Abstract
Twin and family studies indicate that the timing of primary tooth eruption is highly heritable, with estimates typically exceeding 80%. To identify variants involved in primary tooth eruption, we performed a population-based genome-wide association study of ‘age at first tooth’ and ‘number of teeth’ using 5998 and 6609 individuals, respectively, from the Avon Longitudinal Study of Parents and Children (ALSPAC) and 5403 individuals from the 1966 Northern Finland Birth Cohort (NFBC1966). We tested 2 446 724 SNPs imputed in both studies. Analyses were controlled for the effect of gestational age, sex and age of measurement. Results from the two studies were combined using fixed effects inverse variance meta-analysis. We identified a total of 15 independent loci, with 10 loci reaching genome-wide significance (P < 5 × 10−8) for ‘age at first tooth’ and 11 loci for ‘number of teeth’. Together, these associations explain 6.06% of the variation in ‘age of first tooth’ and 4.76% of the variation in ‘number of teeth’. The identified loci included eight previously unidentified loci, some containing genes known to play a role in tooth and other developmental pathways, including an SNP in the protein-coding region of BMP4 (rs17563, P = 9.080 × 10−17). Three of these loci, containing the genes HMGA2, AJUBA and ADK, also showed evidence of association with craniofacial distances, particularly those indexing facial width. Our results suggest that the genome-wide association approach is a powerful strategy for detecting variants involved in tooth eruption, and potentially craniofacial growth and more generally organ development.
INTRODUCTION
Primary tooth eruption is a complex and highly regulated process through which primary teeth enter the mouth and become visible. Prior to eruption, mononuclear cells move to the dental follicle and fuse to form osteoclasts. These osteoclasts subsequently resorb alveolar bone and in doing so form an eruption pathway through which the primary dentition can then emerge (1).
Twin studies have provided insight into the genetic control of primary tooth eruption during childhood. The ‘Dental Development and Oral Health of Australian Twins and Their Families’ was a longitudinal study of 98 sets of twins of European ancestry aged between 1 and 3 years of age that aimed to assess the degree to which variation in tooth eruption was due to genetic factors. Although there was no statistically significant difference in eruption times between zygosity and the sexes, there was strong genetic control with regard to the timing of primary incisor eruption with an estimated heritability of ∼ 82 to 94% in males, and 71 to 96% in females (2).
The majority of current knowledge regarding the genetics of tooth eruption and tooth development has been acquired from studies involving transgenic mice and other model organisms, including fish and reptiles, as well as from clinical genetic studies of humans with congenital disorders in which dental abnormalities are a feature. For example, studies in mice have implicated a host of signalling pathways as being critical in proper tooth eruption and development, including those involving the gene families Bmp, Eda, Fgf, Shh and Wnt, among others (3–5). These pathways are integrated at several stages of the tooth development process and the network appears to be highly conserved evolutionarily across species (4). Disruption of these pathways typically results in severe aberrations of dentition, including tooth agenesis or arrest in the early stages of tooth development (3).
Population-based genome-wide association studies (GWASs) of tooth eruption in children have the capacity to provide complementary information to these studies, by identifying common genetic variation which is associated with non-pathological differences in the timing of tooth eruption between individuals. Loci implicated by GWASs may not necessarily be the same as those that have been identified in molecular studies or be associated with abnormalities, but rather may reflect variation in genes important in more subtle aspects of tooth development, including differences in the timing of tooth eruption or perhaps even genetic variation important in more generalized aspects of growth and development.
In a previous genome-wide meta-analysis of primary tooth eruption, we identified five loci associated with ‘age at first tooth’ and ‘number of teeth’ at 1 year of age at genome-wide levels of significance, and a further five at suggestive levels of significance (6). Many of these loci contained genes previously implicated in tooth or other organ development. A more recent GWAS of secondary tooth eruption identified two of the same loci as well as two others containing the genes ADK and CACNA1S/TMEM9 (7). What was particularly striking about both studies was the number of loci displaying large effect sizes. Typically, GWASs of quantitative traits require tens of thousands of individuals to identify common variants of small effect. However, the tooth eruption phenotype appears to be influenced by some loci of comparably large effect (i.e. >1% of the phenotypic variance), implying that the genome-wide study of primary tooth eruption might be a powerful strategy not only at detecting variants involved in dentition, but also SNPs that may exert pleiotropic actions on other aspects of growth and development.
In order to identify novel variants involved in primary tooth eruption, we doubled the size of our previous population-based genome-wide association meta-analysis, increasing our sample to include 5998 and 6609 individuals from the Avon Longitudinal Study of Parents and Children (ALSPAC) for ‘age at first tooth’ and ‘number of teeth’, and a further 5403 individuals from the 1966 Northern Finland Birth Cohort (NFBC1966). SNPs that met the criteria for genome-wide significance (P < 5 × 10−8) were then assessed for association with other related phenotypes, including measures of craniofacial shape and size, secondary tooth eruption, and height. The aim of our study was to (i) identify novel genetic loci associated with tooth eruption, and (ii) to investigate whether variants associated with tooth development exhibited pleiotropic effects on growth in general. Specifically, we examined the relationship between tooth-associated loci and eruption of secondary teeth, height, craniofacial size and shape, as well as possible relationships between known height-associated loci and tooth eruption.
RESULTS
A total of 2 446 724 SNPs common to both studies were tested for association with ‘age at first tooth’ and ‘number of teeth at one year’. All analyses were adjusted for gestational age, sex and age, where appropriate (see Materials and Methods). Results from the two studies were combined using fixed effects inverse variance meta-analysis, where effect size estimates are weighted according to the inverse of their standard errors. Q–Q plots indicated little inflation of the test statistics in the individual cohorts and for the meta-analysis overall (‘Age at first tooth’ LAMBDA ALSPAC = 1.04; ‘Age at first tooth’LAMBDA NFBC1966 = 1.05; LAMBDA META = 1.07; ‘Number of teeth’: LAMBDA ALSPAC = 1.02; LAMBDA NFBC1966 = 1.04; LAMBDA META = 1.06) (Supplementary Material, Fig. S1). The genomic inflation factor λ is well known to increase with sample size; we, therefore, also calculated λ1000 values (8) for the ‘Age at first tooth’ (λ1000 = 1.01) and ‘Number of teeth’ (λ1000 = 1.00) meta-analyses. Both values are consistent with little latent population stratification or other systematic biases affecting our results.
We identified 10 loci reaching genome-wide significance (P < 5 × 10−8) for ‘age at first tooth’ and a further 11 loci for ‘number of teeth’, giving a total of 15 independent loci (Fig. 1). The full GWAS results corresponding to Figure 1 are available from the Human Molecular Genetics website. Table 1 shows the top-ranking SNPs for each phenotype at each locus. Eight of these loci are novel associations; the top SNPs at these loci are rs17563 (BMP4), rs10740993 (CACNB2), rs4937076 (CDON), rs1799922 (CALU/OPN1SW), rs997154 (AJUBA/C14orf93), rs7924176 (ADK), rs412000 (TEX14/RAD51C) and rs9316505 (DLEU7). Four of the loci identified confirm previously reported genes/regions (6) (KCNJ2, MSRB3, IGF2BP1 and EDA). Furthermore, we detected genome-wide significance for the variant rs17101923 in the HMGA2 region (‘number of teeth’ P = 1.1 × 10−10, Table 1), rs10932688 in the 2q35 region and the rs6568401 variant in the 6q21 region, which were identified at suggestive levels of significance in a previous study (6). We also note that SNPs at the RAD51L1 locus reported as genome-wide significant for association with ‘number of teeth’ in Pillas et al. (6) did not meet the 5 × 10−8 threshold in this study, although there was still suggestive evidence for association at this locus [‘Age at first tooth’ (rs17105278): P = 2.1 × 10−6; ‘Number of teeth’(rs1956529): P = 6.4 × 10−7].
Figure 1.
