Summary
Molecular mechanisms of the mammalian circadian clock have been studied primarily by genetic perturbation and behavioral analysis. Here, we used bioluminescence imaging to monitor Per2 gene expression in tissues and cells from clock mutant mice. We discovered that Per1 and Cry1 are required for sustained rhythms in peripheral tissues and cells, and in neurons dissociated from the suprachiasmatic nuclei (SCN). Per2 is also required for sustained rhythms, whereas Cry2 and Per3 deficiencies cause only period length defects. However, oscillator network interactions in the SCN can compensate for Per1 or Cry1 deficiency, preserving sustained rhythmicity in mutant SCN slices and behavior. Thus, behavior does not necessarily reflect cell-autonomous clock phenotypes. Our studies reveal previously unappreciated requirements for Per1, Per2, and Cry1 in sustaining cellular circadian rhythmicity and demonstrate that SCN intercellular coupling is essential not only to synchronize component cellular oscillators but also for robustness against genetic perturbations.
Introduction
In mammals, the circadian timing system is organized in a hierarchy of multiple oscillators (Reppert and Weaver, 2002 and Lowrey and Takahashi, 2004). At the organismal level, the suprachiasmatic nuclei (SCN) of the anterior hypothalamus comprise the central pacemaker at the top of the hierarchy, integrating light information and coordinating peripheral oscillators throughout the body. Peripheral clocks, in turn, directly regulate many local rhythms (Kornmann et al., 2007), and overt rhythms in physiology and behavior likely feed back to the SCN through hypothalamic integration (Buijs and Kalsbeek, 2001). At the tissue level, individual cells within the SCN are synchronized to form a coherent oscillator through intercellular coupling (Aton and Herzog, 2005). Within cells, the clockwork consists of a core feedback loop in which BMAL1 and CLOCK drive expression of the Per and Cry genes; the PER and CRY repressor proteins in turn feed back to inhibit the transcription of their own genes (Reppert and Weaver, 2002 and Lowrey and Takahashi, 2004).
The most common approach to characterizing the clockwork has involved genetic perturbation followed by behavioral and molecular assays (Lowrey and Takahashi, 2004 and Takahashi, 2004). Though these assays have been instrumental in advancing our understanding of the basic clockwork, they do not take into sufficient consideration the hierarchical nature of the clock system. First of all, locomotor activity reflects a behavioral output downstream of SCN function, far removed from the intracellular molecular oscillations themselves. Wheel-running is a complex rhythmic output confounded by association with feeding, phenotypic variability, and pleiotropy of the underlying gene mutation (Bucan and Abel, 2002, Sato et al., 2004 and Lowrey and Takahashi, 2004). Second, because of intercellular synchronization at the tissue level, previous studies may not have revealed the intrinsic properties of individual cellular oscillators. Third, because of SCN-to-periphery synchronization and the hierarchical dominance of the SCN, molecular phenotypes determined from peripheral tissues in vivo are strongly influenced by the state of the SCN oscillator (Pando et al., 2002) rather than reporting tissue-autonomous properties of peripheral oscillators. Furthermore, previous molecular assays were relatively brief and were lacking in temporal resolution, typically measuring gene expression with only 4 hr resolution for 1-2 cycles. In summary, most previous characterizations of clock phenotypes do not report molecular details of clock operation, reveal system-level complexities, or distinguish between SCN and peripheral oscillators.
In order to test the roles of clock components more directly, we crossed circadian clock gene knockout mice with the mPer2::Luciferase fusion (mPer2Luc) knockin reporter line and examined the persistence and dynamics of molecular circadian rhythms by real-time bioluminescence measurements of tissue explants and dissociated cells (Yoo et al., 2004 and Welsh et al., 2004). We focused our analyses on the negative limb of the core clockwork, the Period (Per) and Cryptochrome (Cry) genes (van der Horst et al., 1999, Zheng et al., 1999, Zheng et al., 2001, Vitaterna et al., 1999, Kume et al., 1999, Shearman et al., 2000, Bae et al., 2001 and Cermakian et al., 2001), where existence of multiple family members provides the potential for functional diversity and redundancy. In this report, we demonstrate that Per1, Per2, and Cry1 are required to sustain circadian rhythms both in peripheral cells and tissues and in uncoupled SCN neurons, whereas Cry2 and Per3 deficiencies only alter circadian period. However, oscillator network interactions uniquely present in the SCN can compensate for genetic defects, preserving rhythms in SCN slices and behavior. These results demonstrate that circadian phenotypes observed in the SCN and in animal behavior are not necessarily cell autonomous.
Results
Per1, Per3, Cry1, and Cry2 Are Individually Dispensable for Sustained mPer2Luc Rhythms in SCN Explants
We crossed Per and Cry knockout mice with the mPer2Luc reporter line and obtained homozygous reporter knockouts. In the mPer2Luc knockin mouse, the transcription of mPer2Luc pre-mRNA is governed by cis-acting elements of the endogenous Per2 locus. The mPer2Luc fusion protein is functional in vivo, as it rescues virtually all phenotypes of Per2 knockout mice and allows for monitoring of molecular circadian rhythmicity in both SCN and peripheral tissues (Yoo et al., 2004).
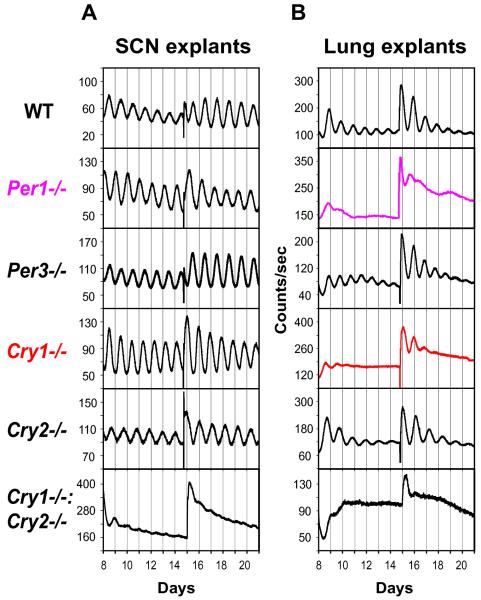
To determine whether wheel-running behavior truly reflects the SCN oscillator, we measured tissue-autonomous mPer2Luc rhythms in SCN explants from various circadian mutant mice and compared the molecular oscillations with locomotor activity patterns. Compared to wild-type (WT) controls, Cry1−/− and Cry2−/− SCN explants displayed rhythms with shorter and longer periods, respectively, while SCN explants from Cry1−/−:Cry2−/− mice were arrhythmic (Figure 1A; Table S1), all consistent with behavioral phenotypes (van der Horst et al., 1999 and Vitaterna et al., 1999).
