Abstract
A primary response to inflammation is an increased survival of the target cell. Several pathways have been identified that promote maintenance of the cell. The principal mechanism for the extended survival is through induction of anti-apoptotic Bcl-2 family proteins. Bcl-2 was the founding member of this family with five additional members, Bcl-XL, Bcl-W, Bcl-B, Bfl-1, and Mcl-1, discovered mostly in hematological malignancies. Another mechanism that could add to cell survival is the Pim kinase pathway. This family of enzymes is associated with Myc-driven transcription, cell cycle regulation, degradation of pro-apoptotic proteins, and protein translation. Chronic lymphocytic leukemia serves as an optimal model to understand the mechanism by which these two protein families provide survival advantage to cells. In addition, since this malignancy is known to be maintained by microenvironment milieu, this further adds advantage to investigate mechanisms by which these pro-survival proteins are induced in presence of stromal support. Multiple mechanisms exists that result in increase in transcript and protein level of anti-apoptotic Bcl-2 family members. Following these inductions, post-translational modifications occur resulting in increased stability of pro-survival proteins, while Pim-mediated phosphorylation inhibits pro-apoptotic protein activity. Furthermore, there is a cross talk between these two (Bcl-2 family proteins and Pim family proteins) pathways that co-operate with each other for CLL cell survival and maintenance. Vigorous efforts are being made to create small molecules that affect these proteins directly or indirectly. Several of these pharmacological inhibitors are in early clinical trials for patients with hematological malignancies.
Keywords: CLL, microenvironment, Bcl-2 family proteins, Pim kinases, survival pathways
Introduction
A primary response to inflammation is a need for cells to exist in an environment that is not conducive to cell survival. To create a favorable setting that promotes cell survival and maintenance, microenvironment provides pro-survival factors and mechanisms. While several pathways have been identified, many of these lead to increased production and stability of Bcl-2 anti-apoptotic family proteins. Because chronic lymphocytic leukemia is a disease where the leukemic lymphocytes survive due to over-expression of Bcl-2 anti-apoptotic proteins, this malignancy serves as an optimal model system to understand and investigate production and maintenance of Bcl-2 anti-apoptotic proteins. In addition, the expression level of these proteins is augmented when cells are growing in microenvironment niches. These systems, CLL primary cells co-cultured with microenvironment cells such as bone marrow stroma cells, nurse-like cells, CD154-expressing cells in presence of IL-4, act as models for malignant cells growing in bone-marrow, spleen, and lymph node, respectively. We review literature and provide some of our work that uses these model systems. Observations obtained with these systems may be applied to other malignancies or diseases.
Chronic lymphocytic leukemia (CLL)
CLL is presently an incurable disease representing the most common form of leukemia in North America and Europe [1–4]. It is a neoplastic disorder, characterized by a gradual accumulation of small, mature B cells with typical B-cell markers CD5, CD19, CD23, and CD20 [5, 6]. Lack of proliferative properties makes these cells inherently quiescent. However, proliferating pool of cells has been described in lymph nodes and in bone marrow that might feed the accumulating pool in the blood [7, 8]. Even though replicationally dormant, the accumulation of leukemic cells both in the bone marrow and the peripheral blood may be due to the numerous parameters such as, intrinsic defects in their apoptotic machinery or dysregulated production of survival signals from their extrinsic microenvironment.
Bcl-2 anti-apoptotic family proteins as pro-survival members in CLL cells
Bcl-2 anti-apoptotic family
The founding member of this ever growing family of proteins is Bcl-2, B-cell leukemia and lymphoma 2 protein. This is a highly conserved protein throughout evolution and in C. elegans, ced-9 has homology to Bcl-2 and it inhibits cell death during worm development [9]. Bcl-2 was discovered by Tsujimoto in Carlo Croce’s laboratory in Non Hodgkin’s lymphoma cell line with chromosomal t(14;18) [10]. With this translocation, the Bcl-2 gene is under the control of IgH enhancer resulting in over production of Bcl-2 transcripts and protein. McDonnell in Korsemeyer’s laboratory elucidated the function of this translocation and protein using Bcl-2 transgenic mice [11]. Bcl-2 has four Bcl-2 homology (BH) domains and a transmembrane domain. Five other members of this family include Bcl-XL [12], Bcl-B [13], Bcl-W [14], Mcl-1 [15], and Bcl-A1 also known as Bfl-1 [16]. The latter two lack BH4 domain. These anti-apoptotic proteins sequester pro-apoptotic counterparts and a balance between the two determines the fate of a cell [17].
Expression in CLL cells
Studies of CLL have established that the survival advantage of CLL lymphocytes is likely due to aberrant over-expression of anti-apoptotic Bcl-2 family proteins [18–20] in general and Bcl-2 and Mcl-1 proteins in particular. Several lines of evidence support this notion. First, the most consistent cytogenetic lesion in CLL is chromosomal deletions of 13q14 [21], resulting in loss of microRNAs (miR-15, miR-16) that down-regulate Bcl-2 mRNA levels [22, 23]. Second, gene ablation studies in mice have shown that Bcl-2, Bcl-XL, and Mcl-1 are essential for lymphocyte survival, with different members of the Bcl-2 family playing prominent roles either during early development or later in adult maintenance of mature T and B-lymphocytes [20, 24, 25]. Third, high levels of Bcl-2 and Mcl-1 mRNAs [19] and proteins [26] have been found in CLL, with levels of Mcl-1 inversely correlated with in vitro response to chemotherapeutic agents [26]. Fourth, high levels of Mcl-1 are observed in relapsed leukemias [27] and are associated with the failure of CLL patients to achieve complete responses (CR) to initial therapy with fludarabine [26]. Fifth, down-regulation of these proteins using antisense oligonucleotides or indirect measures to reduce Mcl-1 protein levels results in CLL cell death in culture [28–34]. Sixth, even in clinic, Bcl-2 antisense therapy has shown promising activity in randomized trials [35]. Seventh, Mcl-1 decline in clinical samples show in vivo activity against CLL [36]. Finally, microenvironments that mimic bone-marrow and lymph nodes further induce these survival proteins or kinases that affect these anti-apoptotic molecules [37–45]. Taken together, these observations establish anti-apoptotic Bcl-2 family proteins as key survival factors for CLL [46].
Microenvironment impact on chronic lymphocytic leukemia
Chronic inflammation and CLL are inter-related in many aspects. The malfunctioning of the immune system helps the first few cancer cells to establish into a full-fledged CLL. Accumulating evidences imply that clinical manifestations and relentless accumulation of CLL cells are frequently due to the progressive infiltration of malignant B lymphocytes in the bone marrow, lymph nodes, and other tissues [38, 47, 48]. In comparison to normal B cells, leukemic cells are rescued from apoptosis by bone marrow stromal cells, signifying the selectivity of microenvironment for malignant cells. These microenvironmental responses are often brought about by the interplay of different chemokines, cytokines, transcriptional factors or post translational modifications.
Chemokines are classified into inflammatory chemokines [49] which are expressed in inflamed tissues and signal for recruitment of neutrophils and homeostatic chemokines that are produced constitutively in distinct tissue microenvironments to sustain traffic of mature lymphocytes in lymphoid and nonlymphoid tissues [50] . Neutrophiles and monocytes migrate to sites of inflammation in response to chemo-attractants, while stromal cell-derived factor-1, (SDF-1) is a haemostatic chemokine that signals through chemoreceptor CXCR4 and play a functional role in B-lymphopoeisis [51–53]. Other chemokines that are important in this context are CCL3 and CCL4 secreted proteins, which are produced by mature hematopoietic cells(reviewed in [54]) and act as potent chemo attractants for monocyte macrophages, dendritic, T, and natural killer cells [54]. Zucchetto et al and others showed that there was a clear cut over expression of transcripts for CCL3 and CCL4 genes upon CD38 triggering, an anti-apoptotic signaling molecule in CLL [55–57], and blocking CCL3 and CCL4 secretion induced apoptosis in CLL cells [44]. SDF-1 and other chemokines appear to form a pro-survival circuitry by regulating leukocyte trafficking, extravagating into sites of tissue inflammation and maintaining extended lymphocyte survival [58–60].
Cytokines are signaling molecules with variety of cellular functions and are key mediators of inflammation or an immune response. Several of them are involved in accelerating inflammation and are present in high levels in CLL patients. They are classified as pro-inflammatory (IL1, IL6, IL15, IL17, IL23 and TNFα [61]-[62]), or anti-inflammatory (IL4, IL10, IL13, transforming growth factor (TGFβ) and interferon (IFNα)) depending on their function in tumorigenesis. One of the primary function for TNFα is to activate the pro-inflammatory NF-κB, while IL-10 suppresses NF-κB, which leads to reduced levels of pro-inflammatory cytokines TNFα, IL6 and IL12 [63]. IL1, IL6 and TNFα activates the Janus kinase, the activator of transcription signaling pathway, or stimulate IL8/CXCL8 chemokine [64] leading to the increased expression of multiple oncogene [65]. Equally, IL2, IL-6, IL7 and IL21 exhibit pro survival effect on CLL [66–68]. Another potential mediator of inflammation induced carcinogenesis is microRNA and the miR-15 and miR-16 on chromosome 13q14 is often deleted in CLL [69]. It has been shown that IL6, a pro-inflammatory cytokine can induce the expression of miR-21, an inflammatory stimuli, in a STAT3-dependent manner [70]. TNFα and IFNβ, can stimulate the expression of miR-155 [71] which is found in increased levels in the bone marrow of leukemic patients and causes hyperproliferation of B-cells [72].