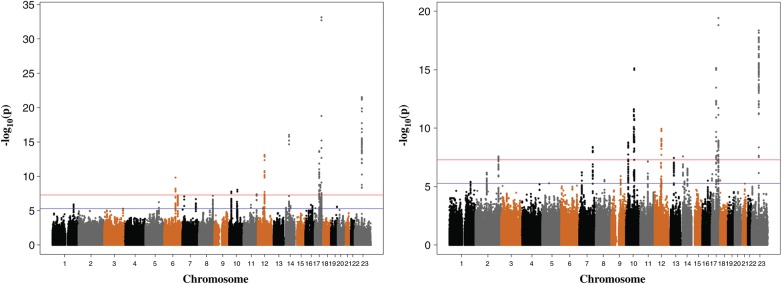
Manhattan plots for meta-analysis of ‘age at first tooth’ and ‘number of teeth’, respectively
Table 1.
Fifteen loci identified at genome-wide significance in meta-analysis of ‘age at first tooth’ or ‘number of teeth’ in ALSPAC and NFBC1966
| Marker | Gene region/locus | Chromosome | Base position | A1/A2 | Effect ALSPAC | SE ALSPAC | % var alspac | P-value ALSPAC | Effect NFBC | SE NFBC | % var nfbc | P-value NFBC | Effect meta | SE meta | P-value meta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at first tooth | |||||||||||||||
| rs10932688 | 2q35 | 2 | 217 571 726 | G/C | −0.107 | 0.048 | 0.05 | 2.7 × 10−2 | −0.106 | 0.037 | 0.13 | 4.0 × 10−3 | −0.106 | 0.029 | 2.9 × 10−4 |
| rs6568401 | 6q21 | 6 | 106 295 511 | C/T | −0.228 | 0.049 | 0.33 | 3.1 × 10−6 | −0.168 | 0.037 | 0.37 | 7.1 × 10−6 | −0.190 | 0.030 | 1.5 × 10−10 |
| rs1799922 | CALU/OPN1SW | 7 | 128 202 431 | T/G | −0.148 | 0.044 | 0.16 | 7.8 × 10−4 | −0.135 | 0.037 | 0.23 | 3.2 × 10−4 | −0.140 | 0.029 | 8.8 × 10−7 |
| rs10740993 | CACNB2 | 10 | 18 482 488 | C/T | −0.175 | 0.043 | 0.24 | 4.7 × 10−5 | −0.118 | 0.035 | 0.19 | 6.7 × 10−4 | −0.141 | 0.0271 | 1.9 × 10−7 |
| rs7924176 | ADK VCL AP3M1 | 10 | 75 965 795 | A/G | −0.261 | 0.043 | 0.58 | 1.2 × 10−9 | −0.081 | 0.037 | 0.06 | 2.5 × 10−2 | −0.167 | 0.028 | 1.8 × 10−8 |
| rs4937076 | CDON | 11 | 125 331 912 | A/G | −0.186 | 0.043 | 0.27 | 1.8 × 10−5 | −0.127 | 0.035 | 0.21 | 3.3 × 10−4 | −0.150 | 0.027 | 4.0 × 10−8 |
| rs12229918 | MSRB3 | 12 | 64 048 325 | C/G | −0.273 | 0.044 | 0.60 | 7.3 × 10−10 | −0.176 | 0.039 | 0.37 | 5.3 × 10−6 | −0.218 | 0.029 | 7.3 × 10−14 |
| rs17101923 | HMGA2 | 12 | 64 624 469 | G/T | −0.282 | 0.053 | 0.44 | 9.99 × 10−8 | −0.170 | 0.041 | 0.30 | 3.5 × 10−5 | −0.212 | 0.033 | 6.3 × 10−11 |
| rs9316505 | DLEU7 | 13 | 50 288 599 | A/G | −0.122 | 0.043 | 0.10 | 4.8 × 10−3 | −0.095 | 0.038 | 0.08 | 1.2 × 10−2 | −0.107 | 0.028 | 1.8 × 10−4 |
| rs997154 | AJUBA/C14orf93 | 14 | 22 534 322 | G/A | −0.132 | 0.051 | 0.08 | 1.0 × 10−2 | −0.142 | 0.038 | 0.23 | 2.2 × 10−4 | 0.138 | 0.031 | 6.9 × 10−6 |
| rs17563 | BMP4 | 14 | 53 487 272 | G/A | −0.339 | 0.043 | 0.98 | 4.9 × 10−15 | −0.160 | 0.038 | 0.29 | 2.9 × 10−5 | −0.239 | 0.029 | 9.1 × 10−17 |
| rs1994969 | IGF2BP1 | 17 | 44 435 430 | T/G | −0.211 | 0.043 | 0.36 | 1.0 × 10−6 | −0.203 | 0.035 | 0.61 | 4.5 × 10−9 | −0.206 | 0.027 | 2.3 × 10−14 |
| rs412000 | TEX14/RAD51C | 17 | 54 064 057 | C/G | −0.752 | 0.0431 | 0.24 | 5.0 × 10−5 | −0.157 | 0.035 | 0.34 | 8.2 × 10−6 | −0.1641 | 0.027 | 1.7 × 10−9 |
| rs8080944 | KCNJ2 KCNJ16 | 17 | 65 697 181 | A/G | −0.378 | 0.045 | 1.14 | 2.8 × 10−17 | −0.317 | 0.036 | 1.45 | 2.0 × 10−18 | −0.341 | 0.028 | 7.6 × 10−34 |
| rs11796357 | FAM155E - EDA | X | 68 581 724 | G/A | −0.290 | 0.041 | 0.81 | 1.1 × 10−12 | −0.222 | 0.033 | 0.85 | 2.0 × 10−11 | −0.250 | 0.026 | 3.1 × 10−22 |
| Number of teeth | |||||||||||||||
| rs10932688 | 2q35 | 2 | 217 571 726 | G/C | 0.118 | 0.035 | 0.09 | 6.4 × 10−4 | 0.173 | 0.038 | 0.39 | 5.8 × 10−6 | 0.143 | 0.026 | 2.5 × 10−8 |
| rs6568401 | 6q21 | 6 | 106 295 511 | C/T | 0.156 | 0.035 | 0.25 | 8.4 × 10−6 | 0.058 | 0.039 | 0.03 | 1.4 × 10−1 | 0.112 | 0.026 | 1.6 × 10−5 |
| rs1799922 | CALU/OPN1SW | 7 | 128 202 431 | T/G | 0.138 | 0.031 | 0.28 | 1.0 × 10−5 | 0.152 | 0.039 | 0.25 | 9.6 × 10−5 | 0.144 | 0.024 | 4.0 × 10−9 |
| rs10740993 | CACNB2 | 10 | 18 482 488 | C/T | 0.132 | 0.031 | 0.26 | 1.7 × 10−5 | 0.153 | 0.036 | 0.3 | 2.2 × 10−5 | 0.141 | 0.023 | 1.7 × 10−9 |
| rs7924176 | ADK VCL AP3M1 | 10 | 75 965 795 | A/G | 0.248 | 0.031 | 0.94 | 8.8 × 10−16 | 0.109 | 0.038 | 0.19 | 3.9 × 10−3 | 0.193 | 0.024 | 7.8 × 10−16 |
| rs4937076 | CDON | 11 | 125 331 912 | A/G | 0.082 | 0.031 | 0.11 | 7.5 × 10−3 | 0.121 | 0.037 | 0.19 | 9.9 × 10−4 | 0.098 | 0.024 | 3.1 × 10−5 |
| rs12229918 | MSRB3 | 12 | 64 048 325 | C/G | 0.144 | 0.032 | 0.30 | 5.3 × 10−6 | 0.158 | 0.041 | 0.21 | 1.1 × 10−4 | 0.149 | 0.025 | 2.3 × 10−9 |
| rs17101923 | HMGA2 | 12 | 64 624 469 | G/T | 0.191 | 0.037 | 0.36 | 3.3 × 10−7 | 0.169 | 0.043 | 0.29 | 7.3 × 10−5 | 0.182 | 0.028 | 1.1 × 10−10 |
| rs9316505 | DLEU7 | 13 | 50 288 599 | A/G | 0.132 | 0.031 | 0.24 | 1.5 × 10−5 | 0.133 | 0.039 | 0.17 | 6.2 × 10−4 | 0.133 | 0.024 | 3.4 × 10−8 |
| rs997154 | AJUBA/C14orf93 | 14 | 22 534 322 | G/A | 0.124 | 0.037 | 0.10 | 7.0 × 10−4 | 0.181 | 0.040 | 0.35 | 5.8 × 10−6 | 0.150 | 0.027 | 2.6 × 10−8 |
| rs17563 | BMP4 | 14 | 53 487 272 | G/A | 0.117 | 0.031 | 0.16 | 1.7 × 10−4 | 0.039 | 0.040 | 0.03 | 3.3 × 10−1 | 0.087 | 0.025 | 3.6 × 10−4 |
| rs1994969 | IGF2BP1 | 17 | 44 435 430 | T/G | 0.189 | 0.031 | 0.50 | 8.5 × 10−10 | 0.190 | 0.036 | 0.54 | 1.6 × 10−7 | 0.190 | 0.024 | 7.2 × 10−16 |
| rs412000 | TEX14/RAD51C | 17 | 54 064 057 | C/G | 0.105 | 0.031 | 0.12 | 7 × 10−4 | 0.098 | 0.037 | 0.10 | 7.4 × 10−3 | 0.102 | 0.024 | 1.6 × 10−5 |
| rs8080944 | KCNJ2 KCNJ16 | 17 | 65 697 181 | A/G | 0.192 | 0.032 | 0.54 | 1.6 × 10−9 | 0.317 | 0.036 | 0.92 | 1.9 × 10−18 | 0.221 | 0.024 | 1.5 × 10−19 |
| rs11796357 | FAM155E - EDA | X | 68 581 724 | G/A | 0.175 | 0.029 | 0.56 | 2.5 × 10−9 | 0.231 | 0.035 | 0.74 | 2.2 × 10−11 | 0.199 | 0.022 | 6.9 × 10−19 |
SNPs showing a genome-wide significance of P < 5 × 10−8 in the meta-analysis. The P-value for each cohort is corrected for gestational age and sex. ALSPAC was also corrected for age at measurement. P-values from the meta-analysis were calculated using a fixed effects inverse variance model. All alleles refer to the forward strand. Positions of SNPs reported correspond to HapMap release II build 36. The effect allele A1 (bold) is defined as the allele associated with faster tooth eruption and an increase in the number of teeth.
