Abstract
Somatic mosaicism caused by in vivo reversion of inherited mutations has been described in several human genetic disorders. Back mutations resulting in restoration of wild-type sequences and second-site mutations leading to compensatory changes have been shown in mosaic individuals. In most cases, however, the precise genetic mechanisms underlying the reversion events have remained unclear, except for the few instances where crossing over or gene conversion have been demonstrated. Here, we report a patient affected with Wiskott–Aldrich syndrome (WAS) caused by a 6-bp insertion (ACGAGG) in the WAS protein gene, which abrogates protein expression. Somatic mosaicism was documented in this patient whose majority of T lymphocytes expressed nearly normal levels of WAS protein. These lymphocytes were found to lack the deleterious mutation and showed a selective growth advantage in vivo. Analysis of the sequence surrounding the mutation site showed that the 6-bp insertion followed a tandem repeat of the same six nucleotides. These findings strongly suggest that DNA polymerase slippage was the cause of the original germ-line insertion mutation in this family and that the same mechanism was responsible for its deletion in one of the propositus T cell progenitors, thus leading to reversion mosaicism.
The Wiskott–Aldrich syndrome (WAS) is an X-linked disorder characterized by thrombocytopenia with small-sized platelets, eczema, and immune deficiency, leading to increased susceptibility to infection from all classes of pathogens (1, 2). Sepsis and severe intracerebral hemorrhage represent frequent causes of death in WAS boys, who also are subjected to malignancies with increased frequency. The disease gene was discovered by positional cloning (3) and encodes the WAS protein (WASP), a 502-aa proline-rich protein, which is constitutively expressed in cytoplasm of all nonerythroid hematopoietic cells (4, 5). Although the definitive function of WASP remains to be determined, this protein has been demonstrated to be involved in signal transduction pathways of important cellular growth factors (e.g., Nck, Fyn, Itk, and Grb2) and in cytoskeleton reorganization events of hematopoietic cells, possibly in response to cellular activation (6).
It has been recently recognized that somatic reversion of inherited mutations occurs in human genetic disorders. Back mutations resulting in restoration of wild-type sequences (7–13) and second-site mutations leading to compensatory changes (14, 15) have been shown in mosaic individuals. If a reverse mutation occurs in a somatic cell during mitosis and the cell with the reverse mutation has a selective growth advantage in vivo, cells carrying the original mutation are expected to be replaced by revertants that have regained wild-type phenotype. This rare event may modify the disease clinical phenotype, leading to atypical and unexpectedly mild presentations. In fact, spontaneous in vivo reversion has been reported in one patient with adenosine deaminase deficiency (7) and one patient with X-linked severe combined immunodeficiency (X-SCID) (8), both of whom showed a progressively mild clinical course because of the selective growth advantage of the revertant lymphocytes.
The molecular mechanism leading to the reversion events has remained unknown in most cases, except for the few cases where crossing over or gene conversion has been shown in compound heterozygous patients (11, 12). DNA polymerase slippage is the most commonly invoked mechanism to explain triplet repeat expansion in human diseases (e.g., Huntington's disease, fragile X syndrome, and Friedreich ataxia) (16). It is well accepted that slippage-type events also can cause small insertion or deletion of tandem repeats (17), and it is therefore possible that in the appropriate genomic context, this mechanism could lead to reversion of a mutation to wild-type sequence (13). In this report, we describe a 43-year-old WAS patient carrying a spontaneous in vivo reversion likely caused by a DNA slippage mechanism. The mutation responsible for the disease in this patient's family is a 6-bp insertion after a tandem microrepeat of the same six nucleotides. In contrast to those of other affected family members, the majority of the proband's T lymphocytes were demonstrated to express WASP and lack the deleterious mutation. In addition, we show evidence of selective advantage of the WASP-expressing (WASP+) T lymphocytes over the WASP-negative (WASP−) ones, which explains the accumulation of the former cells. Finally, our patient has shown clinical improvement over the years, which suggests the revertant T cells having contributed to the modification of his previously severe clinical phenotype.
Materials and Methods
Case Presentation.
