Abstract
Ability to modulate fluorescence of optical probes can be used to enhance signal-to-noise ratio for imaging within highly autofluorescent environments, such as intact tissues and living organisms. Here we report two phytochrome-based photoactivatable near-infrared fluorescent proteins, named PAiRFP1 and PAiRFP2. PAiRFPs utilize heme-derived biliverdin, ubiquitous in mammalian tissues, as the chromophore. Initially weakly fluorescent PAiRFPs undergo photoconversion into a highly fluorescent state with excitation/emission at 690 nm/717 nm following a brief irradiation with far-red light. After photoactivation, PAiRFPs slowly revert back to initial state, enabling multiple photoactivation-relaxation cycles. Low-temperature optical spectroscopy reveals several intermediates involved in PAiRFP photocycles, which all differ from that of the bacteriophytochrome precursor. PAiRFPs can be photoactivated in a spatially selective manner in mouse tissues, and optical modulation of their fluorescence allows for substantial contrast enhancement, making PAiRFPs advantageous over permanently fluorescent probes for in vivo imaging conditions of high autofluorescence and low signal levels.
Bacterial phytochrome photoreceptors (BphPs) are natural red/far-red water-soluble photoreceptors and belong to the phytochrome superfamily, which also include photoreceptors from plants, fungi, and cyanobacteria1–3. Phytochromes share common domain architecture, which consists of a photosensory core domain (PCD) and an output effector module usually represented by histidine kinase. In turn, PCD is formed by three distinct domains, PAS (Per-ARNT-Sim), GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA transcriptional activator) and PHY (phytochrome-specific) connected with α-helix linkers4–6. All chromophore-protein interactions occur in the PCD, which is mainly responsible for the spectral and photochemical properties of phytochromes.
A light-sensing moiety in all phytochromes is comprised of linear tetrapyrrole bilins2. Specifically, BphPs utilize biliverdin IXα (BV) as a chromophore, which is the product of oxidation of heme by heme oxygenase7. BphP apoprotein autocatalytically and covalently attach BV via thioester bond to a cysteine side chain at the N-terminus of the PAS domain2, 8. BphPs are reversibly photoconverted between two stable states, termed Pr and Pfr, respectively by “red” (peaked at 680–710 nm) and “far-red” (peaked at 740–760 nm) light. Upon absorbance of “red” light, Pr state photoconverts into the Pfr state and vice versa, while illumination of the Pfr state with “far-red” light results in the appearance of Pr state. Once generated by light illumination, the photoconverted Pr and Pfr states revert back into their respective ground state either relatively slowly and non-photochemically, by means of the process called dark reversion, or rapidly, upon illumination with “far-red”/”red” light3–5. BphPs adopting Pfr ground state are called bathy BphPs9–11.
The photoconversion of phytochromes is a multistep process typically consisting of an initial photoreaction followed by a number of the dark relaxation steps12, 13. The photoinduced Pr→Pfr and Pfr→Pr conversions were shown to proceed via distinct pathways involving different metastable intermediates (see reviews for details4, 5). The first intermediates formed after Pr and Pfr photoconversion are ascribed as lumi-R and lumi-F, respectively14. After these photoproducts undergo dark relaxation into intermediates designated by the prefix meta. For instance, meta-Ra and meta-Rb are relaxations of lumi-R, whereas meta-Fa and meta-Fb are relaxations of lumi-F (Supplementary Fig. S1)14. Intermediates differ from each other in their absorption spectra and can be stabilized at certain low temperatures in water buffers15–19.
Compared to genetically encoded probes of blue, green, or red fluorescent probes, near-infrared (NIR) fluorescent proteins (FPs) are superior for deep-tissue and whole-body imaging of small mammals20, 21. This is due to low tissue autofluorescence and light scattering and minimal absorbance of melanin, water and hemoglobin in the NIR spectral region of ~650–900 nm, which is termed NIR “optical window” of mammalian tissue22. BphPs possess properties that make them potential templates for engineering NIR FPs. First, BphPs can be readily expressed in mammalian cells and tissues, requiring no enzymes or exogenous cofactors20, 21, 23. Second, BphPs exhibit absorbance and fluorescence spectra, which lie within the NIR “optical window” and are red-shifted relative to other types of phytochromes3 and their fluorescent derivatives24–26. Recently the PAS-GAF domains of several BphPs have been developed into NIR FPs, such as IFP1.4 and iRFP, and applied to whole-body imaging20, 21, 23. To further extend this set of genetically coded fluorescent tools for imaging in NIR “optical window” we designed photoactivatable (PA) NIR FPs using one of the recently proposed strategies of BphP-based NIR FP engineering27.
The capability to control spectral properties of PA FPs with light of specific wavelength and intensity allows for optical labeling and tracking of proteins, organelles, and living cells in a spatiotemporal manner, which is not possible with conventional FPs28. In addition, PA FPs can improve the achievable signal-to-background ratio, thus, allowing higher resolution in samples containing substantial autofluorescence background29. In this paper, we apply molecular evolution to a bathy BphP from Agrobacterium tumefaciens C58, called AtBphP2 (refs.8, 10, 30, 31) and develop two PA NIR FPs, named PAiRFP1 and PAiRFP2. We then study PAiRFPs photoconversion mechanisms using a low temperature UV-visible spectroscopy. Furthermore, we utilize PAiRFPs for in vivo fluorescence imaging and demonstrate spatially selective photoactivation of PAiRFPs in small regions of tumors in living mice. Lastly, an optical modulation of PAiRFPs fluorescent states allows us to substantially enhance the imaging contrast in early stage tumors in vivo, resulting in the signal-to-background ratio values several-fold higher than for permanently fluorescent iRFP.
RESULTS
Development of PAiRFPs
We found that the Pfr state of AtBphP2 protein truncated following PCD (termed AtPCD, amino acids 1–502 of AtBphP2) can be photoconverted into the Pr state using both 660 and 750 nm light. Similar results were reported for the full length AtBphP2 protein10, 32. However, the Pr state of the AtPCD protein is unstable and undergoes rapid dark reversion into Pfr with a half-time of 11 s (Supplementary Fig. S2)33. To stabilize the Pr state and enhance its fluorescence without interfering the Pfr→Pr photoconversion, a gene encoding AtPCD was subjected to several rounds of directed molecular evolution. Each round started from generation of library of mutants using random mutagenesis. The obtained library was co-expressed with heme oxygenase HO-1 in E. coli and screened using a fluorescence activated cell sorter (FACS), followed by selection of the appropriate clones on Petri dishes using a fluorescence stereomicroscope equipped with 660 nm and 750 nm light-emitting diodes (LED) arrays. After the first round we have identified a weakly photoactivatable mutant, called At/660-I#1, having 5 substitutions compared to AtPCD. The At/660-I#1 mutant, initially in the non-fluorescent Pfr state, underwent almost complete photoconversion into the Pr state under both 660 nm and 750 nm illumination and after photoconvertion reverted back into the initial state with a half-time of 4.1 min (Supplementary Table S1 and Fig. S3).
Consequently, At/660-I#1 was used as a template for random mutagenesis, aimed to select PA NIR FPs with increased photoactivation contrast, enhanced brightness, blocked Pr→Pfr photoconversion and slowed or disabled dark reversion. In the subsequent rounds of molecular evolution we used 660 nm photoactivation light as opposed to 750 nm light, for screening of PA NIR FP desirable variants. 660 nm light was advantageous due to the wide availability of light sources emitting at 700 nm and shorter wavelengths such as LEDs and arc lamp sources. The latter are often equipped with 700 nm cut-off filters in a light path which blocks light at 750 nm. Additionally, high power 660 nm light could be used to select clones that do not exhibit Pr→Pfr photoconversion under exposure to ~660 nm lower power probing light, which is used to excite fluorescence of the Pr state. As the result we found two variants with slightly different phenotypes, designated PAiRFP1 and PAiRFP2. PAiRFP1 exhibited higher photoactivation contrast than PAiRFP2 but 4-fold faster dark reversion. To understand an effect of the introduced mutations we characterized spectral and biochemical properties of several mutants found during the molecular evolution of the AtPCD protein (Supplementary Table S1, Figs. S4 and S5). The introduced mutations mainly resulted in the increase of brightness and photoactivation contrast and in the decrease of dark reversion rates.