In addition to chemokines and cytokines, the key mediators of inflammation induced cancer include activation of transcription factors. There are a wide range of transcriptional factors that bind to the promoter region of target genes and activate transcription of these oncogenes. Even though MYC involved chromosomal translocations are characteristics of human Burkitt’s lymphoma, recent studies show that MYC rearrangements are associated with complex cytogenetic abnormalities and poor prognosis in CLL [73]. STAT proteins originally shown to exhibit as latent cytoplasmic transcription factors now known to be constitutively phosphorylated on Ser727 and activate transcription in CLL cells [74].
Nuclear factor of κB (NF-κB), that is constitutively activated in CLL [75, 76], is a key mediator of inflammation-induced carcinogenesis [77] and is strongly attributed to interactions of the malignant cells with the bone marrow microenvironment [78]. NFκB activates the transcription of target genes, including inflammation-related genes (e.g. cytokines and chemokines, TNFα, p53 and Bcl-2, Bcl-XL) [79]. In this respect, Taylor et al proposed a model where they illustrate that integrated effects of three microenvironmental signals (CD40 receptor ligation, IL-4 receptor stimulation and the interaction of integrin ligand VCAM-1 with its receptor) work in concert to increase the mRNA and protein levels of Bcl-XL and promote Bcl-XL-Bax binding to suppress drug-induced apoptosis in B lymphoma cells [80]. NF-κB activation could be triggered by members of the TNF super family; for eg. BAFF and APRIL support CLL B-cell survival through activation of the canonical NF-κB pathway [81], or Tcl-1 could activate NF-κB by interacting with p300/cyclic adenosine monophosphate response element binding protein [82].
Signaling pathways, including those involving NF-κB, MAPK (mitogen-activated protein kinase) and PI3K (phosphoinositide 3-kinase) have been shown to be key regulators of inflammatory cell survival and apoptosis in vitro. Manipulation of such pathways in vivo has indicated that they also play a role in the resolution of inflammation [83]. ERK inhibitor is shown to enhance the resolution of inflammation while BAX inhibitor delayed the inflammation resolution [84, 85].
In conjunction with transcriptional regulation, post translational modifications of Bcl-2 family proteins further affect longevity of anti- and pro- apoptotic proteins [40]. Bcl-2 protein undergoes phosphorylation at sites Thr56, Thr69, Ser70, Thr74 and Ser87 in response to different stimuli. Bcl-2 phosphorylation at Ser70 has been found necessary for its potent anti-apoptotic function [86–89] and associate with poor survival in AML [90, 91]. Bcl-2 phosphorylation regulates its interaction with BAX, BIM or BID [91] and protects Bcl-2 from proteasomal degradation and increase stabilization [92, 93].
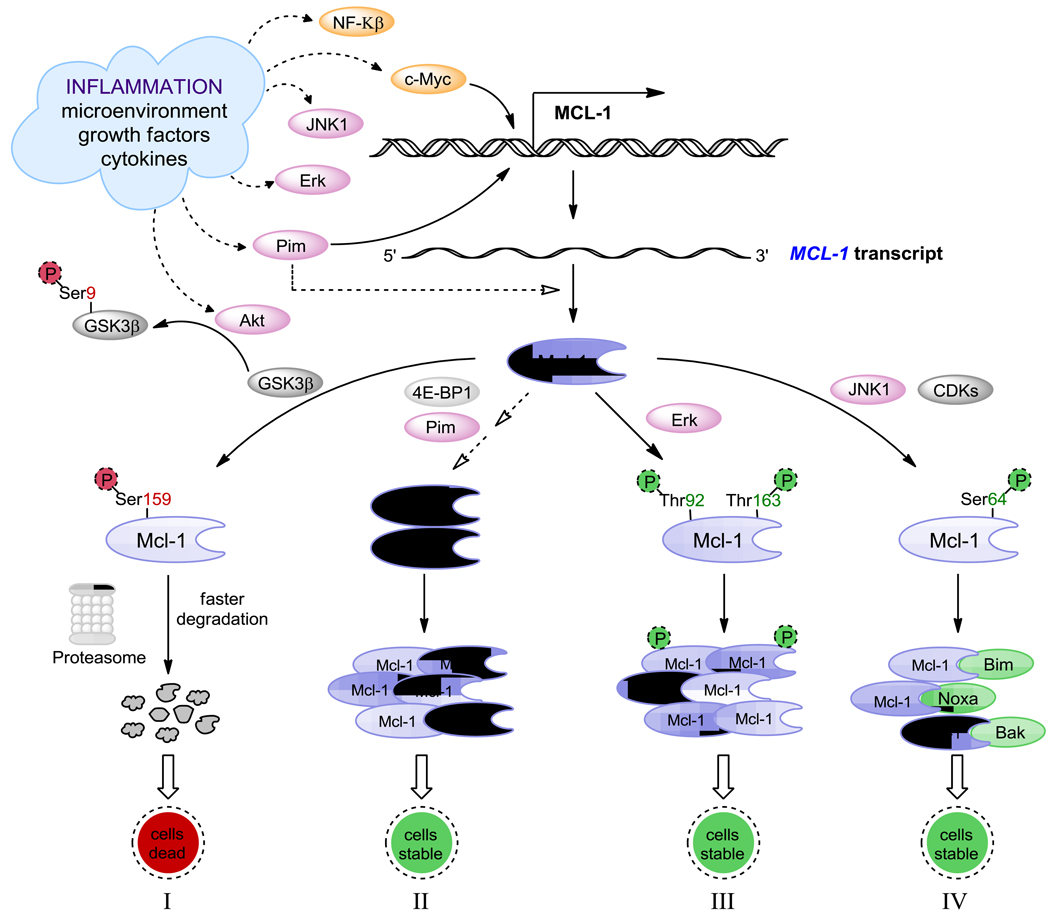
Another important anti-apoptotic Bcl-2 family protein, Mcl-1 has an extended amino terminal PEST region [15] and AU rich elements [94, 95] in the 3’ – untranslated region, which are responsible for its relatively short half life, and it has two caspase cleavage sites D127 and D157 for faster degradation [96]. However, it has several mechanisms for increased production and maintenance of protein, which are depicted in part in Figure 1. Activation of c-Myc and NF-κβ induce transcription of Mcl-1. It is also rapidly transcribed via a PI3K/AKT-dependent pathway, resulting in its increased expression during myeloid differentiation and cytokine stimulation [97–99], and it is phosphorylated upon oxidative stress and cytokine withdrawal [100, 101]. Increased transcript levels lead to higher protein levels and proteins such as Pim kinase (induced by microenvironment) and 4E-BP1 assist in this pathway (Figure 1, pathway II). Phosphorylation at Thr163, the conserved MAP kinase/ERK site located within PEST region, slows Mcl-1 protein turnover (Figure 1, pathway III) [102], but may prime the GSK-3β mediated phosphorylation at Ser159 that leads to Mcl-1 stabilization (Figure 1, pathway I) [103]. Microenvironment-mediated phosphorylation by Akt inactivates GSK-3β resulting in inhibition of proteasomal degradation of Mcl-1. Microenvironment factors also stimulate JNK pathways, and JNK1 phosphorylates Mcl-1 at Ser64 resulting in increased Mcl-1 anti-apoptotic function and binding and sequestering pro-apoptotic proteins (Figure 1, pathway IV) [104, 105]. Finally Mcl-1 has a selective ubiquitinating and deubiquitinating enzymes, MULE [106] and USP9X [107] respectively. In contrast to Mcl-1, Bcl-2 is a long-lived protein [108].
Figure 1.
Microenvironment-mediated transcription, translation, and post-translational modifications of Mcl-1 anti-apoptotic protein. (I) Accelerated degradation of Mcl-1 protein following phosphorylation at Ser159 by GSK-3. (II) Increased Mcl-1 protein levels from Pim/c-Myc driven gene expression. (III) Mcl-1 protein stabilization via phosphorylation at Thr92 and Thr163 by ERK. (IV) Increased Mcl-1 anti-apoptotic function mediated by phosphorylation at Ser64 by JNK/CDKs resulting in increased binding and sequestering of pro-apoptotic proteins.
Phosphorylation of pro-apoptotic protein Bad via PI3K/AKT and Pim kinase pathways leads to its premature degradation and survival of cancer cells. This will be further discussed in the Pim kinase section of this review. Taken together, the extracellular signals from cytokines and chemokines, the contribution of transcriptional factors and post-translational modifications on anti-apoptotic proteins ultimately form a complex network to deliver microenvironmental support to the malignant cells.