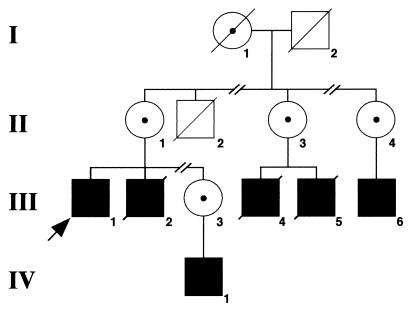
Fig. 1 shows a pedigree of the proband's family whose history began at the age of 10 months with encephalitis. Between the ages of 2 and 5 years, he had recurrent easy bruising, eczema, and recurrent otitis media. At age 5, it was noted that his younger brother had petechiae and thrombocytopenia. The patient's platelet count was then tested and found to be in the range of 13,000 to 20,000/mm3. A clinical diagnosis of WAS was made, and the patient underwent an elective splenectomy, leading to correction of platelet numbers and size. Shortly after splenectomy, the patient suffered from pneumococcal meningitis, from which he recovered. Frequent upper respiratory and/or ear infections and continued eczema are described until the age of 12, when the patient was hospitalized for vasculitic rash, thrombocytopenia, and an illness resembling rheumatoid arthritis with concurrent dysgammaglobulinemia and nephritis. The same year, he developed pneumococcal meningitis and sepsis, which were successfully treated. One month later, another episode of pneumococcal meningitis occurred. At age 16, the patient developed a right mastoiditis. This clinical history is consistent with severe WAS phenotype (score of 5) (18). Since his 20s, the patient has been relatively well, with complaints of sinusitis episodes responding to antibiotic treatment. The patient is now 43 years old and has been free of serious illnesses for the past 20 years.
Figure 1.
Simplified pedigree of the proband's family. Solid squares represent affected individuals; diagonal lines indicate deceased subjects. Carrier status of female subjects is indicated by a dot.
A maternal uncle (II-2) developed petechiae early after birth and died at 6 months of age from unspecified causes. The proband's brother (III-2) had severe WAS phenotype including thrombocytopenia, infections (pneumococcal meningitis, Pneumocystis carinii pneumonia), arthritis, and vasculitis and died of renal failure at the age of 33 years. Two cousins (III-4 and III-5) also had severe WAS symptoms and died from pulmonary hemorrhage at 2.5 years of age and from lymphoma at the age of 18 years, respectively. A third affected cousin (III-6, age 15 years) has a history of thrombocytopenia, eczema, and molluscum contagiosum, and a nephew (IV-1, age 9 months) has thrombocytopenia and eczema.
Cell Preparations.
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll–Hypaque (Mediatech, Washington, DC) gradient centrifugation from the proband, his family members, and normal controls. Granulocytes were recovered from the pellet of the gradient after lysis of erythrocytes. To obtain activated T lymphocytes, PBMC were cultured in the presence of 100 ng/ml anti-CD3 (OKT3; Ortho Diagnostics), 5 μg/ml anti-CD28 mAb (PharMingen), and 100 units/ml recombinant human IL-2 (gift of C. Reynolds, National Cancer Institute) for 3 days in RPMI 1640 medium (Life Technologies, Grand Island, NY) containing 10% FBS and antibiotics (R-10). Thereafter, cells were maintained in R-10 medium supplemented with 100 units/ml recombinant IL-2 for 9 days.
Western Blot Analysis of WASP.
PBMC were lysed in buffer containing 300 mM NaCl/50 mM Tris⋅HCl/2 mM EDTA/0.5% Triton-X/2.5 μM p-nitrophenyl p′-guanidino-benzoate (both from Sigma), 10 μg/ml aprotinin (ICN), and 10 μg/ml leupeptin (Calbiochem). Protein concentration was determined by using the BCA protein assay (Pierce). Twenty micrograms of protein was boiled, subjected to 8% SDS/PAGE, and electrotransferred onto nylon membranes (Immobilon-P, Millipore). The detection of WASP was performed as described (19).
Flow Cytometric Analysis of WASP.
Three-color immunofluorescence analysis was used to study cell-surface antigens and intracellular WASP expression simultaneously. Detection of WASP was performed as described with minor modifications (20). Briefly, PBMC were stained for cell-surface antigens before cell membrane permeabilization using the following mAbs: Cy-chrome-conjugated anti-CD3 (PharMingen); TRI-color labeled anti-CD8 and anti-CD56 (Caltag, South San Francisco, CA); FITC-conjugated anti-CD4 (Caltag); and FITC-conjugated anti-CD45RA, anti-CD45R0, and anti-CD20 (PharMingen). After washing, cells were treated with Cytofix/Cytoperm solution from CytoStain kit (PharMingen) at 4°C for 20 min. After washing twice, they were incubated with 1:200 diluted anti-WASP (3F3-A5) mAb (5) or purified mouse IgG1 (PharMingen) at 4°C for 20 min. Cells were then reacted with biotin-conjugated anti-mouse IgG1 at 4°C for 20 min, followed by further incubation with phycoerythrin-conjugated streptavidin (both from PharMingen). Stained cells were analyzed with a FACScan [fluorescence-activated cell sorter (FACS)] flow cytometer using CELLQUEST software (Becton Dickinson). WASP expression of monocytes was evaluated in CD3-negative cells among monocytic cells determined by a distribution pattern in the forward and side scatter. Because anti-WASP mAb (3F3-A5) belongs to the mouse IgG1 subclass, all antibodies for surface staining were chosen from mouse IgG2 subclass to avoid crossreaction.