Characterization of purified PAiRFPs
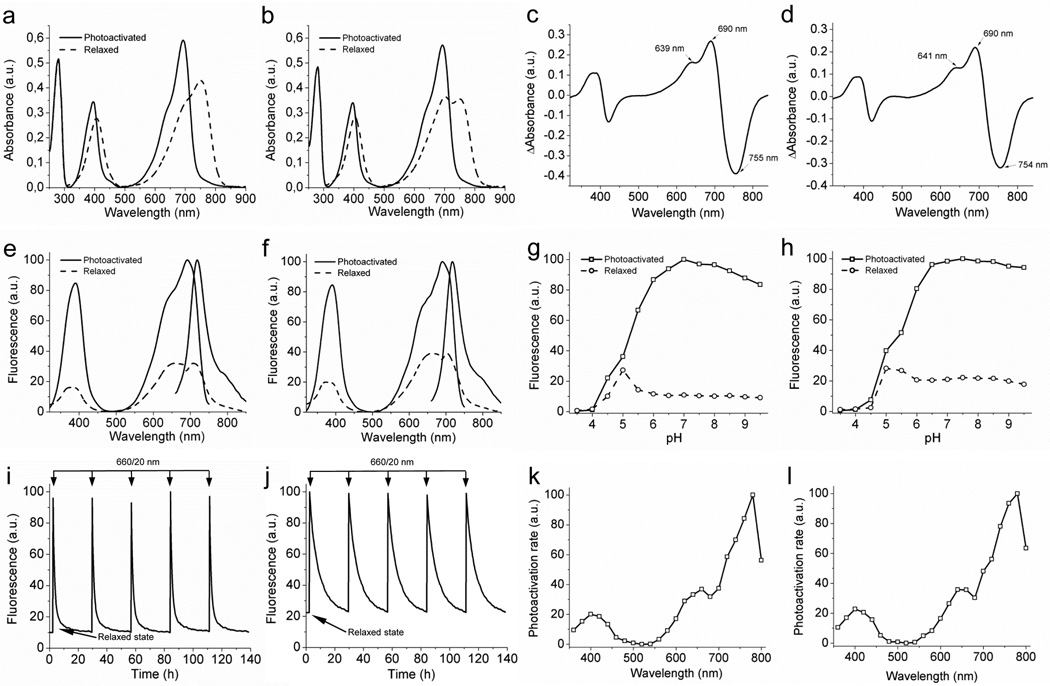
We further compared properties of PAiRFP1 and PAiRFP2 to those of the AtPCD template and common permanently fluorescent iRFP20. Both PAiRFPs could exist in two stable interconvertable states, which we denoted as the relaxed and photoactivated states. Characteristics of PAiRFP1 and PAiRFP are summarized in Table 1 and Fig. 1. The difference absorbance spectra for PAiRFPs demonstrated the characteristic phytochrome signature8, 33–35 with maxima at ~690 nm and ~755 nm for the Pr and Pfr forms, respectively (Fig. 1c,d), similar to those of parental AtPCD33 (Supplementary Fig. S6). After photoactivation, both PAiRFPs slowly reverted back to the relaxed state with the half-times of 58 and 233 min, respectively, enabling multiple photoactivation-relaxation cycles without substantial changes in fluorescence brightness, relaxation rates, and photoactivation contrast (Fig. 1i,j).
Table 1.
Spectral and photochemical characteristics of PAiRFPs in comparison with AtPCD and iRFP.
| Protein | AtPCD | PAiRFP1 | PAiRFP2 | iRFP | |||
|---|---|---|---|---|---|---|---|
| relaxed | photo- converted |
relaxed | photo- activated |
relaxed | photo- activated |
||
| Absorbance maximum, nm | 758 | 699 | 407/751 | 396/693 | 403/702/748 | 396/693 | 692 |
| Extinction coefficient, M−1cm−1 | 27,400 | 23,900 | 48,700 | 67,100 | 39,500 | 63,600 | 85,000 |
| Excitation maximum, nm | none | 703a | 373/659 | 390/690 | 383/657 | 389/692 | 690 |
| Emission maximum, nm | none | 713a | 703 | 717 | 708 | 719 | 713 |
| Quantum yield, % | none | 0.13a | ND | 4.8±0.1 | ND | 4.7±0.1 | 5.9 |
| Relative brightness, % | none | ND | ND | 64 | ND | 60 | 100 |
| Contrast, fold | 1.8b | 9.0 | 5.9 | n.a. | |||
| pKa/pKb | 4.8c | 4.6; 5.6 | 5.2 | 4.7 | 5.4 | 4.0 | |
| Half-time of dark reversion, min | 0.18 | 58 | 233 | n.a. | |||
| Photobleachi ng half-timed, min | n.d. | 2.9 | 3.9 | n.d. | |||
Data from the ref46;
Measured by absorbance changes at 758 nm;
Measured for Pfr by absorbance changes at 758 nm (Supplementary Fig. S7);
Measured in Met-1 mammalian cells stably expressing PAiRFPs variants and normalized to absorbance spectra and extinction coefficients of the proteins, spectrum of a 200 W arc lamp and transmission of a 665/45 nm photobleaching filter. n.a., not applicable. n.d., not determined.
Figure 1. Spectral and biochemical properties of PAiRFP1 and PAiRFP2 in vitro.
Absorbance spectra of the relaxed (dashed line) and photoactivated (solid line) PAiRFP1 (a) and PAiRFP2 (b). Difference spectra “relaxed” minus “photoactivated” for PAiRFP1 (c) and PAiRFP2 (d). Fluorescence spectra of the relaxed (dashed line) and photoactivated (solid line) PAiRFP1 (e) and PAiRFP2 (f). Equilibrium pH dependence for fluorescence of the relaxed (circles) and photoactivated (squares) PAiRFP1 (g) and PAiRFP2 (h). Fluorescence of the relaxed form was normalized to the maximal fluorescence of the photoactivated form. Time courses of the PAiRFP1 (i) and PAiRFP2 (j) fluorescence during multiple cycles of photoactivation and subsequent relaxation in the dark. After purification, the protein was briefly photoactivated with 660 nm light (26 mW/cm2). After the protein reached the relaxed state in the dark, it was briefly photoactivated again with 660 nm light (26 mW/cm2). Fluorescence in the photoactivated state was normalized to 100%. Spectral dependence of the maximal photoactivation rate for PAiRFP1 (k) and PAiRFP2 (l). The maximal values of the photoactivation rates were normalized to the light power in each wavelength band.
Next, photoactivation properties of PAiRFPs were studied in more detail. Both PAiRFPs had a similar spectral dependence of the photoactivation rates with maximal efficiencies of photoactivation observed at 780 nm (Fig. 1k,l). To estimate minimal light intensities required for complete photoactivation of PAiRFPs we used 660 nm light from 12 µW/cm2 to 26 mW/cm2. The complete photoactivation of both PAiRFPs was achieved at the light power of ~2 mW/cm2.
