamic acid residue as part of the active site of ribonuclease T1. J. Biol. Chem. 242, 4682–4690 [PubMed] [Google Scholar]
- 24).Blake C.C., Koenig D.F., Mair G.A., North A.C., Phillips D.C., Sarma V.R. (1965) Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 angstrom resolution. Nature 206, 757–761 [DOI] [PubMed] [Google Scholar]
- 25).Wyckoff H., Hardman K., Allewell N., Inagami T., Johnson L., Richards F. (1967) The structure of ribonuclease-S at 3.5 Å resolution. J. Biol. Chem. 242, 3984–3988 [PubMed] [Google Scholar]
- 26).Takahashi K. (1968) The reaction of phenylglyoxal with arginine residues in proteins. J. Biol. Chem. 243, 6171–6179 [PubMed] [Google Scholar]
- 27).Takahashi K. (1977) The reactions of phenylglyoxal and related reagents with amino acids. J. Biochem. 81, 395–402 [DOI] [PubMed] [Google Scholar]
- 28).Takahashi K. (1977) Further studies on the reactions of phenylglyoxal and related reagents with proteins. J. Biochem. 81, 403–414 [DOI] [PubMed] [Google Scholar]
- 29).Takahashi K. (1970) The structure and function of ribonuclease T1. XI. Modification of the single arginine residue in ribonuclease T1 by phenylglyoxal and glyoxal. J. Biochem. 68, 659–664 [DOI] [PubMed] [Google Scholar]
- 30).Takahashi K. (1976) Specific modification of arginine residues in proteins with ninhydrin. J. Biochem. 80, 1173–1176 [DOI] [PubMed] [Google Scholar]
- 31).Takahashi K. (1970) The structure and function of ribonuclease T1. IX. Photooxidation of ribonuclease T1 in the presence of rose bengal. J. Biochem. 67, 833–839 [DOI] [PubMed] [Google Scholar]
- 32).Takahashi K. (1970) The structure and function of ribonuclease T1. X. Reactions of iodoacetate, iodoacetamide and related alkylating reagents with ribonuclease T1. J. Biochem. 68, 517–527 [DOI] [PubMed] [Google Scholar]
- 33).Takahashi K. (1971) The structure and function of ribonuclease T1. XII. Further studies on rose bengal-catalyzed photooxidation of ribonuclease T1—Identification of a critical histidine residue. J. Biochem. 69, 331–338 [DOI] [PubMed] [Google Scholar]
- 34).Takahashi K. (1973) Evidence for the implication of histidines-40 and -92 in the active site of ribonuclease T1. J. Biochem. 74, 1279–1282 [DOI] [PubMed] [Google Scholar]
- 35).Takahashi K. (1976) The structure and function of ribonuclease T1. XXI. Modification of histidine residues in ribonuclease T1 with iodoacetamide. J. Biochem. 80, 1267–1275 [DOI] [PubMed] [Google Scholar]
- 36).Inagaki F., Kawano Y., Shimada I., Takahashi K., Miyazawa T. (1981) Nuclear magnetic resonance study on the microenvironments of histidine residues of ribonuclease T1 and carboxymethylated ribonuclease T1. J. Biochem. 89, 1185–1195 [PubMed] [Google Scholar]
- 37).Takahashi K. (1972) The structure and function of ribonuclease T1. XVIII. Gel filtration studies on the interaction of ribonuclease T1 with substrate analogs. J. Biochem. 72, 1469–1481 [DOI] [PubMed] [Google Scholar]
- 38).Heinemann U., Saenger W. (1982) Specific protein-nucleic acid recognition in ribonuclease T1-2′-guanylic acid complex: an X-ray study. Nature 299, 27–31 [DOI] [PubMed] [Google Scholar]
- 39).Ishikawa K., Suzuki E., Tanokura M., Takahashi K. (1996) Crystal structure of ribonuclease T1 carboxymethylated at Glu58 in complex with 2′-GMP. Biochemistry 35, 8329–8334 [DOI] [PubMed] [Google Scholar]
- 40).Hatano K., Kojima M., Suzuki E., Tanokura M., Takahashi K. (2003) Determination of the NMR structure of Gln25-Ribonuclease T1. Biol. Chem. 384, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 41).Tanokura M., Miyano H., Suzuki E., Takahashi K. (1993) Effects of carboxymethylation on the conformation of the complex of ribonuclease T1 and guanosine 2′-monophosphate. Bioimages 1, 125–128 [Google Scholar]
- 42).Nishikawa S., Morioka H., Fukushima K., Tanaka T., Uesugi S., Ohtsuka E., Ikehara M. (1986) Modification of Glu 58, an amino acid of the active center of ribonuclease T1, to Gln and Asp. Biochem. Biophys. Res. Commun. 138, 789–794 [DOI] [PubMed] [Google Scholar]
- 43).Nishikawa S., Morioka H., Kim H.J., Fuchimura K., Tanaka T., Uesugi S., Hakoshima T., Tomita K., Ohtsuka E., Ikehara M. (1987) Two histidine residues are essential for ribonuclease T1 activity as in the case of ribonuclease A. Biochemistry 26, 8620–8624 [DOI] [PubMed] [Google Scholar]
- 44).Steyaert J., Hallenga K., Wyns L., Stanssens P. (1990) Histidine-40 of ribonuclease T1 acts as base catalyst when the true catalytic base, glutamic acid-58, is replaced by alanine. Biochemistry 29, 9064–9072 [DOI] [PubMed] [Google Scholar]
- 45).Zegers I., Verhelst P., Choe H.W., Steyaert J., Heinemann U., Saenger W., Wyns L. (1992) Role of histidine-40 in ribonuclease T1 catalysis: three-dimensional structures of the partially active His40Lys mutant. Biochemistry 31, 11317–11325 [DOI] [PubMed] [Google Scholar]
- 46).Steyaert J., Wyns L. (1993) Functional interactions among the His40, Glu58 and His92 catalysts of ribonuclease T1 as studied by double and triple mutants. J. Mol. Biol. 229, 770–781 [DOI] [PubMed] [Google Scholar]
- 47).Pletinckx J., Steyaert J., Zegers I., Choe H.W., Heinemann U., Wyns L. (1994) Crystallographic study of Glu58Ala RNase 2′-guanosine monophosphate at 1.9-Å resolution. Biochemistry 33, 1654–1662 [DOI] [PubMed] [Google Scholar]
