Abstract
Lung cancer is the most lethal carcinoma worldwide. Mutations of p53, inactivation of p16INK4a, and overexpression of cyclins E, A and B are independently associated with poor prognoses of patients, while the prognostic value of cyclin D1 or RB expression is inconclusive. Cyclin D binding myb-like protein 1 (Dmp1) encodes a DNA binding protein that receives signals from oncogenic Ras and functions as a tumor suppressor by activating the Arf-p53 pathway. Dmp1 has been shown to be haplo-insufficient for tumor suppression in mouse models including K-ras-mediated lung carcinogenesis. The human DMP1 gene is located on chromosome 7q21, and our recent results revealed that the hDMP1 gene is deleted, but not mutated or silenced, in approximately 40 % of human non-small-cell lung carcinomas. These cases typically retained wild-type ARF and p53 and expressed very low levels of the hDMP1 protein. Thus, hDMP1 loss could be a novel diagnostic marker for non-small-cell lung carcinomas.
Keywords: ARF, DMP1l, haploid insufficiency, immunohistochemistryl, LOHl, loss of heterozygosity, lung cancer, p16INK4a, p53, Ras, tumor-suppressor gene
Lung cancer is the second most common human malignancy regardless of ethnic origin or sex [1]. In the USA, there are approximately 215,000 new patients and 162,000 deaths per year due to lung cancer, accounting for approximately 30% of total cancer deaths [1]. Novel anticancer therapies including novel cytotoxic agents and molecular-targeted reagents are developed each year, but the prognosis for lung cancer patients is still extremely poor, with overall 5-year survival of approximately 15% [1–4]. Lung cancer is categorized into two major histopathological groups: non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC). Approximately 80% of human lung cancers are NSCLC and they are further classified into adenocarcinomas, squamous cell carcinomas and large-cell carcinomas [3]. NSCLC and SCLC show striking differences in histopathologic characteristics that can be explained by the differential patterns of genetic alterations found in both tumor types [3,5–8]. The diagnostic and prognostic values of molecules that are involved in normal and malignant cell cycles have been extensively studied for human lung cancer in the past 20 years. In this review, we will briefly discuss the diagnostic values of known markers for cell cycle regulators in human NSCLC and then we will focus on the roles of Dmp1 in lung carcinogenesis and its possible diagnostic value.
Physiological cell cycle regulators
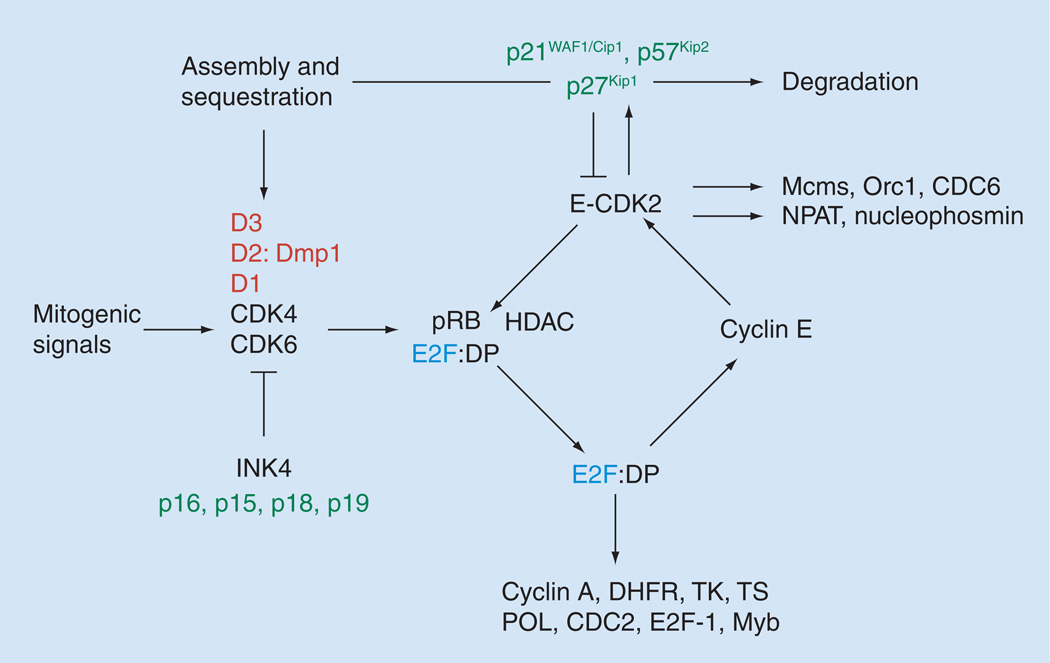
In nontransformed cells, cell cycle division is regulated in an ordered, securely regulated process involving multiple checkpoints that respond to extracellular growth signals, cell size and DNA integrity [9–12]. The replication of DNA occurs in the S phase and segregation of the chromosomes into daughter progeny occurs in the M phase (mitosis). There are two ‘gap’ phases in the mammalian cell cycle, named G1 and G2. During the G1 phase, cells prepare for DNA synthesis and, during G2, cells prepare for mitosis [9–12]. Cyclin/CDK complexes are formed during distinct phases of the cell cycle and are specifically involved in the phosphorylation of target proteins, including pocket proteins (RB, p107 and p130) (Figure 1). Mammalian G1 cyclins D and E mediate progression through the G1/S phases. Three D-type cyclins exist (cyclin D1, D2 and D3), which are expressed differently in various cell lineages, with most cells expressing cyclin D3 and either D1 or D2 (Figure 1). E-type cyclins (cyclins E1 and E2) are expressed during late G1 to the end of S phase of the cell cycle. The activity of cyclin E plays critical roles in the passage of cells through the restriction point, which marks an irreversible point for cells to complete the rest of the cell division cycle. Expression of cyclin E is regulated at the level of gene transcription mainly by E2F proteins and by its degradation via the proteasome pathway. Cyclin E binds and activates the kinase CDK2 to phosphorylate pocket proteins and initiate a cascade of events that leads to the expression of S phase-specific genes (Figure 1) [9–13]. Aside from this specific function as a regulator of S phase entry, cyclin E plays distinct roles in the initiation of DNA replication, the control of genomic stability and the duplication of the centrosome. Mitotic cyclins A and B mediate progression through the S/G2 to M phases. Cyclin A2 is expressed in proliferating somatic cells, while cyclin A1 is specifically detected in the testis and early embryogenesis. Cyclin B1 plays general roles in M phase progression, while cyclin B2 has a special function in Golgi remodeling during mitosis. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero.
Figure 1. Restriction point control and the G1–S transition.
D-type cyclins are induced as delayed early responses to mitogenic signals. Among the three D-cyclins, only cyclin D1 is Ras-responsive. Dmp1 is a novel transcription factor that was isolated in a yeast two-hybrid screen with cyclin D2 as bait. The cyclin D/CDK 4/6; p21CIP1/p27KIP1 complexes assemble, sequestering CIP/KIP proteins from cyclin E–CDK2. Cyclin D- and E- dependent kinases phosphorylate the HDACs and the RB, resulting in release and activation of E2F transcription factors, which are necessary for the transcription of genes required for S phase progression. The targets of E2Fs include DHFR, TK, TS, POL, CDC2, E2F1, cyclin E and cyclin A, creating a positive feedback loop at the G1–S boundary. This will eventually cause irreversible restriction point transition to the S phase. Cyclin E–CDK2 opposes the inhibitory effect of p27KIP1 by phosphorylating it. This allows cyclin A–CDK2 and cyclin E–CDK2 to start S phase. E2Fs also target c-Myb, B-Myb (activation) and Dmp1 (repression). Other cyclin E–CDK substrates include Mcms, Orc1 and CDC6, all of which assemble into pre-initiation complexes to start DNA replication. Cyclin E-CDK2 also phosphorylates p220NPAT and nucleophosmin. As cells age, p16INK4a is induced, which inhibits CDK 4/6, causing release and degradation of D-type cyclins and redistribution of p21CIP1/p27KIP proteins to cyclin E–CDK2.
DHFR: Dihydrofolate reductase; Dmp1: Cyclin D-binding myb-like protein 1; DP: Dimerization partner, E2F dimerization partner; CDC: Cell division cycle; CDK: Cyclin-dependent kinase; HDAC: Histone deacetylase; Mcm: Minichromosome maintenance; NPAT: Nuclear protein, ataxia-telangiectasia locus; Orc1: Origin recognition complex 1; POL: DNA polymerase α; RB: Retinoblastoma protein; TK: Thymidylate kinase; TS: Thymidine synthase.
The product of the retinoblastoma susceptibility gene (RB), plays a central role in the G1–S transition (Figure 1) [11,12]. In its unphosphorylated state, RB prevents progression from G1 to S phase by binding the key transcription factor, E2Fs1–3/DP-1 [11–13]. Once the RB protein is phosphorylated by the cyclin D/Cdk complex, E2F is released, thus allowing transcription of a battery of genes that regulate DNA synthesis. The p107/p130 proteins are required for the repression of distinct sets of genes, potentially due to their selective interactions with E2F4 and E2F5 that are engaged at specific promoter elements [13]. In addition to the regulation of E2F–responsive genes, pocket proteins contribute to silencing of genes in cells that are undergoing senescence or terminal differentiation. Pocket proteins also affect the G1–S transition through E2F–independent mechanisms, such as by inhibiting CDK2 or stabilizing p27KIP1 and these mechanisms have been implicated in the control of G0 exit, DNA replication and genomic re-replication [11–13].