Figure 1.
Per1 and Cry1 Are Required for Sustained mPer2Luc Rhythms in Peripheral Tissue Explants. Representative records of tissue-autonomous mPer2Luc bioluminescence rhythms in SCN explants (A) and lung explants (B) from Per and Cry knockout mice. Tissue explants were dissected (day 0) and immediately cultured in explant medium (EM) for recording. Data are shown beginning immediately following a change to fresh EM (day 8); another medium change occurred at day 15.
We also detected persistent rhythms in both Per1−/− and Per3−/− SCN explants (Figure 1A). While Per3−/− SCN explants displayed a slightly shorter mean period than WT, Per1−/− SCN exhibited a period similar to WT (Table S1), again consistent with behavioral phenotypes (Bae et al., 2001 and Zheng et al., 2001). Thus, locomotor activity rhythms generally reflected the molecular oscillations in SCN explants, confirming the validity of the real-time bioluminescence approach in studying circadian mutants. Our results demonstrate that Per1, Per3, Cry1, and Cry2 are all individually dispensable for persistent rhythmicity in cultured SCN tissue.
Per1 and Cry1 Are Required for Sustained mPer2Luc Rhythms in Peripheral Tissue Explants
Given that the molecular clock components in peripheral oscillators appear to be similar to those in the SCN, we expected that circadian phenotypes of clock gene knockouts in the periphery would resemble those in the SCN. We found that, similar to behavioral and SCN explant phenotypes, tissue explants of lung, liver, and cornea from WT mice showed persistent rhythms; those from Cry1−/−:Cry2−/− mice were arrhythmic; and Cry2−/− tissue explants displayed persistent rhythms with longer periods than WT (Figures 1B and S1; Table S1). Gradual damping observed in these peripheral tissue rhythms is consistent with progressive cellular desynchrony due to period variation among cells, as previously demonstrated in fibroblasts (Welsh et al., 2004 and Nagoshi et al., 2004). Surprisingly, however, Cry1−/− lung explants exhibited much less persistent or absent rhythms, indicative of a profound disruption of intrinsic clock function (Figure 1B). The less persistent rhythms were also observed in Cry1−/− liver and cornea (Figure S1). Although a medium change could briefly restart oscillations in some samples, the revived rhythms were transient and had unstable periods. Interestingly, this requirement for Cry1 for persistent rhythms is reminiscent of the role of dCry in Drosophila (Krishnan et al., 2001). Deletion of Per1 also severely compromised the precision and persistence of the oscillator in lung explants (Figure 1B). Lung explants from Per3−/− mice displayed persistent rhythms with a shorter mean period than in WT (Table S1). Taken together, these findings reveal a previously unrecognized critical requirement for Per1 and Cry1 in sustaining robust rhythms in tissue-autonomous peripheral oscillators (Bae et al., 2001, Zheng et al., 2001 and Cermakian et al., 2001). In contrast, Cry2 and Per3 are clearly dispensable, functioning only to modulate period length.
Per1 and Cry1 Are Required for Sustained mPer2Luc Rhythms in Dissociated Fibroblast Cultures
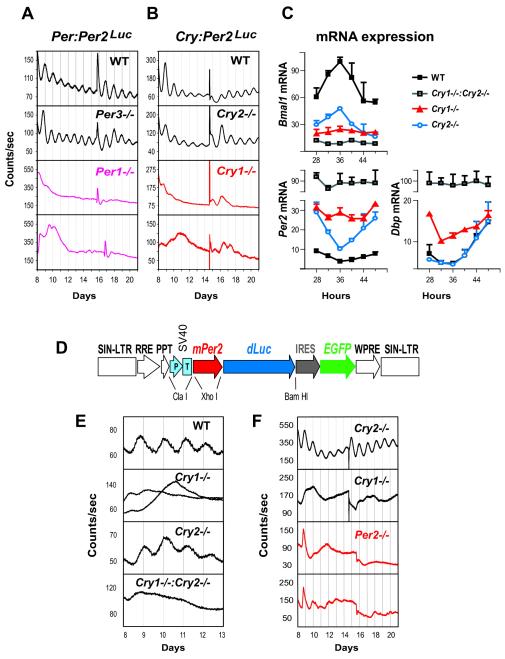
To examine peripheral circadian oscillators in the absence of potential intercellular synchronization (Welsh et al., 2004 and Nagoshi et al., 2004), we assessed the effects of clock gene deletions in cultures of dissociated primary fibroblasts. As expected, WT fibroblasts showed persistent mPer2Luc rhythms (Figures 2A and 2B). Per3−/− and Cry2−/− fibroblasts showed persistent rhythms with short and long periods, respectively. In contrast, both Per1−/− and Cry1−/− fibroblasts were largely arrhythmic (Figures 2A and 2B, bottom two panels). Thus, the respective functions of clock components in dissociated fibroblasts appear to be comparable to those in all the peripheral tissues examined.
Figure 2.
Per1, Per2, and Cry1 Are Required for Sustained mPer2Luc Rhythms in Dissociated Fibroblasts (A and B) Representative records of mPer2Luc bioluminescence rhythms in populations of primary fibroblasts derived from Per1 and Per3 knockout mice (A) and Cry1 and Cry2 knockout mice (B). Cells were first grown to confluence and then changed to EM for recording (day 0). Data are shown beginning immediately following a change to fresh EM (day 8), and another medium change occurred near the middle of each record. Two traces are shown for Per1−/− and for Cry1−/− fibroblasts.(C) Temporal mRNA expression profiles of Per2, Dbp, and Bmal1 in immortalized fibroblasts. Expression was analyzed at 4 hr intervals by reverse transcription and quantitative PCR. Values are expressed as a percent of maximum expression for each gene. Error bars represent standard deviation (SD) of expression levels from two independent cell lines for each genotype. Each PCR reaction was repeated three times, and results were confirmed in two independent time courses.(D) Lentiviral mPer2-dLuc clock reporter construct. The vector contains the mouse Per2 promoter, the coding region dLuc, and an IRES-mediated EGFP coding sequence as detailed in Supplemental Data. This entire DNA cassette is flanked by the long terminal repeats (LTRs) of a lentiviral packaging vector.(E and F) Circadian bioluminescence recordings from primary fibroblasts (E) and immortalized fibroblasts (F) transduced with the mPer2-dLuc reporter. Two traces are shown for Per2−/− fibroblasts.