- 48).Kojima M., Tanokura M., Murakami T., Takahashi K. (1994) Role of glutamic acid-58 of ribonuclease T1 in enzyme action as studied by molecular dynamics simulation of aspartic acid-58 and alanine-58 mutants. Bioimages 2, 53–57 [Google Scholar]
- 49).Hartley R.W. (1980) Homology between prokaryotic and eukaryotic ribonucleases. J. Mol. Evol. 15, 355–358 [DOI] [PubMed] [Google Scholar]
- 50).Kojima M., Mizukoshi T., Miyano H., Suzuki E., Tanokura M., Takahashi K. (1994) Thermal stabilization of ribonuclease T1 by carboxymethylation at Glu-58 as revealed by 1H nuclear magnetic resonance spectroscopy. FEBS Lett. 351, 389–392 [DOI] [PubMed] [Google Scholar]
- 51).Matsuura M., Shimotakahara S., Sakuma C., Tashiro M., Shindo H., Mochizuki K., Yamagishi A., Kojima M., Takahashi K. (2004) Thermal unfolding of ribonuclease T1 studied by multi-dimensional NMR spectroscopy. Biol. Chem. 385, 1157–1164 [DOI] [PubMed] [Google Scholar]
- 52).Takahashi K., Hashimoto J. (1988) The amino acid sequence of ribonuclease U1, a guanine-specific ribonuclease from the fungus Ustilago sphaerogena. J. Biochem. 103, 313–320 [DOI] [PubMed] [Google Scholar]
- 53).Takahashi K. (1988) The amino acid sequence of ribonuclease N1, a guanine-specific ribonuclease from the fungus Neurospora crassa. J. Biochem. 104, 375–382 [DOI] [PubMed] [Google Scholar]
- 54).Takahashi K. (1985) A revision and confirmation of the amino acid sequence of ribonuclease T1. J. Biochem. 98, 815–817 [DOI] [PubMed] [Google Scholar]
- 55).Hashimoto J., Takahashi K., Uchida T. (1973) Photooxidation of ribonuclease U1. J. Biochem. 73, 13–22 [PubMed] [Google Scholar]
- 56).Hashimoto J., Takahashi K. (1977) Chemical modifications of ribonuclease U1. J. Biochem. 81, 1175–1180 [DOI] [PubMed] [Google Scholar]
- 57).Yoshida H. (2001) The ribonuclease T1 family. Methods Enzymol. 341, 28–41 [DOI] [PubMed] [Google Scholar]
- 58).Foltmann B. (1981) Gastric proteinases—structure, function, evolution and mechanism of action. Essays Biochem. 17, 52–84 [PubMed] [Google Scholar]
- 59).Rajagopalan T.G., Stein W.H., Moore S. (1966) The inactivation of pepsin by diazoacetylnorleucine methyl ester. J. Biol. Chem. 241, 4295–4297 [PubMed] [Google Scholar]
- 60).Hofmann T. (1974) Structure, function, and evolution of acid proteases. Adv. Chem. Ser. 136, 146–185 [Google Scholar]
- 61).Chen K.C.S., Tang J. (1972) Amino acid sequence around the epoxide-reactive residues in pepsin. J. Biol. Chem. 247, 2566–2574 [PubMed] [Google Scholar]
- 62).Chang W.-J., Takahashi K. (1973) The structure and function of acid proteases. II. Inactivation of bovine rennin by acid protease-specific inhibitors. J. Biochem. 74, 231–237 [PubMed] [Google Scholar]
- 63).Takahashi K., Mizobe F., Chang W.-J. (1972) Inactivation of acid proteases from Rhizopus chinensis, Aspergillus saitoi and Mucor pusillus, and calf rennin by diazoacetylnorleucine methyl ester. J. Biochem. 71, 161–164 [DOI] [PubMed] [Google Scholar]
- 64).Mizobe F., Takahashi K., Ando T. (1973) The structure and function of acid proteases. I. Specific inactivation of an acid protease form Rhizopus chinensis by diazoacetyl-dl-norleucine methyl ester. J. Biochem. 73, 61–68 [PubMed] [Google Scholar]
- 65).Takahashi K., Chang W.-J. (1973) Specific chemical modifications of acid proteases in the presence and absence of pepstatin. J. Biochem. 73, 675–677 [DOI] [PubMed] [Google Scholar]
- 66).Takahashi K., Chang W.-J., Ko J.-S. (1974) Specific inhibition of acid proteases form brain, kidney, skeletal muscle, and insectivorous plants by diazoacetyl-dl-norleucine methyl ester and by pepstatin. J. Biochem. 76, 897–899 [PubMed] [Google Scholar]
- 67).Takahashi K., Chang W.-J., Arima K. (1976) The structure and function of acid proteases. IV. Inactivation of the acid protease from Mucor pussilus by acid protease-specific inhibitors. J. Biochem. 80, 61–67 [DOI] [PubMed] [Google Scholar]
- 68).Takahashi K., Chang W.-J. (1976) The structure and function of acid proteases. V. Comparative studies on the specific inhibition of acid proteases by diazoacetyl-dl-norleucine methyl ester, 1,2-epoxy-3-(p-nitrophenoxy)propane and pepstatin. J. Biochem. 80, 497–506 [DOI] [PubMed] [Google Scholar]
- 69).Chang W.-J., Takahashi K. (1974) The structure and function of acid proteases. III. Isolation and characterization of the active-site peptides from bovine rennin. J. Biochem. 76, 467–474 [DOI] [PubMed] [Google Scholar]
- 70).Nakamura S., Takahashi K. (1978) The structure and function of acid proteases. IX. Isolation and amino acid sequences of the peptides containing the active site aspartyl residues reactive with diazoacetyl-dl-norleucine methyl ester and 1,2-epoxy-3-(p-nitrophenoxy) propane in Rhizopus chinensis acid protease. J. Biochem. 84, 1593–1600 [DOI] [PubMed] [Google Scholar]
- 71).Ito H., Hirono T., Morita Y., Nemoto Y., Kim Y.-T., Takahashi K. (2008) Novel peptide-based pepsin inhibitors containing an epoxide group. J. Enzyme Inhib. Med. Chem. 23, 352–356 [DOI] [PubMed] [Google Scholar]
- 72).James M.N.G., Hsu I.-N., Delbaere L.T.J. (1977) Mechanism of acid protease catalysis based on the crystal structure of penicillopepsin. Nature 267, 808–813 [DOI] [PubMed] [Google Scholar]
- 73).Kageyama T., Takahashi K. (1976) Pepsinogens and pepsins from gastric mucosa of Japanese monkey. Purification and characterization. J. Biochem. 79, 455–468 [DOI] [PubMed] [Google Scholar]