The CIP/KIP (p21CIP1, p27KIP1 and p57KIP2) and INK4 families (p16INK4a, p15INK4b, p18INK4c and p19INK4d) represent two distinct families of CDK inhibitors that share no primary sequence similarity in spite of their binding to common targets, CDK4 and CDK6 (Figure 1) [14,15]. The binding mode and CDK specificity are different between these two families of inhibitors. While p21CIP1, p27KIP1 and p57KIP2 bind to and form ternary complexes with cyclin D/CDK4 or CDK6, cyclin E/CDK2, cyclin A/CDK2, cyclinA/CDC2 and cyclin B/CDC2, the INK4 proteins bind exclusively to, and form tight binary complexes with, CDK4 and CDK6. Moreover, the expression pattern of each CDK inhibitor gene is differentially regulated by distinct antiproliferative signals and does not appear to be coordinated in most cases. For instance, p53 directly binds and activates the p21CIP1 promoter while pRB represses p16INK4a transcription [15]. TGF-β treatment stimulates the transcription of p15INK4b, but not p16INK4a or p14ARF although these three genes are located on the same genomic locus 9p21 in humans [15]. The transcription of p18INK4c or p19INK4d is not affected by these antiproliferative stimuli. These distinct transcriptional regulations in response to different antiproliferative signals together with their tissue-and developmental stage-specific expression patterns, established the concept that different CDK inhibitors are regulated by different growth inhibitory pathways, as in the case of sequential cyclin expression and CDK activation. Therefore, alterations in any one of these cell cycle regulatory proteins could lead to failure of cell cycle arrest, which will eventually contribute to neoplastic transformation of cells.
Prognostic values of the retinoblastoma susceptibility gene in human NSCLC
Inactivation of RB (by truncation, gene deletion, nonsense mutation or splicing alterations), together with loss of the wild-type RB allele, have been demonstrated in lung cancers, with protein abnormalities detected in approximately 90% of SCLC and 15–30% of NSCLC [16,17]. Whether the absence of RB expression is associated with poor prognosis in NSCLC is controversial. A study conducted by immunohistochemical detection of pRB in more than 100 patients with stage I and II NSCLC showed that the median survival was 32 months for patients with RB-positive tumors and 18 months for individuals in whom expression of RB protein was absent or altered [18]. However, later studies failed to show an independent prognostic value of RB status in NSCLC [19,20]. Nonetheless, it was reported that pRB; p53 combined status was a predictive factor of overall survival [18,21]. Patients with pRB(−); p53(+) tumors had a median survival of only 12 months, whereas those with pRB(+); p53(−) tumors had a median survival of over 40 months [18,21]. Zagorvski et al. studied the roles of RB loss in tumorigenic proliferation and sensitivity to chemotherapeutics in NSCLC cells [22]. Downregulation of RB by shRNA led to a proliferative advantage in vitro and aggressive tumorigenic growth in xenograft models with increased chemosensitivity. However, this response was transient and a durable response was dependent on prolonged chemotherapeutic administration [22]. They concluded that although RB loss enhances sensitivity of NSCLC cells to chemotherapeutic agents, efficient and sustainable response was highly dependent on the specific therapeutic regimen in addition to the molecular environment [22]. So far, no correlation between the RB status and patients’ survival has been reported in SCLC, possibly because there are very few patients with SCLC with intact RB [17,23].
Impact of cyclins & CDK inhibitors in NSCLC
Upregulation of the cyclin D1 proto-oncogene is known to play key roles in G1–S progression of the cell cycle as described earlier. An increase in this gene’s expression permits loss of G1 restriction point integrity. The impact of cyclin D1 overexpression in NSCLC is again a topic of debate [24,25]. Of the four main prognostic studies of cyclin D1 in NSCLC, two of them showed improved survival, whereas the other two showed shorter survival. In a study with 106 patients with stages I and II of NSCLC, cyclin D1 expression was associated with shorter survival and the cumulative survival rate of cyclin D1(+), p16INK4a(−) patients was significantly lower than that of cyclin D1(−), p16INK4a(+) patients (logrank test, p = 0.0004; Wilcoxon test, p = 0.0002) [24]. In contrast to cyclin D1, over-expression of cyclin E, cyclin A or cyclin B has been reproducibly associated with shorter survival among stage I–IIIA NSCLC patients undergoing curative surgical resection [25].
The prognostic value of expression of CDK inhibitor has also been examined. In two studies that adequately controlled for disease stage, p21CIP1 expression was associated with improved survival [25]. Studies evaluating the effect of p27KIP1 expression have also demonstrated a favorable effect on lung cancer survival in NSCLC with p27KIP1 expression [25]. Among the four INK4 family proteins, the impact of lung cancer patients’ survival has been studied exclusively on p16INK4a. The absence of p16INK4a protein expression as detected by immunohistochemistry or Western blotting has reproducibly shown shorter survival, although two of seven studies did not reach statistically significant differences [25]. Additionally, Kratzke et al. reported an inverse correlation between pRB and p16INK4a expression in 65% of NSCLC cases (p = 0.00019) [26]. The observation that lack of p16INK4a expression was associated with a worse prognosis was consistent with the increased incidence of p16INK4a mutations observed in metastatic NSCLC. Other studies have also reported p16INK4a mutations with advanced stage (stage III and IV) in NSCLC [27]. The frequency of deletions of the p15INK4b gene was 12% (four of 34 cases) and no point mutations in the p15INK4b gene were detected in the NSCLC [28]. For the p18INK4c gene, no abnormality was detected in human NSCLC [28]. Alterations of p19INK4d or p57KIP2 have not been reported in human lung cancer. In summary, loss of expression of the inhibitors p16INK4a, p27KIP1 and p21CIP1 and/or overexpression of the cyclins A, E and B1 predict a poor prognosis of NSCLC patients after surgery [25]. Conversely, the impact of the expression of pRB and cyclin D1 on patients’ survival has not been determined in human NSCLC.
Involvement of the ARF–p53 pathway in NSCLC
The p53 tumor-suppressor gene has been reported to be mutated in approximately 50% of all human cancers. p53 responds to a variety of stress signaling including DNA damage, overexpression of oncoproteins and metabolic limitations to regulate a battery of target genes that induce cell cycle arrest, apoptosis, DNA repair and metabolism [29]. The importance of p53 mutations in the pathogenesis of human lung carcinoma is very well established. Since wild-type p53 has a very short half-life (10–20 min), it is usually undetectable by standard immunostaining of normal tissues. By contrast, most mutant p53 proteins have prolonged half-lives, thus allowing visualization of the protein by immunohistochemistry. The significance of the p53 protein expression on the prognosis of NSCLC patients has been extensively studied by many different groups [27,30–39]. Approximately half of the studies found an increased risk for shorter survival with p53 expression, while high p53 expression had no effect, or was associated with favorable disease outcome, in the other half of studies. This controversy is, at least in part, due to the methodological differences in the detection of p53 proteins in lung cancer (i.e., differences in the antibodies or protocols for immunohistochemistry and/or in different criteria for the grading of p53-positive signals in tissues).
In good contrast to the controversial studies with p53 protein expression, genetic analyses of p53 have consistently demonstrated that NSCLC with mutated p53 had an adverse effect on the survival of patients with NSCLC [27,38–50]. Most genetic analyses have been conducted by single-strand conformation polymorphism for screening followed by nucleotide sequencing or p53 GeneChip® assay [42]. One report demonstrated that p53 mutations at exons 7 and 8 were the most predictive for poor clinical outcome [40], while another group reported that p53 mutations in exon 5 were associated with poor prognosis of NSCLC patients [49]. Although the results were different depending on the patient population and the methods they used, p53 mutations detected by molecular genetic analyses are generally a more reliable predictor of poor outcome than p53 protein overexpression in patients with stage I–IIIA NSCLC.
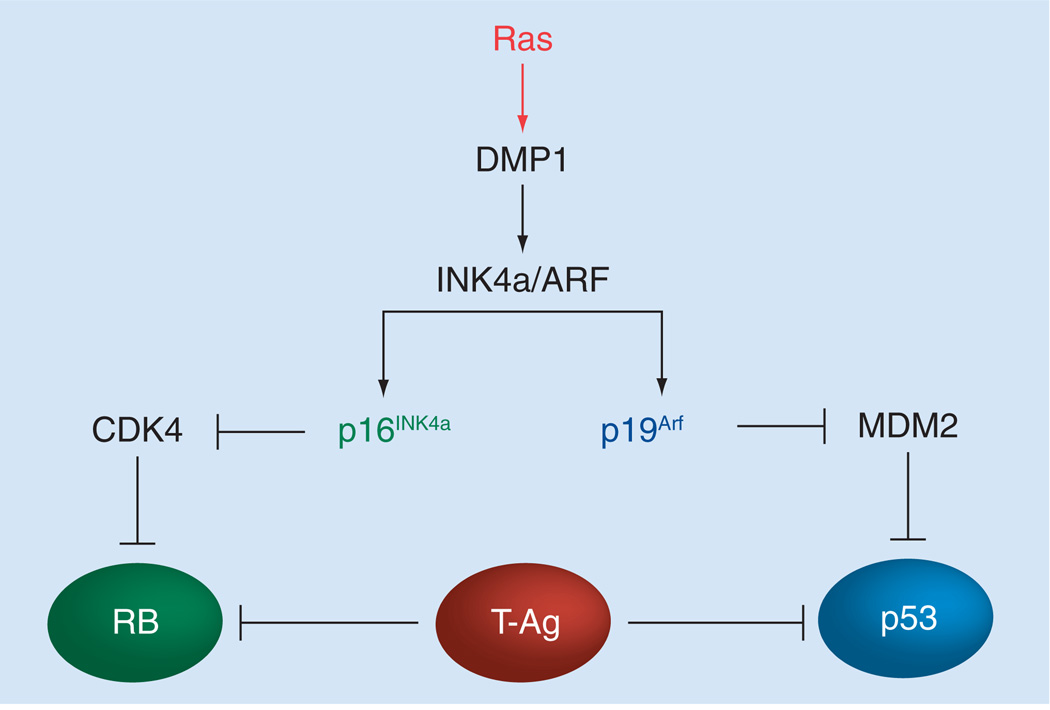
The activity of p53 is positively regulated by p14ARF (p19Arf in mice) in response to oncogenic stress (Figures 2 & 3) [51–53]. p14ARF is an alternative reading frame gene product generated from the INK4a/ARF locus which also encodes the cyclin-dependent kinase inhibitor p16INK4a [54]. p14ARF directly binds to Hdm2, thereby stabilizing and activating p53, whereas p16INK4a binds to cyclin-dependent kinase 4 to inhibit Rb phosphorylation (Figure 2) [51–55]. Since this single genetic locus encodes two independent tumor-suppressor proteins that regulate the p53 and the RB pathways, it is very frequently (~40%) disrupted in human cancer [56]. The ARF induction by potentially harmful growth-promoting signals forces early-stage cancer cells to undergo p53-dependent and p53-independent cell cycle arrest or apoptosis, providing a powerful mode of tumor suppression [51–53]. The Arf promoter is activated by latent oncogenic signals in vivo [57] and thus Arf-null mice are highly prone to spontaneous tumor development [58]. p19Arf (or p14ARF) interacts with nucleophosmin, E2F1, DP1 and numerous other proteins, showing the p53-independent functions of Arf [53]. In human lung cancers, p14ARF is more frequently inactivated in SCLC (~65%) than in NSCLC (~20%) [6]. Promoter hypermethylation of ARF has been reported in approximately 10% of NSCLC, but is much less frequent than that of p16INK4a (~40%) on the same locus [6]. Point mutations for ARF are very rare in human NSCLC.