Mutation Analysis.
DNA was extracted from blood samples with the QIAamp DNA blood mini kit and from a skin biopsy specimen with the DNeasy tissue kit (both from Qiagen, Valencia, CA). Each exon of WASP gene was amplified from genomic DNA using specific primers described (21). Direct sequencing was performed by using the Applied Biosystems Prism BigDye terminator cycle sequencing kit on an Applied Biosystems 310 automated sequencer (Perkin–Elmer).
Complementarity-Determining Region 3 (CDR3) Size Distribution Analysis.
Total RNA was extracted from PBMC with Trizol reagent (Life Technologies), and first-strand cDNA was generated from 2 μg total RNA with oligo(dT) using the RETROscript kit (Ambion, Austin, TX). CDR3 size distribution assay was performed as described with minor modifications (22). Briefly, each T cell receptor (TCR) Vβ fragment was amplified with one of the 24 Vβ-specific primers and a 6-fluorescein phosphoramidite (6-FAM)-labeled Cβ primer recognizing both Cβ1 and Cβ2. Thirty-five cycles of amplification (94°C for 1 min, 55°C for 1 min, and 72°C for 1 min) were used. One microliter of 1:10 diluted PCR products was mixed with 12 μl of deionized formamide and 0.5 μl of size standard and heated at 95°C for 3 min. The size distribution of each fluorescent PCR product was determined by electrophoresis in the Performance Optimized Polymer 4 on the Applied Biosystems 310 automated sequencer, and data were analyzed by genescan software (Perkin–Elmer). The TCR Vβ 10 and Vβ 19 pseudogenes were excluded from the analysis.
TCR/CD3 Down-Regulation Assay.
PBMC (5 × 105) were incubated with anti-CD3 mAb (Leu4; Becton Dickinson) for 30 min on ice. After washing, cells were incubated with biotinylated goat anti-mouse Ab (5 μg/ml, Biosource International, Camarillo, CA) for 30 min on ice and warmed at 37°C for 60 min to allow down-regulation of CD3. Cells were then transferred back to ice, washed, and stained with phycoerythrin-conjugated streptavidin and FITC-conjugated anti-CD2 mAb (Becton Dickinson). After fixation in 2% paraformaldehyde, CD2+ T cells were gated, and CD3 expression was analyzed by FACS.
Cell Proliferation Assay.
PBMC from the proband, a typical WAS patient, and a healthy control were seeded at 1 × 105 cells per well in flat-bottom 96-well plates in a final volume of 200 μl of R-10 medium and cultured for 72 h with 10 ng/ml OKT3 alone or together with 5 μg/ml anti-CD28 mAb. Cells were pulsed with [3H]thymidine (1 μCi per well) for the final 6 h and harvested onto glass fiber filters. Thymidine incorporation into cellular DNA was evaluated by using a scintillation counter (Betaplate; Wallac, Gaithersburg, MD).
Results
Mosaicism of WASP Expression in T Cells.
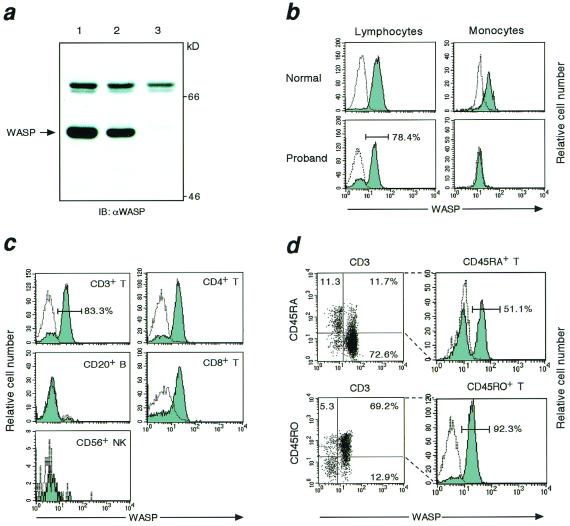
We first analyzed WASP expression of PBMC by Western blot technique. As shown in Fig. 2a, normal PBMC showed the presence of a ≈59-kDa WASP band, which was not detectable in PBMC from affected individual III-6. In contrast, the proband's PBMC expressed significant levels of WASP. To determine what populations of PBMC were responsible for the expression, flow cytometric analysis of WASP was performed. When whole PBMC including lymphocytes and monocytes were gated, WASP was detected in about 60% of cells (data not shown). Although the proband exhibited negligible expression of WASP in monocytes, flow cytometric analysis clearly demonstrated mosaic pattern of WASP expression in lymphocytes, indicating that they consisted of WASP+ and WASP− cells (Fig. 2b). As shown in Fig. 2c, WASP+ cells were found to account for the majority of CD3+ T cells but were absent among CD20+ B cells and CD56+ NK cells. The CD4/CD8 ratio in the proband was 0.98, and both CD4+ and CD8+ T cells were involved equally in the mosaicism for WASP expression. In contrast, a markedly different pattern of WASP expression was found among CD45RA+ “naïve” and CD45R0+ “memory” T cells. The majority of memory T lymphocytes expressed WASP, whereas WASP+ cells were detected only in about half of the naïve T cells (Fig. 2d).