CRY1 Plays a More Prominent Repressor Role than CRY2
To address the molecular basis for differential effects of Cry1 and Cry2 deletions on circadian rhythms, we used quantitative PCR to measure mRNA rhythms of Bmal1, Per2, and Dbp in fibroblast cultures derived from WT, Cry1−/−, Cry2−/−, and Cry1−/−:Cry2−/− mice. As expected, Per2 and Bmal1 showed rhythmic but nearly antiphasic expression in WT fibroblasts, and the highest Per2 and Dbp RNA levels were detected in Cry1−/−:Cry2−/− cells lacking both repressors (Figure 2C). Importantly, the relief of Per2 or Dbp transcriptional repression in Cry1−/− cells was greater than in Cry2−/− cells, and Per2 RNA levels in Cry1−/− fibroblasts exhibited weak and unstable rhythms (Figure 2C), similar to results from a previous study (Yagita et al., 2001). These results indicate that CRY1 plays a more prominent repressor role than CRY2.
The greater importance of CRY1 for clock function is supported by two lines of behavioral observations: Cry1+/-:Cry2−/− mice display more persistent free-running activity rhythms than Cry1−/−:Cry2+/- animals (van der Horst et al., 1999), and Per2−/−:Cry1−/− mice are completely arrhythmic, whereas Per2−/−:Cry2−/− mice are rhythmic (Oster et al., 2002). It is known that Cry1 is strongly rhythmic in most tissues, while Cry2 has only weak rhythms (Kume et al., 1999), which might partly explain their differential importance for clock function. Alternatively, CRY1 protein level may be higher than that of CRY2, or CRY1 may be a stronger biochemical repressor than CRY2.
Per2 Is Required for Sustained mPer2Luc Rhythms in Dissociated Fibroblast Cultures
Given the prominent role of Per2 in SCN-controlled circadian behavior (Bae et al., 2001 and Zheng et al., 2001), we sought to determine whether it also plays a critical role in the periphery. However, a Per2−/−:mPer2Luc mouse line cannot be generated because the knockin reporter construct codes for a functional mPER2Luc fusion protein. As an alternative approach, we engineered a lentivirus-based mPer2-dLuc clock reporter (Figure 2D). Like the mPer2Luc knockin reporter, the lentiviral mPer2-dLuc construct reported persistent circadian rhythms in both primary (Figure 2E) and immortalized WT fibroblasts (data not shown). Using this approach, the phenotypes of Cry1 or Cry2 deficiency in fibroblasts were independently confirmed (Figures 2E and 2F). We found that Per2−/−:mPer2-dLuc fibroblast cultures either displayed unstable rhythms of low amplitude or were arrhythmic, similar to Cry1−/− fibroblasts (Figure 2F, bottom two panels). Thus, it appears that Per1 and Per2 are not functionally redundant, and both are required to generate robust circadian oscillations in fibroblasts. Alternatively, Per1 and Per2 functions might be overlapping, and their protein-expression levels might be too low in Per1−/− or Per2−/− fibroblasts.
Per1 and Cry1 Are Required for Sustained mPer2Luc Rhythms in Dissociated Individual Fibroblasts
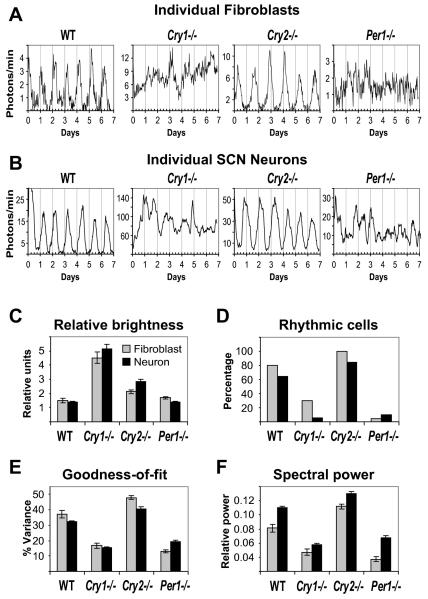
Arrhythmicity in peripheral tissues at the cell population level could arise from single-cell oscillators that are either arrhythmic or poorly synchronized. Thus, to determine whether Per1 and Cry1 are required to sustain rhythmicity in individual cells, we imaged mPer2Luc bioluminescence from dissociated fibroblasts at the single-cell level. We observed that Cry1−/− and Cry2−/− fibroblasts were significantly brighter than WT (p ≤ 0.0025, n = 20 cells per genotype; Figure 3C), consistent with the known repressor functions of CRY1 and CRY2. Cry1−/− cells were also brighter than Cry2−/− cells (p < 0.0001), which supports results from mRNA expression analysis (Figure 2C).
Figure 3.
Per1 and Cry1 Are Required for Cell-Autonomous mPer2Luc Rhythms in Fibroblasts and SCN Neurons (A and B) Representative bioluminescence intensity patterns for fibroblasts (A) and SCN neurons (B) derived from WT, Cry1−/−, Cry2−/−, and Per1−/− mice. Imaging began immediately following a change to fresh EM (day 0). Most WT and Cry2−/− cells were rhythmic, while most Cry1−/− and Per1−/− cells were arrhythmic. See Figure 4 and Figure 5 for more individual cell rhythms.(C) Single-cell brightness comparisons across genotype. SCN neurons were brighter than fibroblasts. The relative unit of brightness was 6.6 photons/min for SCN neurons and 1 photon/min for fibroblasts. Values are presented as mean ± standard error of the mean (SEM).(D-F) Summary of single-cell analysis. Most individual WT and Cry2−/− fibroblasts and SCN neurons were rhythmic as determined using FFT spectral power, whereas most Per1−/− and Cry1−/− cells were either arrhythmic or showed weak or transient rhythms (D). Overall, Per1−/− and Cry1−/− cells had significantly reduced goodness-of-fit (E) and spectral power (FFT-RelPower; F), compared to WT and Cry2−/−. In (E) and (F), values are presented as mean ± SEM.