Photoactivation of PAiRFPs at low temperature
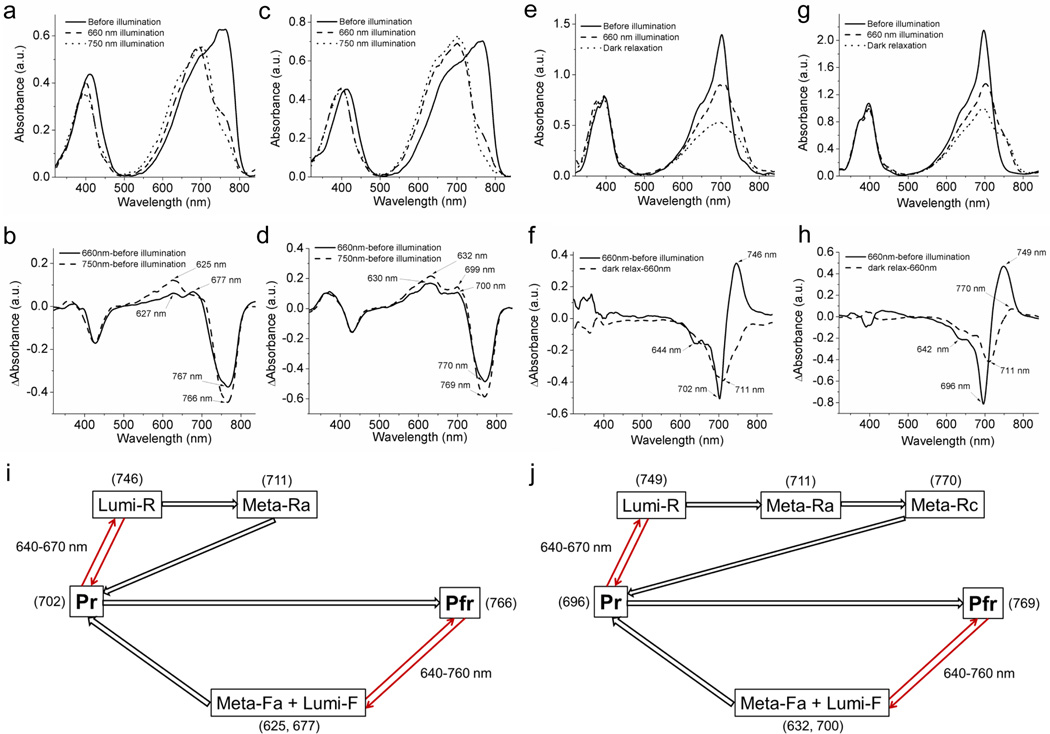
We used low-temperature UV-visible spectroscopy to study PAiRFP photoconversion photoproducts and dark relaxation intermediates photoconversion under 660 and 750 nm light irradiation. After complete relaxation, PAiRFP proteins were flash frozen in liquid nitrogen and subsequently irradiated with 660 nm or 750 nm light. After the brief irradiation (about 10 sec) the absorption spectra displayed significant changes, which possibly indicated accumulation of an intermediate state (Fig. 2a,b). PAiRFP1 absorbance spectra recorded after 660 nm or 750 nm illumination had slightly different profiles and exhibited maxima at 693 and 704 nm (Fig. 2a). The difference PAiRFP1 spectra indicated formation of at least two overlapping absorption bands in the “red” region (Fig. 2b). For PAiRFP2 illumination with 660 or 750 nm light yielded very similar absorbance spectra peaked at 700 nm with the only difference in the “red” shoulder of the 660 nm illuminated sample (Fig. 2c). In the “red” region, the difference PAiRFP2 spectra consisted of at least two overlapping absorption bands as well, with maxima around 630 and 700 nm. Under the same conditions, AtPCD also demonstrated substantial changes in absorbance spectra (Supplementary Fig. S8). It should be noted that the absorbance bands of phytochromes usually shift to longer wavelength upon decreasing the temperature16, 17.
Figure 2. Spectroscopy of intermediate products of PAiRFP1 and PAiRPF2 photoconversion.
(a) Absorbance spectra of relaxed PAiRFP1 measured at 77 K before illumination (solid line), after 10 s of 660 nm illumination (dashed line), and after 10 s of 750 nm illumination (dotted line). (b) Difference spectra for phototransformation of relaxed PAiRFP1 at 77 K: “660 nm illumination” minus “before illumination” (solid line), and “750 nm illumination” minus “before illumination” (dashed line). (c) Absorbance spectra of relaxed PAiRFP2 measured at 77 K before illumination (solid line), after 10 s of 660 nm illumination (dashed line) and after 10 s of 750 nm illumination (dotted line). (d) Difference spectra for phototransformation of relaxed PAiRFP2 at 77 K: “660 nm illumination” minus “before illumination” (solid line), and “750 nm illumination” minus “before illumination” (dashed line). (e) Absorbance spectra of photoactivated PAiRFP1 measured before illumination at 77 K (solid line), and after 10 s of 660 nm illumination at 77 K (dashed line) and subsequent dark relaxation at 245 K (dotted line). (f) Difference spectra for phototransformation of photoactivated PAiRFP1 at low temperature: “660 nm illumination” minus “before illumination” (solid line), and “dark relaxation” minus “660 nm illumination” (dashed line). (g) Absorbance spectra of photoactivated PAiRFP2 measured before illumination at 77 K (solid line), and after 10 s of 660 nm illumination at 77 K (dashed line) and subsequent dark relaxation at 245 K (dotted line). (h) Difference spectra for phototransformation of photoactivated PAiRFP2 at low temperature: “660 nm illumination” minus “before illumination” (solid line) and “dark relaxation” minus “660 nm illumination” (dashed line). (i, j) Schematic representation of the proposed photocylces for PAiRFP1 (i) and PAiRFP2 (j). Values in the brackets denote wavelength of difference spectrum peaks. The indicated absorbance maxima of the intermediates, measured at low temperatures, can be different at room temperature where they likely will shift by 10–15 nm to shorter wavelengths, as was observed for wild-type phytochromes.16, 17 Arrows indicate photoreaction (thin red) or dark relaxation (wide black).
It has been shown that illumination of the Pr state of the wild-type phytochromes at temperatures below 170 K results in the formation of the lumi-R photoproduct, which can further undergo dark relaxation into the meta-Ra and meta-Rc intermediates12–14, 16. Indeed, after brief 660 nm irradiation of flash-frozen photoactivated PAiRFPs at 77 K we observed formation of a pronounced shoulder at ~746–749 nm, which could be ascribed to the lumi-R state (Fig. 2e–h). The subsequent dark relaxation of 660 nm illuminated samples at 245 K resulted in a noticeable decrease of this absorbance band (Fig. 2e,g), which, upon thawing, completely reverted back into the band corresponding to photoactivated PAiRFPs. Overall, these spectroscopic data suggest the following photocycles for PAiRFPs (Fig. 2i,j) (see also Supplementary Note 1).
Behavior of PAiRFPs in cultured cells
To characterize PAiRFPs in mammalian cells we transiently transfected HeLa cells with plasmids encoding non-targeted PAiRFPs. The cells also co-expressed mEGFP for the purpose of normalizing transfection efficiency. No exogenous heme oxygenase was co-expressed. The cells were analyzed using epifluorescence microscopy and flow cytometry (Fig. 3).
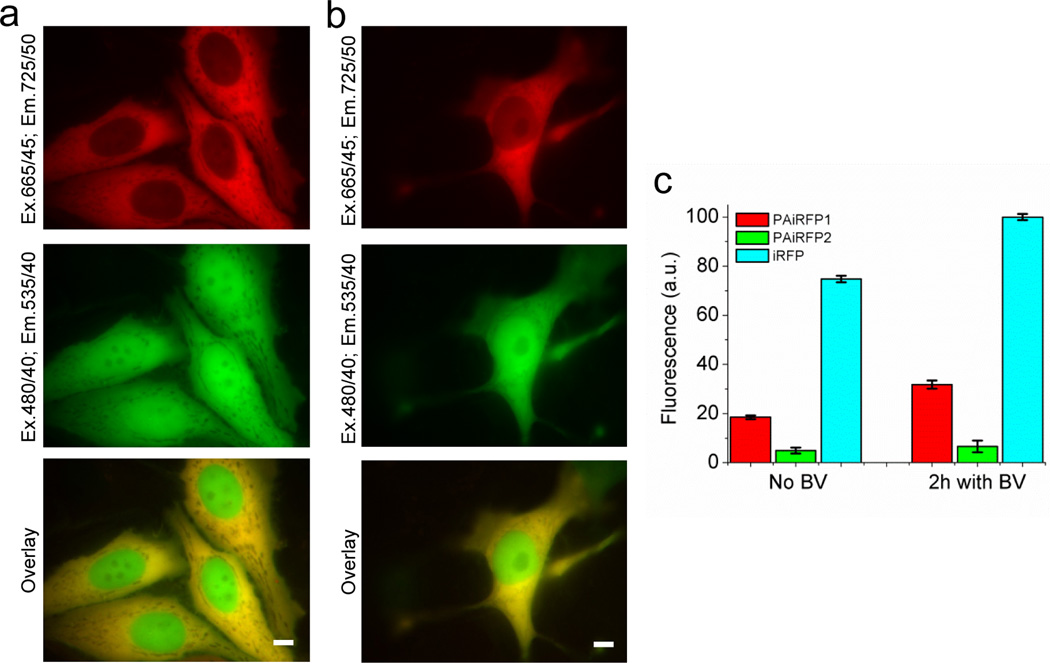
Figure 3. Properties of PAiRFP1 and PAiRFP2 in cultured mammalian cells.
Fluorescence images of transiently transfected HeLa cells co-expressing PAiRFP1 (a) and PAiRFP2 (b) with EGFP. Scale bar, 10 µm. (c) Mean near-infrared fluorescence intensity of the double-positive, transiently transfected HeLa cells normalized to transfection efficiency (EGFP signal), absorbance of the respective protein at 640 nm and overlap of the fluorescence spectrum of the respective protein with the transmission of the emission filter. Near-infrared fluorescence intensities were analyzed using flow cytometer with 640 nm excitation laser. Error bars, s.d. (n = 3).
