Abstract
In humans and experimental animals, protein-enriched diets are beneficial for weight management, muscle development, managing early-stage insulin resistance and overall health. Previous studies have shown that in mice consuming a high fat diet, whey protein isolate (WPI) reduced hepatosteatosis and insulin resistance due in part to an increase in basal metabolic rate. In the current study, we examined the ability of WPI to increase energy metabolism in mouse brain. Female C57BL/6J mice were fed a normal AIN-93M diet for 12 weeks, with (WPI group) or without (Control group) 100 g WPI/L drinking water. In WPI mice compared to controls, the oxidative stress biomarkers malondialdehyde and 4-hydroxyalkenals were 40% lower in brain homogenates, and the production of hydrogen peroxide and superoxide were 25-35% less in brain mitochondria. Brain mitochondria from WPI mice remained coupled, and exhibited higher rates of respiration with proportionately greater levels of cytochromes a+a3 and c+c1. These results suggested that WPI treatment increased the number or improved the function of brain mitochondria. qRT-PCR revealed that the gene encoding a master regulator of mitochondrial activity and biogenesis, Pgc-1alpha (peroxisome proliferator-activated receptor-gamma coactivator-1alpha) was elevated 2.2-fold, as were the PGC-1alpha downstream genes, Tfam (mitochondrial transcription factor A), Gabpa/Nrf-2a (GA-binding protein alpha/nuclear respiratory factor-2a), and Cox-6a1 (cytochrome oxidase-6a1). Each of these genes had twice the levels of transcript in brain tissue from WPI mice, relative to controls. There was no change in the expression of the housekeeping gene B2mg (beta-2 microglobulin). We conclude that dietary whey protein decreases oxidative stress and increases mitochondrial activity in mouse brain. Dietary supplementation with WPI may be a useful clinical intervention to treat conditions associated with oxidative stress or diminished mitochondrial activity in the brain.
Keywords: brain, gene expression, mitochondria, mouse, oxidative stress, whey protein
Introduction
Due to the high energy demand in brain tissue, neurons have a high mitochondrial density and central nervous system (CNS) activities depend heavily on proper mitochondrial function. CNS mitochondrial dysfunction results in lower energy production, increased oxidative stress and apoptosis, factors related to neurodegenerative diseases, such as Parkinson's, Alzheimer's, Huntington's diseases, as well as amyotrophic lateral sclerosis [1;2]. Impaired mitochondrial function has also been linked to psychotic illnesses, including schizophrenia [3] and bipolar disorder [4], as well as senile dementia and diminished longevity [5;6]. Clinical symptoms in various types of mitochondrial dysfunction are often very general, such that identifying mitochondrial dysfunction in the etiology of such disorders may be quite difficult [1]. Since mitochondria are important in maintaining CNS health, strategies to improve mitochondrial function would have clinical relevance.
Although protein-deficient diets result in impaired brain development and activity [7], there is little information on how a protein-enriched diet may influence biochemical and physiological parameters that impact on CNS function. We have previously shown that dairy protein dietary supplementation, in the form of whey protein isolate (WPI) increased mitochondrial activity in mouse female liver [8]. We extended that study herein, to determine whether dietary WPI might increase mitochondrial activity in other tissues, such as brain. In the current study, we focused on biochemical and genetic parameters related to oxidative stress, respiration and the expression of genes associated with mitochondrial function and biogenesis.
Materials and Methods
Animals and treatment
All experiments involving mice were conducted in accordance with the National Institutes of Health standards for care and use of experimental animals as stated in Principles of Laboratory Animal Care (NIH Publication No. 85-23, revised 1985; http://grants1.nih.gov/grants/olaw/references/phspol.htm), and the University of Cincinnati Institutional Animal Care and Use Committee. Female C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME, USA), 10 weeks of age, were group-housed, maintained on a 12-h light/dark cycle, and had ad libitum access to water and chow (AIN-93M, Research Diets, New Brunswick, NJ). After 10 d acclimation, mice were assigned groups (4 mice/group) such that the average initial body weights for groups were as close as possible. Mice were allowed either water (Control group) or water supplemented with 100 g WPI/L (WPI group). The estrous cycle was not monitored. The precise compositions of the control and WPI diets, as well as body weights, food consumption, fluid intake and energy balance during experimental period, have been described [8]. A formulation of WPI (Bioplex Nutrition, Blaine, WA) was chosen due to its high purity, containing over 90% protein with no lipids, sugars or additives, and low salt content. Body weight, food consumption and fluid intake were measured weekly. After 12 weeks, mice were fasted for 4 h, killed by CO2 asphyxiation, and brain tissue harvested.