Figure 2.
Analysis of WASP expression. (a) Western blot analysis of WASP was performed using lysates of PBMC obtained from normal (lane 1), the proband (lane 2), and affected individual III-6 (lane 3). The arrow indicates the position of the WASP band. (b–d) Flow cytometric analysis of WASP. PBMC were stained with mAbs for cell-surface antigens, fixed, and permeabilized. Cells were then reacted with anti-WASP mAb (solid histogram) or a control Ab (open histogram) and further incubated with biotin-conjugated anti-mouse IgG1 followed by streptavidin-phycoerythrin. WASP expression in lymphocytes and monocytes obtained from normal control and the proband is presented (b). WASP expression in CD3+ T, CD20+ B, CD56+ NK, CD4+ T, and CD8+ T cell subsets from the proband is shown (c). The dot plot of a two-color immunofluorescence profile of CD45 isoforms and WASP expression in the proband's CD3+ T cells is presented (d, Left). A second gate is set for the upper quadrants to include CD45RA+CD3+ or CD45R0+CD3+ cells, and WASP expression in each cell population is shown (d, Right).
Absence of Mutation in WASP+ T Cells.
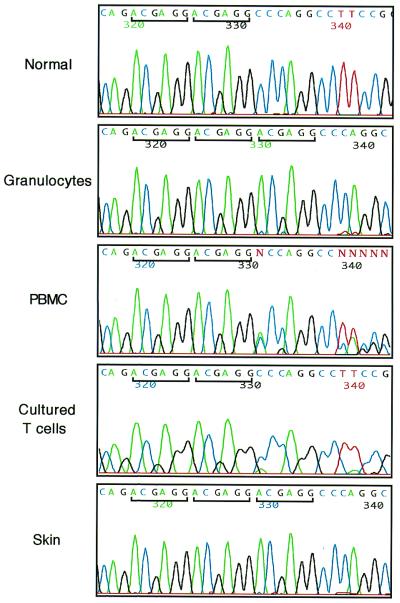
The mutation causing WAS in the proband's family was previously described as a 6-bp ACGAGG insertion at nucleotide 434 in exon 4 of the WASP gene (23). A review of the WASP gene sequence surrounding the mutation showed that the insertion follows a tandem microrepeat of the same ACGAGG sequence. To determine whether the same mutation was present in WASP+ T cells, direct genomic sequence analysis was performed with various samples isolated from the proband. We found the 6-bp insertion in DNA from the proband's granulocytes, whereas DNA from his PBMC showed coexistence of wild-type and mutated sequences. Importantly, the mutation was not detectable in DNA obtained from lymphocytes cultured in the presence of anti-CD3, anti-CD28, and IL-2 (Fig. 3). By flow cytometry analysis, more than 90% of these activated T cells expressed WASP, and 99% of them were CD45R0+ (data not shown). We also analyzed DNA from a proband's skin biopsy specimen and found the presence of the 6-bp insertion mutation (Fig. 3). No other mutations in the WASP gene were found in any of the samples studied. In addition, mutation analysis demonstrated the presence of the insertion in PBMC from affected individuals III-6 and IV-1 and showed that female individuals II-1, II-3, II-4, and III-3 were heterozygous carriers (data not shown). Analysis of 100 normal chromosomes showed that the ACGAGG tandem microrepeat is highly conserved in the general populations and was never followed by a third ACGAGG repeat. To rule out the possibility that the proband's T cell population expressing WASP was derived from the engraftment of T cells from his mother or others, we performed a standard molecular study of HLA typing at the A, B, Cw, DR, and DQ loci of granulocytes (WASP−) and cultured T cells (WASP+) and found that they showed identical HLA type (data not shown).
Figure 3.
Mutational analysis of WASP gene. The sequence of exon 4 of the WASP gene was amplified from DNA extracted from normal PBMC and the proband's granulocytes, PBMC, cultured T cells, and skin. Direct sequencing was performed using an automated sequencer. A bar highlights the 6-bp repeat sequence.
Skewed CDR3 Size Pattern in T Cells.