Fluorescence signal of PAiRFPs was evenly distributed within the cytosol of living HeLa cells without aggregation or nonspecific localization (Fig. 3a,b). The PAiRFP proteins were excluded from the nucleus possibly due to the size (56.6 kDa ×2 for a dimer). The effective brightness of PAiRFP1 and PAiRFP2 in cells was 25% and 7% relative to iRFP, respectively. Treatment with 25 µM of exogenous BV for 2 h increased the brightness of PAiRFP1 cells by 1.7-fold and PAiRFP2 and iRFP cells by 1.3-fold (Fig. 2c). Photobleaching half-times for PAiRFP1 and PAiRFP2 in cells were 2.9 min and 3.9 min, respectively (Table 1). Both PAiRFPs demonstrated irreversible photobleaching under arc lamp illumination on a wide-field microscope (Supplementary Fig. S9). Based on these photobleaching curves, there is no photochromism (i.e., reversible phototransformation between two states with distinct spectral properties) for either PAiRFP.
Imaging of PAiRFPs in live mice
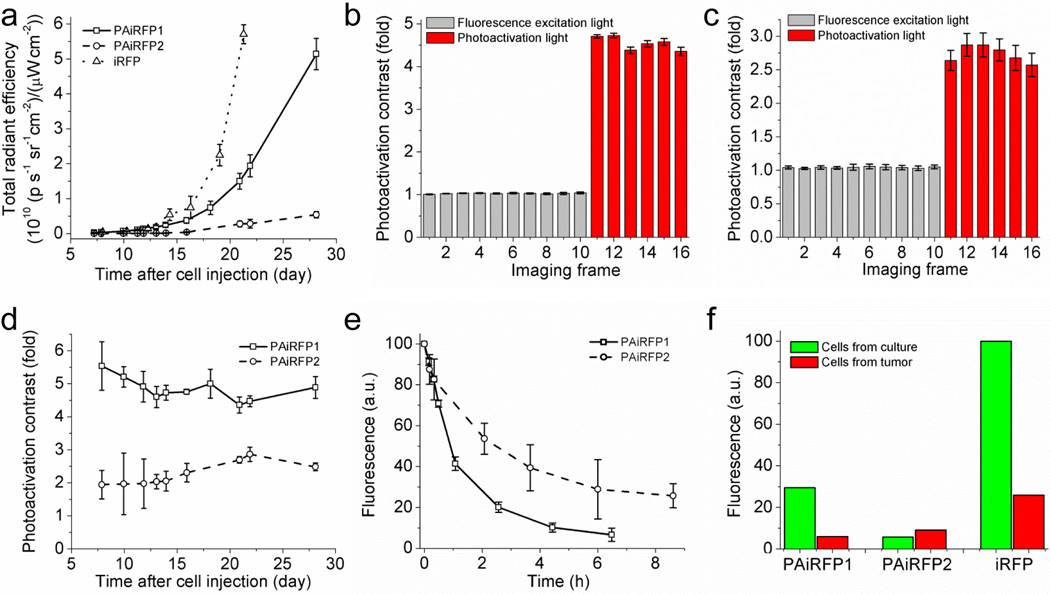
To examine applicability of PAiRFPs for in vivo imaging, we used a xenograft mouse model. Preclonal mixtures of rat adenocarcinoma MTLn3 cells stably expressing PAiRFPs were injected into the mammary glands of immunocompromised mice. MTLn3 cells expressing iRFP were injected into separate animals as a control. After cell transplantation, mammary tumors were allowed to grow over a four week period and imaged using an IVIS Spectrum instrument. Fluorescent signal of both photoactivated PAiRFPs and iRFP were first detected one week after MTLn3 injection using the 675/30 and 720/20 nm excitation/emission filters. Monitoring the fluorescence signals allowed us to plot growth curves for the tumors (Fig. 4a). For the first two weeks after cell injections, fluorescence brightness of the PAiRFP1 and iRFP expressing tumors was similar, where PAiRFP2 brightness was about 4.5% of that of the iRFP tumor. After two weeks, brightness of the PAiRFP1 and PAiRFP2 tumors was approximately 40% and 8% of the iRFP tumor.
Figure 4. Properties of PAiRFP1 and PAiRFP2 in mouse tumor model.
(a) Tumor growth curves plotted based on increase of fluorescence brightness of PAiRFP1 (solid line), PAiRFP2 (dashed line), and iRFP (dotted line). Fluorescence brightness of PAiRFP1 and PAiRFP2 were measured after photoactivation with 660 nm 26 mW/cm2 light. Photoactivation kinetics of the PAiRFP1 (b) and PAiRFP2 (c) expressing tumors in mice. The mice were imaged with 675/30 nm 6.4 µW/cm2 excitation light (2 s exposure time per frame) before (frames 1–10; gray columns) and after repetitive 5 s irradiation with 660 nm 26 mW/cm2 LED photoactivation light per each frame (frames 11–16; red columns). See Supplementary Figs. S10 and S11 for the respective images of mice. (d) Photoactivation contrast of PAiRFP1 (squares) and PAiRFP2 (circles) for different tumor sizes. (e) Kinetics of the dark reversion of PAiRFP1 (squares) and PAiRFP2 (circles) after complete photoactivation. (f) Mean fluorescence intensity of the cells isolated from PAiRFP1, PAiRFP2, and iRFP expressing tumors five weeks after injection, in comparison with the respective preclonal cell mixtures growing in culture. Fluorescence was analyzed using flow cytometer with 640 nm excitation laser. Fluorescence intensities were normalized to absorbance of the respective protein at 640 nm and overlap of the fluorescence spectrum of the respective protein with the transmission of the emission filter. Error bars, s.d. (n = 3).
To establish optimal conditions for complete photoactivation of PAiRFPs in vivo, we measured photoactivation kinetics of a tumor under illumination used for fluorescence read out and light irradiation used for photoactivation. 675/30 nm light with 6.4 µW/cm2 power used for fluorescence excitation in the IVIS Spectrum did not photoactivate PAiRFPs, allowing prolonged imaging of a non-photoactivated tumor (Fig. 4b,c, imaging frames 1 to 10; Supplementary Figs. S10 and S11). Here and below the light powers are indicated on the samples. However, 5 s illumination with 660 nm light of 26 mW/cm2 led to almost complete photoactivation of the whole tumor (Fig. 4b,c, imaging frames 11 to 16). For complete in vivo photoactivation, we further used 10 s illumination with 660 nm light of 26 mW/cm2. The photoactivation contrast of PAiRFPs measured during development of the tumors showed no significant dependence on their size and total brightness (Fig. 4d). The values of photoactivation contrast were in the range of 4.4±0.3 to 5.5±0.7 for PAiRFP1 and 1.9±0.4 to 2.9±0.2 for PAiRFP2. After photoactivation, fluorescence of PAiRFP1 and PAiRFP2 in tumors decreased with the half-times of 55 and 155 min, respectively (Fig. 4e).
Post-mortem flow cytometry analysis of PAiRFP1 and iRFP expressing tumor cells demonstrated a similar ratio between negative and fluorescent cells (~6.1% and ~6.6% positive cells for PAiRFP1 and iRFP, respectively) that resembles the previously reported results for iRFP20. Mean fluorescence intensities of the PAiRFP1 and iRFP positive cells were 20% and 26% relative to respective preclonal MTLn3 mixtures growing in culture (Fig. 4f). In the case of PAiRFP2, the ratio between negative and fluorescent cells isolated from a tumor was 0.72%. Interestingly, mean fluorescence intensity of the PAiRFP2 cells from the tumor was 1.6-fold higher than preclonal mixed MTLn3 cells growing in culture. Overall, in vivo data suggested that PAiRFPs are suitable for long-term expression in mammals.