Mitochondrial preparation and respiration
Each brain was homogenized in ice-cold isolation buffer consisting of 70 mM sucrose, 225 mM mannitol, 1 mM EGTA and 5 mM HEPES, pH 7.4. Nonsynaptic brain mitochondria were prepared by centrifugation through a Percoll (Amersham Biosciences, Piscataway, NJ) gradient, as described [9]. The mitochondrial pellet was suspended in respiratory buffer consisting of 70 mM sucrose, 220 mM mannitol, 0.5 mM EDTA, 2.5 mM KH2PO4, 2.5 mM MgCl2, 0.1% recrystallized bovine serum albumin, and 2 mM HEPES, pH 7.4 [10]. Respiration was measured polarographically with a Clark-type oxygen electrode (Oxytherm electrode, Hansatech Instruments, Norfolk, England). After determining state 2 respiration in the presence of 6 mM succinate (Succ), or 3 mM glutamate + 3 mM malate (G/M), state 3 respiration was determined following the addition of 0.4 mM ADP. Respiratory Control Index was calculated as the ratio of rates, state 3/state 2.
Products of lipid peroxidation and reactive oxygen
The tissue content of malondialdehyde and 4-hydroxyalkenals, products of lipid peroxidation, were estimated using the colorimetric probe 1-methyl-2-phenylindole (BIOXYTECH LPO-586™, OXIS Health Products, Inc., Portland, OR), following the procedure described by the manufacturer. H2O2 was monitored at 37°C with freshly-prepared mitochondria as 5 μM luminol (5-amino-2,3-dihydro-1,4-phthalazinedione) chemiluminescence that was inhibited by 500 U/ml catalase, utilizing the substrates G/M or Succ, under ADP-limited conditions (state 2) [10]. Superoxide production was monitored as 20 μM lucigenin (bis-N-methylacridinium) chemiluminescence that was inhibited by 5 μM of the superoxide dismutase mimetic Mn(III)tetrakis(1-methyl-4-pyridyl)porphyrin pentachloride [10].
Other assays
Brain reduced glutathione (GSH) levels [11] and cytochromes a+a3 and c+c1 [12] were quantified as described. Protein was measured by the bicinchoninic acid method (Pierce Chemical Co.; Rockford, IL).
RNA isolation, cDNA synthesis and quantitative real-time PCR (qRT-PCR)
Each entire brain was removed and quickly rinsed in ice-cold RNase-free PBS, flash-frozen in liquid N2 and stored at −80°C. Frozen tissue was pulverized and ~50 mg was homogenized in Tri Reagent™ (Molecular Research Center, Inc., Cincinnati, OH) and RNA was isolated from the homogenate per the manufacturer's protocol. Total RNA quantity and quality were assessed by 260 nm absorbance and absorbance ratios of 260/280 nm and 260/230 nm ratios, respectively, utilizing Agilent Bioanalyzer/Nanodrop analysis (Agilent 2100 Bioanalyzer). RNA samples (n=4 per treatment group) with 260/280 nm absorbance ratios ≥ 2.0 were used for qRT-PCR. cDNA synthesis was performed using an RNA-to-cDNA kit (Verso cDNA synthesis kit, #AB1453, Thermo Fisher Scientific, Waltham, MA). Synthesis of cDNA was carried out using 0.5 μg total RNA. qRT-PCR was performed on an MJ Opticon (Bio-Rad, Hercules, CA) using SYBR Green 2X RT-PCR master mix (Bio-Rad) with a total reaction volume of 20 μL. Thermocycling parameters were: 95°C for 3 min followed by 40 cycles of 95°C for 30 s, 55°C for 60 s, 72°C for 60 s. Primer sequences and amplicon size are given in Table 1. Primer sequences were confirmed using a basic local alignment search tool (PRIMER-BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi). All reactions were performed in duplicate, and gene expression values were calculated using the difference in target gene expression relative to 18S rRNA utilizing the 2–ΔΔct method [13].