Targeting Bcl-2 survival pathways
The recent advancement in understanding the primary reasons for extended survival of CLL cells had proposed many strategies to inhibit anti-apoptotic proteins in this tumor model. One key approach is to employ synthetic BH3 mimetics to modulate the function of Bcl-2 [109]. Bcl-2 inhibitors are synthesized in a manner that they mimic the structure and/or function of BH3 domain of the pro-apoptotic proteins and competitively bind to anti-apoptotic proteins with greater affinity to displace the sequestered pro-apoptotic proteins.
Obatoclax, an indole bipyrrole compound was specifically designed to inhibit all relevant anti-apoptotic members of the Bcl-2 family (Bcl-2, Bcl-XL, Mcl-1, Bcl-W, Bcl2-A1, and Bcl-B) with an IC50 of 1–7 µM [110] . Preclinical studies with obatoclax revealed promising results in multiple myeloma [111], acute myeloid leukemia [112] and mantle cell lymphoma [113] and solid tumors [114, 115]. It is first in its class of BH3 mimetics to enter clinical trials [116].
Polyphenol compound gossypol is a natural product BH3 mimetic, designed to inhibit all 3 major anti-apoptotic proteins, Bcl-2, Bcl-XL and Mcl-1 with Ki values of 35 nM , 600 nM and 25 nM respectively[109]. Our investigations using CLL primary cells revealed that gossypol, a BH3-mimetic, induces caspase independent apoptosis; required Bax recruitment to the mitochondrial membrane to trigger cytochrome c and AIF release into the cytosol [117]. Although gossypol was tested clinically [118]-[119], mainly this compound has been considered a lead compound for the development of a new class of antineoplastic agents.
AT-101 is ℓ-isomer of gossypol, developed as more potent, less toxic compound compared to gossypol with added oral bio-availability. Similar to gossypol, AT-101 binds to the BH3 motif of all major anti-apoptotic proteins, besides with greater affinity than the parent compound (e.g., 230, 570, and 130 nM for Bcl-2, Bcl-XL, and Mcl-1, respectively) [120]. Extensive studies with single agent AT-101 in CLL demonstrated promising results [120, 121] and its combination with monoclonal antibody rituximab exhibited synergy [122]. Additionally, AT-101 by far overcame the protective effect conferred by nurse like cells in CLL [120] or bone-marrow stroma cells [121]. Phase I study of single agent AT-101 in advanced malignancies [123] and an open-label phase II study to evaluate the safety and efficacy in combination with rituximab in patients with relapsed or refractory CLL [124] have been closed, however, phase I randomized study in combination with chemotherapy or chemoradiotherapy in solid tumors are ongoing.
The ABT-737, is a small molecule Bcl-2 inhibitor, designed to mimic BH3 domain of BAD protein [125]. In comparison to other Bcl-2 inhibitors, ABT-737 displays high potency, with superior affinity to Bcl-2, Bcl-XL, and Bcl-W (Ki, <1 nmol/L for all three proteins) [126]. Unlike other pan Bcl-2 inhibitors, ABT-737 has a constraint that it does not bind to other members in the Bcl-2 family, such as Mcl-1 or A1 [127]. In recent years, ABT-737 has been studied widely in many types of cancer systems. This compound exhibits anti-tumor activity in broad range of tumor models including solid tumors [126] and hematological malignancies [128–130]. Mechanistically, this compound primarily induces apoptosis through mitochondrial pathway, the hallmark feature of intrinsic apoptotic pathway by disrupting Bcl-2/Bax association [128] or dissociating Bim from Bcl-2 [131]. Thus, ABT-737 classically represents the function of a BH3 only protein either by activating pro-apoptotic Bcl-2 proteins (Bak/Bax) or by inhibiting anti-apoptotic protein functions (Bcl-2/Bcl-XL). In CLL cells, ABT-737 showed nanomolar efficacy (EC50 of 4.5 ± 2.2 nM) [129] in displacing Bim from Bcl-2 to promote cell death, signifying Bcl-2 complexed to Bim is the critical target for ABT-737 in CLL [129].
Pim Kinases as pro-survival members in CLL cells
Pim kinase family
While targeting Bcl-2 family proteins is one strategy, the other is to identify Pim kinases as pro survival members of CLL cells. Proviral integration of murine leukemia viral (Pim) kinases were discovered in 1984 [132], and are anti-apoptotic serine/threonine kinases. Increased levels of these proteins have been implicated in transcription regulation, cell survival, tumorigenesis/leukemogenesis and resistance to cytotoxic agents (reviewed by Amaravadi [133]), and Pim kinases have been identified to be over-expressed in leukemias (reviewed by Shah[134]). Three Pim kinases have been identified to date (Pim-1, -2 and -3), each with variant isoforms of expressed protein due to alternative translational start sites [135]. At the amino acid level, there is substantial homology between Pim-1 and Pim-2 (53%) [136] and Pim-3 (69%) [137]. The three proteins have overlapping functions and compensate for one another and Pim-1 knockout experiments identified high frequency of activation of Pim-2 [138].
Function
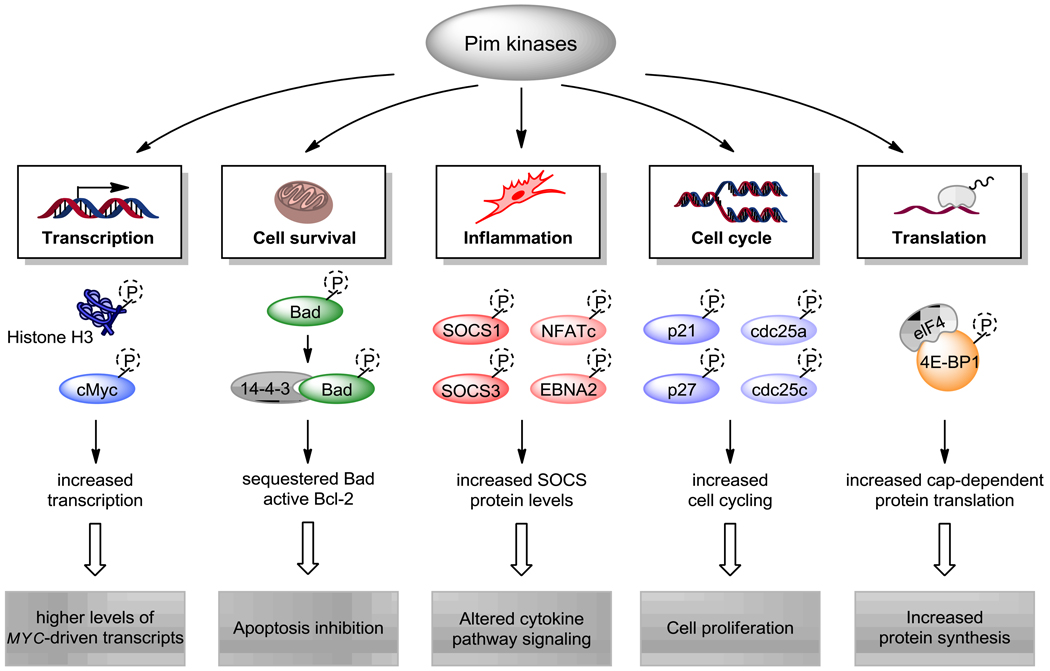
Pim kinases are constitutively active and their activity affects a number of cellular processes [139–141]. An ever-increasing number of proteins have been identified as phosphorylation targets by Pim-1, and a selection of targets and their functions are shown in Figure 2. Phosphorylation of Histone H3 at Ser10 by Pim-1 is necessary for MYC-dependent transcriptional activation and oncogenic transformation.[142] In addition, c-Myc phosphorylation at Ser62 by Pim kinases results in the stabilization of c-Myc protein.[143] These actions support the generation of oncogene transcripts and the sustenance of oncogenic protein levels. Many of c-Myc-target oncogenes have short transcript half-lives and thus inhibition of Pim kinase activity would reduce the expression levels of these oncoproteins. MCL-1 is one such oncogene (Figure 1, pathway II) , and disruption of Pim function may be instrumental in killing cancer cells whose survival is dependent on such oncogenes (reviewed in [144]).
Figure 2.
Pim kinase targets and their roles in inflammation and cancer. Pim kinases phosphorylate a number of substrates such as histone H3 and c-Myc, which activate Myc-driven transcription. Apoptotic protein targets of Pim kinases include pro-apoptotic Bad protein, and Bad phosphorylation allows binding with scaffold protein 14-3-3 and the inhibition of apoptosis. In the inflammatory response, Pim phosphorylation of various targets results in stabilization of SOCS proteins, which leads to cytokine pathway signaling. Cell cycle (p21, p27, cdc25a and cdc25c) protein phosphorylation removes cell cycle checkpoints and induces proliferation. Cap-dependent protein translation is increased by phosphorylation of 4E-BP1, a Pim kinase target. Collectively, phosphorylation of these Pim kinase targets results in increased transcription, protein synthesis, inflammatory response, cell survival and proliferation.