Figure 2. Regulation of the RB and p53 pathways by proteins encoded from the INK4a/ARF locus and DMP1.
The INK4a/ARF locus located on human chromosome 9p21 encodes two tumor-suppressor genes, namely p16INK4a and p14ARF. p16INK4a binds to cyclin-dependent kinase 4 to inhibit RB phosphorylation, while p14ARF (p19Arf in mice) directly binds to Hdm2 (Mdm2 in mice), thereby stabilizing and activating p53 [51–55]. Since the single genetic locus encodes two independent tumor-suppressor proteins that regulate the RB and the p53 pathways, the locus is very frequently disrupted in human cancer [56]. Dmp1 directly binds to the Arf promoter and activates its gene expression. Since high-affinity Dmp1-binding sites are also found in the genomic locus between exon 1β and exon 1α, Dmp1 may stimulate p16INK4a transcription. SV40 T antigen binds to both RB and p53 to neutralize their tumor-suppressor activity.
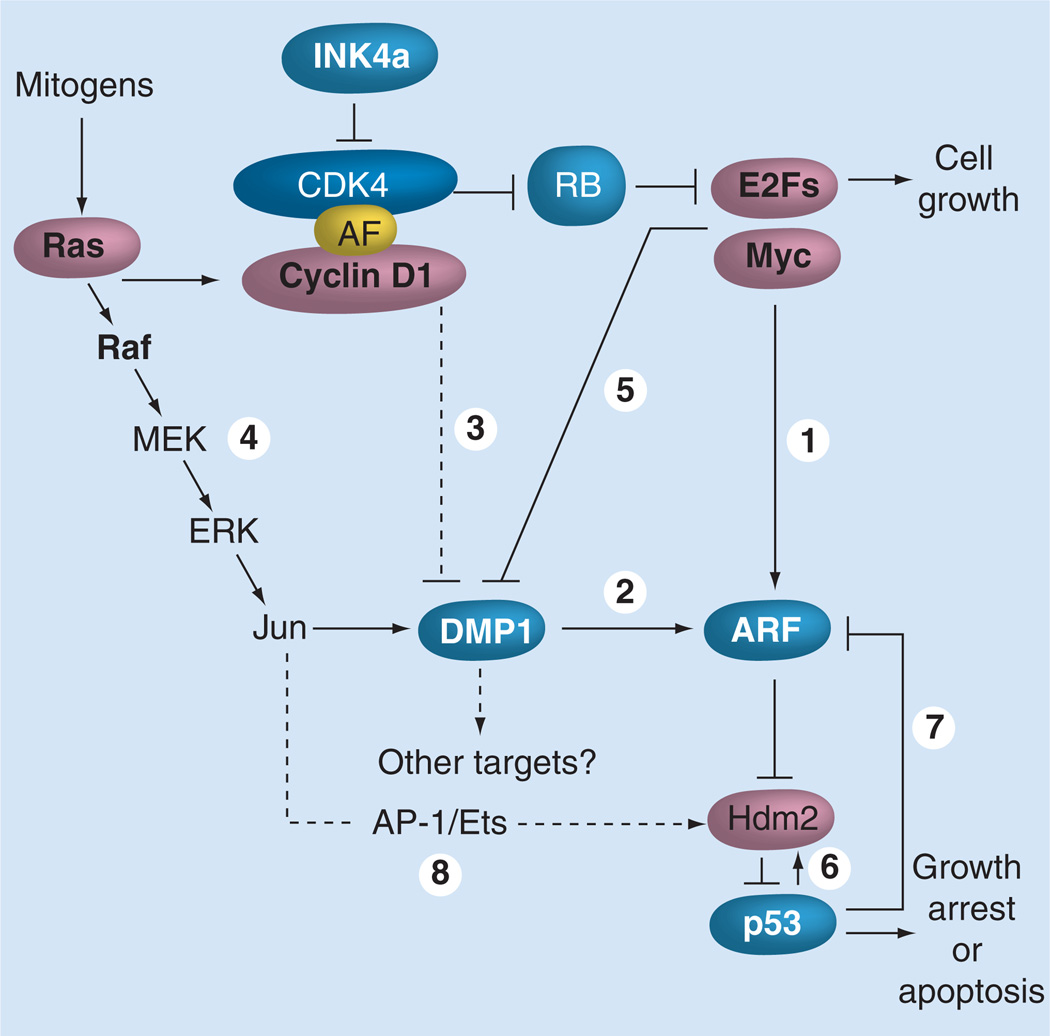
Figure 3. Signaling pathways involving the Dmp1 transcription factor.
p19Arf is induced by potentially oncogenic signals stemming from overexpression of oncogenes such as c-Myc, E2F1 and activated Ras (1). This Arf induction quenches inappropriate mitogenic signaling by diverting incipient cancer cells to undergo p53-dependent and -independent growth arrest or cell death. Dmp1 is unique in that it directly binds and activates the Arf promoter and induces cell cycle arrest in an Arf-dependent fashion (2). Both Dmp1-null and heterozygous mice show hypersensitivity to develop tumors in response to carcinogen DMBA and γ-irradiation. This phenotype could be explained by the inactivation of the Arf–Mdm2–p53 pathway in the absence of the functional Dmp1 protein, although it is possible that Dmp1 has other targets than Arf. D-type cyclins inhibit Dmp1’s transcriptional activity in a CDK-independent fashion when E2Fs do not bind to the same promoter; however, D-cyclins cooperate with Dmp1 to activate the Arf promoter (3). The Dmp1 promoter is efficiently activated by the oncogenic Ras–Raf–MEK–ERK–Jun pathway (4), but is repressed by overexpressed c-Myc, E2Fs and by physiological mitogenic signaling (5) [72,73]. Our study shows that the induction of Arf by oncogenic Ras is dependent on Dmp1 [72]. The Dmp1–Arf pathway is inhibited by NF-κB proteins in response to genotoxic stress signaling. Both Mdm2 and Hdm2 are direct targets of p53 (6), and both human and murine Arf promoters are repressed by p53 (7) [59–61]. It was reported that the Hdm2 promoter is also responsive to oncogenic Ras signaling (8), which can antagonize the Arf induction by the Dmp1 pathway. However, the Arf induction by Ras eventually overrides the Mdm2 activity in normal cells, which undergo p53-dependent cell cycle arrest. AF: Assembly factor.
The prognostic value of p14ARF has rarely been studied in human NSCLC. Wang et al. made a striking discovery that overexpression of p53 is associated with low expression of Hdm2 (p < 0.001) and high expression of p14ARF (p = 0.001) [62]. The overexpressed p53 proteins detected in their study were considered to be mutant p53 since wild-type p53 increases the Hdm2 levels by transactivation of the Hdm2 (and also Mdm2) promoter and repression of the p14ARF promoter (Figure 3) [59–61]. Both overexpression of p53 and absence of Hdm2 expression were associated with squamous cell carcinoma, advanced stages and shorter survival of NSCLC patients (all p < 0.05), suggesting that disruption of the ARF–Hdm2–p53 pathway is important in the pathogenesis and outcome of NSCLC [62].
Novel transcription factor Dmp1 is a regulator of the ARF–p53 pathway
Among known Arf activators, cyclin D-binding myb-like protein-1 (Dmp1), also called cyclin D-binding myb-like transcription factor 1 (Dmtf1), is a unique tumor suppressor [63–71]. Dmp1 was originally isolated in a yeast two-hybrid screen of a murine T-lymphocyte library with cyclin D2 as bait (Figure 1) [63]. Importantly, Dmp1 directly binds to the Arf promoter to activate its expression, thereby inducing p53-dependent cell cycle arrest (Figures 2 & 3) [64,65]. Dmp1 also binds to and activates the CD13/aminopeptidase N promoter through interaction with the c-Myb protein, suggesting its role in hematopoietic cell differentiation [66]. Dmp1-null mice are prone to spontaneous tumor development, which was accelerated when the animals were neonatally treated with ionizing radiation or dimethylbenzanthracene [67,68]. Although Dmp1-knockout mice develop a broad spectrum of epithelial and non-epithelial tumors, lung adenomas/adenocarcinomas were the most frequently found tumors in Dmp1-null and Dmp1-heterozygous mice (Figures 4A & 4B). The wild-type Dmp1 allele is very often retained and expressed in tumors arising from Dmp1+/− mice, demonstrating a typical haplo-insufficiency for tumor suppression, although the molecular mechanisms are not clear [68,69]. Tumors from Eμ-Myc; Dmp1−/− or Dmp1+/− mice rarely show mutations, deletions, or silencing of p19Arf or p53, suggesting that Dmp1 is a critical regulator of the ARF–p53 tumor-suppressor pathway in living animals [68,69]. We have recently characterized the Dmp1 promoter [70–74]. The Dmp1 promoter is activated by the oncogenic Ras–Raf–MEK–ERK–Jun pathway. It is well known that continuous oncogenic Ras activation upregulates p19Arf and induces p53-dependent cell cycle arrest. Our results demonstrated that the induction of Arf by mutant Ras was Dmp1-dependent (Figures 2 & 3) [72]. On the other hand, the Dmp1 promoter is repressed by overexpression of E2Fs and also by physiological mitogenic signaling [73]. Thus, Dmp1 is a marker of cells that have exited from the cell cycle [73]. Our most recent study shows that the Dmp1 promoter is repressed by genotoxic stimuli (daunomycin, doxorubicin or UVC) that activate NF-κB through phosphorylation of the p65 subunit, and that the repression of the Arf promoter by genotoxic stress was Dmp1-dependent [74]. Thus, Dmp1 is a sensor to convey some forms of oncogenic and nononcogenic stress to the ARF–p53 pathway (Figure 3).