Table 1.
qRT-PCR parameters for expression of genes involved in mitochondrial biogenesis.
| Gene Name | GeneSymbol (Amplicon, bp) | Forward primer (5′-) Reverse primer (5′-) |
mRNA gene product |
|---|---|---|---|
| Transcription factor A-mitochondrial | Tfam (144) | F5′-CCGAAGTGTTTTTCCAGCAT R5′-CAGGGCTGCAATTTTCCTAA |
NM_009360.4 |
| Mitofusin-2 | Mfn2 (263) | F5′-GCTGGGACAGTGATGGTCTT R5′-CAGATACAGGCTCTCCCTGG |
NM_133201.2 |
| Cytochrome oxidase-6a1 | Cox6a1 (115) | F5′-TCAACGTGTTCCTCAAGTCGC R5′-AGGGTATGGTTACCGTCTCCC |
NM_007748.3 |
| GA- binding protein (Nuclear respiratory factor-2a) | Gabpa (Nrf2a) (129) | F5′-CTCCCGCTACACCGACTAC R5′-TCTGACCATTGTTTCCTGTTCTG |
NM_008065.2 |
| Peroxisome proliferator-activated receptor γ coactivator-1α | Pgc1a (134) | F5′-TATGGAGTGACATAGAGTGTGCT R5′-CCACTTCAATCCACCCAGAAAG |
NM_027710.1 |
| Beta-2 microglobulin | B2MG (106) | F5′-CTGATACATACGCCTGCAGAGTTAA R5′-ATGAATCTTCAGAGCATCATGAT | NM_009735.3 |
| 18S rRNA | 18S rRNA (201) | F5′-CGGCTACCACATCCAAGGAA R5′-GCTGGAATTACCGCGGCT |
- |
Statistics
Data obtained from 4 brains were used for data analysis. Statistical significance of the differences between group sample mean values was determined by a one-way ANOVA to determine that the data was normally distributed, followed by the Student-Newman-Keuls test for pair-wise comparison of means, utilizing SPSS Statistical Analysis software (SPSS Inc., Chicago, IL).
Results
Oxidative stress and GSH status
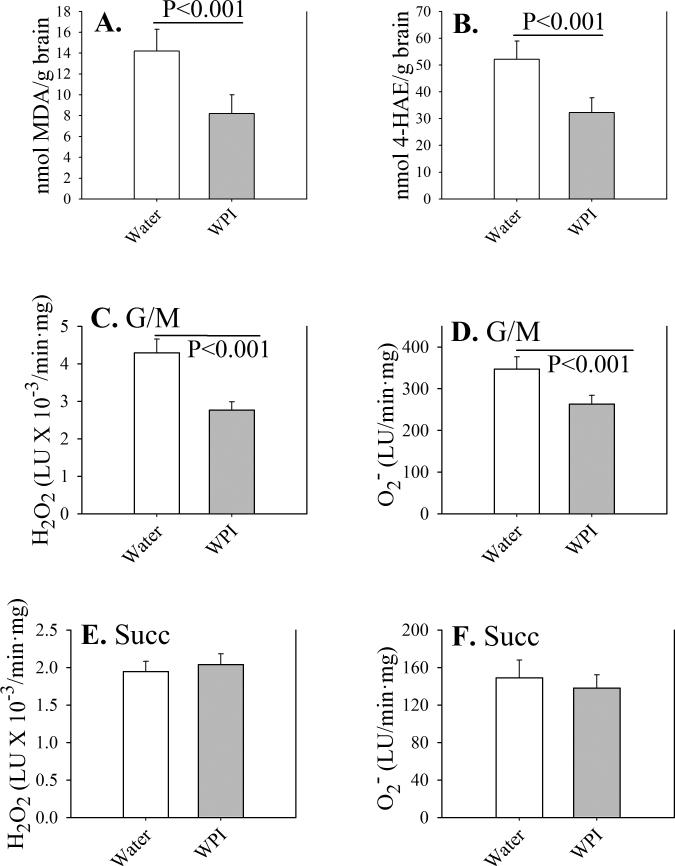
After 12 weeks of treatment, brain tissue from WPI contained lower levels of the products of lipid peroxidation, malondialdehyde (Fig. 1A) and 4-hydroxyalkenals (Fig. 1B). These results may be due in part to lower rates of production of ROS hydrogen peroxide (Fig. 1C) and superoxide (Fig. 1D) by brain mitochondria supplied with NADH-linked complex 1 substrates glutamate+malate (G/M). When mitochondria were supplied with the FADH-linked complex 2 substrate succinate (Succ), rates of ROS production were lower than with G/M, and not affected by WPI dietary supplementation (Figs. 1E and 1F). These results suggest that complex 1 is the primary source of mitochondrial reactive oxygen in the brain. Differing levels of ROS or products of lipid peroxidation in brain were not due to altered levels of GSH, which were the same for control mice (2.0 ± 0.2 μmol/g tissue) and WPI mice (2.1 ± 0.2 μmol/g tissue).