To investigate the TCR Vβ diversity of the proband's T lymphocytes, we performed CDR3 size distribution analysis. Consistent with previous reports (22), the majority of Vβ subfamilies (≈93%) exhibited a Gaussian curve with 6–9 peaks in normal controls, reflecting polyclonal Vβ repertoire (Fig. 4a). All 24 different Vβ segments were amplified from proband's cDNA, and the general pattern of TCR Vβ usage based on gel images did not significantly differ from that of normal controls (data not shown). However, most of the Vβ subfamilies amplified from the proband's cells exhibited a skewed CDR3 size pattern; Vβ subfamilies 5.1, 15, and 21 showed a monoclonal spike, and subfamilies 1, 2, 3, 5.2, 6, 7, 8, 11, 12, 13.2, 16, 17, 22, 23, and 24 demonstrated an oligoclonal pattern (Fig. 4b). These results are consistent with a highly restricted TCR repertoire in the proband.
Figure 4.
CDR3 size distribution of TCR Vβ. Each TCR Vβ fragment was amplified from cDNA with one of the 24 Vβ-specific primers. The size distribution of PCR products was determined by an automated sequencer and GENESCAN software. (a) Normal control; (b) the proband.
Restoration of T Cell Function.
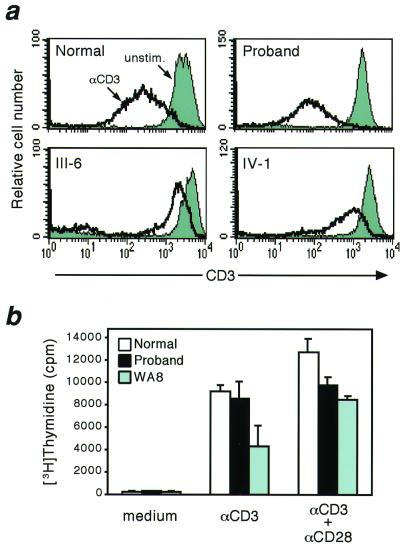
T lymphocytes from WAS patients and WASP knockout mice show abnormal proliferation in response to anti-CD3 stimulation (24, 25). In addition, down-regulation of TCR/CD3 expression after engagement appears to require reorganization of actin cytoskeleton and is defective in lymphocytes from WASP knockout mice (25). To evaluate the functionality of the proband's T lymphocytes, we therefore tested for their responses to anti-CD3 in terms of down-regulation of surface CD3 expression and cell proliferation. As shown in Fig. 5a, the level of CD3 expression on CD2+ T lymphocytes was significantly decreased 1 h after CD3 crosslinking in normal controls, whereas it was severely impaired in T cells from individuals III-6 and IV-1. In contrast, T lymphocytes from the proband showed normal CD3 down-regulation. In addition and in contrast to what was observed in cells from a typical WAS patient (WA8), the proband's PBMC were consistently able to respond to anti-CD3 at levels comparable to normal. The combined stimulation with anti-CD3 and anti-CD28 further improved, but did not fully normalize, the proliferative responses of proband's PBMC that were again significantly higher than in the typical WAS patient (Fig. 5b). Taken together, these results indicate that T lymphocytes present in the proband's peripheral blood (80% of which express WAS and are presumably revertants) are functionally competent.
Figure 5.
(a) Analysis of TCR/CD3 down-regulation. PBMC from normal control, the proband, and affected individuals III-6 and IV-1 were incubated with anti-CD3 mAb, washed, and further reacted with biotinylated goat anti-mouse Ab at 37°C for 60 min. Cells were then washed and stained with streptavidin and anti-CD2 mAb. After fixation, CD3 expression on CD2+ T cells was analyzed by FACS. Solid and open histogram represent CD3 expression in cells unstimulated or treated for 60 min, respectively. (b) Cell proliferation assay. PBMC from normal control, the proband, and WA8, a patient with typical WAS and with a clinical score of 5, were cultured for 72 h with anti-CD3 mAb alone or together with anti-CD28 mAb. DNA incorporation of [3H]thymidine was evaluated. Each bar represents the mean ± SD of triplicate cultures.
Discussion
Revertant mosaicism has been described in a variety of genetic disorders, including tyrosinemia type 1 (10), epidermolysis bullosa (11, 14), Fanconi anemia (12, 13, 15), and primary immunodeficiency syndromes, such as adenosine deaminase deficiency, X-SCID, and WAS (7–9). We report on a 43-year-old WAS patient with somatic mosaicism caused by the reversion of a 6-bp insertion to the normal WASP coding sequence. Such a reversion was observed only in DNA from the patient's T lymphocytes and leads to reconstitution of WASP expression that is detectable in the majority of his circulating T cells. Sequencing analysis of the WASP gene in the general population excluded the possibility that the 6-bp insertion could be a functional polymorphism. Molecular HLA typing demonstrated that the T lymphocytes carrying the normal WASP sequence and granulocytes obtained from the proband were identical, thus ruling out the possibility that normal allogeneic T cells were acquired in utero from the maternal circulation or postnatally from blood products.