Each SNP that reached genome-wide significance explained only a small fraction of the overall phenotypic variation in ‘age at first tooth’ (0.05–1.14%, ALSPAC; 0.06–1.45%, NFBC1966) and ‘number of teeth’ (0.09–0.94%, ALSPAC; 0.03–0.92%, NFBC1966). Pooling together the effects of the top SNPs at the genome-wide significant loci (Table 1) into a single allelic score explained 6.06% of the overall phenotypic variation in ‘age at first tooth’ and 4.76% of the variation in ‘number of teeth’. We also report loci displaying suggestive levels of association (Supplementary Material, Tables S1 and S2), 5 × 10−6 > P > 5 × 10−8, which included SNPs in the TMEM9 region that were reported as genome-wide significant in the study of secondary dentition by Geller et al. (7).
Supplementary Material, Figures S2 and S3 show LocusZoom plots of regression analyses for ‘age at first tooth’ and ‘number of teeth’, respectively, at each genome-wide significant locus after meta-analysis (9). For most loci, there appeared to be evidence of secondary signals independent of the lead SNP at the locus. To quantify the evidence for independent secondary signals, we first calculated the effective number of statistical tests in each region using Nyholt's procedure (10). For each locus, we estimated the threshold for a family-wise error rate of 5% by dividing alpha = 0.05 by the corresponding number of effective tests in that region, and used this threshold for declaring a secondary signal as significant. These thresholds as well as the strongest P-value in each region after conditioning on the lead SNP are presented in Supplementary Material, Table S3. These analyses showed that there were likely to be independent secondary signals at rs11077486 (KCNJ2 KCNJ160), rs2520397 (FAM155E–EDA), rs1951867 (BMP4), rs1472259 (HMGA2) and rs8069452 (IGF2BP1) for ‘age at first tooth’ and rs9788982 (KCNJ2 KCNJ160), rs2804391 (FAM155E–EDA), rs1458991 (BMP4), rs9894411 (IGF2BP1), rs1976274 (MSRB3) and rs1472259 (HMGA2) for ‘number of teeth’ (Supplementary Material, Table S3).
We next investigated whether the SNPs at our top loci have pleiotropic effects, specifically whether they are associated with both primary tooth and craniofacial development. A recent GWAS investigated the genetic determinants of 54 measures of craniofacial shape and size recorded in ALSPAC (Supplementary Material, Fig. S4) (11). We used these data to test for association between the top SNPs at genome-wide significance and each of the 54 measures of craniofacial development. Because of the large number of correlated craniofacial phenotypes analysed, and consequently the large number of statistical tests performed, we calculated empirical P-values for each SNP permuting each genotype against the 54 phenotypes. This procedure is less conservative than a Bonferroni correction (which assumes that the phenotypes are independent) and ensures that the correlation between phenotypes is properly accounted for in the multiple testing correction. Empirical P-values were calculated for each SNP and those with P < 0.05 were declared significant (Table 2). Using this procedure, we identified three SNPs, which were associated with 10 of the 54 craniofacial measures. Specifically, the SNP rs17101923 (HMGA2) was associated with measurements indexing the width of the upper region of the face and nose (Table 2 and Supplementary Material, Fig. S4). Alleles that were associated with increased face width were associated with increased number of teeth and earlier tooth eruption. The rs7924176 marker (ADK) was also associated with measures indexing the width of the nose. Alleles that predisposed to earlier tooth eruption were also associated with a wider nose. Furthermore, rs997154 (AJUBA) was associated with an increase in height and prominence of the mid-brow.
Table 2.
Association results for SNPs that met P < 0.05 after permutation in the analysis of craniofacial size and shape
| Traita | Marker | Gene/locus | Alleles | Frequency of A1 | RSQR | Effect | SE | P-value | Permuted P-value |
|---|---|---|---|---|---|---|---|---|---|
| Width of eye region | |||||||||
| psL-psR: left-to-right palpebrale superius distance | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.11 | 0.028 | 0.00011 | 0.0042 |
| piL-piR: left-to-right palpebrale inferius distance | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.109 | 0.028 | 0.00012 | 0.0049 |
| exR.yz: distance of the right exocanthion from the YZ plane | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.099 | 0.028 | 0.00043 | 0.018 |
| enL.yz: distance of the left endocanthion from the YZ plane | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.096 | 0.028 | 0.00067 | 0.025 |
| enL-enR: left-to-right endocanthion distance | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.094 | 0.028 | 0.00074 | 0.028 |
| exL-exR: left-to-right exocanthion distance | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.093 | 0.028 | 0.00087 | 0.033 |
| Width of lower part of nose | |||||||||
| prn-alL: pronasale to left alare distance | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.08 | 0.025 | 0.0013 | 0.044 |
| sn-alL: subnasale to left alare | rs7924176 | ADK VCL AP3M1 | A/G | 0.58 | 0.9714 | 0.071 | 0.022 | 0.0013 | 0.049 |
| alL-alR: left-to-right alare distance | rs17101923 | HMGA2 | G/T | 0.78 | 0.9645 | 0.078 | 0.024 | 0.0014 | 0.048 |
| Height and prominence of the mid-brow | |||||||||
| g-men: glabella to mid-endocanthion distance | rs997154 | AJUBA | G/A | 0.23 | 0.9698 | 0.104 | 0.027 | 0.00016 | 0.0069 |
All alleles are on the forward strand. The effect allele is displayed in bold font and in each case is also the allele associated with increased the number of teeth at 12 months. Freq1 is the allele frequency of the effect allele.
aSee Supplementary Material Figure S4 for additional information on landmark positions.