Fig. 1. Oxidative stress parameters in mouse brain.
Control mice received plain drinking water (open bars) and WPI mice received WPI protein in drinking water (shaded bars) for 12 weeks. Levels of malondialdehyde (MDA; panel A) and 4-hydroxyalkenals (4-HAE; panel B) were determined in whole brain homogenates. Reactive oxygen production (H2O2, panel C; or superoxide, panel D) were determined in brain mitochondria under state 2 (ADP-limited) conditions, utilizing 3 mM glutamate + 3 mM malate (G/M) as substrates. For panels E and F, 6 mM succinate (Succ) was the substrate. Reactive oxygen production was normalized to protein.
Data are shown as mean values ± SE (n=4 mice).
P-values are indicated for mean value differences between WPI mice and controls consuming plain water.
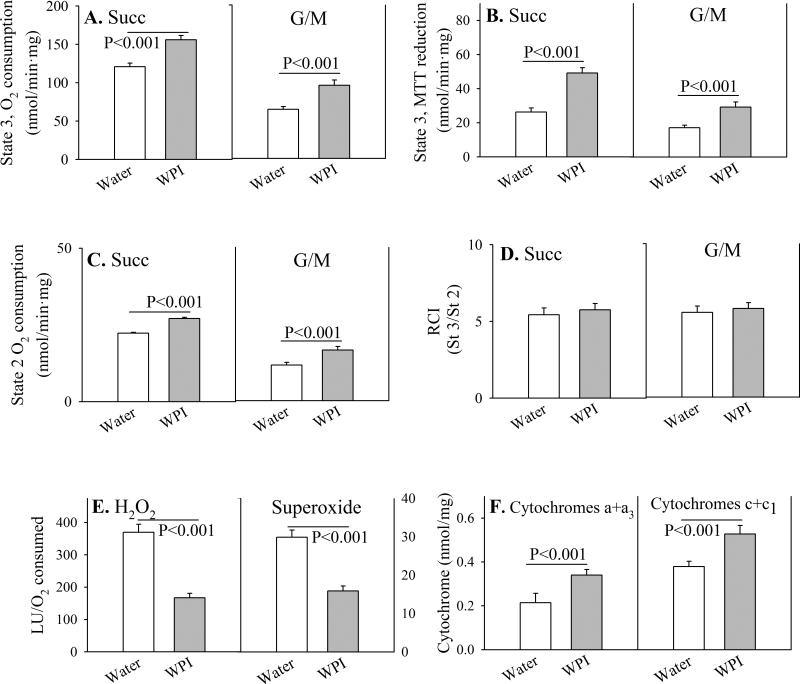
Brain mitochondrial respiration
In order to determine how WPI reduces ROS production in mouse brain mitochondria, we examined mitochondrial respiration in greater detail. For both state 3 respiration (Figs. 2A and 2B), and state 2 (ADP-limited) (Fig. 2C), mitochondrial rates of O2 consumption with the NADH-dependent substrates G/M are less than half the rates utilizing the FAD-dependent substrate Succ, indicating that the NADH dehydrogenase complex 1 is rate-limiting for brain mitochondrial respiration. Under every condition, states 2 and 3 respiration change proportionally, such that energy coupling, calculated as the Respiratory Control Index, is the same with G/M as with Succ (Fig. 2D). Since mitochondrial complex 1 appears to be the major source of brain reactive oxygen production, we calculated ROS production on an oxygen consumption basis. Brain mitochondria from mice consuming WPI generated about half of the amount of H2O2 and superoxide from O2 than mitochondria from mice drinking plain water (Fig. 2E).