The survival advantage provided by Pim kinases is not limited to driving the expression of MYC target genes, but also via the disruption of apoptotic signaling pathways. PIM1 has been shown to efficiently cooperate BCL2 genes in lymphomagenesis,[145] and the simultaneous upregulation of Pim-1 and anti-apoptotic protein A1 is required for Bcr-Abl-induced leukemogenesis [146]. Pim kinases are involved with the regulation of pro- and anti-apoptotic Bcl-2 family proteins, and one of the targets of Pim kinase phosphorylation is pro-apoptotic Bad protein. All three Pim kinases phosphorylate Bad with varying specificities for the different phosphorylation sites (Ser112, Ser136 and Ser155) [139, 147], however the predominant target is the Ser112 “gatekeeper” site [148]. Bad protein phosphorylation at Ser112 by Pim-1 enhances phosphorylation at Ser136 by other kinases such as Akt[148]. Phosphorylation of Bad protein allows binding to 14-3-3 scaffold protein, which prevents 14-3-3 binding to anti-apoptotic Bcl-2 or Bcl-XL proteins and promotes cell survival [149, 150].
Regarding cell cycle regulation, Pim kinases have been shown to phosphorylate cell cycle proteins such as cyclin-dependent kinase (CDK) inhibitors p21 and p27, and phosphatases cdc25a and cdc25c. Pim-1 function on p21 may contribute to tumorigenesis in part via stabilization of p21 and subsequent promoting cell proliferation. Thr145 site on p21 is a Pim-1 target, and its phosphorylation results in the stabilization of p21 protein and disruption of the association between proliferating cell nuclear antigen (PCNA) and p21 [151]. p27 is also a CDK inhibitor of abnormal cell cycle progression, and Pim kinases promote cell cycle progression and tumorigenesis via the down-regulation of p27 expression at both the mRNA and protein level [152]. Pim-mediated suppression of p27 transcription is via the phosphorylation and inactivation of forkhead transcription factors, FoxO1a and FoxO3a. All three Pim kinases can phosphorylate p27 at Thr157 and Thr198, which as with Bad protein, allows binding to 14-3-3 protein resulting in nuclear export and proteasome-dependent degradation. Similarly, both cdc25a and cdc25c are substrates of Pim kinases, and phosphorylation of these cell cycle phosphatase positively regulate cell cycling [153, 154]. In addition to cell cycle effects, Pim kinases may contribute to proliferation via regulation of protein translation. Pim-2 phosphorylates translation repressor protein 4E-BP1 at Ser65, which is the site required for inhibition of binding to eIF4E and formation of the active translational initiation complex [135, 155, 156]. Phosphorylation at Ser65 also inhibits 4E-BP1 pro-apoptotic activity [157], and thus Pim kinase induction of cell survival signaling may be in part through 4E-BP1.
Degradation
Pim kinases interact and phosphorylate SOCS1 and SOCS3 [158, 159], which are negative regulators of STAT-driven Pim expression. The stability and function of Pim-1 is further regulated in part by phosphatase PP2A [160] and heat-shock proteins (HSPs) and through the ubiquitin-proteosome pathway [141, 161]. Through interaction with PP2A, dephosphorylation of Pim-1 allows for ubiquitinylation and protein degradation of Pim-1 [160]. With respect to HSPs, in PIM1-transfected BAF-B03 cells, an interleukin 3 (IL-3)-dependent murine pro-B-cell line, both HSP90α and HSP90β were identified to interact with Pim-1. Furthermore, treatment with HSP90 inhibitor, geldanamycin, resulted in the reduction in both Pim-1 kinase activity and protein stability [161]. Pim-1 is tagged with ubiquitin by HSP70 for degradation by the proteosome, whereas HSP90 binding of Pim-1 provides protection from proteosome degradation [141].
Hypoxic conditions can also alter Pim-1 expression and degradation, and hypoxia increases Pim-1 in several cancer cell lines in a hypoxia-inducible factor-1alpha-independent manner [162]. In pancreatic cancer cell line PCI-43, hypoxic conditions result in the stabilization of Pim-1 via prevention of its ubiquitin-mediated proteasomal degradation [163]. This increase in Pim-1 protein contributes to solid tumor progression and hypoxia-induced chemoresistance, and thus Pim-1 may act as a survival factor under hypoxia.
Microenvironment impact on Pim kinase expression
Pim kinases are transcriptionally regulated and are targets of a variety of growth factor-dependent transcriptional responses [135, 164]. Stromal cell co-culture results in STAT5 activation and increased expression of PIM1 transcript [165], and IL-5 enhances gene expression of MYC, and PIM-1 in activated B cells [166]. In leukemic cells, previous studies in our laboratory and other groups have shown that Pim-2 RNA and protein levels are higher in primary CLL cells compared with normal lymphocytes [31, 167]. The mechanism of increased Pim-2 expression in CLL is unknown, but its increased expression may provide a survival advantage to CLL cells. Microenvironment factors may play a key role in upregulation of Pim kinases and interestingly, Pim kinases themselves may provide a feedback loop as recent advances have shed light on Pim kinase function on microenvironment signaling. CXCR4 is phoshorylated by Pim-1 at Ser339, a site known to be essential for normal receptor recycling [168].
The proposal of CLL primary cells co-cultured with microenvironment cells as a model system for inflammation is supported by investigations in various other hematopoietic cell types that are involved with inflammation. In eosinophils, Pim kinases are involved with inflammation induced by allergic response. Allergic inflammation is characterized by elevated eosinophil numbers and by the increased production of the cytokines IL-5 and GM-CSF, which control several eosinophil functions, including the suppression of apoptosis. Treatment of human eosinophils with IL-5 or GM-CSF results in the activation of STAT3 and STAT5 signaling, and as described earlier, Pim-1 is a target gene of STAT5. This signaling activation induced the protein expression of cyclin D3 and Pim-1, and Pim-1 action is linked to the suppression of eosinophil apoptosis by these cytokines [169]. In basophils, the contribution of these cells in allergic disease depends on their persistence at sites of inflammation (reviewed in [170]) Basophils have a considerably longer spontaneous life span than neutrophils and eosinophils [171, 172], and IL-3 protects basophils from apoptosis through the upregulation of the protein expression of a number of anti-apoptotic proteins including cIAP2, Mcl-1, and Bcl-XL and Pim-1, which is correlated with enhanced cell survival [171].
Targeting Pim kinase pathways
The therapeutic strategy of Pim kinase inhibitors are to block critical signaling pathways involved with cell survival or proliferation to induce cell death in malignant cells. Pim kinase overexpression in malignant cells renders them more susceptible to Pim kinase inhibition, and thus may also be a method to limit toxicity to normal healthy cells. There are currently a number of Pim kinase inhibitors at the preclinical development stage, including a recent report on SGI-1776 from our laboratory [31]. Our studies demonstrated that SGI-1776 was preferentially cytotoxic in towards primary CLL cells compared with normal lymphocytes from healthy donors, and resulted in the decrease of both Mcl-1 protein and transcript level in treated cells. Other Pim inhibitors from both academic institutions [173–175] and pharmaceutical companies are also under development (reviewed in [176]) with varying levels of specificity towards Pim kinases. There has also been a recent report describing the development of monoclonal antibodies against cell surface Pim-1 [177]. The use of Pim kinase inhibitors is in an early stage in development, however, may be a valuable new class of therapeutics for the treatment of cancers.