We also looked up the top SNP from each of the 15 genome-wide significant loci in a previous analysis of secondary dentition and found that 7 were at least nominally associated (P < 0.05) with the number of permanent teeth between 6 and 14 years old (Supplementary Material, Table S4) (7). For the three loci (i.e. HMGA2, BMP4, MSRB3) associated with ‘age at first tooth’ at genome-wide significance, the allele associated with earlier primary tooth eruption was also associated with a greater number of permanent teeth. Furthermore at the four loci (ADK/VCL/AP3M1, 2q35, CACNB2, 6q21) associated with ‘number of primary teeth’, the allele associated with a greater number of teeth at 1 year was also the allele associated with greater number of permanent teeth (6–14 years).
In order to explore the connection between known height-associated SNPs and teeth phenotypes more deeply, we took 180 robustly associated height variants from the Lango Allen et al. (12) Giant Consortium meta-analysis and examined the degree to which these SNPs were associated with tooth eruption (Supplementary Material, Table S5). Several height-associated SNPs showed strong evidence of association with tooth eruption in the expected direction (i.e. the height increaser allele was associated with faster tooth eruption/more teeth), including rs1351394 in HMGA2 (‘Age at first tooth’: P = 5.3 × 10−7; ‘Number of teeth’: P = 1.0 × 10−9), rs12534093 in IGF2BP3 (‘Age at first tooth’: P = 0.0026; ‘Number of teeth’: P = 2.7 × 10−5), rs1490384 near C6orf173 (‘Age at first tooth’: P = 1.0 × 10−7; ‘Number of teeth’: P = 0.12) and rs1570106 in RAD51L1 (‘Age at first tooth’: P = 0.00012; ‘Number of teeth’: 2.3 × 10−6). Overall, however, the number of height-associated SNPs for which the height increaser allele had a positive effect on faster tooth eruption was not greater than expected by chance (‘Age at first tooth’: 89/180 SNPs in the expected direction P = 0.94; ‘Number of teeth’: 92/180 SNPs in the expected direction P = 0.71). Likewise, a weighted allelic score of height-associated SNPs did not significantly predict the age at first tooth or the number of teeth (‘Age at first tooth’: PMETA = 0.18; ‘Number of teeth’: PMETA = 0.44).
We also regressed height at 17 years in ALSPAC and at 31 years in NFBC1966 on an allelic score constructed from the genome-wide significant SNPs for ‘Age at first tooth’ and ‘Number of teeth’ listed in Table 1. Allelic scores for ‘Age at first tooth’ (PMETA = 0.0012) and ‘Number of teeth’ (PMETA = 9.8 × 10−4) showed moderate evidence of association with height; however, the associations appeared to be driven largely by variants in HMGA2 and BMP4. After these SNPS were removed from the construction of the scores, the evidence for association attenuated markedly (‘Age at first tooth’ PMETA = 0.11; ‘Number of teeth’ PMETA = 0.04).
Finally, we conducted a pathway analysis using the ALLIGATOR software (13). In the pathway analyses, an SNP association P-value threshold of 0.005 gave the most significant over-representation of genes in pathways in the ‘age at first tooth’ GWAS (see Materials and Methods and Supplementary Material, Table S6a). The top 20 pathways (of the 2276 considered) from this analysis are shown in Table 3; 11 of these pathways had pathway association P-values <0.001 and the P-value associated with this degree of overrepresentation is 0.004 (Supplementary Material, Table S6A). However, none of the association P-value thresholds applied to the ‘number of teeth’ GWAS resulted in a significant over-representation of pathways (Supplementary Material, Table S6B). In Discussion, we focus on the results from the ‘age at first tooth’ GWAS.
Table 3.
Pathway analysis
| Pathway | Number of significant genes in pathway | Total number of genes in pathway | Expected number of genes on list | P-value | Study-wide P-value | E hits/study |
|---|---|---|---|---|---|---|
| Signalling events mediated by focal adhesion kinase | 70 | 567 | 47.11 | <10−4 | 0.088 | 0.13 |
| Trail signalling pathway | 71 | 591 | 47.01 | <10−4 | 0.088 | 0.13 |
| p53 pathway | 22 | 160 | 9.24 | <10−4 | 0.088 | 0.13 |
| Signalling events mediated by the hedgehog family | 13 | 51 | 4.78 | <10−4 | 0.088 | 0.13 |
| Class I PI3K signalling events | 66 | 544 | 43.87 | 10−4 | 0.1285 | 0.19 |
| Syndecan-1-mediated signalling events | 72 | 597 | 49.57 | 2.0 × 10−4 | 0.17 | 0.26 |
| Glypican pathway | 92 | 808 | 68.2 | 4.0 × 10−4 | 0.2465 | 0.42 |
| Hedgehog signalling pathway (KEGG) | 11 | 54 | 3.68 | 5.0 × 10−4 | 0.2795 | 0.51 |
| Signalling events mediated by hepatocyte growth factor receptor (c-Met) | 71 | 585 | 49.13 | 6.0 × 10−4 | 0.3135 | 0.59 |
| Class I PI3K signalling events mediated by AKT | 54 | 457 | 35.99 | 7.0 × 10−4 | 0.3485 | 0.66 |
| Glypican 1 network | 79 | 685 | 57.33 | 8.0 × 10−4 | 0.3845 | 0.76 |
| Endothelins | 49 | 364 | 33.3 | 0.0011 | 0.4665 | 1.03 |
| EGF receptor (ErbB1) signalling pathway | 79 | 697 | 59.01 | 0.0019 | 0.6565 | 1.8 |
| Internalization of ErbB1 | 79 | 697 | 59.01 | 0.0019 | 0.6565 | 1.8 |
| ErbB1 downstream signalling | 79 | 697 | 59.01 | 0.0019 | 0.6565 | 1.8 |
| Integrins in angiogenesis | 13 | 63 | 5.7 | 0.002 | 0.671 | 1.89 |
| Prostate cancer (KEGG) | 14 | 79 | 6.4 | 0.002 | 0.671 | 1.89 |
| Downstream signalling in naïve CD8+ T cells | 9 | 42 | 3.87 | 0.0027 | 0.758 | 2.54 |
| 1 and 2 methylninaphthalene degradation (KEGG) | 3 | 7 | 0.31 | 0.0027 | 0.758 | 2.54 |
| Immunoregulatory interactions between A lymphoid and a non-lymphoid cell | 6 | 34 | 1.59 | 0.003 | 0.7875 | 2.81 |
Results of top 20 pathways from the ALIGATOR analyses of the ‘age at first tooth’ GWAS. P-value threshold of 0.005. There were 1358 genes significant at this threshold from a total of 19 887 genes included in our analysis. All pathways presented are from the NCI pathway interaction database unless stated.
DISCUSSION
We report genetic variants at 15 loci associated with primary tooth eruption at genome-wide significant levels, including 8 novel variants within or near the following genes: BMP4, CACNB2, CDON, CALU/OPN1SW, AJUBA, DLEU7, TEX14/RAD51C and ADK. We confirm association with six loci previously associated with primary tooth eruption (KCNJ2/KCNJ16, EDA, IGF2BP1, MSRB3, Chr6q21, 2q35) (6). The SNPs from the ADK and 2q35 associations have also been previously associated with secondary tooth eruption (7).
Two genes identified in this study that have been implicated repeatedly in animal and human models of tooth development are BMP4 and EDA. BMP4 is a member of the transforming growth factor beta-1 superfamily of secretory signalling molecules that play essential roles in embryonic development, including mesoderm induction, tooth development, limb formation, bone induction and fracture repair (14). Mutations in BMP4 can cause eye, brain and digit developmental anomalies (14). BMP4 is expressed early in tooth development and has an expression profile which coincides with the shift of odontogenic potential from the epithelium to the mesenchyme during development of the tooth bud (15). Recent data suggest that BMP4 signalling suppresses tooth developmental inhibitors in the tooth mesenchyme, including Dkk2 and Osr2, and synergizes with Msx1 to activate mesenchymal odontogenic potential for tooth morphogenesis and sequential tooth formation (16). Given BMP4's important role in tooth development, it is perhaps not surprising that SNPs at this locus also associate with timing of tooth eruption. Interestingly, variants within BMP4 have previously been associated with Parkinson's disease (17) and colorectal cancer in other GWASs (18) suggesting pleiotropic actions of this gene.