Fig. 2. Parameters of respiration in mouse brain mitochondria.
Control mice received plain drinking water (open bars) or WPI protein in drinking water (shaded bars) for 12 weeks. Respiration was measured in mitochondria isolated from mouse brain, measured under State 3 (substrate + 0.4 mM ADP; Panels A and B) and State 2 (substrate alone, ADP-limited; Panel C) conditions. The substrates were 3 mM glutamate + 3 mM malate (G/M), or 6 mM succinate (Succ). The Respiratory Control Index (RCI) as an estimate of energy coupling was calculated as the ratio of state 3/state 2 respiration (Panel D). The production of reactive oxygen metabolites H2O2 and O2, relative to oxygen consumption under state 2 conditions, is shown in Panel E. Levels of mitochondrial cytochromes a+a3 and c+c1 are shown in Panel F.
Data are shown as mean values ± SE (n=4 mice).
P-values are indicated for mean value differences between WPI mice and controls consuming plain water.
The increase in oxygen consumption in the WPI mouse brain mitochondria, in the absence of uncoupling, suggested that mitochondria may be increased in numbers. This possibility is consistent with the amounts of the mitochondrial structural components, cytochromes a+a3 and c+c1, which were higher in mice consuming WPI, and proportional to the increase in oxygen consumption with both complex 1 and complex 2 substrates (Fig. 2F).
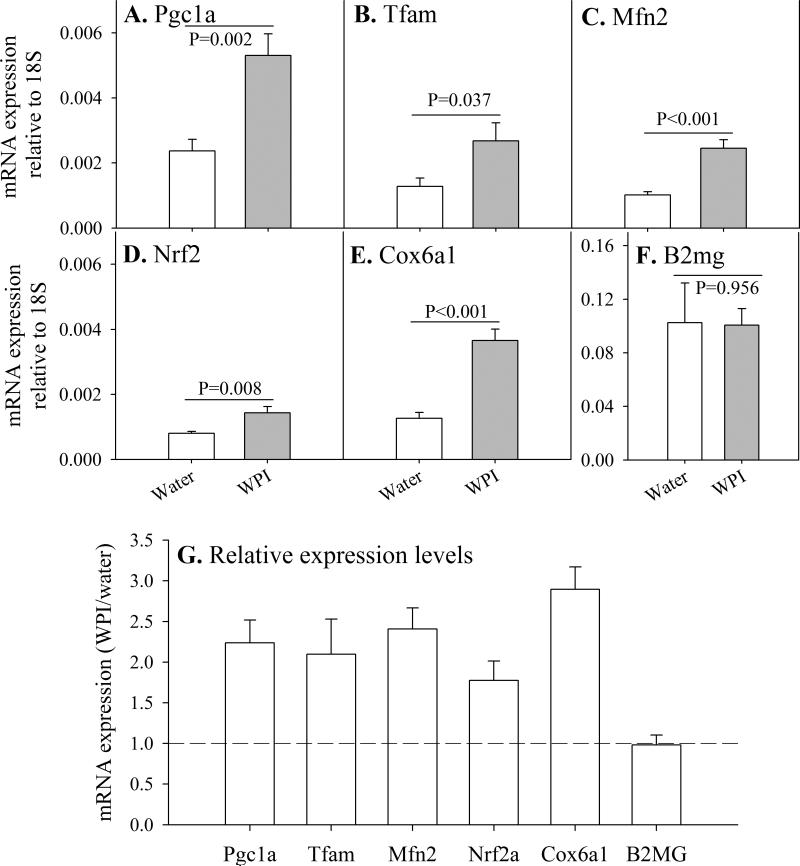
Genes related to mitochondrial activity and biogenesis
Since WPI improves mitochondrial function and oxidative metabolism, we examined how WPI affected some of the genetic regulators and components of mitochondrial activity. Utilizing qRT-PCR, we compared transcriptional levels of genes relative to that of 18S rRNA. Several genes controlling mitochondrial activity and biogenesis are up-regulated in mouse brain by WPI dietary supplementation (Fig. 3 A-F, with fold-changes shown in 3G). These include genes controlling mitochondrial biogenesis [transcriptional coactivator (Pgc1a, 2.2-fold), mitochondrial replication and repair (Tfam, 2.1-fold), mitochondrial remodeling (Mfn2, 2.4-fold), mitochondrial gene transcription (Gabpa/Nrf2a, 1.8-fold)], and a structural component gene (Cox6a1, 2.9-fold). In contrast, there was no change in transcriptional levels of B2mg, a gene that encodes beta-2 microglobulin, a component of the major histocompatibility complex class 1 which is not considered to be involved in energy metabolism or mitochondrial function.