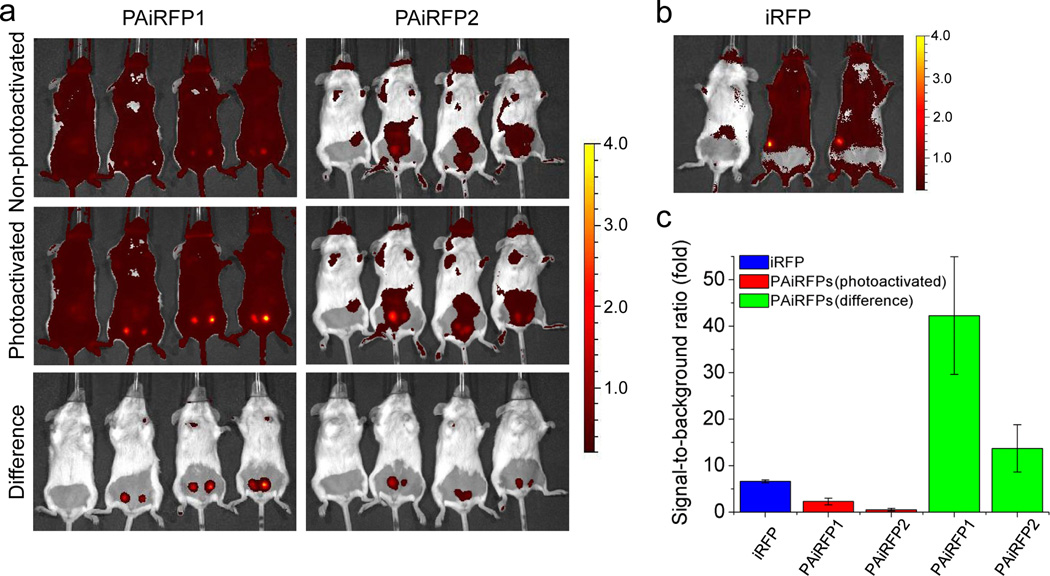
The ability to easily change the PAiRFPs fluorescence signal from the relaxed state to the fluorescent state by photoactivation enables simple image processing, which results in a substantially enhanced fluorescence contrast. To estimate contrast enhancement, we performed in vivo imaging of mice bearing PAiRFPs and control iRFP-expressing tumors at early stages of approximately 10 days after the MTLn3 cells injection. Subtracting images captured before photoactivation from the images captured after photoactivation resulted in the 18-fold and 26-fold increase in the signal-to-background ratios for the PAiRFP1 and PAiRFP2 tumors, respectively, relative to the respective unprocessed images (Fig. 5a). Similar procedure is not possible for permanently fluorescent iRFP because its fluorescence signal remains constant (Fig. 5b). Therefore, the signal-to-background ratio values for PAiRFP1 and PAiRFP2 images after this contrast enhancement procedure were 6.5-fold and 2.1-fold higher than that for iRFP (Fig. 5c).
Figure 5. Contrast enhancement of in vivo imaging using PAiRFP1 and PAiRFP2.
(a) Overlay of the representative light and fluorescent images of a control mouse (left on each image) and the mice bearing a 10 day old PAiRFP1 expressing tumor (left column) and a 13 day old PAiRFP2 expressing tumor (right column) before (upper row) and after photoactivation (middle row), Overlay of the representative light and fluorescent images obtained by subtracting of the “non-photoactivated” image from the “photoactivated” are shown in the lower row. The color bar indicates the fluorescence radiant efficiency, multiplied by 108. (b) Overlay of the representative light and fluorescent images of a control mice (left) and the mice bearing a 10 day old iRFP expressing tumor. The color bar indicates the fluorescence radiant efficiency, multiplied by 108. (c) The signal-to-background ratio values calculated for the respective images in (a) and (b) panels. Error bars, s.d. (n = 6).
Selective photoactivation of PAiRFPs in vivo
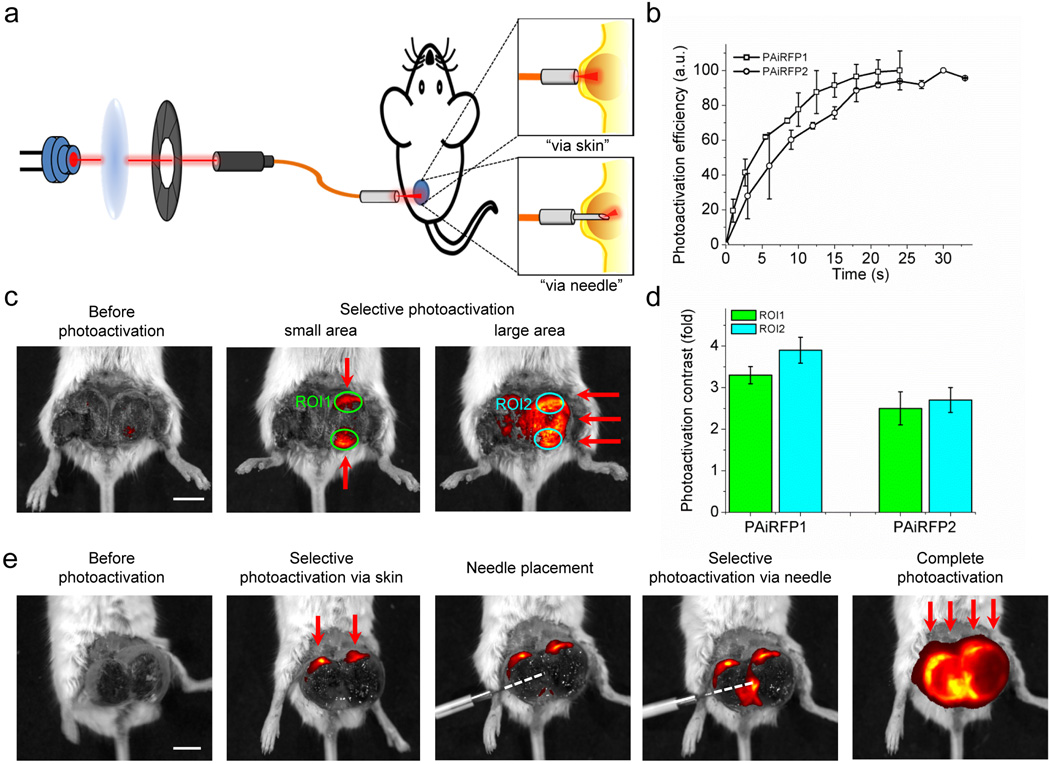
Low light intensity required to photoactivate PAiRFPs prompted us to perform spatially selective photoactivation within an organ in a mouse. We built a setup capable of delivering a focused beam of light of specific power to small areas of a tumor. The setup consisted of a 660 nm laser diode, a focusing lens, a mechanical shutter, a patch cable and a fiber optic cannula. We tested two methods of the selective photoactivation, dubbed “via skin” and “via needle” (Fig. 6a).
Figure 6. Selective photoactivation of PAiRFP1 and PAiRFP2 in mouse tumor model.
(a) Scheme of an optical device consisting of a 660 nm laser diode, a focusing lens, a mechanical shutter and a patch cable and a fiber optic cannula. Device is used for “via skin” and “via needle” selective photoactivation of the PAiRFP1 and PAiRFP2 expressing tumors in living mice. (b) Kinetics of via skin selective photoactivation of the PAiRFP1 (square) and PAiRFP2 (circle) expressing tumors with 660 nm laser beam of 1.5 mW total power. Photoactivation efficiency was calculated as (ROIt−ROI0)/ROImax ×100, where ROIt, ROI0 and ROImax were total radiant efficiencies of the respective areas after t seconds of illumination, before illumination and after complete photoactivation, respectively. (c) Overlay of the representative light and fluorescent images of a mouse bearing a four-week old MTLn3 tumor expressing PAiRFP1 before photoactivation (left image), after selective photoactivation of two small areas (middle image) and after complete photoactivation of the whole tumor (right image). Red arrows indicate areas where the light beam was applied. ROI, region of interest. (d) Average photoactivation contrast of the PAiRFP1 and PAiRFP2 expressing tumors in ROI1 and ROI2, represented on Figure 4c. Average photoactivation contrast values were obtained from 9 and 4 ROIs of PAiRFP1 and PAiRFP2 tumors, respectively. (e) Overlay of the representative light and fluorescent images of a mouse bearing a five-week old MTLn3 tumor expressing PAiRFP2 before photoactivation (first image), after selective via skin photoactivation of two small areas (second image), with subsequent placement of the needle into the tumor (third image), selective photoactivation via needle (fourth image) and complete photoactivation of the tumors (fifth image). Red arrows indicate areas photoactivated via skin. White dashed line indicates path of the needle inside of the tumor. Error bars, s.d. (n = 3). Scale bars, 1 cm.
For determining optimal photoactivation conditions, we initially measured kinetics of the “via skin” photoactivation, which were used for all further experiments (Fig. 6b). Total light power at the exit of the patch cable was ~1.5 mW. Half-times of selective photoactivation of PAiRFP1 and PAiRFP2 in tumors were 4.1±0.4 s and 7±2 s, respectively. We photoactivated two spots within the same tumor with an average photoactivation contrast in the selected regions of interest (ROIs) of 3.3±0.2 and 2.5±0.4 fold for PAiRFP1 and PAiRFP2 expressing tumors respectively (ROI1 in Fig. 6c,d). After photoactivation of the whole tumor, average contrast in the selected ROIs was 3.9±0.3 and 2.7±0.3 fold for PAiRFP1 and PAiRFP2, respectively (ROI2 in Fig. 6c,d).