Conclusion
The pathway similarities between CLL cultured in the microenvironment and inflammation leads to speculation of whether BH3-mimetics or Pim kinase inhibitors may have anti-inflammatory properties. Furthermore, because there is a cross-talk between these two pathways, a combination of two could be explored. Since prevention is a favorable option for inflammation, these agents could also be investigated for such potentials.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Byrd JC, Stilgenbauer S, Flinn IW. Chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2004:163–183. doi: 10.1182/asheducation-2004.1.163. [DOI] [PubMed] [Google Scholar]
- 2.D'Arena G, Di Renzo N, Brugiatelli M, Vigliotti ML, Keating MJ. Biological and clinical heterogeneity of B-cell chronic lymphocytic leukemia. Leuk Lymphoma. 2003;44:223–228. doi: 10.1080/1042819021000035756. [DOI] [PubMed] [Google Scholar]
- 3.Keating MJ. Management of chronic lymphocytic leukemia: a changing field. Rev Clin Exp Hematol. 2002;6:350–365. doi: 10.1046/j.1468-0734.2002.00303.x. discussion 449–50. [DOI] [PubMed] [Google Scholar]
- 4.Keating MJ, Chiorazzi N, Messmer B, Damle RN, Allen SL, Rai KR, et al. Biology and treatment of chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2003:153–175. doi: 10.1182/asheducation-2003.1.153. [DOI] [PubMed] [Google Scholar]
- 5.Bannerji R, Byrd JC. Update on the biology of chronic lymphocytic leukemia. Curr Opin Oncol. 2000;12:22–29. doi: 10.1097/00001622-200001000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Keating MJ. Chronic lymphocytic leukemia. Semin Oncol. 1999;26:107–114. [PubMed] [Google Scholar]
- 7.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 8.Decker T, Hipp S, Ringshausen I, Bogner C, Oelsner M, Schneller F, et al. Rapamycin- induced G1 arrest in cycling B-CLL cells is associated with reduced expression of cyclin D3, cyclin E, cyclin A, and survivin. Blood. 2003;101:278–285. doi: 10.1182/blood-2002-01-0189. [DOI] [PubMed] [Google Scholar]
- 9.Hengartner MO, Horvitz HRC. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 10.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 11.McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- 12.Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 13.Ke N, Godzik A, Reed JC. Bcl-B, a novel Bcl-2 family member that differentially binds and regulates Bax and Bak. J Biol Chem. 2001;276:12481–12484. doi: 10.1074/jbc.C000871200. [DOI] [PubMed] [Google Scholar]
- 14.Gibson L, Holmgreen SP, Huang DC, Bernard O, Copeland NG, Jenkins NA, et al. bcl-w, a novel member of the bcl-2 family, promotes cell survival. Oncogene. 1996;13:665–675. [PubMed] [Google Scholar]
- 15.Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci U S A. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin EY, Orlofsky A, Berger MS, Prystowsky MB. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J Immunol. 1993;151:1979–1988. [PubMed] [Google Scholar]
- 17.Reed JC. Bcl-2-family proteins and hematologic malignancies: history and future prospects. Blood. 2008;111:3322–3330. doi: 10.1182/blood-2007-09-078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krajewski S, Bodrug S, Krajewska M, Shabaik A, Gascoyne R, Berean K, et al. Immunohistochemical analysis of Mcl-1 protein in human tissues. Differential regulation of Mcl-1 and Bcl-2 protein production suggests a unique role for Mcl-1 in control of programmed cell death in vivo. Am J Pathol. 1995;146:1309–1319. [PMC free article] [PubMed] [Google Scholar]
- 19.Gottardi D, Alfarano A, De Leo AM, Stacchini A, Aragno M, Rigo A, et al. In leukaemic CD5+ B cells the expression of BCL-2 gene family is shifted toward protection from apoptosis. Br J Haematol. 1996;94:612–618. doi: 10.1046/j.1365-2141.1996.d01-1856.x. [DOI] [PubMed] [Google Scholar]
- 20.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 21.Bullrich F, Veronese ML, Kitada S, Jurlander J, Caligiuri MA, Reed JC, et al. Minimal region of loss at 13q14 in B-cell chronic lymphocytic leukemia. Blood. 1996;88:3109–3115. [PubMed] [Google Scholar]
- 22.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 23.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motoyama N, Wang F, Roth KA, Sawa H, Nakayama K, Negishi I, et al. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 25.Veis DJ, Sorenson CM, Shutter JR, Korsmeyer SJ. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 1993;75:229–240. doi: 10.1016/0092-8674(93)80065-m. [DOI] [PubMed] [Google Scholar]
- 26.Kitada S, Andersen J, Akar S, Zapata JM, Takayama S, Krajewski S, et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with In vitro and In vivo chemoresponses. Blood. 1998;91:3379–3389. [PubMed] [Google Scholar]
- 27.Kaufmann SH, Karp JE, Svingen PA, Krajewski S, Burke PJ, Gore SD, et al. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998;91:991–1000. [PubMed] [Google Scholar]
- 28.Derenne S, Monia B, Dean NM, Taylor JK, Rapp MJ, Harousseau JL, et al. Antisense strategy shows that Mcl-1 rather than Bcl-2 or Bcl-x(L) is an essential survival protein of human myeloma cells. Blood. 2002;100:194–199. doi: 10.1182/blood.v100.1.194. [DOI] [PubMed] [Google Scholar]
- 29.Balakrishnan K, Stellrecht CM, Genini D, Ayres M, Wierda WG, Keating MJ, et al. Cell death of bioenergetically compromised and transcriptionally challenged CLL lymphocytes by chlorinated ATP. Blood. 2005;105:4455–4462. doi: 10.1182/blood-2004-05-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balakrishnan K, Wierda WG, Keating MJ, Gandhi V. Mechanisms of cell death of chronic lymphocytic leukemia lymphocytes by RNA-directed agent, 8-NH2-adenosine. Clin Cancer Res. 2005;11:6745–6752. doi: 10.1158/1078-0432.CCR-05-0553. [DOI] [PubMed] [Google Scholar]
- 31.Chen LS, Redkar S, Bearss D, Wierda WG, Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009;114:4150–4157. doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R, Keating MJ, Gandhi V, Plunkett W. Transcription inhibition by flavopiridol: mechanism of chronic lymphocytic leukemia cell death. Blood. 2005;106:2513–2519. doi: 10.1182/blood-2005-04-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glassman AB, Hayes KJ. The value of fluorescence in situ hybridization in the diagnosis and prognosis of chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2005;158:88–91. doi: 10.1016/j.cancergencyto.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Kitada S, Zapata JM, Andreeff M, Reed JC. Protein kinase inhibitors flavopiridol and 7- hydroxy-staurosporine down-regulate antiapoptosis proteins in B-cell chronic lymphocytic leukemia. Blood. 2000;96:393–397. [PubMed] [Google Scholar]
- 35.O'Brien S, Moore JO, Boyd TE, Larratt LM, Skotnicki A, Koziner B, et al. Randomized phase III trial of fludarabine plus cyclophosphamide with or without oblimersen sodium (Bcl-2 antisense) in patients with relapsed or refractory chronic lymphocytic leukemia. J Clin Oncol. 2007;25:1114–1120. doi: 10.1200/JCO.2006.07.1191. [DOI] [PubMed] [Google Scholar]
- 36.Byrd JC, Kitada S, Flinn IW, Aron JL, Pearson M, Lucas D, et al. The mechanism of tumor cell clearance by rituximab in vivo in patients with B-cell chronic lymphocytic leukemia: evidence of caspase activation and apoptosis induction. Blood. 2002;99:1038–1043. doi: 10.1182/blood.v99.3.1038. [DOI] [PubMed] [Google Scholar]
- 37.Balakrishnan K, Burger JA, Quiroga MP, Henneberg M, Ayres ML, Wierda WG, et al. Influence of bone marrow stromal microenvironment on forodesine-induced response in CLL primary cells. Blood. 2010 doi: 10.1182/blood-2009-10-246199. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burger JA, Tsukada N, Burger M, Zvaifler NJ, Dell'Aquila M, Kipps TJ. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood. 2000;96:2655–2663. [PubMed] [Google Scholar]
- 39.Ding Q, Huo L, Yang JY, Xia W, Wei Y, Liao Y, et al. Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res. 2008;68:6109–6117. doi: 10.1158/0008-5472.CAN-08-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandhi V, Balakrishnan K, Chen LS. Mcl-1: the 1 in CLL. Blood. 2008;112:3538–3540. doi: 10.1182/blood-2008-07-170241. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh AK, Secreto CR, Knox TR, Ding W, Mukhopadhyay D, Kay NE. Circulating microvesicles in B-cell chronic lymphocytic leukemia can stimulate marrow stromal cells: implications for disease progression. Blood. 2010;115:1755–1764. doi: 10.1182/blood-2009-09-242719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurtova AV, Balakrishnan K, Chen R, Ding W, Schnabl S, Quiroga MP, et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood. 2009;114:4441–4450. doi: 10.1182/blood-2009-07-233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen IM, Kitada S, Leoni LM, Zapata JM, Karras JG, Tsukada N, et al. Protection of CLL B cells by a follicular dendritic cell line is dependent on induction of Mcl-1. Blood. 2002;100:1795–1801. [PubMed] [Google Scholar]