Fig. 3. Transcriptional activation of genes associated with mitochondrial activity and biogenesis in mouse brain.
Control and WPI mice were treated for 12 weeks. Expression levels for genes involved in mitochondrial activity and biogenesis are show relative to 18S rRNA (A-F). Relative change in levels of gene expression due to WPI inclusion in drinking water is shown in panel G.
Data are shown as mean values ± SE (n=4 mice).
P-values are indicated for differences in mean value between WPI mice and controls consuming plain water.
Discussion
Neurological diseases and psychological illnesses are often associated with a loss of neuronal function and even neuronal death. Human and animal models of CNS dysfunction have often been associated with both oxidative stress [14] and mitochondrial dysfunction [15]. Mitochondria are major sources of such intracellular ROS as superoxide and hydrogen peroxide. The production of these oxidants requires a relatively reduced electron transport chain, which in turn depends on maintaining a membrane potential of approximately –180 mV [16;17]. Such a membrane potential produces redox cycling with generation of reactive oxygen, especially at energy coupling sites associated with complexes 1 and 3. In the present study, WPI dietary supplementation in female mice reduced oxidative stress and produced a proportional increase in respiration and respiratory chain components, with no change in energy coupling. We were attempting to determine whether improved mitochondrial function in mouse female liver shown in a previous study [8], could be applied to other organs, such as brain. Neither the study design nor the results should be construed to suggest that there is any gender specificity in our findings.
Mitochondria are composed primarily of nuclear-encoded components and a limited number of components encoded by mitochondrial DNA. Mitochondrial biogenesis, therefore, requires a coordinated expression of both nuclear and mitochondrial genes involved in the synthesis and assembly of these organelles [18]. Nuclear genes encoding transcription and regulatory proteins, considered to be major contributors to the function and biogenesis of mitochondria, were evaluated in this study.
An important finding in this study was the transcriptional increase in PGC-1α, a transcriptional coactivator that responds to metabolic and environmental triggers with a coordinated transcriptional cellular response resulting in mitochondrial biogenesis [19-21]. PGC-1α is an inducible coactivator of the nuclear respiratory factors NRF1 and GABPA/NRF2 [19], as well as MFN2 [22]. In turn, NRF1 activates such downstream targets as TFAM, a mitochondrial gene transcription factor required for maintaining mtDNA copy number and preventing deficiencies in respiration [23]. GABPA/NRF2 is part of the GABP transcription factor complex that activates the expression of many nuclear genes encoding mitochondrial respiratory components, such as cytochrome oxidase subunits such as Cox6a1 [24;25]. GABPA/NRF2 regulates many genes involved in mtDNA transcription and replication [26], and acts in concert with the orphan receptor ERRα to activate additional transcription factors such as PPARα and PPARγ, involved in fatty acid metabolism [19]. Therefore, WPI-triggered elevation of PGC-1α may be sufficient to increase the levels of both nuclear and mitochondrial transcription factors and associated genes central to mitochondrial biogenesis [26].