- 74).Kageyama T., Takahashi K. (1976) Pepsinogen C and pepsin C from gastric mucosa of Japanese monkey. J. Biochem. 80, 983–992 [DOI] [PubMed] [Google Scholar]
- 75).Kageyama T., Takahashi K. (1977) The carbohydrate moiety of Japanese monkey pepsinogen—its composition and site of attachment to protein. Biochem. Biophys. Res. Commun. 74, 789–795 [DOI] [PubMed] [Google Scholar]
- 76).Kageyama T., Takahashi K. (1978) Monkey pepsinogens and pepsins. III. Carbohydrate moiety of Japanese monkey pepsinogens and the amino acid sequence around the site of its attachment to protein. J. Biochem. 84, 771–778 [DOI] [PubMed] [Google Scholar]
- 77).Kageyama T., Takahashi K. (1980) Monkey pepsinogens and pepsins. IV. The amino acid sequence of the activation peptide segment of Japanese monkey pepsinogen. J. Biochem. 88, 9–16 [PubMed] [Google Scholar]
- 78).Kageyama T., Takahashi K. (1980) Monkey pepsinogens and pepsins. V. Purification, characterization, and amino-terminal sequence determination of crab-eating monkey pepsinogens and pepsins. J. Biochem. 88, 635–645 [DOI] [PubMed] [Google Scholar]
- 79).Kageyama T., Takahashi K. (1980) Isolation of an activation intermediate and determination of the amino acid sequence of the activation segment of human pepsinogen A. J. Biochem. 88, 571–582 [DOI] [PubMed] [Google Scholar]
- 80).Kageyama T., Moriyama A., Takahashi K. (1983) Purification and characterization of pepsinogens and pepsins from asiatic black bear, and amino acid sequence determination of the NH2-terminal 60 residues of the major pepsinogen. J. Biochem. 94, 1557–1567 [PubMed] [Google Scholar]
- 81).Kageyama T., Takahashi K. (1984) Rabbit pepsinogens. Purification, characterization, analysis of the conversion process to pepsin, and determination of the NH2-terminal amino acid sequence. Eur. J. Biochem. 141, 261–269 [DOI] [PubMed] [Google Scholar]
- 82).Kageyama T., Takahashi K. (1985) Monkey pepsinogens and pepsins. VII. Analysis of the activation process and determination of the NH2-terminal 60-residue sequence of Japanese monkey progastricsin, and molecular evolution of pepsinogens. J. Biochem. 97, 1235–1246 [DOI] [PubMed] [Google Scholar]
- 83).Kageyama T., Takahashi K. (1986) The complete amino acid sequence of monkey pepsinogen A. J. Biol. Chem. 261, 4395–4405 [PubMed] [Google Scholar]
- 84).Kageyama T., Takahashi K. (1986) The complete amino acid sequence of monkey progastricsin. J. Biol. Chem. 261, 4406–4419 [PubMed] [Google Scholar]
- 85).Ichihara Y., Sogawa K., Takahashi K. (1982) Rat gastric prepepsinogen: in vitro synthesis and partial amino-terminal signal sequence. J. Biochem. 92, 603–606 [DOI] [PubMed] [Google Scholar]
- 86).Ichihara Y., Sogawa K., Takahashi K. (1985) Isolation of human, swine, and rat prepepsinogens and calf preprochymosin, and determination of the primary structures of their NH2-terminal signal sequences. J. Biochem. 98, 483–492 [DOI] [PubMed] [Google Scholar]
- 87).Ong E.B., Perlmann G.E. (1968) The amino-terminal sequence of porcine pepsinogen. J. Biol. Chem. 246, 6104–6109 [PubMed] [Google Scholar]
- 88).Pedersen V.B., Foltmann B. (1973) The amino acid sequence of a hitherto unobserved segment from porcine pepsinogen preceding the N-terminus of pepsin. FEBS Lett. 35, 255–256 [DOI] [PubMed] [Google Scholar]
- 89).Tang J., Sepulveda P., Marciniszyn J., Jr., Chen K.C., Huang W.Y., Tao N., Liu D., Lanier J.P. (1973) Amino-acid sequence of porcine pepsin. Proc. Natl. Acad. Sci. U.S.A. 70, 3437–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Tsukagoshi N., Ando Y., Tomita Y., Uchida R., Takemura T., Sasaki T., Yamagata H., Udaka S., Ichihara Y., Takahashi K. (1988) Nucleotide sequence and expression in Escherichia coli of cDNA of swine pepsinogen: involvement of the amino-terminal portion of the activation peptide segment in restoration of the functional protein. Gene 65, 285–292 [DOI] [PubMed] [Google Scholar]
- 91).Moriyama A., Takahashi K. (1980) Studies on the distribution of acid proteases in primate lungs and other tissues by diethylaminoethyl-cellulose chromatography. J. Biochem. 87, 737–743 [DOI] [PubMed] [Google Scholar]
- 92).Moriyama A., Takahashi K. (1980) Cathepsins D from rhesus monkey lung. Purification and characterization. J. Biochem. 88, 619–633 [DOI] [PubMed] [Google Scholar]
- 93).Moriyama A., Kageyama K., Takahashi K. (1983) Identification of monkey lung procathepsin D-II as a pepsinogen-C-like acid protease zymogen. Eur. J. Biochem. 132, 687–692 [DOI] [PubMed] [Google Scholar]
- 94).Moriyama A., Kageyama T., Takahashi K., Sasaki M. (1985) Purification of Japanese monkey prostate acid protease zymogen and its identification as a pepsinogen c-like zymogen. J. Biochem. 98, 1255–1261 [DOI] [PubMed] [Google Scholar]
- 95).Kageyama T., Takahashi K. (1980) A cathepsin D-like acid proteinase from human gastric mucosa. Purification and characterization. J. Biochem. 87, 725–735 [DOI] [PubMed] [Google Scholar]
- 96).Tang, J.N. (2013) Pepsin A. Handbook of Proteolytic Enzymes, 3rd ed. (eds. Rawlings, N.D. and Salvesen, G.). Vol. 1, Elsevier Academic Press, London, pp. 27–35. [Google Scholar]
- 97).Kageyama T., Takahashi K. (1982) Monkey pepsinogens and pepsins. VI. One-step activation of Japanese monkey pepsinogen to pepsin. J. Biochem. 92, 1179–1188 [DOI] [PubMed] [Google Scholar]