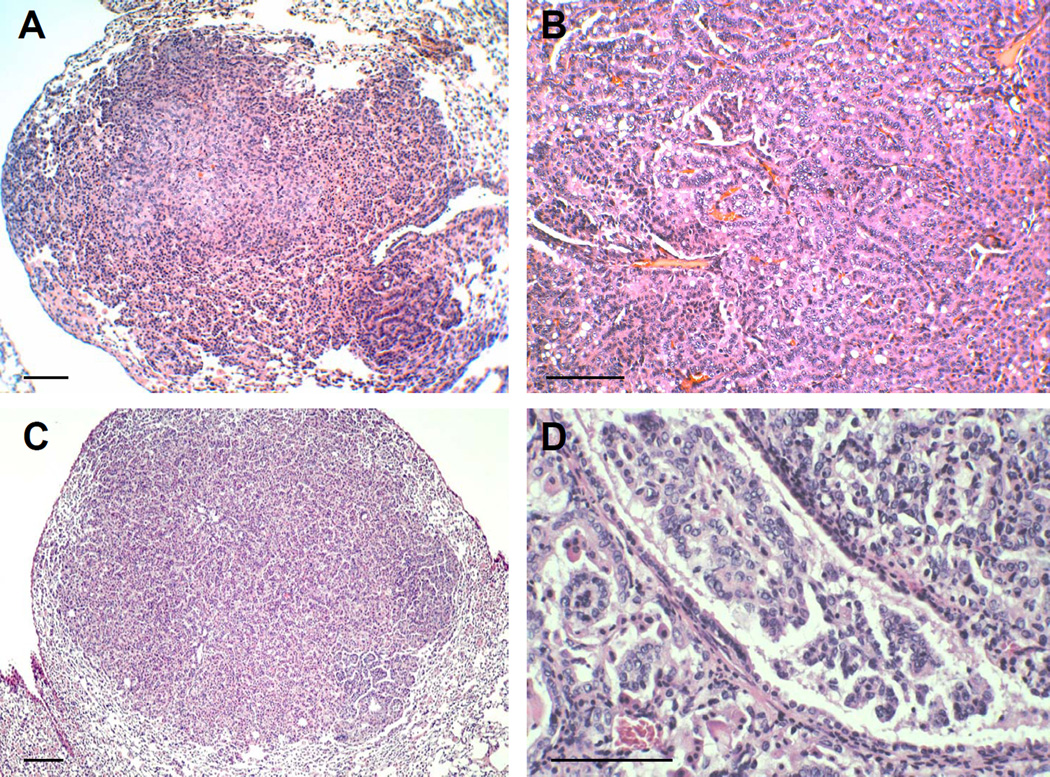
Figure 4. Lung tumors found in Dmp1 deficient mice.
(A) Spontaneous lung adenoma found in an untreated Dmp1-null mouse (60-weeks old). (B) Lung adenocarcinoma found in a DMBA-treated Dmp1-null mouse (40-weeks old). (C) Lung adenoma observed in a wild-type K-rasLA1/+ mouse (50-weeks old). (D) Invasive lung adenocarcinoma found in a Dmp1+/−; K-rasLA1 mouse (35-weeks old). The scale bar in A–D is 100 µM.
Roles of Dmp1 in K-rasLA medicated lung cancer development
Dmp1-null mice were crossed with K-rasLA mice to demonstrate the interactions between Dmp1-loss and oncogenic K-ras activation in vivo [75]. K-rasLA1/+ and K-rasLA2/+ are unique mouse models of lung cancer where the K-ras gene is controlled by its own promoter and is activated during spontaneous recombination events in the whole animal [76]. We found that the survival of K-rasLA mice was significantly shortened in both Dmp1+/− and Dmp1−/− mice, with little difference between the two cohorts [75]. The lung tumor cells from Dmp1+/−, K-rasLA mice expressed Dmp1 mRNA and protein in most cases, clearly demonstrating the haploid-insufficiency of Dmp1 in lung cancer suppression in these mice models. However, K-rasLA lung tumors are different from Eμ-Myc lymphomas because bi-allelic Arf deletion or Mdm2 overexpression was not found in any tumors regardless of the genotype of Dmp1 [75]. Moreover, none of the known Ink4a/Arf repressors, such as Bmi1, Twist, Tbx2, Tbx3 and Pokemon, were overexpressed in K-rasLA lung tumors, ruling out the possibility of the contribution of these Ink4a/Arf modulators for K-ras-induced lung tumor development [75,77–81]. Approximately 40% of lung tumors from wild-type K-rasLA mice showed mutations of the p53 gene, recapitulating the molecular genetic alterations of p53 in human NSCLC [75]. Interestingly, p53 mutations were rarely found in lung tumors from Dmp1+/−, Dmp1−/−, K-rasLA mice; thus, it was assumed that Dmp1-deletions might have similar effects to p53 mutations. In fact, we have found that tumors present in Dmp1+/−, Dmp1−/− , K-rasLA mice tended to show malignant features of carcinomas, such as intravascular and/or intrabronchial invasion (Figures 4C & 4D) [75]. Moreover, the Dmp1+/−, Dmp1−/−, K-rasLA group frequently developed types of tumors other than lung carcinomas [75]. Of note, the Ink4a/Arf locus is rarely inactivated by homozygous gene deletion or silencing in K-rasLA lung tumors [75,76]. Thus, Dmp1-deletion and p53 mutations play major roles in the development of K-rasLA lung carcinomas.
Human DMP1 is a critical tumor suppressor in human lung cancer
The human DMP1 (hDMP1) gene is located on human chromosome 7q21. The 7q21–31 region has been reported to be a hot locus of genomic DNA deletion in human carcinomas and hematopoietic malignancies [82–84]. Bodner et al. studied the copy numbers of the hDMP1 locus by FISH analysis in leukemic samples with chromosome 7q abnormalities. The results demonstrated that one allele of the hDMP1 locus was invariably deleted in tumor cells with 7q alterations, suggesting that the hDMP1 locus was critically involved in 7q–leukemias [84]. Later, Tschan et al. characterized the hDMP1 splicing variants, hDMP1α, β and γ [85]. The β- and γ splicing isoforms do not bind to DNA since they lack most of the DNA-binding domain of DMP1 [85]. The full-length hDMP1α is equivalent to full-length murine Dmp1, which directly binds to the Arf promoter and positively regulates the p19Arf–p53 pathway. Interestingly, Tschan et al. showed that these variant isoforms are specifically expressed in immature hematopoietic cells and that hDMP1β inhibited CD13/aminopeptidase N promoter transactivation by hDMP1α [85]. Notably, stable and ectopic overexpression of hDMP1β efficiently blocked phorbol-12-myristate-13-acetate-induced terminal differentiation of U937 cells to macrophages, which resulted in maintenance of proliferation [85]. Therefore, in humans, the hDMP1 α isoform has tumor-suppressor activity and the β and γ proteins are regarded as dominant negative isoforms for hDMP1α [85].
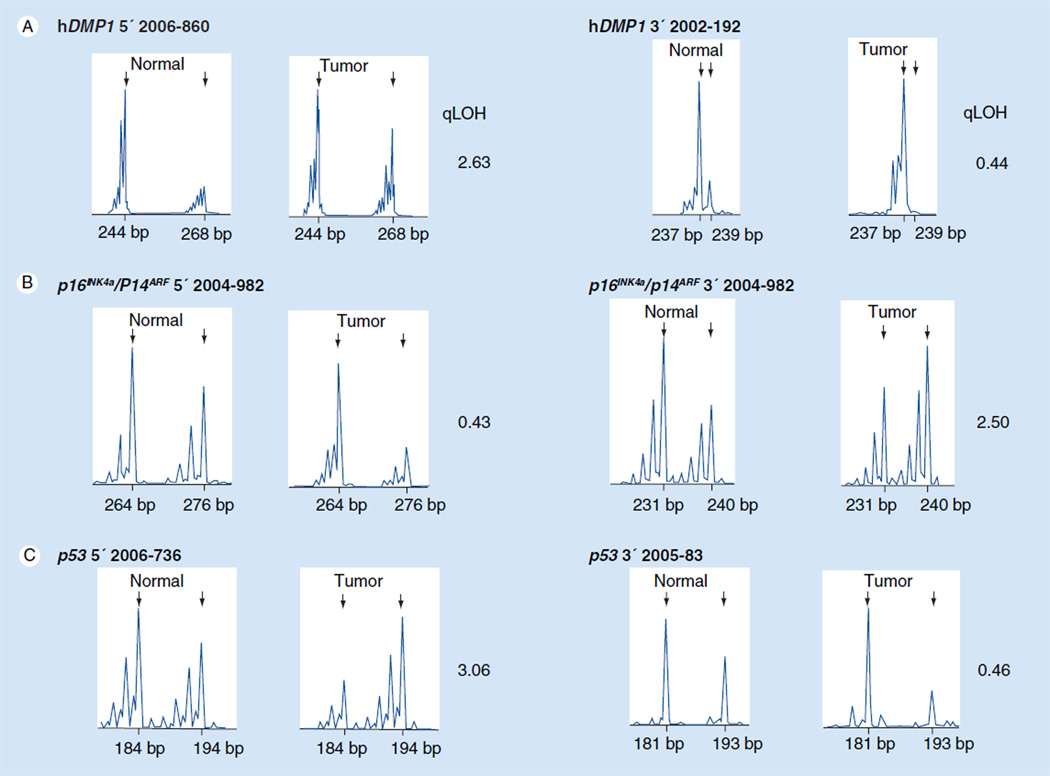
Previous studies have shown differential involvement of the INK4a/ARF, RB and the p53 locus in human lung cancers. For instance, RB is inactivated in approximately 90% of human NSCLC, while p16INK4a is deleted and/or promoter silenced in more than 50% of NSCLC. Promoter hypermethylation or deletion of ARF is relatively rare in NSCLC; however, ARF is inactivated in approximately 65% of human SCLC [6]. The p53 gene is mutated in 90% of SCLC and in 50% of NSCLC [6]. In order to demonstrate the involvement of hDMP1 in human lung cancer, we have recently conducted genomic DNA deletion analyses of hDMP1, INK4a/ARF and p53 by loss of heterozygosity (LOH) assays in more than 50 cases of human NSCLC (total 51 patients: 33 cases of lung adenocarcinoma, 16 cases of squamous cell carcinoma and two cases of adenosquamous carcinoma) [75]. This is the first report of human cancer analysis for hDMP1. LOH of hDMP1 was found in approximately 35% (average of two different sets of primers; 41% if we use relaxed criteria) of NSCLC (Figure 5) [75]. LOH for the INK4a/ARF or p53 locus was also found in 30–50% of the same samples (Figure 5) [75]. Interestingly, LOH of the hDMP1 locus and that of the INK4a/ARF or p53 locus occurred in a mutually exclusive fashion (p = 0.0035 for hDMP1 vs INK4a/ARF; p = 0.027 for hDMP1 vs p53), consistent with our hypothesis that hemizygous deletion of hDMP1 may be inactivating the ARF–p53 pathway in human NSCLC [75]. Of note, the LOH for INK4a/ARF and that of p53 were overlapping at a higher frequency than random (p = 0.0045), possibly because the INK4a/ARF locus regulates both RB and p53 pathways and because p14ARF has p53-independent function for tumor suppression.