For “via needle” photoactivation, a light fiber was protected with a syringe needle for its insertion into a tissue. To test this method of photoactivation, two spots in the PAiRFP2 expressing tumor were photoactivated via skin, as described above. Subsequently, the needle with the optical fiber was inserted into the middle of the tumor and illumination was applied for 5–6 s (Fig. 6e). Total light power at the exit of the needle was ~0.5–1.0 mW. The fiber optic cannula with the needle inserted in the tissue can be easily disconnected from the optical setup and the animal can be imaged. After imaging, the cannula is connected back to the device allowing further illumination of the same spot to achieve a desirable level of photoactivation. Combining the “via skin” and “via needle” light irradiation techniques allowed rapid and selective photoactivation of several spots in the tumor tissue.
Discussion
We employed molecular evolution of a bathy BphP named AtBphP2 for the purpose of developing dark-to-NIR PA FPs. As a result, we have found two variants, named PAiRFP1 and PAiRFP2, with distinctive biochemical characteristics (Table 1 and Fig. 1). We have introduced 15 and 24 amino acid substitutions into AtPCD to engineer PAiRFP1 and PAiRPF2 respectively. Spectral and photochemical properties of light-active proteins are mainly determined by interactions between the chromophore and its immediate environment. We proposed the possible effects of the newly introduced amino acid residues in close proximity to the chromophore in PAiRFPs through analysis of the spectral and biochemical properties of the AtPCD variants as well as mutagenesis and structural data available for other BphPs. We aligned the amino acid sequence of the AtBphP2 PCD with the bathy BphP PaBphP sequence from Pseudomonas aeruginosa34, which share 37% sequence identity (Supplementary Fig. S12), to suggest the possible location of the introduced mutations in the PAiRFPs tertiary structures. Next, the available crystal structures of PaBphP-PCD in its dark-adapted Pfr state and the Pr state were used to locate the residue of interest11, 36. Analysis of the alignment revealed that 12 out of 15 substitutions of PAiRPF1 reside in the GAF domain with the remaining 3 in the PHY domain. PAiRFP2 contains 3 mutations in the PAS domain, 11 in the GAF domain, and 10 in the PHY domain. In addition, according to the alignment, only 3 mutations (M163L, V244F, A276V) of PAiRFP1 and 4 mutations (M163L, S243N, V244F, A276V) of PAiRFP2 reside in the chromophore binding pocket (Supplementary Fig. S12). In Supplementary Note 1 we proposed possible effects of the M163L, V244F, and A276V mutations on the PAiRFPs photophysical and photochemical properties based on the properties of others bacteriophytochromes, such as DrBphP, PaBphP, and their mutants, and mutants generated during molecular evolution of AtPCD. To determine the roles of all other introduced mutations, further structural, spectroscopic and mutational analysis of PAiRFPs is required. Overall, we conclude that the novel mutations in the PAiRFPs are mainly responsible for (i) altering sensitivity to “red” versus “far-red” light, (ii) disabling the Pr→Pfr photoconversion, (iii) decreasing dark reversion rates, (iv) increasing the extinction coefficient and quantum yield of the Pr state, and (v) increasing the photoactivation contrast. The introduced mutations did not cause substantial shift of the absorbance and fluorescence spectra: parental AtPCD and its variants exhibit the similar absorbance and fluorescence maxima (Supplementary Table S1).
In the relaxed and photoactivated states, PAiRFP1 has absorbance maxima at 756 nm and 692 nm, respectively, hence the relaxed and photoactivated states of PAiRFP1 correspond to the Pfr and Pr states, respectively. In the relaxed state, PAiRFPs exhibit residual fluorescence, which is blue-shifted relative to the photoactivated state (Table 1). This fluorescence can arise from a state other than Pr, which probably exists in a thermal equilibrium with the Pfr state. Interestingly, the excitation maximum of this state corresponds to the 660 nm local maximum of the spectral dependence of photoactivation for PAiRFPs (Table 1 and Fig. 1e,f,k.l). Possibly, illumination of this state with 660 nm light can result in the formation of Pr and is an intermediate in the Pfr→Pr photoconversion pathway.
We utilized low temperature and UV-visible spectroscopy to study mechanism of PAiRFPs photoconversion. To identify the intermediates involved in the PAiRFPs photocycle, we compared the calculated differences spectra for PAiRFPs photoconversion (Fig. 2) with the ones available for wild-type phytochromes14–18 (see Supplementary Note 2). These suggest that photoconversion of PAiRFPs proceeds via intermediates similar to that of wild-type phytochromes which, we believe, are involved in the alternation of BV conformations and re-armament of protein moieties around BV (Fig. 2i,j). Thus, photoactivation of PAiRFPs does not seem to involve chemical modifications of the BV chromophore except its protonation-deprotonation.
Although PAiRFPs in mammalian cells are less bright than iRFP, their brightness is sufficient for imaging cells in culture and in live mice. The noticeable difference in PAiRFPs brightness in vitro and in mammalian cells can be possible explained by a lower affinity to BV and/or instability of PAiRFP holoproteins, as it has been suggested for IFP1.4 (ref21). However, imaging with PAiRFPs does not require BV administration, as endogenous BV in mammalian cells is sufficient for formation of the fluorescent apoprotein. Using the xenograft tumor mouse model we showed applicability of PAiRFPs for whole-body imaging. Tumor growth dynamics and post-mortem analysis of tumor cells expressing PAiRFPs suggest that the proteins are suitable for long-term expression in mammals. Additionally, photoactivation of PAiRFPs is rapid, easy and allows simultaneous tissue photolabeling in several mice using an LED array. The described “via skin” and “via needle” photoactivation setups allow precise and gradual photoactivation of a selected region of interest. Photoactivation contrast does not depend on the size and brightness of the expressing tissue, making PAiRFPs reliable probes for semi-quantitative in vivo imaging.
The ability to change the PAiRFPs fluorescence signal by photoactivation has allowed to substantially increase the signal-to-background ratio values of the in vivo images, thus resulting in an order of magnitude higher fluorescence contrast (Fig. 5). In spite of the PAiRFP2 expressing tumor was about 10-fold less bright after photoactivation than the iRFP expressing tumor, the processed signal-to-background ratio for PAiRFP2 was more than twice higher than for iRFP. The possibility to subtract the PAiRFPs images before and after photoactivation makes these proteins advantageous over permanently fluorescent NIR FPs for imaging in vivo when there are typically high autofluorescence levels. This approach is particularly important for imaging of small organs and tissues such as early tumors, which provide low fluorescence signals due to the small number of fluorescent cells. Potentially, multiple photoactivation – dark relaxation cycles of PAiRFPs may be useful to allow unambiguous assignment of the signals to PAiRFPs and to improve spatial resolution of in vivo imaging.
PAiRFPs extend the set of available and widely used photoactivatable FPs derived from GFP-like proteins28, 37. However, PAiRFPs possess several features distinct of the GFP-like PAFPs. First, both excitation and emission fluorescence spectra of PAiRFPs lie within the NIR “optical window” and are more than 50 nm red-shifted relative to the most red-shifted photoswitchable GFP-like protein, PSmOrange38. Second, PAiRFPs are photoactivated by NIR light, whereas the most of the GFP-like PAFPs are activated by UV-violet light, with the exception of a few reversibly photoswitchable RFPs, which are photoconverted by orange light39. Third, the light power required for PAiRFP photoactivation is orders of magnitude lower than that required for GFP-like PAFP photoactivation (Supplementary Table S2). For comparison, the photoconvertion energy density required for the PSmOrange38 and PATagRFP40 photoconvertion is ~5,000-fold higher than that for PAiRFPs. The latter can be explained by the distinct molecular mechanisms of chromophore transformations involved in photoactivation. Phototransformation of phytochromes and their derivatives only involves isomerization and protonation-deprotonation of the chromophore4, 5, whereas photoconversion of GFP-like FPs involve chemical modification of the chromophores, which requires higher activation energy37, 41. PAiRFPs can be completely photoactivated in vivo within seconds whereas in vivo photoactivation of PSmOrange takes more than a half of hour38. Lastly, in vivo photoactivation of PAiRFPs does not require sophisticated equipment, in contrast to, for example, Dendra PAFP photoactivation42. These features make PAiRFPs advantageous over the GFP-like PAFPs for in vivo photolabeling and imaging cells, tissues, and whole mammals.