The elevated expression levels of Pgc1a and its downstream transcriptional target genes Gabpa/Nrf2, Tfam, and Mfn2, and Cox6a1 suggest that WPI may promote mitochondrial biogenesis, mediated largely through increased expression and activity of Pgc1α. Interestingly, transcriptional targets for the PGC-1α protein include not only Gabpa/Nrf2, Tfam and Ppara, but the Pgc1a gene itself [19;21;27;28]. Activation of PGC-1α occurs primarily through deacetylation of lysine residues of NAD-dependent silent information regulator T1 (Sirt1), and by phosphorylation of ser/thr residues by AMP-activated protein kinase (AMPK). While mitochondrial biogenesis may be triggered through PGC-1α by such environmental factors as nutrient deprivation, cold exposure and exercise [24], these variables are not involved in the present study and, therefore, would not be involved in PGC-1α gene activation resulting from dietary WPI. However, WPI supplies substrates for the citric acid cycle and for electron transport, through its constituent amino acids. Catabolism, transamination and deamination of every constitutive amino acid in WPI produce acetyl Co-A and citric acid cycle intermediates as products of intermediary metabolism. We speculate that the increase in substrate availability would result in an increase in mitochondrial metabolism and an activation of PGC-1α through rapid metabolism of NADH and an increase in cellular NAD+/NADH ratios [29].
Conclusions
We have shown that WPI dietary supplementation resulted in a decrease in oxidative stress and increased mitochondrial respiration and mitochondrial structural components in mouse brain. These changes were accompanied by a transcriptional increase in the gene encoding the master regulator of mitochondrial activity and biogenesis, PGC-1α, and increases in associated downstream genes, Tfam, Mfn2, Gabpa/Nrf2a, and Cox6a1. Should these results be applicable to humans, dietary WPI supplementation could prove clinically useful to reduce oxidative stress in the brain, and increase brain mitochondrial activity.
Highlights.
Dietary whey protein isolate (WPI) decreases oxidative stress in mouse brain.
Dietary WPI increases mitochondrial activity in mouse brain.
Dietary WPI increases gene expression associated with mitochondrial biogenesis.
Clinical evaluation for WPI treating certain neurological disorders is warranted.
Acknowledgements
This study was supported in part by NIEHS Center for Environmental Genetics Grant P30 ES06096.
Abbreviations
- B2MG
beta-2 microglobulin
- CNS
central nervous system
- COX-6A1
cytochrome oxidase-6a1
- ERR
estrogen-related receptor
- G/M
glutamate + malate
- GABPA/NRF-2A
GA-binding protein alpha (mouse)/nuclear respiratory factor-2a (human)
- MFN2
mitofusin-2
- PGC-1a
peroxisome proliferator-activated receptor-gamma coactivator-1alpha
- PPAR
peroxisome proliferator-activated receptor
- qRT-PCR
quantitative real-time polymerase chain reaction
- ROS
reactive oxygen species
- Succ
succinate
- TFAM
mitochondrial transcription factor A
- WPI
whey protein isolate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship contributions:
Conception and design: HGS, MBG
Acquisition of data: MK, HGS
Analysis and interpretation of data: HGS, MK
Drafting and editing: MBG, MK, HGS
Final approval: HGS, MBG
Conflict of interest statement:
The authors have nothing to disclose regarding funding or conflicts of interest regarding this manuscript.
References
- 1.Federico A, Cardaioli E, Da PP, Formichi P, Gallus GN, Radi E. Mitochondria, oxidative stress and neurodegeneration. J. Neurol. Sci. 2012;322:254–262. doi: 10.1016/j.jns.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 2.Filosto M, Scarpelli M, Cotelli MS, Vielmi V, Todeschini A, Gregorelli V, Tonin P, Tomelleri G, Padovani A. The role of mitochondria in neurodegenerative diseases. J. Neurol. 2011;258:1763–1774. doi: 10.1007/s00415-011-6104-z. [DOI] [PubMed] [Google Scholar]
- 3.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, Karp NA, Hester S, Tkachev D, Mimmack ML, Yolken RH, Webster MJ, Torrey EF, Bahn S. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatry. 2004;9:684–697. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 4.Andreazza AC, Shao L, Wang JF, Young LT. Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder. Arch. Gen. Psychiatry. 2010;67:360–368. doi: 10.1001/archgenpsychiatry.2010.22. [DOI] [PubMed] [Google Scholar]