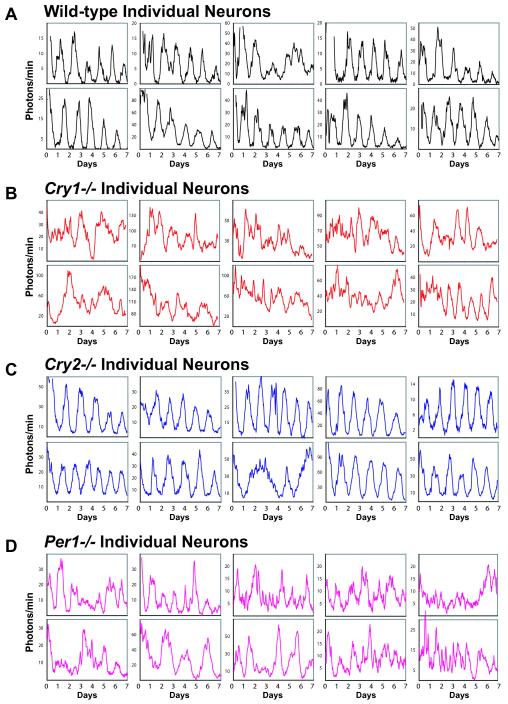
Dissociated WT and Cry2−/− cells generally exhibited persistent rhythms with high amplitude. Rhythmicity was detected in 80% of WT fibroblasts (n = 16/20) and 100% of Cry2−/− fibroblasts (n = 20/20). Interestingly, Cry2−/− fibroblasts exhibited higher rhythm amplitudes than WT cells (FFT-RelPower; p < 0.0001). Of the rhythmic cells, the mean period was 24.63 hr in WT and significantly longer (28.79 hr) in Cry2−/− fibroblasts (p < 0.0001; Table S1). In striking contrast, most Per1−/− and Cry1−/− cells were arrhythmic (Figures 3A, 3D, and 4). A few cells showed weak or transient rhythms, including 5% of Per1−/− fibroblasts (n = 1/20) and 30% of Cry1−/− fibroblasts (n = 6/20). Overall, compared to WT, Per1−/− and Cry1−/− cells had much weaker rhythms as measured by either goodness-of-fit of a fitted sine wave (p < 0.0001; Figure 3E) or spectral power (FFT-RelPower; p < 0.0001; Figure 3F). These intrinsically compromised rhythms were of low amplitude, of variable period, or persistent for only a few cycles (Figure 4). Similar patterns were also observed in individual cells of Cry1−/− liver slice cultures (Figure S2). Thus, in peripheral tissues, single Per1−/− or Cry1−/− cells cannot sustain circadian oscillations.
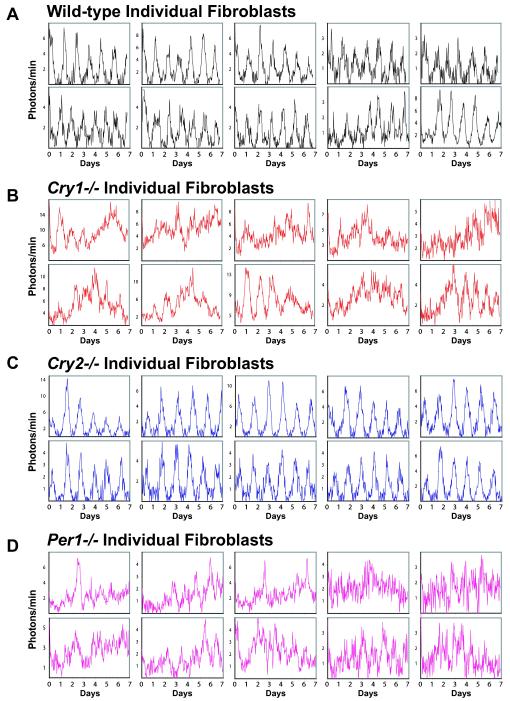
Figure 4.
mPer2Luc Bioluminescence Patterns of Individual Fibroblasts Ten single-cell fibroblast rhythms representative of each genotype are presented for (A) WT, (B) Cry1−/−, (C) Cry2−/−, and (D) Per1−/−. Imaging began immediately following a change to fresh EM (day 0). These results show that both Cry1 and Per1 are required for sustained rhythmicity in the fibroblasts.
Per1 and Cry1 Are Required for Sustained mPer2Luc Rhythms in Dissociated Individual SCN Neurons
Robust rhythmicity in Per1−/− or Cry1−/− SCN slices might be explained either by a distinct intracellular clock mechanism (different from peripheral oscillators) or by intercellular coupling and mutual reinforcement of compromised clock cells. To distinguish between these two possibilities, we decided to uncouple SCN neurons to examine cell-autonomous rhythmicity. Uncoupling of SCN cellular oscillators has been demonstrated by mechanical dissociation into single cells (Welsh et al., 1995 and Herzog et al., 1998) or in slice preparations using tetrodotoxin (Yamaguchi et al., 2003) or genetic disruption of VIP signaling (Aton et al., 2005 and Maywood et al., 2006). However, the latter two approaches cause damping of individual cell rhythms, confounding interpretation of mutant phenotypes. In this study, we employed mechanical dissociation.
To address whether the overt bioluminescence expression patterns observed at the SCN tissue level are cell autonomous, we imaged mPer2Luc bioluminescence from dissociated SCN neurons at the single-cell level. At least 100 individual neurons were analyzed for each genotype. Overall, SCN neurons were nearly 7-fold brighter than fibroblasts, indicating a much higher average level of PER2 expression in the SCN (Figure 3C). Just as in fibroblasts, SCN neurons of Cry1−/− and Cry2−/− genotypes were significantly brighter than WT (p < 0.0001), and Cry1−/− cells were also brighter than Cry2−/− cells (p < 0.0001).
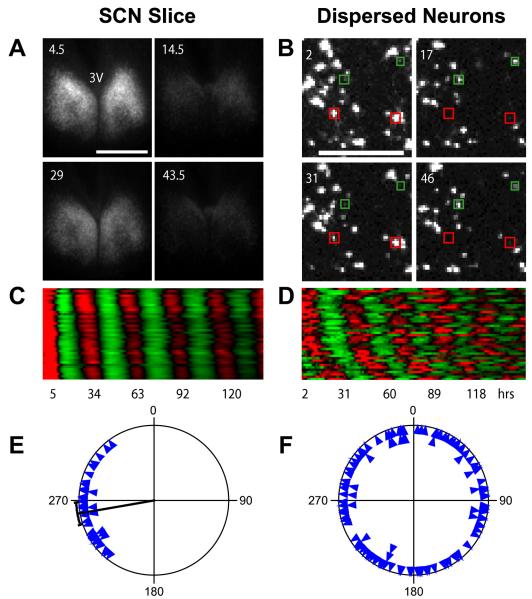
Unlike the stable and synchronized bioluminescence rhythms in neurons within WT and Cry2−/− SCN slices (Figures 6A, 6C, and 6E), dispersed SCN neurons of these genotypes drifted out of phase from one another by the end of the recording; their end phases were randomly distributed (Figures 6B, 6D, and 6F), indicating a lack of functional coupling. Even among cells that were close together (<500 μm apart), closer cells did not tend to have more similar phases (r = 0.04 and p > 0.4 for both WT and Cry2−/−), ruling out any measurable local coupling.
Figure 6.