The demonstration of the same 6-bp insertion mutation in other affected members of the family excludes the possibility that the mosaicism observed in the proband was derived from a postzygotic somatic mutation of the normal maternal X chromosome at the level of a lymphoid progenitor. A spontaneous in vivo reversion event, therefore, seems to be the most likely explanation for the appearance of genetically normal T lymphocytes in this individual who was found to have a normal XY male karyotype, thus not allowing for gene conversion or crossing over mechanisms to be responsible for the correction of the mutation at the WASP locus (Xp11.23). Reversions of single point mutations to the original wild-type sequence have been reported for adenosine deaminase deficiency, X-SCID, and WAS (7–9); however, the mechanism underlying these back mutations remains undefined. Our findings differ from previous reports on two counts. First, the correction of the mutation in the present case is sustained by the precise deletion of a 6-bp fragment, an occurrence previously unreported. Second, we suggest that both original and reverse mutations may have been caused by DNA polymerase slippage, a mechanism that has been invoked to explain insertions and deletion of DNA repeats both in germ-line and somatic cells (16, 17, 26). The presence of a high GC content 6-bp tandem repeat (ACGAGG) may have induced slippage and mispairing of the nascent and template strand during DNA replication. The occurrence of a misalignment of the 3′ end of the nascent DNA strand backward in the germline of a family ancestor would explain the 6-bp insertion, whereas a forward slippage event in a somatic cell of the proband would account for the reversion mosaicism and make this case an example of a back mutation as a result of DNA polymerase slippage.
WASP+ cells were only detectable by FACS among CD3+ T lymphocytes and not among granulocytes, monocytes, or B or NK cells. CD4+ and CD8+ T cells contributed equally to the mosaicism, and multiple TCR Vβ families were expressed by the patient's T lymphocytes that contained more than 80% WASP+ cells. Taken together, these findings suggest that the reversion of the mutation occurred in a T cell progenitor at a stage before CD4/CD8 lineage commitment and TCRβ rearrangement. Alternatively, the reversion may have occurred in a more primitive hematopoietic progenitor, and the presence of normal numbers of normal-size platelets in the proband's peripheral blood could be interpreted as the result of a reversion event at the level of a progenitor common to myeloid and lymphoid lineages (pluripotent stem cell). However, the proband's monocytes are WASP− by FACS analysis (Fig. 2b), and his granulocytes carry the WASP mutation (Fig. 3), thus showing the absence of detectable revertants among these myeloid cell types. The somatic mosaicism could be evident only among T lymphocytes because revertant cells had selective advantage among these and not other cell lineages (e.g., myeloid, B, or NK cells) differentiating from the corrected stem cell. X-chromosome inactivation studies performed in WAS carrier females, however, have demonstrated skewed inactivation in all hematopoietic lineages, as well as in granulocyte-macrophage colony-forming units and CD34+ progenitors (27–30). These findings indicate that revertant cells should have selective advantage within all lineages and suggest that a committed T cell progenitor is the most likely element where the reversion originated. For these reasons, although we have not tested the proband's platelets for WASP expression, we believe that their presence in normal numbers and size is the likely consequence of the splenectomy, rather than the reversion of the WASP mutation at the level of the pluripotent stem cell.
There is clear evidence of in vivo selective advantage of T lymphocytes able to express WASP in the proband. Because of the rarity of reverse mutation events, we assume that all T lymphocytes present in the proband's circulation and expressing WASP derived from a single revertant progenitor. Our flow cytometry studies showed that about half of CD45RA+ “naïve” circulating T cells stained positive for WASP expression, thus suggesting that, although T cell progenitors carrying the original mutation continued to differentiate into T lymphocytes unable to express WASP, at the time of analysis, half of the “naïve” T lymphocytes present in the proband's peripheral blood derived a single revertant cell. Even more significant was the finding that more than 90% of CD45R0+ “memory” T cells were WASP+. Altogether, these observations indicate that WASP+ cells had selective growth advantage during thymic development and the establishment of immunological memory. Although the precise mechanism underlying the selective advantage of WASP+ cells over WASP− counterparts is not clear, it is known that T cells from WAS patients have defective proliferative responses (24) and higher susceptibility to cell death by spontaneous apoptosis as compared with normal T lymphocytes (31, 32), and it is therefore conceivable that they are outperformed by WASP+ revertant T cells in the various steps of development, differentiation, and immune surveillance.