- 5.Calabrese V, Scapagnini G, Giuffrida Stella AM, Bates TE, Clark JB. Mitochondrial involvement in brain function and dysfunction: relevance to aging, neurodegenerative disorders and longevity. Neurochem. Res. 2001;26:739–764. doi: 10.1023/a:1010955807739. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti S, Munshi S, Banerjee K, Thakurta IG, Sinha M, Bagh MB. Mitochondrial Dysfunction during Brain Aging: Role of Oxidative Stress and Modulation by Antioxidant Supplementation. Aging Dis. 2011;2:242–256. [PMC free article] [PubMed] [Google Scholar]
- 7.Habibulla M, Krishnan H. Retardation of brain myelination by malnourishment and feeding low protein irradiated diet in rats. Adv. Exp. Med. Biol. 1978;100:179–186. doi: 10.1007/978-1-4684-2514-7_13. [DOI] [PubMed] [Google Scholar]
- 8.Shertzer HG, Woods SE, Krishan M, Genter MB, Pearson KJ. Dietary whey protein lowers the risk for metabolic disease in mice fed a high-fat diet. J. Nutr. 2011;141:582–587. doi: 10.3945/jn.110.133736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrasekaran K, Hazelton JL, Wang Y, Fiskum G, Kristian T. Neuron-specific conditional expression of a mitochondrially targeted fluorescent protein in mice. J. Neurosci. 2006;26:13123–13127. doi: 10.1523/JNEUROSCI.4191-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senft AP, Dalton TP, Nebert DW, Genter MB, Puga A, Hutchinson RJ, Kerzee JK, Uno S, Shertzer HG. Mitochondrial reactive oxygen production is dependent on the aromatic hydrocarbon receptor. Free Radic. Biol. Med. 2002;33:1268–1278. doi: 10.1016/s0891-5849(02)01014-6. [DOI] [PubMed] [Google Scholar]
- 11.Senft AP, Dalton TP, Shertzer HG. Determining glutathione and glutathione disulfide using the fluorescence probe o-phthalaldehyde. Anal. Biochem. 2000;280:80–86. doi: 10.1006/abio.2000.4498. [DOI] [PubMed] [Google Scholar]
- 12.Shertzer HG, Cascarano J. Mitochondrial alterations in heart, liver, and kidney of altitude-acclimated rats. Am. J. Physiol. 1972;223:632–636. doi: 10.1152/ajplegacy.1972.223.3.632. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi S, Abramov AY. Mechanism of oxidative stress in neurodegeneration. Oxid. Med. Cell Longev. 2012;2012:428010. doi: 10.1155/2012/428010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marazziti D, Baroni S, Picchetti M, Landi P, Silvestri S, Vatteroni E, Catena DM. Psychiatric disorders and mitochondrial dysfunctions. Eur. Rev. Med. Pharmacol. Sci. 2012;16:270–275. [PubMed] [Google Scholar]
- 16.Shen D, Dalton TP, Nebert DW, Shertzer HG. Glutathione redox state regulates mitochondrial reactive oxygen production. J. Biol. Chem. 2005;280:25305–25312. doi: 10.1074/jbc.M500095200. [DOI] [PubMed] [Google Scholar]
- 17.Shertzer HG, Genter MB, Shen D, Nebert DW, Chen Y, Dalton TP. TCDD decreases ATP levels and increases reactive oxygen production through changes in mitochondrial F(0)F(1)-ATP synthase and ubiquinone. Toxicol. Appl. Pharmacol. 2006;217:363–374. doi: 10.1016/j.taap.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J. Cell Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 19.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarpulla RC. Nucleus-encoded regulators of mitochondrial function: Integration of respiratory chain expression, nutrient sensing and metabolic stress. Biochim. Biophys. Acta. 2012;1819:1088–1097. doi: 10.1016/j.bbagrm.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piantadosi CA, Suliman HB. Redox Regulation of Mitochondrial Biogenesis. Free Radic. Biol. Med. 2012;53:2043–2053. doi: 10.1016/j.freeradbiomed.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowell RM, Talati P, Blake KR, Meador-Woodruff JH, Russell JW. Identification of novel targets for PGC-1alpha and histone deacetylase inhibitors in neuroblastoma cells. Biochem. Biophys. Res. Commun. 2009;379:578–582. doi: 10.1016/j.bbrc.2008.12.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 24.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 25.Ristevski S, O'Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol. Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruni F, Polosa PL, Gadaleta MN, Cantatore P, Roberti M. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J. Biol. Chem. 2010;285:3939–3948. doi: 10.1074/jbc.M109.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 28.Mootha VK, Handschin C, Arlow D, Xie X, St PJ, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]