Network Interaction Synchronizes Cellular Oscillators in Cry2−/− SCN (A) Bioluminescence images of a Cry2−/− SCN slice culture at peak and trough phases, showing stable and synchronized oscillations. Numbers indicate hours after start of imaging. 3V indicates third ventricle. Scale bar is 500 μm.(B) Bioluminescence images of dissociated individual Cry2−/− SCN neurons showing cell-autonomous, desynchronized oscillations. Note that the cells highlighted with red and green are antiphasic. Numbers and scale bar are as in (A). See also Figure 5 and Movie S1.(C and D) Raster plots of bioluminescence intensity of individual Cry2−/− neurons in SCN slice in (A) and from dispersed culture in (B). Forty cells are presented in each plot, and each horizontal line represents a single cell. Values above and below the mean are shown in red and green, respectively.(E and F) Circadian phase plots of rhythmic Cry2−/− neurons within the SCN slice in (A) and from the dispersal culture in (B). Each blue triangle represents the phase of one cell at the end of a 6-7 day experiment, where 0° = phase of the fitted peak of the rhythm. The radial line indicates the average phase, and the arc indicates the 95% confidence interval for mean phase. Rayleigh test: p < 0.0001 (n = 40) for neurons in the SCN slice; p = 0.97 (n = 121) for dissociated neurons. Most dissociated Cry2−/− SCN neurons expressed rhythmic patterns of bioluminescence that were initially partly synchronized by medium change but then gradually desynchronized due to varying intrinsic periods in the absence of functional intercellular coupling (B, D, and F), whereas cells in the slice culture were synchronized across the SCN and throughout the course of the experiment (A, C, and E).
Dissociated WT and Cry2−/− SCN neurons generally exhibited persistent rhythms with high amplitude (Figure 5; Movie S1). Rhythmicity was detected in 64% of WT SCN neurons (n = 106/165) and 84% of Cry2−/− SCN neurons (n = 136/162; Figure 3D). The mean circadian period in Cry2−/− cells was significantly longer than in WT (p = 0.0017; Table S1). Interestingly, just as in fibroblasts, Cry2−/− neurons displayed higher rhythm amplitudes than WT cells (FFT-RelPower; p < 0.0001).
Figure 5.
mPer2Luc Bioluminescence Patterns of Individual SCN Neurons Ten single SCN neuron rhythms representative of each genotype are presented for (A) WT, (B) Cry1−/−, (C) Cry2−/−, and (D) Per1−/−. Imaging began immediately following a change to fresh EM (day 0). These results show that both Cry1 and Per1 are required for sustained rhythmicity in SCN neurons. See Movies S1 and S2 for dynamic views of Cry2−/− and Cry1−/− neurons, respectively.
In striking contrast to WT and Cry2−/− neurons, most Per1−/− and Cry1−/− neurons were arrhythmic (Figures 3B, 3D, and 5; Movie S2). Overall, these cells had much weaker rhythms as measured by either goodness-of-fit of a fitted sine wave (p < 0.0001; Figure 3E) or spectral power (FFT-RelPower; p < 0.0001; Figure 3F). A few individual cells showed weak or transient rhythms, including 10% of Per1−/− SCN neurons (n = 12/125) and 6% of Cry1−/− SCN neurons (n = 7/118). Although these rhythms were statistically detectable, most were of low amplitude, of variable period, or persistent for only a few cycles (Figures 5B and 5D; Movie S2); as a result, a reliable period length could not be determined.
SCN Oscillator Network Interactions Compensate for Per1 and Cry1 Deficiency
How can the SCN, but not the examined peripheral tissues, compensate for the loss of Per1 or Cry1? A higher abundance of circadian clock components in the SCN might contribute to the robustness of the SCN oscillator, but that is clearly insufficient to sustain rhythmicity in dissociated SCN cells. So, robustness of the Per1−/− and Cry1−/− SCN is not a single-cell characteristic but depends upon a coupled network of oscillator cells. The observation that most WT SCN neurons within cultured explants express synchronized circadian rhythms, whereas individual neurons in dispersal are not synchronized (Figure 6; Welsh et al., 1995, Liu et al., 1997, Herzog et al., 1998, Herzog et al., 2004, Yamaguchi et al., 2003 and Aton and Herzog, 2005), implies that the robust rhythmicity of the SCN ensemble is attained by oscillator coupling interactions. This notion is supported by mouse chimera experiments with WT and Clock mutant embryos, showing that both period averaging and coherence of circadian behavior can occur at the SCN and organismal levels (Low-Zeddies and Takahashi, 2001).
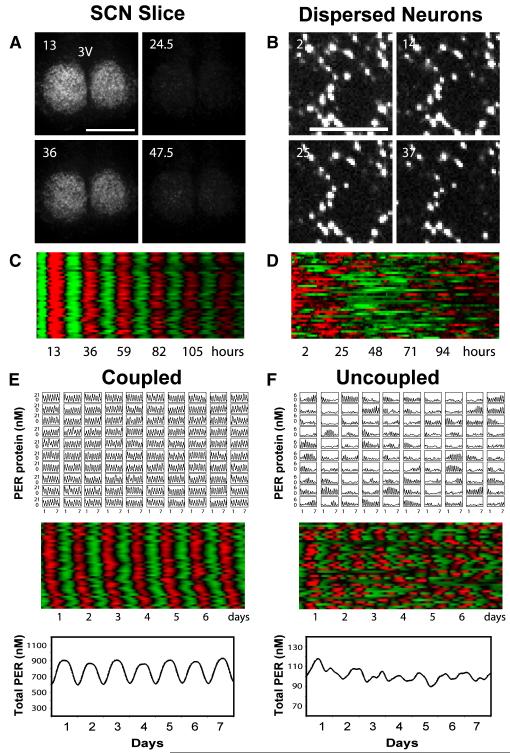
Our results demonstrate that coupling among SCN neurons in slices and in vivo was able not only to synchronize rhythmic cells (Figure 6; Movie S1) but also to stabilize and synchronize the transient rhythms in intrinsically compromised cellular oscillators such as those in Cry1−/− SCN (Figures 7A-7D; Movie S2). In this context, the transient or weak rhythms present in a small percentage of Per1−/− and Cry1−/− neurons must play a crucial role, as no obvious circadian rhythmicity was detected in Cry1−/−:Cry2−/− SCN slices (Figure 1A).
Figure 7.