- 44.Quiroga MP, Balakrishnan K, Kurtova AV, Sivina M, Keating MJ, Wierda WG, et al. B- cell antigen receptor signaling enhances chronic lymphocytic leukemia cell migration and survival: specific targeting with a novel spleen tyrosine kinase inhibitor, R406. Blood. 2009;114:1029–1037. doi: 10.1182/blood-2009-03-212837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukada N, Burger JA, Zvaifler NJ, Kipps TJ. Distinctive features of "nurselike" cells that differentiate in the context of chronic lymphocytic leukemia. Blood. 2002;99:1030–1037. doi: 10.1182/blood.v99.3.1030. [DOI] [PubMed] [Google Scholar]
- 46.Pepper C, Lin TT, Pratt G, Hewamana S, Brennan P, Hiller L, et al. Mcl-1 expression has in vitro and in vivo significance in chronic lymphocytic leukemia and is associated with other poor prognostic markers. Blood. 2008;112:3807–3817. doi: 10.1182/blood-2008-05-157131. [DOI] [PubMed] [Google Scholar]
- 47.Panayiotidis P, Jones D, Ganeshaguru K, Foroni L, Hoffbrand AV. Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocytic leukaemia cells in vitro. Br J Haematol. 1996;92:97–103. doi: 10.1046/j.1365-2141.1996.00305.x. [DOI] [PubMed] [Google Scholar]
- 48.Caligaris-Cappio F. Role of the microenvironment in chronic lymphocytic leukaemia. Br J Haematol. 2003;123:380–388. doi: 10.1046/j.1365-2141.2003.04679.x. [DOI] [PubMed] [Google Scholar]
- 49.Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123–128. doi: 10.1038/84219. [DOI] [PubMed] [Google Scholar]
- 50.Kim CH, Broxmeyer HE. Chemokines: signal lamps for trafficking of T and B cells for development and effector function. J Leukoc Biol. 1999;65:6–15. doi: 10.1002/jlb.65.1.6. [DOI] [PubMed] [Google Scholar]
- 51.Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc Natl Acad Sci U S A. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loetscher P, Moser B, Baggiolini M. Chemokines and their receptors in lymphocyte traffic and HIV infection. Adv Immunol. 2000;74:127–180. doi: 10.1016/s0065-2776(08)60910-4. [DOI] [PubMed] [Google Scholar]
- 54.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 55.Deaglio S, Capobianco A, Bergui L, Durig J, Morabito F, Duhrsen U, et al. CD38 is a signaling molecule in B-cell chronic lymphocytic leukemia cells. Blood. 2003;102:2146–2155. doi: 10.1182/blood-2003-03-0989. [DOI] [PubMed] [Google Scholar]
- 56.Zucchetto A, Benedetti D, Tripodo C, Bomben R, Dal Bo M, Marconi D, et al. CD38/CD31, the CCL3 and CCL4 chemokines, and CD49d/vascular cell adhesion molecule-1 are interchained by sequential events sustaining chronic lymphocytic leukemia cell survival. Cancer Res. 2009;69:4001–4009. doi: 10.1158/0008-5472.CAN-08-4173. [DOI] [PubMed] [Google Scholar]
- 57.Deaglio S, Vaisitti T, Bergui L, Bonello L, Horenstein AL, Tamagnone L, et al. CD38 and CD100 lead a network of surface receptors relaying positive signals for B-CLL growth and survival. Blood. 2005;105:3042–3050. doi: 10.1182/blood-2004-10-3873. [DOI] [PubMed] [Google Scholar]
- 58.Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94:3658–3667. [PubMed] [Google Scholar]
- 59.Mohle R, Failenschmid C, Bautz F, Kanz L. Overexpression of the chemokine receptor CXCR4 in B cell chronic lymphocytic leukemia is associated with increased functional response to stromal cell-derived factor-1 (SDF-1) Leukemia. 1999;13:1954–1959. doi: 10.1038/sj.leu.2401602. [DOI] [PubMed] [Google Scholar]
- 60.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 61.Kast RE, Altschuler EL. Anti-apoptosis function of TNF-alpha in chronic lymphocytic leukemia: lessons from Crohn's disease and the therapeutic potential of bupropion to lower TNF-alpha. Arch Immunol Ther Exp (Warsz) 2005;53:143–147. [PubMed] [Google Scholar]
- 62.Waage A, Espevik T. TNF receptors in chronic lymphocytic leukemia. Leuk Lymphoma. 1994;13:41–46. doi: 10.3109/10428199409051650. [DOI] [PubMed] [Google Scholar]
- 63.Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 65.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 66.Sasson SC, Smith S, Seddiki N, Zaunders JJ, Bryant A, Koelsch KK, et al. IL-7 receptor is expressed on adult pre-B-cell acute lymphoblastic leukemia and other B-cell derived neoplasms and correlates with expression of proliferation and survival markers. Cytokine. 50:58–68. doi: 10.1016/j.cyto.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 67.Gowda A, Ramanunni A, Cheney C, Rozewski D, Kindsvogel W, Lehman A, et al. Differential effects of IL-2 and IL-21 on expansion of the CD4+ CD25+ Foxp3+ T regulatory cells with redundant roles in natural killer cell mediated antibody dependent cellular cytotoxicity in chronic lymphocytic leukemia. MAbs. 2:35–41. doi: 10.4161/mabs.2.1.10561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai R, O'Brien S, Maushouri T, Rogers A, Kantarjian H, Keating M, et al. Prognostic value of plasma interleukin-6 levels in patients with chronic lymphocytic leukemia. Cancer. 2002;95:1071–1075. doi: 10.1002/cncr.10772. [DOI] [PubMed] [Google Scholar]
- 69.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 71.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 72.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huh YO, Lin KI, Vega F, Schlette E, Yin CC, Keating MJ, et al. MYC translocation in chronic lymphocytic leukaemia is associated with increased prolymphocytes and a poor prognosis. Br J Haematol. 2008;142:36–44. doi: 10.1111/j.1365-2141.2008.07152.x. [DOI] [PubMed] [Google Scholar]
- 74.Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, et al. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood. 115:2852–2863. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cuni S, Perez-Aciego P, Perez-Chacon G, Vargas JA, Sanchez A, Martin-Saavedra FM, et al. A sustained activation of PI3K/NF-kappaB pathway is critical for the survival of chronic lymphocytic leukemia B cells. Leukemia. 2004;18:1391–1400. doi: 10.1038/sj.leu.2403398. [DOI] [PubMed] [Google Scholar]
- 76.Furman RR, Asgary Z, Mascarenhas JO, Liou HC, Schattner EJ. Modulation of NF-kappa B activity and apoptosis in chronic lymphocytic leukemia B cells. J Immunol. 2000;164:2200–2206. doi: 10.4049/jimmunol.164.4.2200. [DOI] [PubMed] [Google Scholar]
- 77.Shen HM, Tergaonkar V. NFkappaB signaling in carcinogenesis and as a potential molecular target for cancer therapy. Apoptosis. 2009;14:348–363. doi: 10.1007/s10495-009-0315-0. [DOI] [PubMed] [Google Scholar]
- 78.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Helbig G, Christopherson KW, 2nd, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, et al. NF-kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278:21631–21638. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 80.Taylor ST, Hickman JA, Dive C. Survival signals within the tumour microenvironment suppress drug-induced apoptosis: lessons learned from B lymphomas. Endocr Relat Cancer. 1999;6:21–23. doi: 10.1677/erc.0.0060021. [DOI] [PubMed] [Google Scholar]
- 81.Endo T, Nishio M, Enzler T, Cottam HB, Fukuda T, James DF, et al. BAFF and APRIL support chronic lymphocytic leukemia B-cell survival through activation of the canonical NF-kappaB pathway. Blood. 2007;109:703–710. doi: 10.1182/blood-2007-04-081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pekarsky Y, Palamarchuk A, Maximov V, Efanov A, Nazaryan N, Santanam U, et al. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci U S A. 2008;105:19643–19648. doi: 10.1073/pnas.0810965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rossi AG, Hallett JM, Sawatzky DA, Teixeira MM, Haslett C. Modulation of granulocyte apoptosis can influence the resolution of inflammation. Biochem Soc Trans. 2007;35:288–291. doi: 10.1042/BST0350288. [DOI] [PubMed] [Google Scholar]
- 84.Hamzaoui K, Hamzaoui A, Zakraoui L, Chabbou A. Expression of Bcl-2 in inflammatory sites from patients with active Behcet's disease. Mediators Inflamm. 1999;8:101–106. doi: 10.1080/09629359990595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sawatzky DA, Willoughby DA, Colville-Nash PR, Rossi AG. The involvement of the apoptosis-modulating proteins ERK 1/2, Bcl-xL and Bax in the resolution of acute inflammation in vivo. Am J Pathol. 2006;168:33–41. doi: 10.2353/ajpath.2006.050058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.May WS, Tyler PG, Ito T, Armstrong DK, Qatsha KA, Davidson NE. Interleukin-3 and bryostatin-1 mediate hyperphosphorylation of BCL2 alpha in association with suppression of apoptosis. J Biol Chem. 1994;269:26865–26870. [PubMed] [Google Scholar]
- 87.Ito T, Deng X, Carr B, May WS. Bcl-2 phosphorylation required for anti-apoptosis function. J Biol Chem. 1997;272:11671–11673. doi: 10.1074/jbc.272.18.11671. [DOI] [PubMed] [Google Scholar]
- 88.Ruvolo PP, Deng X, May WS. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia. 2001;15:515–522. doi: 10.1038/sj.leu.2402090. [DOI] [PubMed] [Google Scholar]
- 89.Perez-Galan P, Roue G, Lopez-Guerra M, Nguyen M, Villamor N, Montserrat E, et al. BCL-2 phosphorylation modulates sensitivity to the BH3 mimetic GX15-070 (Obatoclax) and reduces its synergistic interaction with bortezomib in chronic lymphocytic leukemia cells. Leukemia. 2008;22:1712–1720. doi: 10.1038/leu.2008.175. [DOI] [PubMed] [Google Scholar]