EDA is a member of the tumour necrosis factor family that signals through a receptor expressed locally in the placodes of all ectodermal appendages as well as in primary and secondary enamel knots (4,19). In humans, mutations in the gene encoding EDA can cause hypohidrotic ectodermal dysplasia-1 (20). This syndrome is characterized by a variety of ectodermal abnormalities, including missing teeth and defects in tooth morphology in that crowns of the remaining teeth lack cusps and are conical in shape (20). The ‘Tabby’ mouse (i.e. EDA null mutant mouse) represents the murine equivalent of hypohidrotic ectodermal dysplasia-1 (21). These mice often lack incisors and third molars and typically express simplified tooth morphology, including missing or fused cusps (22). Conversely, mice that overexpress EDA in the epithelium develop an extra tooth in front of the molars (23). The EDA locus was implicated in our previous GWA study of tooth eruption in humans (6). Our results confirm that SNPs at this locus are also associated with subtle effects on tooth development, including alterations in the timing of tooth eruption.
CACNB2 (rs10740993) is a member of the voltage-gated calcium channel superfamily, and the third ion channel gene to be implicated in tooth eruption (24). Mutations in CACNB2 have been implicated in a form of Brugada syndrome, a genetic disease characterized by electrocardiogram abnormalities (25). Variants in the gene have also been associated with hypertension, systolic and diastolic blood pressure in GWASs (26). Interestingly, the top SNP from the present study is in LD (r2 > 0.7) with an SNP associated with blood pressure from the Ehret et al. study; the allele associated with earlier tooth eruption is on the same haplotype as the allele associated with lower blood pressure.
Variants in DLEU7 have also been associated with height in three GWASs (27–29), although the LD between the topmost SNPs from these studies and the topmost SNP from the present study is low (r2 < 0.01), suggesting that the underlying signals are independent of each other. Transcription factors of DLEU7 are known to have roles in cell proliferation and differentiation (30).
CDON (rs4937076) is involved in muscle cell differentiation and cell adhesion. This gene is part of a cell-surface receptor complex that mediates cell–cell interactions (31). Cell adhesion molecules have been implicated in several processes, including cell migration, growth control and tumour genesis. Cole and Krauss (32) generated mice lacking CDON, 60% of which failed to survive beyond weaning at postpartum day 21. CDON−/− mice displayed the hallmark facial defects associated with microforms of holoprosencephaly, including lack of or solitary central maxillary incisors.
CALU (rs1799922) is a calcium-binding protein found in the endoplasmic reticulum. It is involved in protein folding and sorting (33). The gene has no known functions related to tooth eruption, and variants within this gene have not been associated with other phenotypes in GWASs.
AJUBA belongs to a group of cell adhesion complexes. It is involved in cell fate determination (34) and is an important regulator of the WNT signalling pathway (35). As well as being associated with the number of teeth at 12 months/15 months, the variant rs997154 was also associated with G-men distance (i.e. distance from the glabella to the mid-endocanthion point), suggesting that this gene might be pleiotropically involved in other aspects of craniofacial development besides dentition.
SNPs at three loci (HMGA2, ADK and AJUBA) showed evidence of association with craniofacial distances, particularly those indexing facial width. The SNP rs17101923 is located in an intron of the gene HMGA2 which is known to contain genetic variants associated with height (29), head circumference (36), intracranial volume (37) and permanent dentition (7). The top variant from our study (rs17101923) is in moderate to high LD with these genetic variants, which could reflect the pleiotropic action on growth, in general, of a single causal variant. Ligon et al. (38) report the case of an 8-year-old boy with a de novo pericentric inversion of chromosome 12 that truncated the HMGA2 gene. The patient exhibited multiple clinical features, including premature dentition, enlarged and supernumerary teeth, as well as macrocephaly, flat supraorbital ridges, widely spaced eyes and prominent alveolar ridges. Our results suggest that common SNPs at this locus can also contribute to normal variation in the timing of tooth eruption and craniofacial distances.
We examined the degree to which known height SNPs were associated with tooth eruption, and similarly whether SNPs associated with tooth eruption explained variance in height. Our analyses suggest there exists a subset of known height-associated variants, including those in the HMGA2, IGF2BP3, C6orf173 and RAD51L1 loci that are also associated with tooth eruption. This may be due to these variants exerting a generalized pleiotropic effect on many aspects of growth. For example, SNPs in HMGA2 have been previously associated with other growth-related phenotypes, including head circumference (36), intracranial volume (37) and birth weight (39) as well as height (12). Likewise, SNPs at the C6orf173 locus have been associated with age at menarche (40). Despite robust associations of a few, the majority of height-related SNPs were not strongly related to tooth eruption. A weighted allelic score of height-associated variants was not strongly related to tooth eruption and there seemed to be little consistency in the direction of allelic effects for 180 height-associated SNPs across height and tooth eruption phenotypes. Likewise, the majority of genome-wide significant tooth eruption SNPs did not appear to be strongly related to height. The exceptions were SNPs in HMGA2 and BMP4, both which appear to have pleiotropic actions that have been discussed previously. Our results suggest that BMP4 is likely to contain novel height-associated variants and could also be followed up in this context.
The RAD51 family of genes encode a strand-transfer protein which is thought to be involved in recombinational repair of DNA damage and in meiotic recombination; variants in two of these genes have been highlighted in this study. A variant near RAD51C was genome-wide significant for tooth eruption; this gene has been implicated in a Fanconi anaemia-like disorder (41) as well as in rare monogenic forms of breast and ovarian cancer (42), but not in tooth development. Further, a variant in RAD51L1 reported in Lango Allen et al. (12) as being associated with height also showed suggestive evidence of association with primary tooth eruption in this study.
The top 20 pathways identified from the pathway analysis are mostly related to growth and/or cancer. The three genes (BMP4, CDON, IGF2BP1) associated with ‘age at first tooth’ in the GWAS meta-analysis are part of the hedgehog-signalling pathway (P = 5 × 10−4), signalling events mediated by the Hedgehog family (P < 10−4) and glypican pathway (P = 4 × 10−4). Hedgehog signalling has been well described in tooth development (43), along with heparan sulphate proteoglycans (44). Growth factors also play a major role in the interaction between dental epithelium and mesenchyme, as well as cell–cell interactions within these tissues during tooth development (45). Several growth factor receptor signalling pathways for the epidermal growth factor receptor and the hepatocyte growth factor receptor were significant in our analysis [signalling events mediated by the hepatocyte growth factor receptor (c-Met) P < 6 × 10−4, EGF receptor (ErbB1) signalling pathway, internalization of ErbB1, ErbB1 downstream signalling, P < 0.0019], which is of interest as their ligands HGF and EGF have been shown to play a role in root development in mice (46,47).
Of the four loci associated with primary tooth development in Pillas et al. (6) and confirmed in this study, three have known developmental functions: KCNJ2, EDA and IGF2BP1. The link between normal development and cancer has been noted previously (48), with both involving shifts between cell proliferation and differentiation. Five of the 15 loci identified by our study have been implicated in cancer. As noted earlier, rare mutations in RAD51C have been implicated in breast and ovarian cancer. Likewise, a variant in BMP4 has been found to be associated with colorectal cancer; also a variant in 2q35 has been found to be associated with breast cancer. In both cases, the reported SNP is in high LD with the lead SNP at the respective locus in our study and was also associated with primary tooth eruption. However, whereas the allele associated with increased risk of colorectal cancer was associated with earlier time to tooth eruption, the allele associated with increased risk of breast cancer was associated with fewer teeth at 12 months. Expression of HMGA2 has been implicated in bladder and lung cancers (49,50). Expression of IGF II mRNA-binding protein produced by IGF2BP1 has been implicated in ovarian cancer (51). Furthermore, the pathway analysis implicated many pathways associated with cancer.