Bioluminescence Imaging and Mathematical Simulations Demonstrate that Intercellular Coupling Can Stabilize and Sustain Circadian mPer2Luc Rhythms in Cry1−/− SCN (A) Bioluminescence images of a representative Cry1−/− SCN slice culture at peak and trough phases, showing stable and synchronized bioluminescence oscillations. Numbers and scale bar are as in Figure 6.(B) Bioluminescence images of dissociated individual Cry1−/− SCN neurons showing cell-autonomous, largely arrhythmic patterns of high bioluminescence intensity. Numbers and scale bar are as in (A). See also Figure 5 and Movie S2.(C and D) Raster plots of bioluminescence intensity of individual Cry1−/− neurons in SCN slice in (A) and from dispersed culture in (B). Plots were constructed as in Figure 6. Dissociated Cry1−/− SCN neurons were largely arrhythmic, whereas cells in the slice culture were highly rhythmic and very tightly synchronized across the SCN.(E and F) Mathematical simulation for coupled oscillators (E) and uncoupled oscillators (F). Time courses of 100 oscillators (single-cell PER protein concentration in 10×10 grid, top), raster plots of 40 oscillators (middle), and the sum (total PER concentration for 100 oscillators, bottom) are presented for each condition.
Mathematical Simulations Demonstrate that Oscillator Coupling Can Compensate for Severely Compromised Single-Cell Oscillators
To complement our in vivo observations, we used mathematical simulations to further explore the significance of coupling. We modified a published computational clock model (Leloup and Goldbeter, 2003) to reproduce the largely arrhythmic behavior of the individual Per1−/− and Cry1−/− neurons by introducing noise to the Bmal1 degradation rate and to the activation threshold of nuclear BMAL1 on Cry transcription. Through the introduction of a simple coupling mechanism (i.e., Per induction dependent on Per mRNA levels in nearby cells), we were able to recover stable, persistent rhythms from the rare, intermittent oscillations observed in uncoupled Per1−/− or Cry1−/− neurons (Figures 7E and 7F). Thus, both in vivo and in silico experiments demonstrate that coupling can restore robust oscillations to severely compromised single-cell oscillators.
Our results, which underscore the importance of intercellular communication in the SCN, serve as an experimental validation of model predictions previously made in the context of a simple synthetic system, where the same general principle was observed (Garcia-Ojalvo et al., 2004). Our results are also consistent with studies of electrical and secretory responses in pancreatic β cells (Smolen et al., 1993), neural oscillators in Drosophila (Stoleru et al., 2005), and the somite segmentation clock in zebrafish and mice (Horikawa et al., 2006 and Masamizu et al., 2006), all showing that oscillator coupling mechanisms are critical for synchronization and maintenance of robust oscillations. However, the systems-level network interactions in the mammalian SCN are likely to be more complex (Low-Zeddies and Takahashi, 2001 and Aton and Herzog, 2005). Given the prevalence of a wide variety of biological oscillators in controlling development and physiology, a general picture is now emerging for the importance of intercellular coupling in maintaining system robustness.
Discussion
The SCN contain an autonomous circadian pacemaker necessary for circadian behavior. Recent studies have established that virtually all peripheral tissues, and even cultured fibroblasts, also contain cell-autonomous, self-sustained circadian oscillators (Balsalobre et al., 1998, Reppert and Weaver, 2002, Yoo et al., 2004, Welsh et al., 2004 and Nagoshi et al., 2004). In this study we uncover critical requirements for Per1, Per2, and Cry1 in sustaining cellular rhythmicity and provide compelling genetic evidence to illuminate the respective contributions of intracellular and intercellular clock mechanisms to the robustness of the clock system.
As single cells are ordinarily capable of functioning as autonomous oscillators, our understanding of clock mechanisms has rested precariously on the assumption that we can divine the roles of molecular clock components by testing behavioral rhythms in mice deficient for particular genes. By examining effects of genetic perturbations at the level of single cells and tissues, as well as behavior, we demonstrate that intercellular mechanisms are in fact essential to the robust operation of cellular circadian clocks (Figure S3). Thus, unlike in peripheral tissues, the SCN circadian clock is more than just the sum of its cells. Our results demonstrate that the integrated SCN circadian pacemaker is qualitatively more robust than its component cellular oscillators. SCN neurons appear to be similar to fibroblasts at the level of individual cells, not only in their capacity to generate persistent circadian oscillations, but also in their requirement for Per1 and Cry1 in sustaining these oscillations. Essentially, the special attributes of the SCN appear to result not from distinctive intracellular clock mechanisms, but rather from specialized intercellular interactions.
Our studies reveal a direct relationship between the coupling of single-cell oscillators in the SCN and robustness of the clock against genetic perturbations. The distribution of period lengths for cultured SCN explants within each genotype was significantly wider than for behavior, and this period dispersion was even more extreme among individual neurons, consistent with previous findings (Herzog et al., 2004). The circadian defects observed in mutant oscillators were clearly more extreme when measured at the tissue and cell levels than in behavioral assays, in agreement with previous observations (Table S1; Liu et al., 1997, Herzog et al., 1998, Nakamura et al., 2002 and Brown et al., 2005). A major challenge in systems neuroscience is to integrate molecular mechanisms with tissue-level organization. Our results show that the SCN, by amplifying and stabilizing unstable (stochastic) component oscillators through coupling interactions, can establish rhythms at the SCN-tissue and behavioral levels that are significantly more reliable (deterministic; Figure S3).
Despite significant experimental effort over the past two decades, how the cellular oscillators in the SCN are synchronized is not well understood. Neurons in the ventral SCN synchronize the dorsal neurons and maintain synchrony of the SCN ensemble (Yamaguchi et al., 2003). Interestingly, long-term exposure to constant light renders mice arrhythmic by disrupting SCN synchrony without impairing oscillator function of individual SCN neurons (Ohta et al., 2005). Several mechanisms, particularly neurochemical synapses and electrical coupling, have been proposed to mediate intercellular synchronization (Aton and Herzog, 2005). In particular, VIP signaling through VPAC2 receptors has been shown to contribute to circadian synchrony (Harmar et al., 2002, Aton et al., 2005 and Maywood et al., 2006). Neural cell adhesion molecule (NCAM) and polysialic acid (PSA) may also play important roles (Shen et al., 1997). In the present work, we demonstrate the necessity of tissue-level coupling for robust operation of the circadian timing system, and future studies should focus on addressing the necessity and sufficiency of specific coupling mechanisms.
Since distinct neuronal types are present in the SCN (e.g., based on neuropeptide content), future studies should also address whether the intermittent rhythmicity we observed in some mutant cells might be associated with one or more subclasses of neurons or whether this rhythmicity is a completely stochastic phenomenon. It is highly likely, however, that in the present study we sampled all known subpopulations of SCN neurons for each genotype (elementary probability calculations, data not shown). As we did not observe even a single Per1−/− or Cry1−/− neuron that maintained circadian rhythmicity for the entire 6-7 day recording, it is extremely unlikely that any known subclass of SCN neurons can maintain normal rhythms despite Per1 or Cry1 deletion.