- 90.Kurinna S, Konopleva M, Palla SL, Chen W, Kornblau S, Contractor R, et al. Bcl2 phosphorylation and active PKC alpha are associated with poor survival in AML. Leukemia. 2006;20:1316–1319. doi: 10.1038/sj.leu.2404248. [DOI] [PubMed] [Google Scholar]
- 91.Bassik MC, Scorrano L, Oakes SA, Pozzan T, Korsmeyer SJ. Phosphorylation of BCL-2 regulates ER Ca2+ homeostasis and apoptosis. EMBO J. 2004;23:1207–1216. doi: 10.1038/sj.emboj.7600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Breitschopf K, Haendeler J, Malchow P, Zeiher AM, Dimmeler S. Posttranslational modification of Bcl-2 facilitates its proteasome-dependent degradation: molecular characterization of the involved signaling pathway. Mol Cell Biol. 2000;20:1886–1896. doi: 10.1128/mcb.20.5.1886-1896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dimmeler S, Breitschopf K, Haendeler J, Zeiher AM. Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J Exp Med. 1999;189:1815–1822. doi: 10.1084/jem.189.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 95.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herrant M, Jacquel A, Marchetti S, Belhacene N, Colosetti P, Luciano F, et al. Cleavage of Mcl-1 by caspases impaired its ability to counteract Bim-induced apoptosis. Oncogene. 2004;23:7863–7873. doi: 10.1038/sj.onc.1208069. [DOI] [PubMed] [Google Scholar]
- 97.Chao JR, Wang JM, Lee SF, Peng HW, Lin YH, Chou CH, et al. mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is one component of the GM-CSF viability response. Mol Cell Biol. 1998;18:4883–4898. doi: 10.1128/mcb.18.8.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jourdan M, Veyrune JL, De Vos J, Redal N, Couderc G, Klein B. A major role for Mcl-1 antiapoptotic protein in the IL-6-induced survival of human myeloma cells. Oncogene. 2003;22:2950–2959. doi: 10.1038/sj.onc.1206423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Domina AM, Smith JH, Craig RW. Myeloid cell leukemia 1 is phosphorylated through two distinct pathways, one associated with extracellular signal-regulated kinase activation and the other with G2/M accumulation or protein phosphatase 1/2A inhibition. J Biol Chem. 2000;275:21688–21694. doi: 10.1074/jbc.M000915200. [DOI] [PubMed] [Google Scholar]
- 101.Inoshita S, Takeda K, Hatai T, Terada Y, Sano M, Hata J, et al. Phosphorylation and inactivation of myeloid cell leukemia 1 by JNK in response to oxidative stress. J Biol Chem. 2002;277:43730–43734. doi: 10.1074/jbc.M207951200. [DOI] [PubMed] [Google Scholar]
- 102.Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23:5301–5315. doi: 10.1038/sj.onc.1207692. [DOI] [PubMed] [Google Scholar]
- 103.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 104.Kobayashi S, Lee SH, Meng XW, Mott JL, Bronk SF, Werneburg NW, et al. Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J Biol Chem. 2007;282:18407–18417. doi: 10.1074/jbc.M610010200. [DOI] [PubMed] [Google Scholar]
- 105.Scupoli MT, Donadelli M, Cioffi F, Rossi M, Perbellini O, Malpeli G, et al. Bone marrow stromal cells and the upregulation of interleukin-8 production in human T-cell acute lymphoblastic leukemia through the CXCL12/CXCR4 axis and the NF-kappaB and JNK/AP-1 pathways. Haematologica. 2008;93:524–532. doi: 10.3324/haematol.12098. [DOI] [PubMed] [Google Scholar]
- 106.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 107.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 108.Merino R, Ding L, Veis DJ, Korsmeyer SJ, Nunez G. Developmental regulation of the Bcl-2 protein and susceptibility to cell death in B lymphocytes. EMBO J. 1994;13:683–691. doi: 10.1002/j.1460-2075.1994.tb06307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kitada S, Leone M, Sareth S, Zhai D, Reed JC, Pellecchia M. Discovery, characterization, and structure-activity relationships studies of proapoptotic polyphenols targeting B-cell lymphocyte/leukemia-2 proteins. Journal of medicinal chemistry. 2003;46:4259–4264. doi: 10.1021/jm030190z. [DOI] [PubMed] [Google Scholar]
- 110.Zhai D, Jin C, Satterthwait AC, Reed JC. Comparison of chemical inhibitors of antiapoptotic Bcl-2-family proteins. Cell death and differentiation. 2006;13:1419–1421. doi: 10.1038/sj.cdd.4401937. [DOI] [PubMed] [Google Scholar]
- 111.Trudel S, Li ZH, Rauw J, Tiedemann RE, Wen XY, Stewart AK. Preclinical studies of the pan-Bcl inhibitor obatoclax (GX015-070) in multiple myeloma. Blood. 2007;109:5430–5438. doi: 10.1182/blood-2006-10-047951. [DOI] [PubMed] [Google Scholar]
- 112.Konopleva M, Watt J, Contractor R, Tsao T, Harris D, Estrov Z, et al. Mechanisms of antileukemic activity of the novel Bcl-2 homology domain-3 mimetic GX15-070 (obatoclax) Cancer Res. 2008;68:3413–3420. doi: 10.1158/0008-5472.CAN-07-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perez-Galan P, Roue G, Villamor N, Campo E, Colomer D. The BH3-mimetic GX15-070 synergizes with bortezomib in mantle cell lymphoma by enhancing Noxa-mediated activation of Bak. Blood. 2007;109:4441–4449. doi: 10.1182/blood-2006-07-034173. [DOI] [PubMed] [Google Scholar]
- 114.Mott JL, Bronk SF, Mesa RA, Kaufmann SH, Gores GJ. BH3-only protein mimetic obatoclax sensitizes cholangiocarcinoma cells to Apo2L/TRAIL-induced apoptosis. Mol Cancer Ther. 2008;7:2339–2347. doi: 10.1158/1535-7163.MCT-08-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Murthy Madiraju SR, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci U S A. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.O'Brien SM, Claxton DF, Crump M, Faderl S, Kipps T, Keating MJ, et al. Phase I study of obatoclax mesylate (GX15-070), a small molecule pan-Bcl-2 family antagonist, in patients with advanced chronic lymphocytic leukemia. Blood. 2009;113:299–305. doi: 10.1182/blood-2008-02-137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Balakrishnan K, Wierda WG, Keating MJ, Gandhi V. Gossypol, a BH3 mimetic, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2008;112:1971–1980. doi: 10.1182/blood-2007-12-126946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stein RC, Joseph AE, Matlin SA, Cunningham DC, Ford HT, Coombes RC. A preliminary clinical study of gossypol in advanced human cancer. Cancer Chemother Pharmacol. 1992;30:480–482. doi: 10.1007/BF00685601. [DOI] [PubMed] [Google Scholar]
- 119.Van Poznak C, Seidman AD, Reidenberg MM, Moasser MM, Sklarin N, Van Zee K, et al. Oral gossypol in the treatment of patients with refractory metastatic breast cancer: a phase I/II clinical trial. Breast Cancer Res Treat. 2001;66:239–248. doi: 10.1023/a:1010686204736. [DOI] [PubMed] [Google Scholar]
- 120.James DFMR, Mosadeghi R, Kipps TJ. AT-101, a pan-inhibitor of Bcl-2 family anti-apoptotic proteins, antagonizes the protective effect conferred by nurse-like cells on primary chronic lymphocytic leukemia cells. Blood. 2006;108:595a. [Google Scholar]
- 121.Balakrishnan K, Burger JA, Wierda WG, Gandhi V. AT-101 induces apoptosis in CLL B cells and overcomes stromal cell-mediated Mcl-1 induction and drug resistance. Blood. 2009;113:149–153. doi: 10.1182/blood-2008-02-138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.James DFPC, Castro JE, Kipps TJ. AT 101, an inhibitor of Bcl-2 family members, is cytotoxic to a heterogeneous group of primary CLL samples and synergistic with rituximab. Blood. 2005;106:835a. [Google Scholar]
- 123.Saleh MPH, Hartung J, Holmlund J, LoBuglio A, Forero A. Phase I trial of AT-101, an orally bioavailable inhibitor of Bcl-2, in patients with advanced malignancies; AACR-NCI-EORTC International Conference; 2005. p. 220. [Google Scholar]
- 124.Castro JELO, Aguillon RA, et al. A phase II, open label study of AT-101 in combination with rituximab in patients with relapsed or refractory chronic lymphocytic leukemia. Blood. 2006;108:803a. [Google Scholar]
- 125.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Molecular cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 126.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 127.van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 129.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. The Journal of clinical investigation. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Del Gaizo Moore V, Schlis KD, Sallan SE, Armstrong SA, Letai A. BCL-2 dependence and ABT-737 sensitivity in acute lymphoblastic leukemia. Blood. 2008;111:2300–2309. doi: 10.1182/blood-2007-06-098012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen S, Dai Y, Pei XY, Grant S. Bim Up-regulation by Histone Deacetylase Inhibitors Mediates Interactions with the Bcl-2 Antagonist ABT-737: Evidence for Distinct Roles for Bcl-2, Bcl-xL and Mcl-1. Molecular and cellular biology. 2009 doi: 10.1128/MCB.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37:141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 133.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shah N, Pang B, Yeoh KG, Thorn S, Chen CS, Lilly MB, et al. Potential roles for the PIM1 kinase in human cancer - a molecular and therapeutic appraisal. Eur J Cancer. 2008;44:2144–2151. doi: 10.1016/j.ejca.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 135.Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17:1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van der Lugt NM, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, et al. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. Embo J. 1995;14:2536–2544. doi: 10.1002/j.1460-2075.1995.tb07251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Konietzko U, Kauselmann G, Scafidi J, Staubli U, Mikkers H, Berns A, et al. Pim kinase expression is induced by LTP stimulation and required for the consolidation of enduring LTP. Embo J. 1999;18:3359–3369. doi: 10.1093/emboj/18.12.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Allen JD, Berns A. Complementation tagging of cooperating oncogenes in knockout mice. Semin Cancer Biol. 1996;7:299–306. doi: 10.1006/scbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- 139.Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur JS. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006;7:1. doi: 10.1186/1471-2121-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qian KC, Wang L, Hickey ER, Studts J, Barringer K, Peng C, et al. Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J Biol Chem. 2005;280:6130–6137. doi: 10.1074/jbc.M409123200. [DOI] [PubMed] [Google Scholar]