The high percentage of WASP+ cells (≈80%) among the circulating lymphocytes offered us the opportunity to analyze the functional effects of the revertant mosaicism. Although all TCR Vβ families were expressed in the proband's T cells, we observed a reduced number of CDR3 fragments, thus indicating that his TCR repertoire had limited complexity. Similar results were obtained from the study of the TCR repertoire of an unusual X-SCID patient carrying a back mutation in the γc gene (33). In vitro functional studies of our patient's lymphocytes, however, showed that TCR/CD3 down-regulation and cell proliferation in response to anti-CD3 antibody were largely normal, demonstrating that the genetic reversion led to restoration of biological defects characteristic of WAS T lymphocytes. Because the clinical phenotype of our patient changed dramatically from a full-blown classical WAS to a much milder immunodeficiency condition starting from his mid-20s, we speculate that the accumulation of WASP+, normally functioning T lymphocytes may have started around that point in time. Spontaneous reversion to normal of inherited mutations in adenosine deaminase deficiency and X-SCID has resulted in mild clinical courses (7, 8), and it seems therefore reasonable that the progressive clinical improvement of our patient is the consequence of his revertant mosaicism. These findings have important implications for gene therapy approaches for WAS, in that they indicate that even the correction of a small number of hematopoietic stem cells with the capability of differentiating into gene-corrected T cell progenitors and mature T lymphocytes may lead to significant clinical benefit. In addition, the fact that the isolated presence of WASP+ T lymphocytes is associated with mild clinical presentation in this patient supports also the feasibility of T cell-directed gene transfer as a potential form of therapy for this disease.
Acknowledgments
We thank Drs. L. Peltonen and D. A. Tagle for insightful discussions, Dr. L. D. Notarangelo for critical review of the manuscript, Dr. Jeanne Meck for karyotype analysis, and Mrs. Jacqueline T. Keller for secretarial assistance. This work was supported in part by Japan Society for the Promotion of Science Research Fellowships for Japanese Biomedical and Behavioral Researchers at the National Institutes of Health (to T.W.) and by National Institutes of Health Grant HD17427 (to H.D.O.).
Abbreviations
- WAS
Wiskott–Aldrich syndrome
- WASP
WAS protein
- X-SCID
X-linked severe combined immunodeficiency
- PBMC
peripheral blood mononuclear cells
- TCR
T cell receptor
- CDR3
complementarity-determining region 3
- FACS
fluorescence-activated cell sorter
References
- 1.Sullivan K E, Mullen C A, Blaese R M, Winkelstein J A. J Pediatr. 1994;125:876–885. doi: 10.1016/s0022-3476(05)82002-5. [DOI] [PubMed] [Google Scholar]
- 2.Ochs H D. Springer Semin Immunopathol. 1998;19:435–458. doi: 10.1007/BF00792601. [DOI] [PubMed] [Google Scholar]
- 3.Derry J M, Ochs H D, Francke U. Cell. 1994;78:635–644. [PubMed] [Google Scholar]
- 4.Parolini O, Berardelli S, Riedl E, Bello-Fernandez C, Strobl H, Majdic O, Knapp W. Blood. 1997;90:70–75. [PubMed] [Google Scholar]
- 5.Stewart D M, Treiber-Held S, Kurman C C, Facchetti F, Notarangelo L D, Nelson D L. J Clin Invest. 1996;97:2627–2634. doi: 10.1172/JCI118712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snapper S B, Rosen F S. Annu Rev Immunol. 1999;17:905–929. doi: 10.1146/annurev.immunol.17.1.905. [DOI] [PubMed] [Google Scholar]
- 7.Hirschhorn R, Yang D R, Puck J M, Huie M L, Jiang C K, Kurlandsky L E. Nat Genet. 1996;13:290–295. doi: 10.1038/ng0796-290. [DOI] [PubMed] [Google Scholar]
- 8.Stephan V, Wahn V, Le Deist F, Dirksen U, Broker B, Muller-Fleckenstein I, Horneff G, Schroten H, Fischer A, de Saint Basile G. N Engl J Med. 1996;335:1563–1567. doi: 10.1056/NEJM199611213352104. [DOI] [PubMed] [Google Scholar]
- 9.Ariga T, Kondoh T, Yamaguchi K, Yamada M, Sasaki S, Nelson D L, Ikeda H, Kobayashi K, Moriuchi H, Sakiyama Y. J Immunol. 2001;166:5245–5249. doi: 10.4049/jimmunol.166.8.5245. [DOI] [PubMed] [Google Scholar]