Figure 5. Representative patterns of LOH for hDMP1 INK4a/ARF and p53 in human non-small-cell lung carcinoma.
Genomic DNA was extracted from non-small-cell lung carcinomas and their normal counterparts and PCR was conducted with 6-FAM-labeled primers that amplify the dinucleotide repeats within (or close to) each locus [75]. The area peaks of the PCR products were quantitated by ABI 3730xl DNA analyzer. The qLOH values were determined through the following equation: qLOH = area peak 1/area peak 2 (normal tissue) divided by area peak 1’/area peak 2’ (tumor tissue). The arrows indicate the peak that was lost in tumor cells. The sample was considered to have LOH when the value was >2.0 or <0.5 [75,98]. Two different sets of primers were used (set 1 sense: 5’-CCCAAAGAAGCCAACCAGAG-3’ and antisense: 5’-GGCAAATGTGGGAGGTAAGG-3’; set 2 sense: 5’-GAGTGAAAGAGAGTGAGACAG-3’ and antisense: 5’-TCTCACTGTCTCGCTCTGTG-3’) to evaluate the LOH for the 3’ region of hDMP1 to increase the chance of finding a polymorphism. (A) LOH analysis of non-small-cell lung cancer with hDMP1 primer sets. (B) LOH analysis of non-small-cell lung cancer with INK4a/ARF primer sets. (C) LOH analysis of NSCLC with p53 primer sets. LOH: Loss of heterozygosity; qLOH: Quantitative LOH.
Importantly, the region that was deleted in human lung cancer was confined to the hDMP1/MGC4175 locus in approximately 80% of the cases that showed LOH for hDMP1. Although it was very difficult to dissect the contribution of hDMP1 deletion and MGC4175 deletion in NSCLC, hDMP1 was considered to be a key player, since MGC4175 encodes a mitochondrial protein that is involved in taxol- and doxorubicin-resistant malignant phenotypes in human cancer cell lines and, therefore, deletion of this gene would result in tumor regression rather than progression. Point mutations, and promoter methylations that inactivate hDMP1 functions, were very rare (<10%) in our NSCLC samples. Importantly, ectopic expression and activation of Dmp1:ER in an ARF+, p53 wild-type lung cancer cell line strongly inhibited the growth of the cells, while other lung cancer cells with deletion for ARF or p53 were relatively resistant to the effects of Dmp1:ER [75]. In summary, our recent study demonstrated that the hDMP1 gene is inactivated in a significant percentage of human NSCLC, especially those which hold the status of wild-type ARF, and p53 and hDMP1 deletion plays a key role in human lung cancer development.
Detection of the hDMP1 protein in human lung cancer
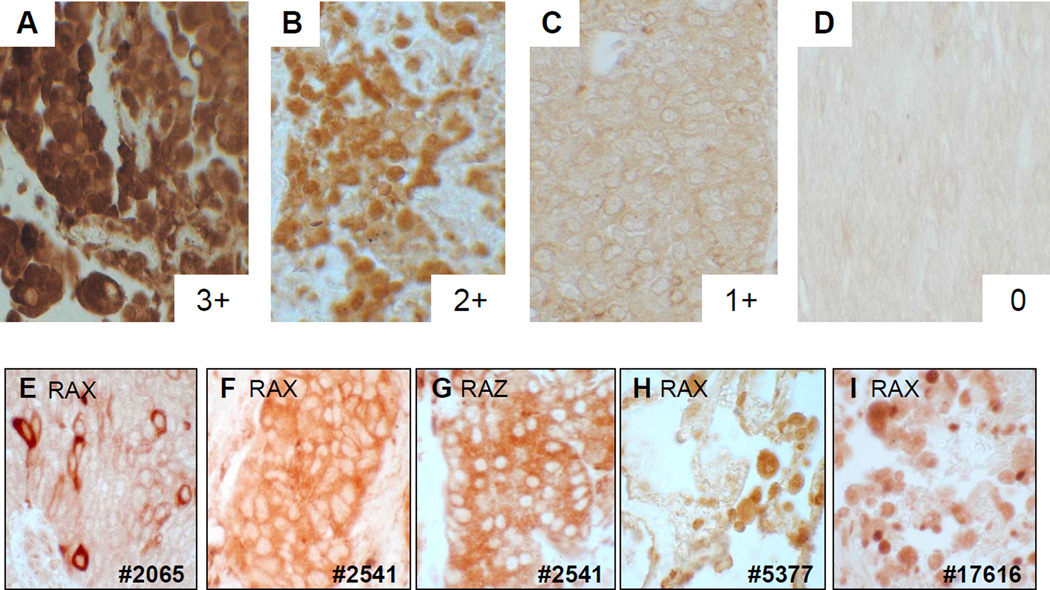
Although Dmp1 (or hDMP1) lacks nuclear localization signals, the endogenous product is localized in the nucleus in normal tissues, NIH 3T3 cells, H460 cells and approximately 30% of lung cancer samples (Figure 6). We found a significant correlation between the intensity of hDMP1 staining in the nucleus and the absence of hDMP1 deletion [75]. However, when we extended our study to approximately 40 NSCLC samples, we noticed that there are many cases where hDMP1 is cytoplasmically localized or localized in both the nucleus and cytoplasm in NSCLC (Figures 6E, F & G). Cytoplasmic localization of the hDMP1 protein was confirmed by immunohistochemistry with two different antibodies to DMP1 in case #2541 (Figure 6). Although the mechanisms of cytoplasmic mislocalization of hDMP1 in cancer cells remain to be determined, there are a few possibilities. One possibility is that lung cancer cells lack binding partner(s) for hDMP1 that normally interact and transport hDMP1 from the cytoplasm to the nucleus. The other possibility is that hDMP1 proteins expressed in tumor cells lack physiological post-translational modification(s) that are essential for their nuclear localization. Physiological cytoplasmic localizations of transcription factors have been reported in repressive E2Fs and NF-κB [86,87]. In repressive E2Fs, the proteins use nuclear localization signals of their binding partners (DPs and pocket proteins) for nuclear transport [86]. The NF-κB dimers are bound by inhibitory IκB molecules and stay in the cytoplasm in unstimulated cells. Since transcriptional activation by NF-κB requires its nuclear translocation, signal-induced degradation of IκB molecules by phosphorylation at serine residues 32 and 36 is considered to be critical in NF-κB activation [87]. Thus, it will be essential to identify physiological hDMP1-binding partners by mass spectrometry analyses and/or binding assays with known molecules to clarify the mechanisms of nuclear localization of DMP1.
Figure 6. Immunohistochemical detection of the hDMP1 protein in human lung cancer.
Pictures (A–D) show the grading of nuclear staining of hDMP1 in different lung cancer cells. It was graded as 3(+), strongly positive; 2(+), positive; 1(+), weakly positive; and 0, negative. Non-small-cell lung cancer samples without LOH for hDMP1 showed significantly stronger signals than LOH(+) samples. (E–I) show abnormal subcellular localization of the hDMP1 protein in lung cancer. (E–G) Immunohistochemical detection of the hDMP1 protein in human non-small-cell lung cancer samples. (H & I) Detection of hDMP1 in normal human lung tissue. The patients’ numbers are listed at the bottom. Paraffin sections were stained with the Dmp1-specific antibody, RAX [73] except for panel (G). Dmp1 antibody to the carboxyl-terminus (RAZ) was used for (G), confirming the cytoplasmic localization of hDMP1 in tumor cells. LOH: Loss of heterozygosity.
Our recent study demonstrated that stimulation of Dmp1:ER with 4-hydroxytamoxifen showed a major shift of the band of Dmp1:ER when the protein translocated from the cytoplasm to the nucleus in H460 cells [75]. These results suggested that post-translational modification(s) may also mediate Dmp1’s nuclear translocation or prevent its nuclear export, the mechanisms of which may be altered in human lung cancer cells.
hDMP1 as a biomarker of NSCLC?