- 141.Shay KP, Wang Z, Xing PX, McKenzie IF, Magnuson NS. Pim-1 kinase stability is regulated by heat shock proteins and the ubiquitin-proteasome pathway. Mol Cancer Res. 2005;3:170–181. doi: 10.1158/1541-7786.MCR-04-0192. [DOI] [PubMed] [Google Scholar]
- 142.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9:932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 143.Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27:4809–4819. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- 144.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 145.Acton D, Domen J, Jacobs H, Vlaar M, Korsmeyer S, Berns A. Collaboration of pim-1 and bcl-2 in lymphomagenesis. Curr Top Microbiol Immunol. 1992;182:293–298. doi: 10.1007/978-3-642-77633-5_36. [DOI] [PubMed] [Google Scholar]
- 146.Nieborowska-Skorska M, Hoser G, Kossev P, Wasik MA, Skorski T. Complementary functions of the antiapoptotic protein A1 and serine/threonine kinase pim-1 in the BCR/ABL-mediated leukemogenesis. Blood. 2002;99:4531–4539. doi: 10.1182/blood.v99.12.4531. [DOI] [PubMed] [Google Scholar]
- 147.Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 2006;66:6741–6747. doi: 10.1158/0008-5472.CAN-05-4272. [DOI] [PubMed] [Google Scholar]
- 148.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571:43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 149.Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Molecular cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 150.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 151.Zhang Y, Wang Z, Magnuson NS. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res. 2007;5:909–922. doi: 10.1158/1541-7786.MCR-06-0388. [DOI] [PubMed] [Google Scholar]
- 152.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 153.Bachmann M, Kosan C, Xing PX, Montenarh M, Hoffmann I, Moroy T. The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int J Biochem Cell Biol. 2006;38:430–443. doi: 10.1016/j.biocel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 154.Mochizuki T, Kitanaka C, Noguchi K, Muramatsu T, Asai A, Kuchino Y. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999;274:18659–18666. doi: 10.1074/jbc.274.26.18659. [DOI] [PubMed] [Google Scholar]
- 155.Gong J, Wang J, Ren K, Liu C, Li B, Shi Y. Serine/threonine kinase Pim-2 promotes liver tumorigenesis induction through mediating survival and preventing apoptosis of liver cell. J Surg Res. 2009;153:17–22. doi: 10.1016/j.jss.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 156.Tamburini J, Green AS, Bardet V, Chapuis N, Park S, Willems L, et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood. 2009;114:1618–1627. doi: 10.1182/blood-2008-10-184515. [DOI] [PubMed] [Google Scholar]
- 157.Li S, Sonenberg N, Gingras AC, Peterson M, Avdulov S, Polunovsky VA, et al. Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol Cell Biol. 2002;22:2853–2861. doi: 10.1128/MCB.22.8.2853-2861.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Peltola KJ, Paukku K, Aho TL, Ruuska M, Silvennoinen O, Koskinen PJ. Pim-1 kinase inhibits STAT5-dependent transcription via its interactions with SOCS1 and SOCS3. Blood. 2004;103:3744–3750. doi: 10.1182/blood-2003-09-3126. [DOI] [PubMed] [Google Scholar]
- 159.Chen XP, Losman JA, Cowan S, Donahue E, Fay S, Vuong BQ, et al. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc Natl Acad Sci U S A. 2002;99:2175–2180. doi: 10.1073/pnas.042035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ma J, Arnold HK, Lilly MB, Sears RC, Kraft AS. Negative regulation of Pim-1 protein kinase levels by the B56beta subunit of PP2A. Oncogene. 2007;26:5145–5153. doi: 10.1038/sj.onc.1210323. [DOI] [PubMed] [Google Scholar]
- 161.Mizuno K, Shirogane T, Shinohara A, Iwamatsu A, Hibi M, Hirano T. Regulation of Pim-1 by Hsp90. Biochem Biophys Res Commun. 2001;281:663–669. doi: 10.1006/bbrc.2001.4405. [DOI] [PubMed] [Google Scholar]
- 162.Chen J, Kobayashi M, Darmanin S, Qiao Y, Gully C, Zhao R, et al. Pim-1 plays a pivotal role in hypoxia-induced chemoresistance. Oncogene. 2009;28:2581–2592. doi: 10.1038/onc.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen J, Kobayashi M, Darmanin S, Qiao Y, Gully C, Zhao R, et al. Hypoxia-mediated up-regulation of Pim-1 contributes to solid tumor formation. Am J Pathol. 2009;175:400–411. doi: 10.2353/ajpath.2009.080972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang Z, Bhattacharya N, Weaver M, Petersen K, Meyer M, Gapter L, et al. Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J Vet Sci. 2001;2:167–179. [PubMed] [Google Scholar]
- 165.Sato AK, Yanai N, Okubo T, Mori KJ, Obinata M. Stromal cells provide signals different from cytokines for STAT5 activation in hematopoietic cells. Cell Struct Funct. 2001;26:95–101. doi: 10.1247/csf.26.95. [DOI] [PubMed] [Google Scholar]
- 166.Horikawa K, Takatsu K. Interleukin-5 regulates genes involved in B-cell terminal maturation. Immunology. 2006;118:497–508. doi: 10.1111/j.1365-2567.2006.02382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cohen AM, Grinblat B, Bessler H, Kristt D, Kremer A, Schwartz A, et al. Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma. 2004;45:951–955. doi: 10.1080/10428190310001641251. [DOI] [PubMed] [Google Scholar]
- 168.Grundler R, Brault L, Gasser C, Bullock AN, Dechow T, Woetzel S, et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J Exp Med. 2009;206:1957–1970. doi: 10.1084/jem.20082074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stout BA, Bates ME, Liu LY, Farrington NN, Bertics PJ. IL-5 and granulocyte-macrophage colony-stimulating factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3 protein expression in human eosinophils. J Immunol. 2004;173:6409–6417. doi: 10.4049/jimmunol.173.10.6409. [DOI] [PubMed] [Google Scholar]
- 170.Siracusa MC, Perrigoue JG, Comeau MR, Artis D. New paradigms in basophil development, regulation and function. Immunol Cell Biol. 2010;88:275–284. doi: 10.1038/icb.2010.1. [DOI] [PubMed] [Google Scholar]
- 171.Didichenko SA, Spiegl N, Brunner T, Dahinden CA. IL-3 induces a Pim1-dependent antiapoptotic pathway in primary human basophils. Blood. 2008;112:3949–3958. doi: 10.1182/blood-2008-04-149419. [DOI] [PubMed] [Google Scholar]
- 172.Zheng X, Karsan A, Duronio V, Chu F, Walker DC, Bai TR, et al. Interleukin-3, but not granulocyte-macrophage colony-stimulating factor and interleukin-5, inhibits apoptosis of human basophils through phosphatidylinositol 3-kinase: requirement of NF-kappaB-dependent and -independent pathways. Immunology. 2002;107:306–315. doi: 10.1046/j.1365-2567.2002.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Beharry Z, Zemskova M, Mahajan S, Zhang F, Ma J, Xia Z, et al. Novel benzylidene-thiazolidine-2,4-diones inhibit Pim protein kinase activity and induce cell cycle arrest in leukemia and prostate cancer cells. Mol Cancer Ther. 2009;8:1473–1483. doi: 10.1158/1535-7163.MCT-08-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pogacic V, Bullock AN, Fedorov O, Filippakopoulos P, Gasser C, Biondi A, et al. Structural analysis identifies imidazo [1,2-b]pyridazines as PIM kinase inhibitors with in vitro antileukemic activity. Cancer Res. 2007;67:6916–6924. doi: 10.1158/0008-5472.CAN-07-0320. [DOI] [PubMed] [Google Scholar]
- 175.Xia Z, Knaak C, Ma J, Beharry ZM, McInnes C, Wang W, et al. Synthesis and evaluation of novel inhibitors of Pim-1 and Pim-2 protein kinases. J Med Chem. 2009;52:74–86. doi: 10.1021/jm800937p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Morwick T. Pim kinase inhibitors: a survey of the patent literature. Expert Opin Ther Pat. 20:193–212. doi: 10.1517/13543770903496442. [DOI] [PubMed] [Google Scholar]
- 177.Hu XF, Li J, Vandervalk S, Wang Z, Magnuson NS, Xing PX. PIM-1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis. J Clin Invest. 2009;119:362–375. doi: 10.1172/JCI33216. [DOI] [PMC free article] [PubMed] [Google Scholar]