METHODS
Mutagenesis and screening of the libraries
An AtBphP2 gene was kindly provided by Eric Giraud. A PCR-amplified BglII-AsuII fragment encoding AtPCD (first 502 amino acids of the AtBphP2 protein with the insertion of the MAGLEYK sequence on the C-terminus) was cloned into the pBAD/His-B vector (Invitrogen). Random mutagenesis was performed with GeneMorph II Random Mutagenesis Kit (Stratagene) using conditions resulting in a mutation frequency up to 16 mutations per 1,000 base pairs. After mutagenesis, a mixture of the mutants was electroporated into LMG194 host cells (Invitrogen) containing the pWA23h plasmid, which facilitates biliverdin synthesis. The pWA23h plasmid contained the rhamnose promoter from the pWA21 plasmid43, Kan resistance, and COLA origin parts from a pCOLADuet-1 plasmid (Novagen). First, the AvrII/PciI fragment containing Kan resistance and COLA origin was PCR amplified 14 from pCOLADuet-1 plasmid and inserted into a pWA21h-AvrIINotI vector. Then, a hmuO gene encoding Bradyrhizobium ORS278 heme oxygenase was PCR amplified from pBAD/HisB-RpBphP2-hmuO plasmid44 and swapped with the EGFP gene in the pWA21-AvrIINotI plasmid.
Typical mutant libraries consisted of about 106–108 independent clones. The LMG194 cells were grown at 37 °C in RM minimal medium supplemented with ampicillin, kanamycin and 100 µM of 5-aminolevulinic acid (ALA). Protein expression in the libraries was induced with 0.002% arabinose and 0.02% rhamnose. The cells were grown up to 18 h at 37°C. Prior to FACS screening, bacterial cells were washed with phosphate buffered saline (PBS) containing 1 mM EDTA and then diluted with PBS to optical density of 0.02 at 600 nm. Cell suspensions were activated using a 750 nm 60 mW/cm2 LED array (20 nm of emission half-width and 1.7 cm diameter of light emitting zone (Marubeni)) or a 660 nm 26 mW/cm2 LED array (20 nm of emission half-width and 17 cm diameter of light emitting zone (90 W UFO LED, Grow Light)). The library photoactivation consisted of ~5 min illumination of the bacterial suspension mixed on ice. After photoactivation the libraries were screened using MoFlo XDP (Beckman Coulter) fluorescence activated cell sorting (FACS) equipped with Argon, Krypton and Argon-Krypton mixed-gas lasers. Typically about 10 sizes of each library were sorted using 647 nm excitation line and 680 nm LP emission filter (Chroma). The brightest collected near-infared bacterial cells were recovered in SOC medium at 37°C for one hour, then grown on LB/ampicillin/kanamycin Petri dishes supplemented with 0.02% arabinose, 0.2% rhamnose, 100 µM ALA and 50 µM FeCl3 overnight at 37°C followed by incubation at 4°C. The dishes were analyzed with Leica MZ16F fluorescence stereomicroscope equipped with the 650/45 nm excitation and 690LP nm emission filters (Chroma) and a CCD camera before and after photoactivation with 750 or 660 nm light for 5–30 s. Approximately 20–50 of the brightest near-infrared clones having the highest contrast after photoactivation, with minimal rate of dark reversion were selected and their DNA was sequenced. A mixture of several selected mutants was then used as a template for the next round of mutagenesis.
Protein characterization in vitro
The AtBphP2 mutants and iRFP with polyhistidine tags on the N-terminus were expressed in LMG194 bacterial cells grown in RM medium supplemented with ampicillin, kanamycin, 0.002% arabinose, 0.02% rhamnose and 100 µM ALA for 15–18 h at 37°C and then for 24 h at 18°C. Proteins were purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s protocol with minor modification. In wash buffer, 400 mM imidazole was substituted with 100 mM EDTA. For spectroscopy, photoactivation of purified proteins was performed with 750 nm or 660 nm light in 50 µl quartz microcuvette (Starna Cells) at room temperature in PBS. Low temperature UV-visible absorption spectra were measured in PBS. The fluorescence spectra and spectral dependence of the maximal photoactivation rate were measured using a FluoroMax-3 spectrofluorometer (Jobin Yvon). Fluorescence spectra of the relaxed PAiRFPs were measured using plate reader SpectraMax-M2 (Molecular Devices). For absorbance measurements a Hitachi U-2000 spectrophotometer was used. Background light scattering was removed by subtracting a fitted λ4 curve from the measured spectrum.
To determine extinction coefficients, we used a direct measurement of protein concentrations with a BCA protein assay kit (Pierce), followed by the calculation of extinction coefficients using a Beer-Lambert-Bouguer equation. For determination of quantum yield, fluorescence signal of purified proteins was compared to that of the equally absorbing iRFP20. pH titrations were done using a series of buffers (100 mM sodium acetate, 300 mM NaCl for pH 2.5–5.0 and 100 mM NaH2PO4, 300 mM NaCl for pH 4.5–9.0).
Mammalian plasmids and cell culture
To construct pPAiRFP1-N1 and pPAiRFP2-N1 plasmids, the respective genes were PCR amplified as BglII-NotI fragments and swapped with the EGFP gene in pEGFP-N1 (Clontech). The pDualCMV-PAiRFP1-EGFP, pDualCMV-PAiRFP2-EGFP and pDualCMV-iRFP-EGFP plasmids for co-expression non-targeted PAiRFPs and iRFP with EGFP were constructed by insertion of the AflII-AflII fragment containing CMV promoter and EGFP gene into the pPAiRFPs-N1 and piRFP-N1 vectors, digested by AflII and dephosphorylated.
Met-1 and HeLa cell lines were grown in DMEM containing 10% FBS, penicillin-streptomycin and 2 mM glutamine (Invitrogen). Cells were cultured in 35 mm glass bottom culture dishes with no. 1 cover glasses (MatTek). MTLn3 rat adenocarcinoma cells were cultured in αMEM (Life Technologies) with 5% FBS and 0.5% penicillin-streptomycin (Life Technologies). Plasmid transfections were performed using an Effectene reagent (Qiagen) according to the manufacturer's protocol. Establishing cell lines and orthotopic injection in mammary glands were performed as previously described45.
Imaging of HeLa cells was performed 48 h after transfection. HeLa cells were imaged using an Olympus IX81 inverted epifluorescence microscope equipped with a 200 W metal-halide arc lamp (Lumen220Pro, standardly equipped with 800 nm cold mirror; Prior), a 60× 1.35 NA oil immersion objective lens (UPlanSApo, Olympus), and standard Cy5.5 filter set (665/45 nm exciter and 725/50 nm emitter) (Chroma).
Photobleaching experiments were performed in Met-1 mammalian cells stably expressing PAiRFPs variants using an Olympus IX81 inverted epifluorescence microscope equipped with the 200 W metal-halide arc lamp (Prior), a 60× 1.35 NA oil immersion objective lens (UPlanSApo, Olympus), and standard Cy5.5 filter set (Chroma). Raw data were normalized to the absorbance spectra and extinction coefficients of the proteins, the spectrum of the 200 W arc lamp and transmission of the 665/45 nm photobleaching filter.
In vivo fluorescence imaging
One million MTLn3 cells stably expressing PAiRFP1, PAiRFP2 or iRFP alone were injected into the mammary gland of SCID/NCr mice (female, 5–7 weeks old) (Taconic), and imaged starting 1 week later using an IVIS Spectrum instrument (PerkinElmer) in epifluorescence mode. The IVIS Spectrum was equipped with the 675/30 nm and 720/20 nm excitation and emission filters for PAiRFPs and iRFP imaging. Belly fur was removed using a depilatory cream. Mice were fed with AIN-93M Maintenance Purified Diet (TestDiet) to reduce intrinsic autofluorescence level. Before imaging, mice were kept in the darkness for 8–10 h. Complete photoactivation of tumors was performed with the 660 nm 26 mW/cm2 LED array (90 W UFO LED, Grow Light). The LED array allowed for simultaneous, evenly distributed illumination of up to 5 mice on the IVIS Spectrum stage. For spatially selective photoactivation the optical setup shown in Fig. 5a was used. The 660 nm laser diode (120 mW), fiber optic cannula (200 and 400 µm core), and optogenetic patch cable (200 and 400 µm core, 50 cm long) were from Thorlabs. Total light power used for “via skin” and “via needle” photoactivation was ~1.5 mW and ~0.5–1.0 mW, respectively.