The diagnostic or prognostic value of hDMP1 has never been tested in the literature. However, our recent study shows that LOH of hDMP1 was typically found mutually exclusively with that of the INK4a/ARF locus or that of p53. Our study has also demonstrated that LOH of INK4a/ARF is often associated with silencing of the p16INK4a or p14ARF promoter, suggesting biallelic inactivation [75]. Our results are consistent with previous reports that showed good correlation between LOH of 9p21 and methylation of p16INK4a promoter in NSCLC [88,89]. We also conducted sequencing analyses of the p53 cDNA in NSCLC when RNA was available. All of the four p53 LOH(+) cases showed mutations for p53 while none of the two p53 LOH(−) cases showed p53 mutations [75], consistent with the results from other groups [90,91]. Therefore, it is possible that NSCLC with hDMP1 deletion existed in the historical group of NSCLC patients without p53 mutation and also in that without p16INK4a alterations. Since previous studies have consistently demonstrated that mutations of p53 or absence of the p16INK4a protein is associated with shorter survival and worse prognosis of patients with NSCLC, LOH of hDMP1 or low expression of the hDMP1 protein in immunohistochemistry might be associated with relatively better prognoses of patients. Nevertheless, we still believe that hDMP1 will a useful bio-marker for human NSCLC and LOH assays should be carried out for genotyping for the following reasons:
There are small numbers of cases of NSCLC where LOH for hDMP1 and that of INK4a/ARF or p53 occurred simultaneously (10–20%) [75]. There are also cases of NSCLC where none of the hDMP1, INK4a/ARF or p53 loci are involved (13% of total) [75]. Thus, some hDMP1 LOH(+) cases might have existed in NSCLC with p53 or p16INK4a alterations.
The primers used for LOH assays of the INK4a/ARF and the p53 loci have been carefully designed by us to accurately evaluate gene deletions for these genomic loci and, thus, our LOH assays are unique. Therefore, although our results show mutually exclusive inactivation of hDMP1 and INK4a/ARF or p53 in the vast majority of NSCLC cases, our results cannot be directly compared with those from historical studies conducted by other groups who used published microsatellite markers located more than 1 Mbp away from the INK4a/ARF or p53 locus.
The INK4a/ARF locus encodes another important tumor suppressor, p14ARF , which has been considered the direct target of hDMP1. We speculate that this is the major reason why LOH for hDMP1 of INK4a/ARF are mutually exclusive in approximately 90% of the cases [75]. Since the prognostic value of p14 ARF inactivation in NSCLC has not been reported in the literature, it is not possible to predict the prognostic value of hDMP1 LOH just from the mutual exclusiveness with the LOH of the INK4a/ARF locus. It is not known whether p16INK4a is a direct target for hDMP1.
We have found increased metastasis of K-rasLA lung tumors in Dmp1-hetrozygous mice [75]. Thus, it is possible that DMP1 regulates other genes that are involved in angiogenesis and/or metastasis of lung cancer cells. These targets will be regulated independently of the ARF-p53 pathway.
Hence, the diagnostic and prognostic values of hDMP1 deletion and its correlation with other biomarkers have to be extensively studied in the near future using lung cancer patients’ samples with known prognostic data.
Expert commentary
Lung cancer has been the leading cause of cancer mortality in the world and, thus, it is the most challenging topic for cancer research. The impact of cell cycle regulators, such as cyclins E,A and B and CDK inhibitors p16INK4a, p21CIP1 and p27KIP1 , in NSCLC have been well established. The prognostic values of p53 mutations as detected by molecular genetic approaches have also been established in human NSCLC, although the impact of p53 detection by immunohistochemistry on patients’ survival has been very controversial. The prognostic significance of p14ARF on clinical stage and/or patients’ survival has not been reported in the literature for NSCLC.
Crossbreeding of K-rasLA1 and K-rasLA2 mice with Dmp1-null mice showed significant acceleration of lung carcinogenesis and shortened survival of K-rasLA mice [75]. Thus our study has established that Dmp1 plays significant roles in the prevention of K-ras-induced lung adenocarcinomas. The survival of Dmp1+/−, K-rasLA and Dmp1−/−, K-rasLA mice were not significantly different and lung tumors from Dmp1+/− mice retained and expressed the wild-type Dmp1 allele as studied by competitive and real-time PCR [75]. Moreover, our immunohistochemical data showed expression of the Dmp1 protein in lung tumors from Dmp1+/− mice [75]. Importantly, lung tumors from Dmp1+/− or Dmp1−/−, K-rasLA mice rarely showed mutations of the p53 gene, which was found in 40% of wild-type K-rasLA lung tumors [75]. Thus, Dmp1 is considered to be a nonclassical, haplo-insufficient tumor suppressor gene which plays a critical role in the Ras–Arf–p53 signaling cascade.
The Dmp1 (or hDMP1) gene is often inactivated by deletion in NSCLC cells with wild-type Arf or p53. Our immunohistochemistry results demonstrate that the hDMP1 protein is significantly downregulated in NSCLC cells that show LOH for the hDMP1 locus. Since NSCLC with p53 mutations have been shown to be associated with shorter survival of patients, it is reasonable to predict that NSCLC samples with LOH for hDMP1 and/or low expression of the hDMP1 protein in immunohistochemistry will have a more favorable outcome in comparison to those with p53 mutations.
Five-year view
Recent studies show improving prediction of drug efficacy and patient survival using molecular biological techniques. Lung cancers, p53 mutations, K-Ras mutations and EGF receptor mutations may become indicators for the success of anticancer therapy and prognosis (reviewed in [92–94]). p53, anti-p53 antibodies, EGF receptor and Ras have been detected in the serum of lung cancer patients. However, routine use for these serum biomarkers for early detection of occupationally derived lung carcinomas is currently controversial [95]. HER2 overexpression has been shown to be a poor prognostic factor [96] and low expression of the excision repair cross-complementation group 1 gene was associated with improved survival within cis-platinum-based chemotherapy for NSCLC [97].
Our study has demonstrated the inactivation of hDMP1 in approximately 40% of human NSCLC. Future studies should focus on the determination of prognostic values of hDMP1 deletion and/or hDMP1 protein expression in NSCLC samples with patients’ data for response to therapy and survival. In addition, the significance and prognostic values for cytoplasmic mislocalization of the hDMP1 protein should be analyzed/evaluated. Cancer-specific splicing alterations of hDMP1 and their relationship with LOH of hDMP1, INK4a/ARF and p53 loci should be studied with a large number of patients’ samples. Since NSCLC cells invariably (>90%) retain one intact hDMP1 allele, hDMP1 gene activation within cancer cells with some naturally occurring or synthetic chemicals will be a possible approach for novel cancer therapy. Indeed, we have recently reported the activation of the Dmp1 promoter by trichostatin A, which is a potent inhibitor of histone deacetylases. We hope that analysis of the hDMP1 gene or proteins will help to plan an individualization of the patient treatment protocols for lung cancer.
Key issues.
Dmp1 is a novel transcription factor that directly binds and activates the Arf promoter and induces Arf-p53-dependent cell cycle arrest in primary cells.
Dmp1 is haplo-insufficient for tumor suppression.
Dmp1-deficient mice are prone to develop lung adenomas/adenocarcinomas.
Oncogenic Ras activates the Dmp1 promoter through the Raf-MEK-ERK-Jun pathway which, in turn, stimulates the Arf promoter to stop cell proliferation.
E2Fs and genotoxic stimuli mediated by NF-κB repress the Dmp1 promoter.
The human DMP1 gene (hDMP1) is located on chromosome 7q21 and is hemizygously deleted in approximately 40% of human non-small-cell lung cancer (NSCLC). This hDMP1 deletion is generally mutually exclusive with deletion, LOH or silencing of INK4a/ARF or p53.
The nuclear hDMP1 protein levels correlate well with the genetic status of hDMP1 in NSCLC.
There are cases where the hDMP1 protein is mislocalized in the cytoplasm of NSCLC cells.
Further study is expected if hDMP1 becomes a novel prognostic factor for the lung cancer and a novel target for gene-induction therapy.
Acknowledgements
We thank Charles Sherr, Martine Roussel, John Cleveland, Linda Shapiro, Martin McMahon, Ali Mallakin, Lauren Matise, Sarah Lagedrost, Robert Kendig and Dana Yancey for collaborative work on Dmp1 projects. We are grateful to Bruce Torbett and Mario Tschan for sharing unpublished data.
Kazushi Inoue is supported by the National Institutes of Health/National Cancer Institute (NIH/NCI) 5R01CA106314, American Cancer Society RSG-07–207–01-MGO and Wake Forest University Golfers against Cancer grant P30CA12197GAC. Donna Frazier was supported by the Ruth L Kirschstein National Research Service Award Institutional Research Training Grant (5T32CA079448, F Torti) from NIH.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this review manuscript.
Contributor Information
Takayuki Sugiyama, The Departments of Pathology & Cancer Biology, Wake Forest University Health Sciences, Medical Center Boulevard, Winston-Salem, NC 27157-0001, USA, Tel.: +1 336 716 6047, Fax: +1 336 716 6757, tsugiyam@wfubmc.edu.
Donna P Frazier, The Departments of Pathology & Cancer Biology, Wake Forest University Health Sciences, Medical Center Boulevard, Winston-Salem, NC 27157-0001, USA, Tel.: +1 336 716 1319, Fax: +1 336 716 6757, dofrazie@wfubmc.edu.
Pankaj Taneja, The Departments of Pathology & Cancer Biology, Wake Forest University Health Sciences, Medical Center Boulevard, Winston-Salem, NC 27157-0001, USA, Tel.: +1 336 716 6047, Fax: +1 336 716 6757, ptaneja@wfubmc.edu.
Rachel L Morgan, The Departments of Pathology & Cancer Biology, Wake Forest University Health Sciences, Medical Center Boulevard, Winston-Salem, NC 27157-0001, USA, Tel.: +1 336 716 1319, Fax: +1 336 716 6757, rachelleighmorgan@gmail.com.
Mark C Willingham, The Departments of Pathology & Cancer Biology, Wake Forest University Health Sciences, Medical Center Boulevard, Winston-Salem, NC 27157-0001, USA, Tel.: +1 336 716 7779, Fax: +1 336 716 6757, mwilling@wfubmc.edu.