- 10.Kvittingen E A, Rootwelt H, Berger R, Brandtzaeg P. J Clin Invest. 1994;94:1657–1661. doi: 10.1172/JCI117509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonkman M F, Scheffer H, Stulp R, Pas H H, Nijenhuis M, Heeres K, Owaribe K, Pulkkinen L, Uitto J. Cell. 1997;88:543–551. doi: 10.1016/s0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- 12.Lo Ten Foe J R, Kwee M L, Rooimans M A, Oostra A B, Veerman A J, van Weel M, Pauli R M, Shahidi N T, Dokal I, Roberts I, et al. Eur J Hum Genet. 1997;5:137–148. [PubMed] [Google Scholar]
- 13.Gregory J J, Jr, Wagner J E, Verlander P C, Levran O, Batish S D, Eide C R, Steffenhagen A, Hirsch B, Auerbach A D. Proc Natl Acad Sci USA. 2001;98:2532–2537. doi: 10.1073/pnas.051609898. . (First Published February 13, 2001; 10.1073/pnas.051609898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darling T N, Yee C, Bauer J W, Hintner H, Yancey K B. J Clin Invest. 1999;103:1371–1377. doi: 10.1172/JCI4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waisfisz Q, Morgan N V, Savino M, de Winter J P, van Berkel C G, Hoatlin M E, Ianzano L, Gibson R A, Arwert F, Savoia A, et al. Nat Genet. 1999;22:379–383. doi: 10.1038/11956. [DOI] [PubMed] [Google Scholar]
- 16.Usdin K, Grabczyk E. Cell Mol Life Sci. 2000;57:914–931. doi: 10.1007/PL00000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levinson G, Gutman G A. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Q, Zhang M, Blaese R M, Derry J M, Junker A, Francke U, Chen S H, Ochs H D. Blood. 1995;86:3797–3804. [PubMed] [Google Scholar]
- 19.Candotti F, Facchetti F, Blanzuoli L, Stewart D M, Nelson D L, Blaese R M. Gene Ther. 1999;6:1170–1174. doi: 10.1038/sj.gt.3300926. [DOI] [PubMed] [Google Scholar]
- 20.Yamada M, Ariga T, Kawamura N, Yamaguchi K, Ohtsu M, Nelson D L, Kondoh T, Kobayashi I, Okano M, Kobayashi K, Sakiyama Y. J Immunol. 2000;165:1119–1122. doi: 10.4049/jimmunol.165.2.1119. [DOI] [PubMed] [Google Scholar]
- 21.Ariga T, Yamada M, Sakiyama Y. Pediatr Res. 1997;41:535–540. doi: 10.1203/00006450-199704000-00013. [DOI] [PubMed] [Google Scholar]
- 22.Zeng W, Nakao S, Takamatsu H, Yachie A, Takami A, Kondo Y, Sugimori N, Yamazaki H, Miura Y, Shiobara S, Matsuda T. Blood. 1999;93:3008–3016. [PubMed] [Google Scholar]
- 23.Kwan S P, Hagemann T L, Radtke B E, Blaese R M, Rosen F S. Proc Natl Acad Sci USA. 1995;92:4706–4710. doi: 10.1073/pnas.92.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molina I J, Sancho J, Terhorst C, Rosen F S, Remold-O'Donnell E. J Immunol. 1993;151:4383–4390. [PubMed] [Google Scholar]
- 25.Zhang J, Shehabeldin A, da Cruz L A, Butler J, Somani A K, McGavin M, Kozieradzki I, dos Santos A O, Nagy A, Grinstein S, et al. J Exp Med. 1999;190:1329–1342. doi: 10.1084/jem.190.9.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efstratiadis A, Posakony J W, Maniatis T, Lawn R M, O'Connell C, Spritz R A, DeRiel J K, Forget B G, Weissman S M, Slightom J L, et al. Cell. 1980;21:653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- 27.Fearon E R, Kohn D B, Winkelstein J A, Vogelstein B, Blaese R M. Blood. 1988;72:1735–1739. [PubMed] [Google Scholar]
- 28.Greer W L, Kwong P C, Peacocke M, Ip P, Rubin L A, Siminovitch K A. Genomics. 1989;4:60–67. doi: 10.1016/0888-7543(89)90315-7. [DOI] [PubMed] [Google Scholar]
- 29.Mantuano E, Candotti F, Giliani S, Parolini O, Lusardi M, Zucchi M, Lanfranchi A, Porta F, Airo P, Albertini A, et al. Immunodeficiency. 1993;4:271–276. [PubMed] [Google Scholar]
- 30.Wengler G, Gorlin J B, Williamson J M, Rosen F S, Bing D H. Blood. 1995;85:2471–2477. [PubMed] [Google Scholar]
- 31.Rawlings S L, Crooks G M, Bockstoce D, Barsky L W, Parkman R, Weinberg K I. Blood. 1999;94:3872–3882. [PubMed] [Google Scholar]
- 32.Rengan R, Ochs H D, Sweet L I, Keil M L, Gunning W T, Lachant N A, Boxer L A, Omann G M. Blood. 2000;95:1283–1292. [PubMed] [Google Scholar]
- 33.Bousso P, Wahn V, Douagi I, Horneff G, Pannetier C, Le Deist F, Zepp F, Niehues T, Kourilsky P, Fischer A, de Saint Basile G. Proc Natl Acad Sci USA. 2000;97:274–278. doi: 10.1073/pnas.97.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]