- 98).Kageyama T., Takahashi K. (1982) Evidence for the occurrence of one-step activation in porcine pepsinogen. Biochem. Biophys. Res. Commun. 107, 1117–1122 [DOI] [PubMed] [Google Scholar]
- 99).Kageyama T., Takahashi K. (1983) Occurrence of two different pathways in the activation of porcine pepsinogen to pepsin. J. Biochem. 93, 743–754 [DOI] [PubMed] [Google Scholar]
- 100).Kageyama T., Takahashi K. (1987) Activation mechanism of monkey and porcine pepsinogens A. Eur. J. Biochem. 165, 483–490 [DOI] [PubMed] [Google Scholar]
- 101).Kageyama T., Ichinose M., Miki K., Athauda S.B.P., Tanji M., Takahashi K. (1989) Difference of activation processes and structure of activation peptides in human pepsinogen A and progastricsin. J. Biochem. 105, 15–22 [DOI] [PubMed] [Google Scholar]
- 102).Takahashi, K. and Kageyama, T. (1985) Multiplicity and intermediates of the activation mechanism of zymogens of gastric aspartic proteinases. Aspartic Proteinases and Their Inhibitors (ed. Kostka, V.). Waltzer de Gruyter & Co., Berlin, New York, pp. 265–282. [Google Scholar]
- 103).Sogawa K., Ichihara Y., Takahashi K. (1981) Molecular cloning of complementary DNA to swine pepsinogen mRNA. J. Biol. Chem. 256, 12561–12565 [PubMed] [Google Scholar]
- 104).Sogawa K., Fujii-Kuriyama Y., Mizukami Y., Ichihara Y., Takahashi K. (1983) Primary structure of human pepsinogen gene. J. Biol. Chem. 258, 5306–5311 [PubMed] [Google Scholar]
- 105).Hayano T., Sogawa K., Ichihara Y., Fujii-Kuriyama Y., Takahashi K. (1986) Close linkage of human chromosomal pepsinogen A genes. Biochem. Biophys. Res. Commun. 138, 289–296 [DOI] [PubMed] [Google Scholar]
- 106).Hayano T., Sogawa K., Ichihara Y., Fujii-Kuriyama Y., Takahashi K. (1988) Primary structure of human Pepsinogen C gene. J. Biol. Chem. 263, 1382–1385 [PubMed] [Google Scholar]
- 107).Takahara K., Fukushige S., Murotsu T., Ichihara Y., Hayano T., Ishihara T., Takahashi K., Matsubara K. (1989) Assignment of human pepsinogen C (PGC) gene to chromosome 6. Cytogenet. Cell Genet. 52, 100–101 [DOI] [PubMed] [Google Scholar]
- 108).Takahashi K. (1992) Gene structures of pepsinogens A and C. Scand. J. Clin. Lab. Invest. 52 (Suppl. 210), 97–110 [PubMed] [Google Scholar]
- 109).Ichihara Y., Sogawa K., Morohashi K., Fujii-Kuriyama Y., Takahashi K. (1986) Nucleotide sequence of a nearly full-length cDNA coding for pepsinogen of rat gastric mucosa. Eur. J. Biochem. 161, 7–12 [DOI] [PubMed] [Google Scholar]
- 110).Ishihara T., Ichihara Y., Hayano T., Katsura I., Sogawa K., Fujii-Kuriyama Y., Takahashi K. (1989) Primary structure and transcriptional regulation of rat pepsinogen C gene. J. Biol. Chem. 264, 10193–10199 [PubMed] [Google Scholar]
- 111).Yakabe Y., Tanji M., Ichinose M., Goto S., Miki K., Kurokawa K., Ito H., Kageyama T., Takahashi K. (1991) Purification, characterization, and amino acid sequences of pepsinogens and pepsins from the esophageal mucosa of bullfrog (Rana catesbeiana). J. Biol. Chem. 266, 22436–22443 [PubMed] [Google Scholar]
- 112).Tanji M., Yakabe E., Kubota K., Kageyama T., Ichinose M., Miki K., Ito H., Takahashi K. (2009) Structural and phylogenetic comparison of three pepsinogens from pacific bluefin tuna: molecular evolution of fish pepsinogens. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 152, 9–19 [DOI] [PubMed] [Google Scholar]
- 113).Ichinose M., Miki K., Furihata C., Tatematsu M., Ichihara Y., Ishihara T., Katsura I., Sogawa K., Fujii-Kuriyama Y., Tanji M., Oka H., Matsushima T., Takahashi K. (1988) DNA methylation and expression of the rat pepsinogen gene in embryonic, adult, and neoplastic tissues. Cancer Res. 48, 1603–1609 [PubMed] [Google Scholar]
- 114).Ichinose M., Miki K., Tatematsu M., Furihata C., Nobuhara M., Ichihara Y., Tanji M., Sogawa K., Fujii-Kuriyama Y., Oka H., Takahashi T., Kageyama T., Takahashi K. (1988) Hypomethylation and expression of pepsinogen A genes in the fundic mucosa of human stomach. Biochem. Biophys. Res. Commun. 151, 275–282 [DOI] [PubMed] [Google Scholar]
- 115).Ichinose M., Miki K., Tatematsu M., Mizuno T., Mutai M., Furihata C., Ichihara Y., Ishihara T., Tanji M., Oka H., Hinohara Y., Takahashi T., Kageyama T., Takahashi K. (1988) Cell-specific hypomethylation of the pepsinogen gene in pepsinogen-producing cells. Biochem. Biophys. Res. Commun. 155, 670–677 [DOI] [PubMed] [Google Scholar]
- 116).Sano J., Miki K., Ichinose M., Kimura M., Kurokawa K., Aida T., Ishizaki M., Asano G., Masugi Y., Wong R.N.S., Takahashi K. (1989) In situ localization of pepsinogens I and II mRNA in human gastric mucosa. Acta Pathol. Jpn. 39, 765–771 [DOI] [PubMed] [Google Scholar]
- 117).Ichinose M., Miki K., Tatematsu M., Furihata C., Matsushima M., Ichihara Y., Tanji M., Konishi T., Obara M., Inoue H., Kurokawa K., Takahashi T., Kageyama T., Takahashi K. (1990) Hydrocortisone-induced enhancement of expression and changes in methylation of pepsinogen genes in stomach mucosa of the developing rat. Biochem. Biophys. Res. Commun. 172, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 118).Ichinose M., Miki K., Wong R.N.S., Tatematsu M., Furihata C., Konishi T., Matsushima M., Tanji M., Sano S., Kurokawa K., Takahashi T., Kageyama T., Tang J.J.N., Takahashi K. (1991) Methylation and expression of human pepsinogen genes in normal tissues and their alteration in stomach cancer. Jpn. J. Cancer Res. 82, 686–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119).Ichinose M., Miki K., Furihata C., Kageyama T., Niwa H., Oka H., Oda T., Matsushima T., Takahashi K. (1982) Radioimmunoassay of group II pepsinogen in human serum. Clin. Chim. Acta 122, 61–69 [DOI] [PubMed] [Google Scholar]