Kazushi Inoue, The Departments of Pathology & Cancer Biology, Wake Forest University Health Sciences, Medical Center Boulevard, Winston-Salem, NC 27157-0001, USA, Tel.: +1 336 716 5863, Fax: +1 336 716 6757, kinoue@wfubmc.edu.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Jemal A, Siegel R, Ward E, et al. Cancer Statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N. Engl J Med. 2004;350(4):379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 3.Sun S, Schiller JH, Spinola M, Minna JD. New molecular targeted therapies for lung cancer. J Clin Invest. 2007;117(10):2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramalingam S, Belani C. Systemic chemotherapy for advanced non-small cell lung cancer: recent advances and future directions. Oncologist. 2008;13(Suppl 1):5–13. doi: 10.1634/theoncologist.13-S1-5. [DOI] [PubMed] [Google Scholar]
- 5.Travis WD. Pathology of lung cancer. Clin. Chest Med. 2002;23(1):65–81. doi: 10.1016/s0272-5231(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 6.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19(6):643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 7.Wistuba I, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin. Oncol. 2001;28(2) Suppl. 4:3–13. [PubMed] [Google Scholar]
- 8.Zochbauer-Muller S, Gazdar AF, Minna JD. Molecular pathogenesis of lung cancer. Annu. Rev. Physiol. 2002;64:681–708. doi: 10.1146/annurev.physiol.64.081501.155828. [DOI] [PubMed] [Google Scholar]
- 9. Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235–246. doi: 10.1016/s0092-8674(03)01075-4.. • Comprehensive review on cell cycle, oncogenes and tumor suppressor genes.
- 10.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 11.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24(17):2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 12.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25(38):5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 13.Taneja P, Frazier DP, Sugiyama T, Lagedrost SJ, Inoue K. Control of cellular physiology by transcription factors E2F and their roles in carcinogenesis. Res. Signp. 2008:179–197. [Google Scholar]
- 14.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 15.Pei XH, Xiong Y. Biochemical and cellular mechanisms of mammalian CDK inhibitors: a few unresolved issues. Oncogene. 2005;24(17):2787–2795. doi: 10.1038/sj.onc.1208611. [DOI] [PubMed] [Google Scholar]
- 16.Salgia R, Skarin AT. Molecular abnormalities in lung cancer. J. Clin. Oncol. 1998;16(3):1207–1217. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- 17.Scambia G, Lovergine S, Masciullo V. RB family members as predictive and prognostic factors in human cancer. Oncogene. 2006;25(38):5302–5308. doi: 10.1038/sj.onc.1209620. [DOI] [PubMed] [Google Scholar]
- 18.Xu HJ, Quinlan DC, Davidson AG, et al. Altered retinoblastoma protein expression and prognosis in early-stage non-small-cell lung carcinoma. J Natl Cancer Inst. 1994;86(9):695–699. doi: 10.1093/jnci/86.9.695. [DOI] [PubMed] [Google Scholar]
- 19.Hommura F, Dosaka-Akita H, Kinoshita I, et al. Predictive value of expression of p16INK4A, retinoblastoma and p53 proteins for the prognosis of non-small-cell lung cancers. Br. J. Cancer. 1999;81(4):696–701. doi: 10.1038/sj.bjc.6690750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JT, Chen YC, Chen CY, Wang YC. Loss of p16 and/or pRb protein expression in NSCLC. An immunohistochemical and prognostic study. Lung Cancer. 2001;31(2–3):163–170. doi: 10.1016/s0169-5002(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 21.Xu HJ, Cagle PT, Hu SX, Li J, Benedict WF. Altered retinoblastoma and p53 protein status in non-small cell carcinoma of the lung: potential synergistic effects on prognosis. Clin. Cancer Res. 1996;2(7):1169–1176. [PubMed] [Google Scholar]
- 22.Zagorski WA, Knudsen ES, Reed MF. Retinoblastoma deficiency increases chemosensitivity in lung cancer. Cancer Res. 2007;67(17):8264–8273. doi: 10.1158/0008-5472.CAN-06-4753. [DOI] [PubMed] [Google Scholar]
- 23.Hensel CH, Hsieh CL, Gazdar AF, et al. Altered structure and expression of the human retinoblastoma susceptibility gene in small cell lung cancer. Cancer Res. 1990;50(10):3067–3072. [PubMed] [Google Scholar]
- 24.Jin M, Inoue S, Umemura T, et al. Cyclin D1, p16 and retinoblastoma gene product expression as a predictor for prognosis in non-small cell lung cancer at stages I and II. Lung Cancer. 2001;34(2):207–218. doi: 10.1016/s0169-5002(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 25.Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin. Cancer Res. 2005;11(11):3974–3986. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- 26.Kratzke RA, Greatens TM, Rubins JB, et al. Rb and p16INK4a expression in resected non-small cell lung tumors. Cancer Res. 1996;56(15):3415–3420. [PubMed] [Google Scholar]
- 27.Nakagawa K, Conrad NK, Williams JP, Johnson BE, Kelley MJ. Mechanism of inactivation of CDKN2 and MTS2 in non-small cell lung cancer and association with advanced stage. Oncogene. 1995;11(9):1843–1851. [PubMed] [Google Scholar]
- 28.Kawamata N, Miller CW, Koeffler HP. Molecular analysis of a family of cyclin-dependent kinase inhibitor genes (p15/MTS2/INK4b and p18/INK4c) in non-small cell lung cancers. Mol. Carcinogen. 1995;14(4):263–268. doi: 10.1002/mc.2940140406. [DOI] [PubMed] [Google Scholar]
- 29.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6(12):909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 30.Pappot H, Francis D, Brunner N, Grondahl-Hansen J, Osterlind K. p53 protein in non-small cell lung cancer as quantitated by enzyme-linked immunosorbent assay: relation to prognosis. Clin. Cancer Res. 1996;2(1):155–160. [PubMed] [Google Scholar]
- 31.Nishio M, Koshikawa T, Kuroishi T, et al. Prognostic significance of abnormal p53 accumulation in primary, resected non-small-cell lung cancers. J Clin. Oncol. 1996;14(2):497–502. doi: 10.1200/JCO.1996.14.2.497. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki M, Nakanishi Y, Kuwano K, Yatsunami J, Takayama K, Hara N. The utility of p53 immunostaining of transbronchial biopsy specimens of lung cancer: p53 overexpression predicts poor prognosis, chemoresistance in advanced non-small cell lung cancer. Clin Cancer Res. 1997;3(7):1195–1200. [PubMed] [Google Scholar]
- 33.Lee JS, Yoon A, Kalapurakal SK, et al. Expression of p53 oncoprotein in non-small-cell lung cancer: a favorable prognostic factor. J.Clin. Oncol. 1995;13(8):1893–1903. doi: 10.1200/JCO.1995.13.8.1893. [DOI] [PubMed] [Google Scholar]
- 34.Quinlan DC, Davidson AG, Summers CL, Warden HE, Doshi HM. Accumulation of p53 protein correlates with a poor prognosis in human lung cancer. Cancer Res. 1992;52(17):4828–4831. [PubMed] [Google Scholar]
- 35.Fujino M, Dosaka-Akita H, Harada M, et al. Prognostic significance of p53 and rasp21 expression in nonsmall cell lung cancer. Cancer. 1995;76(12):2457–2463. doi: 10.1002/1097-0142(19951215)76:12<2457::aid-cncr2820761209>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 36.Tormanen U, Eerola AK, Rainio P, et al. Enhanced apoptosis predicts shortened survival in non-small cell lung carcinoma. Cancer Res. 1995;55(23):5595–5602. [PubMed] [Google Scholar]
- 37.Passlick B, Izbicki JR, Riethmuller G, Pantel K. p53 in non-small-cell lung cancer. J. Natl Cancer Inst. 1994;86(10):801–803. doi: 10.1093/jnci/86.10.801. [DOI] [PubMed] [Google Scholar]
- 38.Ebina M, Steinberg SM, Mulshine JL, Linnoila RI. Relationship of p53 overexpression and up-regulation of proliferating cell nuclear antigen with the clinical course of non-small cell lung cancer. Cancer Res. 1994;54(9):2496–2503. [PubMed] [Google Scholar]
- 39.Carbone DP, Mitsudomi T, Chiba I, et al. p53 immunostaining positivity is associated with reduced survival and is imperfectly correlated with gene mutations in resected non-small cell lung cancer. A preliminary report of LCSG 871. Chest. 1994;106(6 Suppl):377S–381S. [PubMed] [Google Scholar]
- 40.Huang C, Taki T, Adachi M, Konishi T, Higashiyama M, Miyake M. Mutations in exon 7 and 8 of p53 as poor prognostic factors in patients with non-small cell lung cancer. Oncogene. 1998;16(19):2469–2477. doi: 10.1038/sj.onc.1201776. [DOI] [PubMed] [Google Scholar]
- 41.Ahrendt SA, Hu Y, Buta M, et al. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. J. Natl Cancer Inst. 2003;95(13):961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- 42.Burke L, Flieder DB, Guinee DG, et al. Prognostic implications of molecular and immunohistochemical profiles of the Rb and p53 cell cycle regulatory pathways in primary non-small cell lung carcinoma. Clin. Cancer Res. 2005;11(1):232–241. [PubMed] [Google Scholar]
- 43.Fukuyama Y, Mitsudomi T, Sugio K, Ishida T, Akazawa K, Sugimachi K. K-ras and p53 mutations are an independent unfavourable prognostic indicator in patients with non-small-cell lung cancer. Br. J. Cancer. 1997;75(8):1125–1130. doi: 10.1038/bjc.1997.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skaug V, Ryberg D, Kure EH, et al. p53 mutations in defined structural and functional domains are related to poor clinical outcome in non-small cell lung cancer patients. Clin. Cancer Res. 2000;6(3):1031–1037. [PubMed] [Google Scholar]
- 45.Hashimoto T, Tokuchi Y, Hayashi M, et al. p53 null mutations undetected by immunohistochemical staining predict a poor outcome with early-stage non-small cell lung carcinomas. Cancer Res. 1999;59(21):5572–5577. [PubMed] [Google Scholar]
- 46.Mitsudomi T, Oyama T, Kusano T, Osaki T, Nakanishi R, Shirakusa T. Mutations of the p53 gene as a predictor of poor prognosis in patients with non-small-cell lung cancer. J. Natl Cancer Inst. 1993;85(24):2018–2023. doi: 10.1093/jnci/85.24.2018. [DOI] [PubMed] [Google Scholar]
- 47.Tomizawa Y, Kohno T, Fujita T, et al. Correlation between the status of the p53 gene and survival in patients with stage I non-small cell lung carcinoma. Oncogene. 1999;18(4):1007–1014. doi: 10.1038/sj.onc.1202384. [DOI] [PubMed] [Google Scholar]
- 48.Laudanski J, Niklinska W, Burzykowski T, Chyczewski L, Niklinski J. Prognostic significance of p53 and bcl-2 abnormalities in operable nonsmall cell lung cancer. Eur. Respir. J. 2001;17(4):660–666. doi: 10.1183/09031936.01.17406600. [DOI] [PubMed] [Google Scholar]
- 49.Vega FJ, Iniesta P, Caldes T, et al. p53 exon 5 mutations as a prognostic indicator of shortened survival in non-small-cell lung cancer. Br. J. Cancer. 1997;76(1):44–51. doi: 10.1038/bjc.1997.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horio Y, Takahashi T, Kuroishi T, et al. Prognostic significance of p53 mutations and 3p deletions in primary resected non-small cell lung cancer. Cancer Res. 1993;53(1):1–4. [PubMed] [Google Scholar]
- 51.Sherr CJ. The INK4a/ARF network in tumor suppression. Nat. Rev. Mol. Cell Biol. 2001;2(10):731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 52.Lowe S, Sherr CJ. Tumor suppression by Ink4a–Arf: progress puzzles. Curr. Opin. Genet. Dev. 2003;13(1):77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 53.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6(9):663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 54.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 55.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127(2):265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor its relatives. Biochim. Biophys. Acta Rev. Cancer. 1998;1378(2):F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 57.Zindy F, Williams RT, Baudino TA, et al. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo . Proc. Natl Acad. Sci. USA. 2003;100(26):15930–15935. doi: 10.1073/pnas.2536808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59(9):2217–2222. [PubMed] [Google Scholar]
- 59.Wu X, Bayle JH, Olson D, Levine AJ. The p53–mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 60.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23(14):2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol. Cell Biol. 1998;18(11):6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang YC, Lin RK, Tan YH, Chen JT, Chen CY, Wang YC. Wild-type p53 overexpression and its correlation with MDM2 and p14ARF alterations: an alternative pathway to non-small-cell lung cancer. J. Clin. Oncol. 2005;23(1):154–164. doi: 10.1200/JCO.2005.03.139. [DOI] [PubMed] [Google Scholar]
- 63. Hirai H, Sherr CJ. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol. Cell Biol. 1996;16(11):6457–6467. doi: 10.1128/mcb.16.11.6457.. • Cloning of the murine Dmp1 cDNA.