Lack of coupling in the periphery explains why intact peripheral tissues were vulnerable to Per1 or Cry1 deletion, but this may actually be an adaptive feature in most circumstances. SCN cells in vivo must synchronize not only to light-dark cycles but also to one another to coordinate circadian behavior. Lack of coupling may allow peripheral oscillators, on the other hand, to anticipate and respond rapidly and flexibly not only to the synchronizing cues emanating from the SCN but also to physiological signals related to feeding and behavior (Hastings and Herzog, 2004).
Remarkably, deletion of Cry2 actually strengthens rhythms; rhythms are weaker when Cry2 is functional. We speculate that by modestly weakening rhythms, CRY2 might enhance responsiveness of the clock to certain physiological signals. Interestingly, a recent stochastic clock model predicted stronger rhythms with Cry2 deletion (Forger and Peskin, 2005). Stochastic, intermittent rhythmicity was also predicted by this model, though rhythms were not predicted to be weaker for Per1−/− or Cry1−/− than for WT. Thus, our results validate model predictions previously overlooked or regarded as model flaws. However, new clock models are needed to accommodate the novel cell-autonomous phenotypes revealed here.
Finally, our findings have important strategic implications for future studies of the circadian clock. As intercellular communication can mask genetic defects in individual clock cells (Figure S3), the molecular mechanisms required for rhythmicity must therefore be studied not only at the organismal and tissue levels but also at the level of single cells. Equally important, because circadian timing is a dynamic process, long-term recordings are required to assess the persistence of circadian rhythmicity. It is interesting to note that the Clock gene was recently shown to be dispensable for locomotor activity rhythms (DeBruyne et al., 2006). Given that dissociated ClockΔ19 mutant SCN neurons appear less robust than those in SCN slices (Herzog et al., 1998 and Nakamura et al., 2002), highly reminiscent of Per1−/− and Cry1−/− cellular phenotypes, it will be interesting to determine whether the effects of Clock deletion are also cell autonomous. Perhaps multioscillator coupling will prove to be a general mechanism for enhancing robustness to a wide variety of genetic and other perturbations.
Experimental Procedures
Animals and Behavioral Analysis
Per mouse lines were obtained from David Weaver at the University of Massachusetts, and Cry lines were obtained from Bert van der Horst at Erasmus University, The Netherlands (van der Horst et al., 1999; Scripps) or from Aziz Sancar and Takeshi Todo (Vitaterna et al., 1999; Northwestern). Knockout mice (Per1−/−, Per3−/−, Cry1−/−, Cry2−/−, and Cry1−/−:Cry2−/−) were bred with mPer2Luc reporter mice to obtain homozygous knockouts harboring the mPer2Luc reporter. Wheel-running assays were performed and analyzed as described previously (Yoo et al., 2004). Behavioral phenotypes of these mice were similar to the respective knockout animals not carrying the reporter (Table S1; van der Horst et al., 1999, Shearman et al., 2000 and Bae et al., 2001). All animal studies were conducted in accordance with the regulations of the Committees on Animal Care and Use at The Scripps Research Institute and Northwestern University.
Cell and Explant Culture
Explants of SCN and peripheral tissues were dissected and cultured in HEPES-buffered serum-free explant medium (EM) containing B-27 and luciferin (Yoo et al., 2004). SCN and liver slices were cut by tissue chopper (Stoelting) to a thickness of 400 μm and cultured on Millicell-CM membrane inserts (Fisher PICMORG50). SCN slices prepared from early postnatal (2-7 days old) or adult animals (1-4 months old) produced highly comparable rhythms for all genotypes.
Primary mouse fibroblasts were generated from tails by a standard enzymatic digestion procedure (Welsh et al., 2004). Fibroblasts that spontaneously overcame replicative senescence (immortalization) were used when indicated. All fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and were grown to confluence prior to bioluminescence recording.
For preparation of SCN neuronal cells, cylindrical punches of unilateral SCN from 2- to 4-day-old pups were made from 400 μm coronal sections using a 20 gauge needle. For each preparation, six mice were used, and the experiment was repeated twice for each genotype. Cells were dissociated using papain and were cultured as previously described (Welsh et al., 1995) except that medium contained 5% FBS instead of rat serum. Cells were maintained in culture for 2-5 weeks before imaging.
Bioluminescence Recording and Data Analysis
Change to fresh EM was sufficient for synchronization, and similar synchronizing effects and cellular phenotypes were seen when peripheral tissue explants or cells were treated with 50% horse serum, forskolin, or dexamethasone (data not shown). After change to fresh EM, culture dishes containing cells or explants were sealed and placed into the LumiCycle luminometer (Actimetrics, Inc.), which was kept inside a standard tissue culture incubator at 36°C. Bioluminescence from each dish was continuously recorded with a photomultiplier tube (PMT) for 70 s at intervals of 10 min. We usually recorded bioluminescence rhythms for 1-2 weeks between medium changes.
Raw data (counts/sec) were plotted against time (days) in culture. For analysis of rhythm parameters, we used the Lumicycle Analysis program (Actimetrics, Inc.). Raw data were baseline fitted, and the baseline-subtracted data were fitted to a sine wave, from which the period was determined. For samples that showed persistent rhythms, goodness-of-fitness >90% was usually achieved. Due to high transient luminescence upon medium change, the first cycle was excluded from rhythm analysis.
Single-Cell Imaging
Single fibroblasts and SCN neurons were studied by bioluminescence imaging (Welsh et al., 2004 and Welsh et al., 2005), repeated twice for each cell type and each genotype. Identical culture conditions were used for all explant, slice, and single-cell bioluminescence experiments except that a higher concentration of luciferin (1 mM) was used for imaging. Culture dishes were sealed and placed on the stage of an inverted microscope (Olympus IX71) in a dark room. A heated lucite chamber around the microscope stage (Solent Scientific, UK) kept the cells at a constant 36°C. Images were collected using an Olympus 4× XLFLUOR (NA 0.28) or UPlanApo (NA 0.16) objective and transmitted to a CCD camera (Spectral Instruments SI800, Tucson, AZ) cooled to −92°C. Signal-to-noise ratio was improved by binning of pixels (8×8 for fibroblasts; 4×4 for dissociated SCN neurons). Images of 29.8 min exposure duration were collected at 30 min intervals for 6-7 days. Integration of bioluminescence over all single cells analyzed or the entire imaging field gave population patterns similar to those measured in the luminometer. The long period of WT neurons (27 hr versus 24.5 hr for behavior or other single-cell studies) is likely due to medium composition (e.g., exclusion of serum; Murakami et al., 1991, Welsh et al., 1995 and Herzog et al., 1998).