- 120).Ichinose M., Miki K., Furihata C., Kageyama T., Hayashi R., Niwa H., Oka H., Matsushima T., Takahashi K. (1982) Radioimmunoassay of serum group I and group II pepsinogens in normal controls and patients with various disorders. Clin. Chim. Acta 126, 183–191 [DOI] [PubMed] [Google Scholar]
- 121).Huang S.C., Miki K., Hirano K., Hayashi Y., Furihata C., Shimizu A., Ichinose M., Oka H., Matsushima T., Takahashi K. (1987) Enzyme-linked immunosorbent assay of serum pepsinogen I. Clin. Chim. Acta 162, 85–96 [DOI] [PubMed] [Google Scholar]
- 122).Miki K., Ichinose M., Shimizu A., Huang S.C., Oka H., Furihata C., Matsushima T., Takahashi K. (1987) Serum pepsinogens as a screening test of extensive chronic gastritis. Gastroenterol. Jpn. 22, 133–141 [DOI] [PubMed] [Google Scholar]
- 123).Miki K., Ichinose M., Kawamura N., Matsushima M., Ahmad H.B., Kimura M., Sano J., Tashiro T., Kakei N., Oka H., Furihata C., Takahashi K. (1989) The significance of low serum pepsinogen levels to detect stomach cancer associated with extensive chronic gastritis in Japanese subjects. Jpn. J. Cancer Res. 80, 111–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124).Miki K., Ichinose M., Kakei N., Yahagi N., Matsushima M., Tsukada S., Ishihama S., Shimizu Y., Suzuki T., Kurokawa K., Takahashi K. (1994) The clinical application of the serum pepsinogen I and II levels as a mass screening method for gastric cancer. Adv. Exp. Med. Biol. 362, 139–143 [DOI] [PubMed] [Google Scholar]
- 125).Miki K. (2011) Gastric cancer screening by combined assay for serum anti-Helicobacter pylori JgG antibody and serum pepsinogen levels—“ABC method.” Proc. Jpn. Acad., Ser. B 87, 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126).Athauda S.B.P., Tanji M., Kageyama T., Takahashi K. (1989) A comparative study on the NH2-terminal amino acid sequences and some other properties of six isozymic forms of human pepsinogens and pepsins. J. Biochem. 106, 920–927 [DOI] [PubMed] [Google Scholar]
- 127).Athauda S.B.P., Nishigai M., Arakawa H., Ikai A., Ukai M., Takahashi K. (2003) Inhibition of human pepsin and gastricsin by α2-macroglobulin. J. Enzyme Inhib Med. Chem. 18, 219–224 [DOI] [PubMed] [Google Scholar]
- 128).Tanji M., Kageyama T., Takahashi K. (1988) Tuna pepsinogens and pepsins: purification, characterization and amino-terminal sequences. Eur. J. Biochem. 177, 251–259 [DOI] [PubMed] [Google Scholar]
- 129).Tanji M., Yakabe E., Kageyama T., Takahashi K. (1996) The primary structure of the major pepsinogen from the gastric mucosa of tuna stomach. J. Biochem. 120, 647–656 [DOI] [PubMed] [Google Scholar]
- 130).Takahashi K., Tanji M., Yakabe E., Hirasawa A., Athauda S.B.P., Kageyama T. (1994) Non-mammalian vertebrate pepsinogens and pepsins: isolation and characterization. Adv. Exp. Med. Biol. 362, 53–65 [DOI] [PubMed] [Google Scholar]
- 131).Hirasawa A., Athauda S.B.P., Takahashi K. (1996) Purification and characterization of turtle pepsinogen and pepsin A. J. Biochem. 120, 407–414 [DOI] [PubMed] [Google Scholar]
- 132).Tanji M., Yakabe E., Kageyama T., Takahashi K. (1996) The primary structure of the major pepsinogen from the gastric mucosa of tuna stomach. J. Biochem. 120, 647–656 [DOI] [PubMed] [Google Scholar]
- 133).Tanji M., Yakabe E., Kageyama K., Yokobori S., Ichinose M., Miki K., Ito H., Takahashi K. (2007) Purification and characterization of pepsinogens from the gastric mucosa of African coelacanth, Latimeria chalumnae, and properties of the major pepsin. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 146, 412–420 [DOI] [PubMed] [Google Scholar]
- 134).Moriyama A., Takahashi K. (1977) The structure and function of acid proteases. VIII. Purification and characterization of cathepsins D from Japanese monkey lung. J. Biochem. 83, 441–451 [DOI] [PubMed] [Google Scholar]
- 135).Tanji M., Kageyama T., Takahashi K. (1991) Occurrence of cathepsin D isozymes with different specificities in monkey skeletal muscle. Biochem. Biophys. Res. Commun. 176, 798–804 [DOI] [PubMed] [Google Scholar]
- 136).Tanji M., Kageyama T., Takahashi K. (1991) Cathepsin D from Japanese monkey skeletal muscle. An efficient and high-yield purification and some properties. Biomed. Res. 12, 339–346 [Google Scholar]
- 137).Athauda S.B.P., Takahashi K. (2002) Cleavage specificities of aspartic proteinases toward oxidized insulin B chain at different pH values. Protein Pept. Lett. 9, 289–294 [DOI] [PubMed] [Google Scholar]
- 138).Athauda S.B.P., Nishigai M., Arakawa H., Ikai A., Takahashi K. (2002) Limited proteolysis of α2-macroglobulin by cathepsin D and structural changes in the inhibitor. Biomed. Res. 23, 199–201 [Google Scholar]
- 139).Athauda S.B.P., Matsuzaki O., Kageyama T., Takahashi K. (1990) Structural evidence for two isozymic forms and the carbohydrate attachment site of human gastric cathepsin E. Biochem. Biophys. Res. Commun. 168, 878–885 [DOI] [PubMed] [Google Scholar]
- 140).Athauda S.B.P., Takahashi T., Kageyama T., Takahashi K. (1991) Autocatalytic processing of procathepsin E to cathepsin E and their structural differences. Biochem. Biophys. Res. Commun. 175, 152–158 [DOI] [PubMed] [Google Scholar]