In summary, we have identified eight new loci affecting primary tooth eruption, which together with previously identified loci explain 6.06% of the variation in ‘age of first tooth’ and 4.76% of the variation in ‘number of teeth’. These estimates compare favourably with larger studies on human height; for example, using a total sample size of 39 509, Gudbjartsson et al. discovered 27 loci associated with human height, which together explained 3.7% of the variation in human height (27). Several of these variants also appear to exhibit pleiotropic actions, including effects on craniofacial development, height and potentially on disease development in later life. Furthermore, we report a number of genes belonging to pathways involved in growth/development and cancer. A thorough understanding of how the functional variants underlying these associations mediate their effects is likely to yield rich rewards not only in terms of understanding tooth eruption and craniofacial development, but also potentially about how disease develops across the life course.
MATERIALS AND METHODS
Participants and phenotypes
Genome-wide association analyses of primary tooth eruption variables were based on data collected from two prospective birth cohorts: ALSPAC and NFBC1966.
ALSPAC
ALSPAC is a population-based birth cohort study consisting of 14 541 women and their children recruited in the county of Avon, UK, in the early 1990s (52). Both mothers and children have been extensively followed from the eighth gestational week onwards using a combination of self-reported questionnaires, medical records and physical examinations. Biological samples including DNA have been collected from the participants. Ethical approval was obtained from the ALSPAC Law and Ethics Committee and relevant local ethics committees, and written informed consent provided by all parents. Tooth eruption phenotypes of the children were derived from questionnaires completed by the mothers and included items regarding the ‘age at first tooth’ (assessed at 15months) and the ‘number of teeth’ in the child's mouth (at 15 months).
NFBC1966
NFBC1966 followed pregnancies with an expected delivery date in the year 1966 in the Oulu and Lapland provinces of Finland (53). A total of 5403 samples were available for analysis from NFBC1966. In NFBC1966, ‘age at first tooth’ and ‘number of teeth’ were gathered by public health professionals during the children's monthly visits to child welfare centres. ‘Age at first tooth’ was recorded as the month of visit at which the first tooth was observed (so that the first tooth could have erupted at any time between the end of the previous month and the recorded month, i.e. ‘interval censoring’). The number of teeth was recorded at 12 months. All aspects of the study were reviewed and approved by the Ethics Committee of the University of Oulu and by the respective local research committees. Participants gave written informed consent to be included in the study.
Genotyping
ALSPAC
In total, 9912 participants were genotyped using the Illumina HumanHap550 quad genome-wide SNP genotyping platform by 23andMe subcontracting the Wellcome Trust Sanger Institute, Cambridge, UK, and the Laboratory Corporation of America, Burlington, NC, USA. Individuals were excluded from analyses on the basis of excessive or minimal heterozygosity, gender mismatch, individual missingness (>3%), cryptic relatedness as measured by identity by descent (genome-wide IBD >10%) and sample duplication. Individuals were assessed for population stratification using multi-dimensional scaling modelling seeded with HapMap Phase II release 22 reference populations. Individuals of non-European ancestry were removed from further analysis. SNPs with a final call rate of <95%, minor allele frequency (MAF) <1% and evidence of departure from Hardy–Weinberg equilibrium (HWE) (P < 5 × 10−7) were also excluded from analyses. After data cleaning, 5998 and 6609 individuals had complete phenotype and genotype data for the analysis of ‘age at first tooth’ and ‘number of teeth’, respectively. Individuals were imputed to HapMap Phase II (Build 36, release 22) using the Markov Chain Haplotyping software (MACH v.1.0.16) (32). X chromosome imputation was carried out on the non-pseudo autosomal region of the X chromosome only, using CEU individuals from HapMap Phase III (release 2). Only SNPs exceeding an rsq imputation quality metric of 0.3 and an MAF of >1% were included in subsequent analyses.
NFBC1966
The Illumina HumanCNV370-Duo DNA Analysis BeadChip was used for genotyping NFBC1966. SNPs were excluded from the analysis if the call rate in the final sample was <95%, if there was a lack of HWE (P < 5 × 10−4) or if the MAF was <1%; more details of genotyping and quality control procedures can be found in Sabatti et al. (53). After quality control, 328 077 SNPs were available for imputation. Imputation was carried out using IMPUTEv1 with CEU haplotypes from HapMap Phase II (release 21) as the reference panel. X chromosome imputation was carried out in the non-pseudo-autosomal region of the X chromosome (54). Only SNPs exceeding an ‘info’ metric of 0.3 and an MAF of >1% were included in subsequent analyses. After data cleaning, 5120 and 4904 individuals had complete phenotype and genotype data for the analysis of ‘age at first tooth’ and ‘number of teeth’, respectively.
Statistical analysis
In order to account for censoring of the data, the association between expected allelic dosage and ‘age at first tooth’ was analysed using parametric survival analysis, with the Gaussian distribution used to model event time. The ALSPAC data were modelled as ‘right censored’, whereas the data in NFBC1966 were modelled as ‘interval censored’. The association between expected allelic dosage and the number of teeth was analysed using proportional odds logistic regression (ordinal regression). Teeth are known to erupt in pairs; hence Poisson regression (which assumes that the events of interest are independent) was not appropriate. Analyses were adjusted for sex (ALSPAC and NFBC1966), gestational age (ALSPAC and NFBC1966) and age of completion (ALSPAC only, all NFBC1966 measurements were recorded at 12 months). In addition, in NFBC1966, the top 10 ancestry-derived principal components were tested for association with the phenotypes and were included in the GWAS of that phenotype if they associated at P < 0.05. This resulted in the inclusion of the second principal component in the NFBC1966 analysis of tooth eruption, and no principal components in the analysis of ‘number of teeth’. Data were analysed using the R software package 2.9.1.
Results from both studies were combined using a fixed effects inverse variance meta-analysis, using the software package METAL (55). This approach weights effect size estimates according to the inverse of their standard errors. Variance explained by each SNP was calculated as 1 − var(res.full)/var(res.null)*100 of the model (proportional odds logistic regression/survival regression) with age at measurement, sex and gestational age (var, variance; res, residuals). To correct for over-fitting each individuals, phenotype was estimated from a model that did not include that individual (6).
In order to investigate the possibility of secondary signals at loci that met the criteria for genome-wide significance (defined as P < 5 × 10−8), conditional regression analyses were performed conditioning on the most strongly associated SNP in each region. We then applied the Nyholt method for multiple testing correction to derive a threshold for determining statistical significance based on the number of SNPs tested and taking into account LD across the region (10). These regions were defined based on locations of nearby recombination hot spots. In absence of these, we defined a region as ±250 kb from the top SNP.
We then investigated whether any of our genome-wide significant loci exerted pleiotropic actions by looking at their association with height, craniofacial shape and size and permanent tooth eruption. In the case of height, we conducted linear regression of height measured at age 17 (in ALSPAC) and age 31 (in NFBC) on genome-wide significant SNPs from Table 1. For craniofacial shape and size, we looked up genome-wide significant SNPs from the present study in the results from a previous GWAS of the 54 variables characterizing different facial features consisting of facial height, width, convexity and prominence of landmark in respect to facial planes (11) (see Supplementary Material, Fig. S4, for a list of distances examined). To account for multiple testing, empirical levels of significance were determined using permutation analyses, where for each SNP, genotype was permuted with respect to the 54 craniofacial variables. In this way, an adjusted P-value could be calculated for each SNP, which took into account the fact that association had been tested across the 54 correlated variables.
Analyses involving secondary tooth eruption were performed using data from the Danish National Birth Cohort (7). The genotype data were derived from two on-going GWASs of preterm birth (56) and obesity (57). The study combined all observations between age 6 and 14 years (starting with the 6th and stopping with the 14th birthday), the time period when eruption of permanent dentition usually occurs. For each visit to the dentist, the total number of permanent teeth (excluding third molars) was recorded, and regressed on age. The resulting residuals were then standardized, and for each individual the mean residual across all available records was used as the phenotype. Genotypes for the two GWASs were imputed separately using MACH (54). The resulting imputed genotypes were analysed separately and meta-analysed with METAL (55).