- 64.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol. Cell Biol. 1998;18(3):1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc. Natl Acad. Sci. USA. 1999;96(7):3993–3998. doi: 10.1073/pnas.96.7.3993.. • Regulation of the Arf promoter by Dmp1.
- 66.Inoue K, Sherr CJ, Shapiro LH. Regulation of the CD13/aminopeptidase N gene by DMP1, a transcription factor antagonized by D-type cyclins. J. Biol. Chem. 1998;273(44):29188–29194. doi: 10.1074/jbc.273.44.29188. [DOI] [PubMed] [Google Scholar]
- 67. Inoue K, Wen R, Rehg JE, et al. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation, and tumorigenesis. Genes Dev. 2000;14(14):1797–1809.. • Creation of Dmp1 knockout mice.
- 68. Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001;15(22):2934–2939. doi: 10.1101/gad.929901.. • Haploid-insufficiency of Dmp1 in tumor suppression.
- 69.Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001;15(22):2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- 70.Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression (Review) Oncogene. 2007;26(30):4329–4335. doi: 10.1038/sj.onc.1210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sugiyama T, Taneja P, Frazier DP, et al. Oncogenic and non-oncogenic signaling pathways that regulate Dmp1 (Dmtf1) Clin. Med. Oncol. 2008;2:1–11. [Google Scholar]
- 72. Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on Dmp1 transcription factor. Mol. Cell Biol. 2005;25(1):220–232. doi: 10.1128/MCB.25.1.220-232.2005.. • Mechanisms of regulation of the Dmp1 promoter by oncogenic Ras signaling.
- 73.Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its repression by E2Fs. Oncogene. 2006;25(59):7703–7713. doi: 10.1038/sj.onc.1209750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taneja P, Mallakin A, Matise LA, Frazier DP, Choudhary M, Inoue K. Repression of Dmp1 and Arf transcription by anthracyclins: critical roles of the NF-κB subunit p65. Oncogene. 2007;26(33):7457–7466. doi: 10.1038/sj.onc.1210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mallakin A, Sugiyama T, Taneja P, et al. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell. 2007;12(4):381–394. doi: 10.1016/j.ccr.2007.08.034.. •• First report on the involvement of hDMP1 in human lung cancer.
- 76.Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogenecauses early onset lung cancer in mice. Nature. 2001;410(6832):1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 77.Maeda T, Hobbs RM, Merghoub T, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433(7023):278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 78.Maestro R, Dei Tos AP, Hamamori Y, et al. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13(17):2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 80.Jacobs JJ, Keblusek P, Robanus-Maandag E, et al. Senescence bypass screen identifies TBX2 , which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat. Genet. 2000;26(3):291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 81.Yang J, Mani S, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 82.Bieche I, Champeme MH, Matifas F, Hacene K, Callahan R, Lidereau R. Loss of heterozygosity on chromosome 7q and aggressive primary breast cancer. Lancet. 1992;339(8786):139–143. doi: 10.1016/0140-6736(92)90208-k. [DOI] [PubMed] [Google Scholar]
- 83.Kerr J, Leary JA, Hurst T, et al. Allelic loss on chromosome 7q in ovarian adenocarcinomas: two critical regions and a rearrangement of the PLANH1 locus. Oncogene. 1996;13(8):1815–1818. [PubMed] [Google Scholar]
- 84. Bodner SM, Naeve CW, Rakestraw KM, et al. Cloning and chromosomal localization of the gene encoding human cyclin D-binding Myb-like protein (hDMP1) Gene. 1999;229(1–2):223–228. doi: 10.1016/s0378-1119(98)00591-5.. • Cloning and chromosomal mapping of the human DMP1 gene.
- 85.Tschan MP, Fischer KM, Fung VS, et al. Alternative splicing of the human cyclin D-binding Myb-like protein (hDMP1) yields a truncated protein isoform that alters macrophage differentiation patterns. J. Biol. Chem. 2003;278(44):42750–42760. doi: 10.1074/jbc.M307067200. [DOI] [PubMed] [Google Scholar]
- 86.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3(1):11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 87.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 88.Awaya H, Takeshima Y, Amatya VJ, et al. Inactivation of the p16 gene by hypermethylation and loss of heterozygosity in adenocarcinoma of the lung. Pathol.Int. 2004;54(7):486–489. doi: 10.1111/j.1440-1827.2004.01655.x. [DOI] [PubMed] [Google Scholar]
- 89.Tanaka R, Wang D, Morishita Y, et al. Loss of function of p16 gene and prognosis of pulmonary adenocarcinoma. Cancer. 2005;103(3):608–615. doi: 10.1002/cncr.20827. [DOI] [PubMed] [Google Scholar]
- 90.Zienolddiny S, Ryberg D, Arab MO, Skaug V, Haugen A. Loss of heterozygosity is related to p53 mutations and smoking in lung cancer. Br. J. Cancer. 2001;84(2):226–231. doi: 10.1054/bjoc.2000.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nelson HH, Wilkojmen M, Marsit CJ, Kelsey KT. TP53 mutation, allelism and survival in non-small cell lung cancer. Carcinogenesis. 2005;26(10):1770–1773. doi: 10.1093/carcin/bgi125. [DOI] [PubMed] [Google Scholar]
- 92.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure lung cancer. Hum. Mutat. 2003;21(3):229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 93.Kim TY, Han SW, Bang YJ. Chasing targets for EGFR tyrosine kinase inhibitors in non-small cell lung cancer: Asian perspectives. Expert Rev. Mol. Diagn. 2007;7(6):821–836. doi: 10.1586/14737159.7.6.821. [DOI] [PubMed] [Google Scholar]
- 94.Stahel RA. Adenocarcinoma, a molecular perspective. Ann. Oncol. 2007;18(Suppl 9):ix147–ix 149. doi: 10.1093/annonc/mdm310. [DOI] [PubMed] [Google Scholar]
- 95.Helmig S, Schneider J. Oncogene tumor-suppressor gene products as serum biomarkers in occupational-derived lung cancer. Expert Rev. Mol. Diagn. 2007;7(5):555–568. doi: 10.1586/14737159.7.5.555. [DOI] [PubMed] [Google Scholar]
- 96.Nakamura H, Kawasaki N, Taguchi M, Kabasawa K. Association of HER-2 overexpression with prognosis in nonsmall cell lung carcinoma: a meta analysis. Cancer. 2005;103(9):1865–1873. doi: 10.1002/cncr.20957. [DOI] [PubMed] [Google Scholar]
- 97.Gray J, Simon G, Bepler G. Molecular predictors of chemotherapy response in non-small-cell lung cancer. Expert Rev. Anticancer Ther. 2007;7(4):545–549. doi: 10.1586/14737140.7.4.545. [DOI] [PubMed] [Google Scholar]
- 98.So CK, Nie Y, Song Y, et al. Loss of heterozygosity and internal tandem duplication mutations of the CBP gene are frequent events in human esophageal squamous cell carcinoma. Clin. Cancer Res. 2004;10:19–27. doi: 10.1158/1078-0432.ccr-03-0160. (1 Pt 1) [DOI] [PubMed] [Google Scholar]