- 141).Athauda S.B.P., Takahashi T., Inoue H., Ichinose M., Takahashi K. (1991) Proteolytic activity and cleavage specificity of cathepsin E at the physiological pH as examined towards the B chain of oxidized insulin. FEBS Lett. 292, 53–56 [DOI] [PubMed] [Google Scholar]
- 142).Kageyama T., Ichinose M., Tsukada S., Miki K., Kurokawa K., Koiwai O., Tanji M., Yakabe E., Athauda S.B.P., Takahashi K. (1992) Gastric procathepsin E and progastricsin from guinea pig. Purification, molecular cloning of cDNAs, and characterization of enzymatic properties, with special reference to procathepsin E. J. Biol. Chem. 267, 16450–16459 [PubMed] [Google Scholar]
- 143).Tsukada S., Ichinose M., Miki K., Tatematsu M., Yonezawa S., Matsushima M., Kakei K., Fukamachi H., Yasugi S., Kurokawa K., Kageyama K., Takahashi K. (1992) Tissue- and cell-specific control of guinea pig cathepsin E gene expression. Biochem. Biophys. Res. Commun. 187, 1401–1408 [DOI] [PubMed] [Google Scholar]
- 144).Athauda S.B.P., Arakawa H., Nishigai M., Takahashi T., Ikai A., Takahashi K. (1993) Inhibition of cathepsin E by α2-macroglobulin and the resulting structural changes in the inhibitor. J. Biochem. 113, 526–530 [DOI] [PubMed] [Google Scholar]
- 145).Athauda S.B.P., Kageyama T., Takahashi T., Inoue H., Ichinose M., Ukai M., Takahashi K. (1994) Isolation and characterization of human gastric procathepsin E and cathepsin E. Adv. Exp. Med. Biol. 362, 201–210 [DOI] [PubMed] [Google Scholar]
- 146).Kageyama T., Ichinose M., Miki K., Moriyama A., Yonezawa Y., Tanji M., Athauda S.B.P., Takahashi K. (1994) Isolation, characterization and structure of procathepsin E and cathepsin E from the gastric mucosa of guinea pig. Adv. Exp. Med. Biol. 362, 211–221 [DOI] [PubMed] [Google Scholar]
- 147).Tsukada S., Ichinose M., Miki K., Kakei K., Matsushima M., Yahagi N., Ishihama S., Shimizu Y., Kido M., Fukamachi H., Kurokawa K., Yonezawa S., Kageyama T., Takahashi K. (1994) Tissue- and cell-specific control of guinea pig cathepsin E gene expression. Adv. Exp. Med. Biol. 362, 349–355 [DOI] [PubMed] [Google Scholar]
- 148).Yonezawa S., Masaki S., Hanai A., Ono T., Sonta S., Hirai H., Ichinose M., Miki K., Takahashi K., Kageyama T. (1998) Cathepsin E gene in mouse. Biomed. Res. 19, 327–334 [Google Scholar]
- 149).Athauda S.B.P., Takahashi K. (2002) Distinct cleavage specificity of human cathepsin E at neutral pH with special preference for Arg-Arg bonds. Protein Pept. Lett. 9, 15–22 [DOI] [PubMed] [Google Scholar]
- 150).Athauda S.B.P., Nomura N., Inoue H., Takahashi K. (2003) Two distinct types of aspartic proteases from the filarial parasite Brugia malayi: molecular cloning and tissue distribution. Biomed. Res. 24, 269–276 [Google Scholar]
- 151).Athauda S.B.P., Ido E., Arakawa H., Nishigai M., Kyushiki H., Yoshinaka Y., Takahashi T., Ikai A., Tang J., Takahashi K. (1993) Entrapment and inhibition of human immunodeficiency virus proteinase by α2-macroglobulin and structural changes in the inhibitor. J. Biochem. 113, 742–746 [DOI] [PubMed] [Google Scholar]
- 152).Kogo H., Takeuchi K., Inoue H., Kihara H., Kojima M., Takahashi K. (2009) Urea-dependent unfolding of HIV-1 protease studied by circular dichroism and small-angle X-ray scattering. Biochim. Biophys. Acta 1794, 70–74 [DOI] [PubMed] [Google Scholar]
- 153).Athauda S.B.P., Yoshioka K., Shiba T., Takahashi K. (2006) Isolation and characterizaton of recombinant Drosophila copia aspartic proteinase. Biochem. J. 399, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154).Takahashi K. (1986) The amino acid sequence of rhizopuspepsin, an aspartic proteinase from Rhizopus chinensis. J. Biol. Chem. 262, 1468–1478 [PubMed] [Google Scholar]
- 155).Takahashi K. (1988) Determination of the amino acid sequences of the two major isozymes of rhizopuspepsin. J. Biochem. 103, 162–167 [DOI] [PubMed] [Google Scholar]
- 156).Lu J.-F., Inoue H., Kimura T., Makabe O., Takahashi K. (1995) Molecular cloning of a cDNA for Proctase B from Aspergillus niger var. macrospours and sequence comparison with other aspergillopepsins I. Biosci. Biotechnol. Biochem. 59, 954–955 [DOI] [PubMed] [Google Scholar]
- 157).Inoue H., Hayashi T., Huang X.-P., Lu J.-F., Athauda S.B.P., Kong K.-H., Yamagata H., Udaka S., Takahashi K. (1996) Heterologous expression and site-directed mutagenesis studies on the activation mechanism and the roles of the basic residues in the prosegment of aspergillopepsinogen I. Eur. J. Biochem. 237, 719–725 [DOI] [PubMed] [Google Scholar]
- 158).Athauda S.B.P., Inoue H., Iwamatsu A., Takahashi K. (1998) Acid proteinase from Nepenthes distillatoria (Badura). Adv. Exp. Med. Biol. 436, 453–458 [DOI] [PubMed] [Google Scholar]
- 159).Athauda S.B.P., Matsumoto K., Rajapakshe S., Kuribayashi M., Kojima M., Kubomura-Yoshida N., Iwamatsu A., Shibata C., Inoue H., Takahashi K. (2004) Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. Biochem. J. 381, 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160).Takahashi K., Athauda S.B.P., Matsumoto K., Rajapakshe S., Kuribayashi M., Kojima M., Kubomura-Yoshida N., Iwamatsu A., Shibata C., Inoue H. (2005) Nepenthesin, a unique member of a novel subfamily of aspartic proteinases: enzymatic and structural characteristics. Curr. Protein Pept. Sci. 6, 513–525 [DOI] [PubMed] [Google Scholar]