The PAiRFPs and iRFP expressing MTLn3 tumors were excised postmortem, chopped into pieces, washed with PBS supplemented with 2% of bovine serum albumin, and subsequently filtered through sieves and a 35 µm filter. FACS analysis was perform with the LSRII cytometer (BD Biosciences) equipped with a 640 nm laser and a 730/45 nm emission filter.
All animal experiments were performed in an AAALAC approved facility using protocols approved by the Albert Einstein College of Medicine Animal Usage Committee.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Giraud for the AtBphP2 gene, J. Zhang and L. Tesfa for the assistance with flow cytometry, Y. Wang and L.-M. Ting for the help with mouse experiments, and G. Filonov for the useful discussions. This work was supported by the grants GM073913, CA164468, and EB013571 from the National Institutes of Health.
Footnotes
Accession codes. Nucleotide sequences for PAiRFP1 and PAiRFP2 have been deposited in NCBI GenBank Nucleotide database under accession codes KF006822 and KF006823, respectively.
AUTHOR CONTRIBUTIONS
K.D.P. developed the proteins and together with F.V.S. characterized them in vitro. K.D.P. characterized the proteins in mammalian cells and in mice. V.V.V. designed the overall project and together with K.D.P. planned experiments, discussed data, and wrote the manuscript.
COMPETING INTEREST STATEMENT
There are no competing financial interests.
References
- 1.van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, "star actors of modern times": a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004;37:13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- 2.Rockwell NC, Lagarias JC. A brief history of phytochromes. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auldridge ME, Forest KT. Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol. 2011;46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 4.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annual review of plant biology. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulijasz AT, Vierstra RD. Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr Opin Plant Biol. 2011;14:498–506. doi: 10.1016/j.pbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Nagatani A. Phytochrome: structural basis for its functions. Curr Opin Plant Biol. 2010;13:565–570. doi: 10.1016/j.pbi.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Dammeyer T, Frankenberg-Dinkel N. Function and distribution of bilin biosynthesis enzymes in photosynthetic organisms. Photochemical & photobiological sciences : Official journal of the European Photochemistry Association and the European Society for Photobiology. 2008;7:1121–1130. doi: 10.1039/b807209b. [DOI] [PubMed] [Google Scholar]
- 8.Lamparter T, Michael N, Mittmann F, Esteban B. Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11628–11633. doi: 10.1073/pnas.152263999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rottwinkel G, Oberpichler I, Lamparter T. Bathy phytochromes in rhizobial soil bacteria. J. Bacteriol. 2010;192:5124–5133. doi: 10.1128/JB.00672-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karniol B, Vierstra RD. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2807–2812. doi: 10.1073/pnas.0437914100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendrick RE, Spruit CJ. Phototransformations of phytochrome. Photochemistry and photobiology. 1977;26:201–214. doi: 10.1111/j.1751-1097.1977.tb07473.x. [DOI] [PubMed] [Google Scholar]
- 13.Rüdiger W, Thümmler F. Phytochrome, the Visual Pigment of Plants. Angewandte Chemie International Edition in English. 1991;30:1216–1228. [Google Scholar]
- 14.Spruit CJP, Kendrick RE, Cooke RJ. Phytochrome intermediates in freeze-dried tissue. Planta. 1975;127:121–132. doi: 10.1007/BF00388373. [DOI] [PubMed] [Google Scholar]
- 15.Borucki B, et al. Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore. The Journal of biological chemistry. 2005;280:34358–34364. doi: 10.1074/jbc.M505493200. [DOI] [PubMed] [Google Scholar]
- 16.Rudiger W, Thummler F. Low temperature spectroscopy of phytochrome: Pr, Pfr and intermediates. Physiologia Plantarum. 1984;60:383–388. [Google Scholar]
- 17.Song P-S, Sarkar HK, Kim I-S, Poff KL. Primary photoprocesses of undegraded phytochrome excited with red and blue light at 77 K. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1981;635:369–382. doi: 10.1016/0005-2728(81)90035-9. [DOI] [PubMed] [Google Scholar]
- 18.Spruit CJP, Kendrick RE. Phototransformation of phytocrome: the characterization of lumi-F and meta-Fa. Photochemistry and photobiology. 1977;26:133–138. doi: 10.1111/j.1751-1097.1977.tb07464.x. [DOI] [PubMed] [Google Scholar]
- 19.Piwowarski P, et al. Light-induced activation of bacterial phytochrome Agp1 monitored by static and time-resolved FTIR spectroscopy. Chemphyschem. 2010;11:1207–1214. doi: 10.1002/cphc.200901008. [DOI] [PubMed] [Google Scholar]
- 20.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konig K. Multiphoton microscopy in life sciences. Journal of microscopy. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 23.Filonov GS, et al. Deep-Tissue Photoacoustic Tomography of a Genetically Encoded Near-Infrared Fluorescent Probe. Angew. Chem. Int. Ed. Engl. 2011;51:1448–1451. doi: 10.1002/anie.201107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer AJ, et al. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry. 2005;44:15203–15215. doi: 10.1021/bi051633z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, et al. Fused-gene approach to photoswitchable and fluorescent biliproteins. Angew Chem Int Ed Engl. 2010;49:5456–5458. doi: 10.1002/anie.201001094. [DOI] [PubMed] [Google Scholar]
- 26.Su YS, Lagarias JC. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chemical Society reviews. 2013;42:3441–3452. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Curr Opin Cell Biol. 2011;23:310–317. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Y, Marriott ME, Petchprayoon C, Marriott G. Optical switch probes and optical lock-in detection (OLID) imaging microscopy: high-contrast fluorescence imaging within living systems. The Biochemical journal. 2011;433:411–422. doi: 10.1042/BJ20100992. [DOI] [PubMed] [Google Scholar]
- 30.Goodner B, et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 31.Wood DW, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 32.Inomata K, et al. Assembly of Agrobacterium phytochromes Agp1 and Agp2 with doubly locked bilin chromophores. Biochemistry. 2009;48:2817–2827. doi: 10.1021/bi802334u. [DOI] [PubMed] [Google Scholar]
- 33.Krieger A, Molina I, Oberpichler I, Michael N, Lamparter T. Spectral properties of phytochrome Agp2 from Agrobacterium tumefaciens are specifically modified by a compound of the cell extract. Journal of photochemistry and photobiology. B, Biology. 2008;93:16–22. doi: 10.1016/j.jphotobiol.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Tasler R, Moises T, Frankenberg-Dinkel N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. The FEBS journal. 2005;272:1927–1936. doi: 10.1111/j.1742-4658.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 35.Wagner JR, et al. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. The Journal of biological chemistry. 2008;283:12212–12226. doi: 10.1074/jbc.M709355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Kuk J, Moffat K. Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15639–15644. doi: 10.1073/pnas.0902178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piatkevich KD, Verkhusha VV. Advances in engineering of fluorescent proteins and photoactivatable proteins with red emission. Curr. Opin. Chem. Biol. 2010;14:23–29. doi: 10.1016/j.cbpa.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subach OM, et al. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nature methods. 2011;8:771–777. doi: 10.1038/nmeth.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subach FV, et al. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem Biol. 2010;17:745–755. doi: 10.1016/j.chembiol.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. Journal of the American Chemical Society. 2010;132:6481–6491. doi: 10.1021/ja100906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subach FV, Piatkevich KD, Verkhusha VV. Directed molecular evolution to design advanced red fluorescent proteins. Nature methods. 2011;8:1019–1026. doi: 10.1038/nmeth.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kedrin D, et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nature methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wegerer A, Sun T, Altenbuchner J. Optimization of an E. coli L-rhamnose-inducible expression vector: test of various genetic module combinations. BMC biotechnology. 2008;8:2. doi: 10.1186/1472-6750-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giraud E, et al. A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. The Journal of biological chemistry. 2005;280:32389–32397. doi: 10.1074/jbc.M506890200. [DOI] [PubMed] [Google Scholar]
- 45.Piatkevich KD, et al. Monomeric red fluorescent proteins with a large Stokes shift. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5369–5374. doi: 10.1073/pnas.0914365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zienicke B, et al. Fluorescence of phytochrome adducts with synthetic locked chromophores. The Journal of biological chemistry. 2011;286:1103–1113. doi: 10.1074/jbc.M110.155143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.