Pathway analyses of the ‘age at first tooth’ and ‘number of teeth’ GWASs were performed using the ALIGATOR method (13). The implementation of ALIGATOR described in Holmans et al. (13) maps genes to gene ontology categories; however, the method is equally applicable to other gene to pathway mappings and we used ALIGATOR to test for the enrichment of significant genes within biological pathways; significant genes are defined by the method as those with one or more SNPs with an association P-value less than a predefined threshold within the gene. We considered 2276 pathways curated by the Broad Institute (http://www.broadinstitute.org/gsea/), as well as pathways from ‘Pathway Commons’ (http://www.pathwaycommons.org/pc/home.do) and additional inflammatory pathways (58,59). All genotyped and imputed SNPs with minor allele frequencies of >0.05 were included in the analyses. The method corrects for variable gene size, and multiple testing of non-independent pathways using permutation. All ALIGATOR analyses used 10 000 simulated replicate gene lists and 2000 simulated replicate studies. We compared results using P-value thresholds for association at 0.005, 0.001 and 0.0005 and 0.0001, and as suggested Holmans et al. (13) report results from the analysis showing the most significant enrichment of pathways.
FUNDING
This work and D.M.E. were supported by a Medical Research Council New Investigator Award (MRC G0800582). G.F. is funded by a Wellcome Trust 4-year PhD studentship in molecular, genetic and life course epidemiology (WT083431MA). The UK Medical Research Council (grant 74882), the Wellcome Trust (grant 076467) and the University of Bristol provide core support ALSPAC. G.F., L.P., J.P.K., N.J.T., G.D.S. and D.M.E. work in a centre that receives funds from the UK Medical Research Council (G0600705) and the University of Bristol. C.J.H. and V.J.W. are funded by European Union's Seventh Framework Programme under EC-GA no. 279185 (EUCLIDS). The research of I.P. is funded through the European Community's Seventh Framework Programme (FP7/2007-2013), ENGAGE project, grant agreement HEALTH-F4-2007-201413. Northern Finland Birth Cohort 1966 (NFBC1966) received financial support from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, 139900/24300796, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu, Finland (75617), the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643), NHLBI grant 5R01HL087679-02 through the STAMPEED programme (1RL1MH083268-01), NIH/NIMH (5R01MH63706:02), ENGAGE project and grant agreement HEALTH-F4-2007-201413, the Medical Research Council, UK (G0500539, G0600705, G0600331, PrevMetSyn/SALVE, PS0476) and the Wellcome Trust (project grant GR069224, WT089549), UK. Replication genotyping was supported in part by MRC grant G0601261, Wellcome Trust grants 085301, 090532 and 083270, and Diabetes UK grants RD08/0003704 and BDA 08/0003775. GOYA is a nested study within The Danish National Birth Cohort, which was established with major funding from the Danish National Research Foundation. Additional support for this cohort has been obtained from the Pharmacy Foundation, the Egmont Foundation, The March of Dimes Birth Defects Foundation, the Augustinus Foundation and the Health Foundation. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Supplementary Material
Acknowledgements
We are extremely grateful to all of the families who took part in this study, the midwives for their help in recruiting and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. For NFBC1966, DNA extractions, sample quality controls, biobank upkeeping and aliquotting were performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki. We thank the late Professor Paula Rantakallio (launch of NFBC1966 and 1986), and Ms Outi Tornwall and Ms Minttu Jussila (DNA biobanking). The authors would like to acknowledge the contribution of the late Academian of Science Leena Peltonen. We thank Dr Hariklia Eleftherohorinou for assembling the pathways. This publication is the work of the authors, and they will serve as guarantors for the contents of the paper.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wise G.E., King G.J. Mechanisms of tooth eruption and orthodontic tooth movement. J. Dent. Res. 2008;87:414–434. doi: 10.1177/154405910808700509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes T.E., Bockmann M.R., Seow K., Gotjamanos T., Gully N., Richards L.C., Townsend G.C. Strong genetic control of emergence of human primary incisors. J. Dent. Res. 2007;86:1160–1165. doi: 10.1177/154405910708601204. [DOI] [PubMed] [Google Scholar]
- 3.Bei M. Molecular genetics of tooth development. Curr. Opin. Genet. Dev. 2009;19:504–510. doi: 10.1016/j.gde.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jernvall J., Thesleff I. Tooth formation and tooth renewal: evolving with the same signals. Development. 2012;139:3487–3497. doi: 10.1242/dev.085084. [DOI] [PubMed] [Google Scholar]
- 5.Jussila M., Thesleff I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb. Perspect. Biol. 2012;4:a008425. doi: 10.1101/cshperspect.a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillas D., Hoggart C.J., Evans D.M., O'Reilly P.F., Sipilä K., Lähdesmäki R., Millwood I.Y., Kaakinen M., Netuveli G., Blane D., et al. Genome-wide association study reveals multiple loci associated with primary tooth development during infancy. PLoS Genet. 2010;6:e1000856. doi: 10.1371/journal.pgen.1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geller F., Feenstra B., Zhang H., Shaffer J.R., Hansen T., Esserlind A.-L., Boyd H.A., Nohr E.A., Timpson N.J., Fatemifar G., et al. Genome-wide association study identifies four loci associated with eruption of permanent teeth. PLoS Genet. 2011;7:e1002275. doi: 10.1371/journal.pgen.1002275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman M.L., Reich D., Penney K.L., McDonald G.J., Mignault A.A., Patterson N., Gabriel S.B., Topol E.J., Smoller J.W., Pato C.N., et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 9.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyholt D.R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paternoster L., Zhurov A.I., Toma A.M., Kemp J.P., St Pourcain B., Timpson N.J., McMahon G., McArdle W., Ring S.M., Smith G.D., et al. Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am. J. Hum. Genet. 2012;90:478–485. doi: 10.1016/j.ajhg.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmans P., Green E.K., Pahwa J.S., Ferreira M.A.R., Purcell S.M., Sklar P., Owen M.J., O'Donovan M.C., Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakrania P., Efthymiou M., Klein J.C., Salt A., Bunyan D.J., Wyatt A., Ponting C.P., Martin A., Williams S., Lindley V., et al. Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am. J. Hum. Genet. 2008;82:304–319. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vainio S., Karavanova I., Jowett A., Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- 16.Jia S., Zhou J., Gao Y., Baek J.-A., Martin J.F., Lan Y., Jiang R. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development. 2013;140:423–432. doi: 10.1242/dev.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simón-Sánchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houlston R.S., Webb E., Broderick P., Pittman A.M., Di Bernardo M.C., Lubbe S., Chandler I., Vijayakrishnan J., Sullivan K., Penegar S., et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikkola M.L. Molecular aspects of hypohidrotic ectodermal dysplasia. Am. J. Med. Genet. A. 2009;149A:2031–2036. doi: 10.1002/ajmg.a.32855. [DOI] [PubMed] [Google Scholar]
- 20.Kere J., Srivastava A.K., Montonen O., Zonana J., Thomas N., Ferguson B., Munoz F., Morgan D., Clarke A., Baybayan P., et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat. Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- 21.Srivastava A.K., Pispa J., Hartung A.J., Du Y., Ezer S., Jenks T., Shimada T., Pekkanen M., Mikkola M.L., Ko M.S.H., et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc. Natl Acad. Sci. USA. 1997;94:13069–13074. doi: 10.1073/pnas.94.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pispa J., Jung H.S., Jernvall J., Kettunen P., Mustonen T., Tabata M.J., Kere J., Thesleff I. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev. Biol. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- 23.Mustonen T., Pispa J., Mikkola M.L., Pummila M., Kangas A.T., Pakkasjärvi L., Jaatinen R., Thesleff I. Stimulation of ectodermal organ development by Ectodysplasin-A1. Dev. Biol. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. [DOI] [PubMed] [Google Scholar]