- 161).Takahashi, K. (2013) Nepenthesin. Handbook of Proteolytic Enzymes, 3rd ed. (eds. Rawlings, N.D. and Salvesen, G.). Vol. 1, Elsevier Academic Press, London, pp. 125–128. [Google Scholar]
- 162).Kubota K., Metoki Y., Athauda S.B.P., Shibata C., Takahashi K. (2010) Stability profiles of nepenthesin in urea and guanidine hydrochloride: comparison with porcine pepsin A. Biosci. Biotechnol. Biochem. 74, 2323–2326 [DOI] [PubMed] [Google Scholar]
- 163).Takahashi K., Matsumoto K., Nishii W., Muramatsu M., Kubota K., Shibata C., Athauda S.B.P. (2009) Comparative studies on the acid proteinase activities in the digestive fluids of Nepenthes, Cephalotus, Dionaea and Drosera. Carniv. Pl. Newslett. 38, 75–82 [Google Scholar]
- 164).Takahashi K., Nishii W., Shibata C. (2012) The digestive fluid of Drosera indica contains a cysteine endopeptidase (“droserain”) similar to dionain from Dionaea muscipula. Carniv. Pl. Newslett. 41, 132–134 [Google Scholar]
- 165).Takahashi K., Niwa H., Yokota N., Inoue H. (2008) Widespread tissue expression of nepenthesin-like aspartic protease genes in Arabidopsis thaliana. Plant Physiol. Biochem. 46, 724–729 [DOI] [PubMed] [Google Scholar]
- 166).Chang W.-J., Horiuchi S., Takahashi K., Yamasaki M., Yamada Y. (1976) The structure and function of acid proteases. VI. Effects of acid protease-specific inhibitors on the acid proteases from Aspergillus niger var. macrosporus. J. Biochem. 80, 975–981 [DOI] [PubMed] [Google Scholar]
- 167).Takahashi K., Tanokura M., Inoue H., Kojima M., Muto Y., Yamasaki M., Makabe O., Kimura T., Takizawa T., Hamaya T., Suzuki E., Miyano H. (1991) Structure and function of a pepstatin-insensitive acid proteinase from Aspergillus niger var. macrosporus. Adv. Exp. Med. Biol. 306, 203–211 [DOI] [PubMed] [Google Scholar]
- 168).Takahashi K. (1995) Proteinase A from Aspergillus niger. Methods Enzymol. 248, 146–155 [DOI] [PubMed] [Google Scholar]
- 169).Takahashi K. (1997) The specificity of peptide bond cleavage of acid proteinase from Aspergillus niger var. macrosporus toward oxidized ribonuclease A. Biosci. Biotechnol. Biochem. 61, 381–383 [DOI] [PubMed] [Google Scholar]
- 170).Takahashi K., Kagami N., Huang X.-P., Kojima M., Inoue H. (1998) Aspergillus niger acid proteinase A. Structure and function. Adv. Exp. Med. Biol. 436, 275–282 [DOI] [PubMed] [Google Scholar]
- 171).Takahashi, K. (2013) Aspergilloglutamic peptidase. Handbook of Proteolytic Enzymes, 3rd ed. (eds. Rawlings, N.D. and Salvesen, G.). Vol. 1, Elsevier Academic Press, London, pp. 307–310. [Google Scholar]
- 172).Fujinaga M., Cherney M.M., Oyama H., Oda K., James M.N.G. (2004) The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum. Proc. Natl. Acad. Sci. U.S.A. 101, 3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173).Pillai B., Cherney M.M., Hiraga K., Takada K., Oda K., James M.N.G. (2007) Crystal structure of scytalidoglutamic peptidase with its first potent inhibitor provides insights into substrate specificity and catalysis. J. Mol. Biol. 365, 343–361 [DOI] [PubMed] [Google Scholar]
- 174).Oda K. (2012) New families of carboxyl peptidases: serine-carboxyl peptidase and glutamic peptidases. J. Biochem. 151, 13–25 [DOI] [PubMed] [Google Scholar]
- 175).Takahashi K., Inoue H., Sakai K., Kohama T., Kitahara S., Takishima K., Tanji M., Athauda S.B.P., Takahashi T., Akanuma H., Mamiya B., Yamasaki M. (1991) The primary structure of Aspergillus niger acid proteinase A. J. Biol. Chem. 266, 19480–19483 [PubMed] [Google Scholar]
- 176).Inoue H., Kimura T., Makabe O., Takahashi K. (1991) The gene and deduced protein sequences of the zymogen of Aspergillus niger acid proteinase. J. Biol. Chem. 266, 19484–19489 [PubMed] [Google Scholar]
- 177).Kubota K., Nishii W., Kojima K., Takahashi K. (2005) Specific inhibition and stabilization of aspergilloglutamic peptidase by the propeptide: identification of critical sequences and residues in the propeptide. J. Biol. Chem. 280, 999–1006 [DOI] [PubMed] [Google Scholar]
- 178).Huang X.-P., Kagami N., Inoue H., Kojima M., Kimura T., Makabe O., Suzuki K., Takahashi K. (2000) Identification of a glutamic acid and an aspartic acid residue essential for catalytic activity of aspergillopepsin II, a non-pepsin type acid proteinase. J. Biol. Chem. 275, 26607–26614 [DOI] [PubMed] [Google Scholar]
- 179).Yabuki Y., Kubota K., Kojima K., Inoue H., Takahashi K. (2004) Identification of a glutamine residue essential for catalytic activity of aspergilloglutamic peptidase by site-directed mutagenesis. FEBS Lett. 569, 161–164 [DOI] [PubMed] [Google Scholar]
- 180).Matsuzaki H., Iwata S., Hamaya T., Takizawa T., Tanokura M., Takahashi K. (1991) Effect of dimethylsulfoxide on the crystallization of Apergillus niger proteinase A. Proc. Jpn. Acad., Ser. B 67, 209–212 [Google Scholar]
- 181).Tanokura M., Matsuzaki H., Iwata S., Nakagawa A., Hamaya T., Takizawa T., Takahashi K. (1992) Crystallization and preliminary X-ray investigation of proteinase A, a non-pepsin-type acid proteinase from Aspergillus niger var. macrosporus. J. Mol. Biol. 223, 373–375 [DOI] [PubMed] [Google Scholar]
- 182).Sasaki H., Nakagawa A., Muramatsu T., Suganuma M., Sawano Y., Kojima M., Kubota K., Takahashi K., Tanokura M. (2004) The three-dimensional structure of aspergilloglutamic peptidase from Aspergillus niger. Proc. Jpn. Acad., Ser. B 80, 435–438 [Google Scholar]