Fibroblast and SCN neuron viability was assessed by cell morphology, and no differences were observed among genotypes. We also measured the stability of average daily mPER2Luc bioluminescence for each cell and compared brightness of bioluminescence averaged over the last 3 days versus the first 3 days of the experiment. From this analysis, there was no indication that mutant genotypes with weaker rhythms (Cry1−/− and Per1−/−) were less viable (data not shown).
Single-Cell Imaging Data Analysis
Bioluminescence images were analyzed using MetaMorph (Molecular Devices) as previously described (Welsh et al., 2004 and Welsh et al., 2005). Cells that were clearly discriminable from adjacent cells and that remained bioluminescent for the entire experiment were selected for analysis.
Luminescence time series were first imported into LumiCycle Analysis (Actimetrics). Due to high initial transients of luminescence, the first 12 hr of data were excluded. A linear baseline was subtracted from raw data (polynomial order = 1). For rhythm analysis, five different procedures were used to detect the presence of circadian rhythmicity for each cell, all of which gave similar results for comparisons across genotype: (1) chi-square periodogram with single major peak (at least 2-fold greater than any minor peak) in the range 20-36 hr, p < 0.01; (2) best-fit sine wave (“Sin fit”) with goodness-of-fit (percent of total variance accounted for by the fitted curve) ≥ 25%; (3) spectral analysis (FFT-RelAmp) with percent of total power (variance) within frequency range corresponding to 20-36 hr periods ≥ 40%; (4) spectral analysis (FFT-RelPower) with percent of total power within a 1 Hz window centered at the peak in this range ≥ 10% for neurons or 6% for fibroblasts; and (5) FFT-NLLS procedure (Plautz et al., 1997) with RelAmp Error ≤ 0.3 for fibroblasts and 0.25 for neurons. Cutoffs were chosen so as to include all clearly rhythmic WT and Cry2−/− cells. We chose the FFT-RelPower criterion to select rhythmic cells for comparisons across genotypes. Comparisons significant by ANOVA (p < 0.05) were further explored by pairwise t tests. Period was defined as the period of the best-fit sine wave. Strength of rhythmicity was defined by spectral analysis (FFT-RelPower) or goodness-of-fit of the fitted sine wave. Other definitions of rhythm strength (see above) gave similar results.
For raster plots, bioluminescence-intensity data were detrended by subtracting a linear baseline, normalized for amplitude, and then color coded, with higher than average red and lower than average green. Plots were constructed using TreeView (Eisen lab, Stanford University). Clustering of circadian phases was evaluated by Rayleigh’s uniformity test and plotted using Oriana (Kovach Computing, UK).
Detailed descriptions of lentiviral reporter construction, mathematical simulation, and quantitative PCR are provided in Supplemental Data.
Supplementary Material
Acknowledgments
We thank D. Weaver, G.T.J. van der Horst, A. Sancar, and T. Todo for Per and Cry knockout mouse lines. We thank Rie Yasuda for providing primary fibroblast preparation protocols, Tom Schultz for FFT-NLLS analysis, and Takato Imaizumi for helpful discussions. This work was supported in part by grants from the NIH (R01 GM074868 and R01 MH051573 to S.A.K., K08 MH067657 to D.K.W., and U01 MH061915 and Silvio O. Conte Center P50 MH074924 to J.S.T.). J.S.T. is an Investigator in the Howard Hughes Medical Institute. This is manuscript #18121-CB of The Scripps Research Institute.
References
- Aton SJ, Herzog ED. Come together, right...now: synchronization of rhythms in a mammalian circadian clock Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin XW, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Brown SA, Fleury-Olela F, Nagoshi E, Hauser C, Juge C, Meier CA, Chicheportiche R, Dayer JM, Albrecht U, Schibler U. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abel T. The mouse: genetics meets behaviour. Nat. Rev. Genet. 2002;3:114–123. doi: 10.1038/nrg728. [DOI] [PubMed] [Google Scholar]
- Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat. Rev. Neurosci. 2001;2:521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. Stochastic simulation of the mammalian circadian clock Proc. Natl. Acad. Sci. USA. 2005;102:321–324. doi: 10.1073/pnas.0408465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ojalvo J, Elowitz MB, Strogatz SH. Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proc. Natl. Acad. Sci. USA. 2004;101:10955–10960. doi: 10.1073/pnas.0307095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen SB, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Herzog ED. Clock genes, oscillators, and cellular networks in the suprachiasmatic nuclei. J. Biol. Rhythms. 2004;19:400–413. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat. Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J. Biol. Rhythms. 2004;19:35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H. Noise-resistant and synchronized oscillation of the segmentation clock. Nature. 2006;441:719–723. doi: 10.1038/nature04861. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. http://dx.doi.org/10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, Hall JC, Hardin PE, Dryer SE. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin XW, Maywood ES, Hastings MH. S.M. Reppert mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Leloup JC, Goldbeter A. Toward a detailed computational model for the mammalian circadian clock. Proc. Natl. Acad. Sci. USA. 2003;100:7051–7056. doi: 10.1073/pnas.1132112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: Period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, Okamura H, Kageyama R. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc. Natl. Acad. Sci. USA. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Murakami N, Takamure M, Takahashi K, Utunomiya K, Kuroda H, Etoh T. Long-term cultured neurons from rat suprachiasmatic nucleus retain the capacity for circadian oscillation of vasopressin release. Brain Res. 1991;545:347–350. doi: 10.1016/0006-8993(91)91312-o. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Nakamura W, Honma S, Shirakawa T, Honma K. Clock mutation lengthens the circadian period without damping rhythms in individual SCN neurons. Nat. Neurosci. 2002;5:399–400. doi: 10.1038/nn843. [DOI] [PubMed] [Google Scholar]
- Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nat. Neurosci. 2005;8:267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- Oster H, Yasui A, van der Horst GT, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 2002;16:2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–117. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Jin XW, Lee CG, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: Subtle effects on circadian clock function. Mol. Cell. Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Watanabe M, Tomasiewicz H, Rutishauser U, Magnuson T, Glass JD. Role of neural cell adhesion molecule and polysialic acid in mouse circadian clock function. J. Neurosci. 1997;17:5221–5229. doi: 10.1523/JNEUROSCI.17-13-05221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen P, Rinzel J, Sherman A. Why pancreatic islets burst but single beta cells do not. The heterogeneity hypothesis Biophys. J. 1993;64:1668–1680. doi: 10.1016/S0006-3495(93)81539-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. Finding new clock components: past and future. J. Biol. Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GTJ, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker APM, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki U, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and Mammalian cells Methods. Enzymol. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, van der Horst GTJ, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng BH, Larkin DW, Albrecht L, Sun ZS, Sage M, Eichele G, Lee CC. A. Bradley The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zheng BH, Albrecht U, Kaasik K, Sage M, Lu WQ, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.