- 24.Taviaux S., Williams M.E., Harpold M.M., Nargeot J., Lory P. Assignment of human genes for β2 and β4 subunits of voltage-dependent Ca2+ channels to chromosomes 10p12 and 2q22-q23. Hum. Genet. 1997;100:151–154. doi: 10.1007/pl00008704. [DOI] [PubMed] [Google Scholar]
- 25.Bayés de Luna A., Brugada J., Baranchuk A., Borggrefe M., Breithardt G., Goldwasser D., Lambiase P., Riera A.P., Garcia-Niebla J., Pastore C., et al. Current electrocardiographic criteria for diagnosis of Brugada pattern: a consensus report. J. Electrocardiol. 2012;45:433–442. doi: 10.1016/j.jelectrocard.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Levy D., Ehret G.B., Rice K., Verwoert G.C., Launer L.J., Dehghan A., Glazer N.L., Morrison A.C., Johnson A.D., Aspelund T., et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 28.Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J.B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A., et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R.B., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammarsund M., Corcoran M.M., Wilson W., Zhu C., Einhorn S., Sangfelt O., Grandér D. Characterization of a novel B-CLL candidate gene – DLEU7 – located in the 13q14 tumor suppressor locus. FEBS Lett. 2004;556:75–80. doi: 10.1016/s0014-5793(03)01371-1. [DOI] [PubMed] [Google Scholar]
- 31.Kang J.-S. CDO: an oncogene-, serum-, and anchorage-regulated member of the Ig/fibronectin type III repeat family. J. Cell Biol. 1997;138:203–213. doi: 10.1083/jcb.138.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole F., Krauss R.S. Microform holoprosencephaly in mice that lack the Ig superfamily member Cdon. Curr. Biol. 2003;13:411–415. doi: 10.1016/s0960-9822(03)00088-5. [DOI] [PubMed] [Google Scholar]
- 33.Yabe D., Taniwaki M., Nakamura T., Kanazawa N., Tashiro K., Honjo T. Human calumenin gene (CALU): cDNA isolation and chromosomal mapping to 7q32. Genomics. 1998;49:331–333. doi: 10.1006/geno.1998.5245. [DOI] [PubMed] [Google Scholar]
- 34.Goyal R.K., Lin P., Kanungo J., Payne A.S., Muslin A.J., Longmore G.D. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol. Cell. Biol. 1999;19:4379–4389. doi: 10.1128/mcb.19.6.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haraguchi K., Ohsugi M., Abe Y., Semba K., Akiyama T., Yamamoto T. Ajuba negatively regulates the Wnt signaling pathway by promoting GSK-3beta-mediated phosphorylation of beta-catenin. Oncogene. 2008;27:274–284. doi: 10.1038/sj.onc.1210644. [DOI] [PubMed] [Google Scholar]
- 36.Taal H.R., St Pourcain B., Thiering E., Das S., Mook-Kanamori D.O., Warrington N.M., Kaakinen M., Kreiner-Møller E., Bradfield J.P., Freathy R.M., et al. Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat. Genet. 2012;44:532–538. doi: 10.1038/ng.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein J.L., Medland S.E., Vasquez A.A., Hibar D.P., Senstad R.E., Winkler A.M., Toro R., Appel K., Bartecek R., Bergmann Ø., et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ligon A.H., Moore S.D.P., Parisi M.A., Mealiffe M.E., Harris D.J., Ferguson H.L., Quade B.J., Morton C.C. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am. J. Hum. Genet. 2005;76:340–348. doi: 10.1086/427565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horikoshi M., Yaghootkar H., Mook-Kanamori D.O., Sovio U., Taal H.R., Hennig B.J., Bradfield J.P., St Pourcain B., Evans D.M., Charoen P., et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elks C.E., Perry J.R.B., Sulem P., Chasman D.I., Franceschini N., He C., Lunetta K.L., Visser J.A., Byrne E.M., Cousminer D.L., et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F., et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 42.Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H., et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 43.Tummers M., Thesleff I. The importance of signal pathway modulation in all aspects of tooth development. J. Exp. Zool. B. 2009;312B:309–319. doi: 10.1002/jez.b.21280. [DOI] [PubMed] [Google Scholar]
- 44.Hikake T., Mori T., Iseki K., Hagino S., Zhang Y., Takagi H., Yokoya S., Wanaka A. Comparison of expression patterns between CREB family transcription factor OASIS and proteoglycan core protein genes during murine tooth development. Anat. Embryol. 2003;206:373–380. doi: 10.1007/s00429-003-0311-z. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y.D., Chen Z., Song Y.Q., Liu C., Chen Y.P. Making a tooth: growth factors, transcription factors, and stem cells. Cell Res. 2005;15:301–316. doi: 10.1038/sj.cr.7290299. [DOI] [PubMed] [Google Scholar]
- 46.Fujiwara N., Akimoto T., Otsu K., Kagiya T., Ishizeki K., Harada H. Reduction of Egf signaling decides transition from crown to root in the development of mouse molars. J. Exp. Zool. B. 2009;312B:486–494. doi: 10.1002/jez.b.21268. [DOI] [PubMed] [Google Scholar]
- 47.Sakuraba H., Fujiwara N., Sasaki-Oikawa A., Sakano M., Tabata Y., Otsu K., Ishizeki K., Harada H. Hepatocyte growth factor stimulates root growth during the development of mouse molar teeth. J. Periodontal Res. 2012;47:81–88. doi: 10.1111/j.1600-0765.2011.01407.x. [DOI] [PubMed] [Google Scholar]
- 48.Nunes F.D., de Almeida F.C., Tucci R., de Sousa S.C.O. Homeobox genes: a molecular link between development and cancer. Pesqui. Odontol. Bras. 2003;17:94–98. doi: 10.1590/s1517-74912003000100018. [DOI] [PubMed] [Google Scholar]
- 49.Di Cello F., Hillion J., Hristov A., Wood L.J., Mukherjee M., Schuldenfrei A., Kowalski J., Bhattacharya R., Ashfaq R., Resar L.M.S. HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang G.L., Zhang L.H., Bo J.J., Hou K.L., Cai X., Chen Y.Y., Li H., Liu D.M., Huang Y.R. Overexpression of HMGA2 in bladder cancer and its association with clinicopathologic features and prognosis HMGA2 as a prognostic marker of bladder cancer. Eur. J. Surg. Oncol. 2011;37:265–271. doi: 10.1016/j.ejso.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Gu L., Shigemasa K., Ohama K. Increased expression of IGF II mRNA-binding protein 1 mRNA is associated with an advanced clinical stage and poor prognosis in patients with ovarian cancer. Int. J. Oncol. 2004;24:671–678. [PubMed] [Google Scholar]
- 52.Golding J., Pembrey M., Jones R. ALSPAC—the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr. Perinat. Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 53.Sabatti C., Service S.K., Hartikainen A.-L., Pouta A., Ripatti S., Brodsky J., Jones C.G., Zaitlen N.A., Varilo T., Kaakinen M., et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornelis M.C., Agrawal A., Cole J.W., Hansel N.N., Barnes K.C., Beaty T.H., Bennett S.N., Bierut L.J., Boerwinkle E., Doheny K.F., et al. The Gene, Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet. Epidemiol. 2010;34:364–372. doi: 10.1002/gepi.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nohr E.A., Timpson N.J., Andersen C.S., Davey Smith G., Olsen J., Sørensen T.I.A. Severe obesity in young women and reproductive health: the Danish National Birth Cohort. PLoS One. 2009;4:e8444. doi: 10.1371/journal.pone.0008444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eleftherohorinou H., Wright V., Hoggart C., Hartikainen A.-L., Jarvelin M.-R., Balding D., Coin L., Levin M. Pathway analysis of GWAS provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS One. 2009;4:e8068. doi: 10.1371/journal.pone.0008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eleftherohorinou H., Hoggart C.J., Wright V.J., Levin M., Coin L.J.M. Pathway-driven gene stability selection of two rheumatoid arthritis GWAS identifies and validates new susceptibility genes in receptor mediated signalling pathways. Hum. Mol. Genet. 2011;20:3494–3506. doi: 10.1093/hmg/ddr248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