- 183).Kataoka Y., Takada K., Oyama H., Tsunemi M., James M.N., Oda K. (2005) Catalytic residues and substrate specificity of scytalidoglutamic peptidase, the first member of the eqolisin in family (G1) of peptidases. FEBS Lett. 579, 2991–2994 [DOI] [PubMed] [Google Scholar]
- 184).Schulz E.C., Ficner R. (2011) Knitting and snipping: chaperones in β-helix folding. Curr. Opin. Struct. Biol. 21, 232–239 [DOI] [PubMed] [Google Scholar]
- 185).Sasaki H., Kubota K., Lee W.C., Ohtsuka J., Kojima M., Iwata S., Nakagawa A., Takahashi K., Tanokura M. (2012) The crystal structure of an intermediate dimer of aspergilloglutamic peptidase that mimics the enzyme-activation product complex produced upon autoproteolysis. J. Biochem. 152, 45–52 [DOI] [PubMed] [Google Scholar]
- 186).Sasaki H., Kojima M., Sawano Y., Kubota K., Suganuma S., Muramatsu T., Takahashi K., Tanokura M. (2005) A proposed catalytic mechanism of aspergilloglutamic peptidase from Aspergillus niger. Proc. Jpn Acad., Ser. B 81, 441–446 [Google Scholar]
- 187).Huang X.-P., Yabuki Y., Kojima M., Inoue H., Takahashi K. (2007) Activation profile of the zymogen of aspergilloglutamic peptidase. Biol. Chem. 388, 129–133 [DOI] [PubMed] [Google Scholar]
- 188).Fukuda H., Takahashi K., Sorai M., Kojima M., Tanokura M., Takahashi K. (1995) Differential scanning calorimetric studies on the thermal unfolding of acid proteinase a from Aspergillus niger at various pHs. Thermochim. Acta 267, 373–378 [Google Scholar]
- 189).Kojima M., Tanokura M., Maeda M., Kimura K., Amemiya Y., Kihara H., Takahashi K. (1999) pH-Dependent unfolding of aspergillopepsin II studied by small-angle X-ray scattering. Biochemistry 39, 1364–1372 [DOI] [PubMed] [Google Scholar]
- 190).Maeda M., Takeuchi K., Kojima M., Tanokura M., Kimura K., Amemiya Y., Kihara H., Takahashi K. (2003) Kinetic studies of unfolding process of aspergillopepsin II by pH-jump methods. Biochem. Biophys. Res. Commun. 301, 745–750 [DOI] [PubMed] [Google Scholar]
- 191).Wlodawer A., Li M., Dauter Z., Gustchina A., Uchida K., Oyama H., Dunn B.M., Oda K. (2001) Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes. Nat. Struct. Biol. 8, 442–446 [DOI] [PubMed] [Google Scholar]
- 192).Wlodawer A., Li M., Gustchina A., Dauter Z., Uchida K., Oyama H., Goldfarb N.E., Dunn B.M., Oda K. (2001) Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase. Biochemistry 40, 15602–15611 [DOI] [PubMed] [Google Scholar]
- 193).Wlodawer A., Gustchina A., Oyama H., Dunn B.M., Oda K. (2003) Structual and enzymatic properties of the sedolisin family of serine-carboxyl peptidases. Acta Biochim. Pol. 50, 81–102 [PubMed] [Google Scholar]
- 194).Nishii, W., Murakami-Murofushi, K. and Takahashi, K. (2013) Physarolisin. Handbook of Proteolytic Enzymes, 3rd ed. (eds. Rawlings, N.D. and Salvesen, G.). Vol. 1, Elsevier Academic Press, London, pp. 3347–3349. [Google Scholar]
- 195).Murakami-Murofushi K., Takahashi T., Minowa Y., Iino S., Takeuchi T., Haruko Kitagaki-Ogawa H., Murofushi H., Takahashi K. (1990) Purification and characterization of a novel intracellular acid proteinase from the plasmodia of a true slime mold Physarum polycephalum. J. Biol. Chem. 265, 19898–19903 [PubMed] [Google Scholar]
- 196).Nishii W., Kubota K., Takahashi K. (2009) The P1 and P1′ residue specificities of physarolisin I, a serine-carboxyl peptidase from the true slime mold Physarum polycephalum. Biosci. Biotechnol. Biochem. 73, 1168–1171 [DOI] [PubMed] [Google Scholar]
- 197).Nishii W., Ueki T., Miyashita R., Kojima M., Kim Y.-T., Sasaki N., Murakami-Murofushi K., Takahashi K. (2003) Structural and enzymatic characterization of physarolisin (formerly physaropepsin) proves that it is a unique serine-carboxyl proteinase. Biochem. Biophys. Res. Commun. 301, 1023–1029 [DOI] [PubMed] [Google Scholar]
- 198).Takahashi K., Oda K. (2008) Structural evidence that scytalidolisin (formerly scytalidopepsin a) is a serine-carboxyl peptidase of the sedolisin family. Biosci. Biotechnol. Biochem. 72, 2239–2242 [DOI] [PubMed] [Google Scholar]
- 199).Benard M., Pallotta D., Pierron G. (1992) Structure and identity of a late-replicating and transcriptionally active gene. Exp. Cell Res. 201, 506–513 [DOI] [PubMed] [Google Scholar]
- 200).Nishii W., Kuriyama H., Takahashi K. (2003) The Physarum polycephalum php gene encodes a unique cold-adapted serine-carboxyl protease, physarolisin II. FEBS Lett. 546, 340–344 [DOI] [PubMed] [Google Scholar]
- 201).Jogl G., Rozovsky S., McDermott A.E., Tong L. (2003) Optimal alignment for enzymatic proton transfer: structure of the Michaelis complex of triosephosphate isomerase at 1.2-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 100, 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202).Oliva G., Fontes M.R., Garratt R.C., Altamirano M.M., Calcagno M.L., Horjales E. (1995) Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1Å resolution. Structure 3, 1323–1332 [DOI] [PubMed] [Google Scholar]
- 203).Hellinga H.W., Evans P.R. (1987) Mutations in the active site of Escherichia coli phosphofructokinase. Nature 327, 437–439 [DOI] [PubMed] [Google Scholar]
- 204).Henrissat B., Davies G. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 205).Vocadlo D.J., Davies G.J., Laine R., Withers S.G. (2001) Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature 412, 835–838 [DOI] [PubMed] [Google Scholar]