Abstract
Transcriptional inactivation of one X chromosome in mammalian female somatic cells leads to condensation of the inactive X chromosome into the heterochromatic sex chromatin, or Barr body. Little is known about the molecular composition and structure of the Barr body or the mechanisms leading to its formation in female nuclei. Because human sera from patients with autoimmune diseases often contain antibodies against a variety of cellular components, we reasoned that some autoimmune sera may contain antibodies against proteins associated with the Barr body. Therefore, we screened autoimmune sera by immunofluorescence of human fibroblasts and identified one serum that immunostained a distinct nuclear structure with a size and nuclear localization consistent with the Barr body. The number of these structures was consistent with the number of Barr bodies expected in diploid female fibroblasts containing two to five X chromosomes. Immunostaining with the serum followed by fluorescence in situ hybridization with a probe against XIST RNA demonstrated that the major fluorescent signal from the autoantibody colocalized with XIST RNA. Further analysis of the serum showed that it stains human metaphase chromosomes and a nuclear structure consistent with the inactive X in female mouse fibroblasts. However, it does not exhibit localization to a Barr body-like structure in female mouse embryonic stem cells or in cells from female mouse E7.5 embryos. The lack of staining of the inactive X in cells from female E7.5 embryos suggests the antigen(s) may be involved in X inactivation at a stage subsequent to initiation of X inactivation. This demonstration of an autoantibody recognizing an antigen(s) associated with the Barr body presents a strategy for identifying molecular components of the Barr body and examining the molecular basis of X inactivation.
During early mammalian female embryogenesis, one of the two transcriptionally active X chromosomes is inactivated in each cell of the embryo (1). The stable inactivation of genes on one of the two X chromosomes in females functionally equalizes the apparent dosage imbalance of X-linked genes between males and females. This chromosome-wide transcriptional silencing is associated with condensation of the inactive X chromosome into the heterochromatic sex chromatin or Barr body, a unique constituent of the female nucleus identified half a century ago (2). The Barr body in female interphase nuclei is characteristically found as a darkly staining nuclear inclusion commonly associated with the nuclear membrane (2). In cells carrying a diploid complement of autosomes, X inactivation and Barr body formation occurs according to the N-1 rule: cells maintain a single active X chromosome and inactivate and condense all remaining X chromosomes (3, 4). However, Barr body formation does not appear to be a requirement for maintaining transcriptional repression of genes on the inactive X because rodent–human somatic cell hybrids containing an inactive human X chromosome do not form Barr bodies but continue to maintain transcriptional silencing of genes on the inactive human X (5).
The molecular mechanisms for establishing and maintaining this unique system of differential gene regulation are not well understood, and currently little is known about the molecular components and structure of the Barr body itself. The Barr body has been examined by electron microscopy (6, 7), and the results indicate the possibility of a special nuclear envelope attachment region for the Barr body. But these studies provide few insights into the possible composition or macromolecular organization of the inactive X chromosome. The Barr body and individual genes on the inactive X have been probed with nucleases, particularly DNase I, to analyze molecular structure. Nick translation assays on female cells after fixation and nicking with DNase I have shown that inactive X chromatin is resistant to nick translation (6, 8). However, analysis of general DNase I sensitivity of the X-linked mouse Hprt and human Pgk genes in unfixed cells (9, 10) showed a much smaller difference in sensitivity between the active and inactive alleles (≤2-fold) than would be expected for highly condensed heterochromatin (i.e., inactive X) versus uncondensed euchromatin (i.e., active X). Recent analysis of the three-dimensional organization of the active and inactive X chromosomes showed the two chromosomes occupy the same volume, but the active X appeared flatter with a larger and fuzzier surface than the inactive X, which appeared rounder in shape with smoother surface structure (11). Two studies have also examined the potential association of the two telomeres and resulting loop structure of the inactive X chromosome (12, 13).
Currently, three macromolecules have been shown to colocalize with the Barr body or inactive X chromosome. Perichromin, a nuclear envelope protein directly or indirectly bound to DNA (14), has been reported to be associated with the Barr body (15). However, its role in X inactivation, if any, is unknown. The XIST gene encodes a large nuclear RNA found to be associated exclusively with the inactive X chromosome by fluorescence in situ hybridization (FISH) (16, 17). This RNA exhibits no conserved and extended ORF (16, 18), is transcribed only from the inactive X chromosome (19, 20), and is essential for normal X inactivation (21, 22). The mechanisms by which the XIST gene and RNA function in X inactivation are currently unresolved. The histone variant macroH2A1.2 also is reported to be concentrated on the inactive X chromosome (23, 24), although association of macroH2A1.2 with the inactive X does not appear to be required for either initiating or maintaining transcriptional repression of genes on the inactive X chromosome (25, 26). Recently, Perche et al. have reported that, in addition to macroH2A, the core histones H2B and H3 also show preferential localization to the Barr body, suggesting that the Barr body may contain a higher density of nucleosomes (27). Conversely, the inactive X chromosome is reported to be deficient in another novel histone variant, termed H2A-Bbd (28). Despite these promising findings, knowledge of the composition and molecular structure of the Barr body, as well as mechanisms of global silencing of genes on the inactive X, remains incomplete.
Antisera from human patients with autoimmune diseases have been used extensively as a tool for studying intracellular structure and function (29, 30). We reasoned that a small subpopulation of autoimmune patients may carry antibodies against one or more components of the Barr body. Further, autoantisera with antibodies against the Barr body then could be used to identify the corresponding Barr body-associated antigen. Therefore, to examine whether or not human autoimmune sera may be useful as probes of Barr body composition and structure, we screened samples of autoantisera by an indirect immunofluorescence assay on male and female fibroblasts and examined the immunostaining patterns for a female-specific staining pattern and colocalization of antibody with the Barr body. Screening of 255 different autoimmune sera identified one serum containing antibodies against the Barr body in both human and mouse fibroblasts.
Materials and Methods
Autoimmune Sera.
Sera were obtained from patients with autoimmune diseases including systemic lupus erythematosus, scleroderma, and mixed connective tissue disease. Each serum sample was assayed at dilutions of 1:20 and 1:200.
Cells and Cell Culture.
The human fibroblast cell lines GM 06111 (46,XX), GM 00254 (47,XXX), GM01415E (48,XXXX), GM05009C (49,XXXXX), and GM00468 (46,XY) were purchased from the NIGMS Human Genetic Mutant Cell Repository and grown according to the recommended conditions. Human–hamster hybrid cell line 8121 contains an inactive human X chromosome in a rodent cell background (31). Male and female mouse embryonic stem (ES) cell lines J1 and 2-1, respectively, were grown on mitotically inactivated mouse embryonic fibroblasts in medium containing leukemia inhibitory factor. Female mouse fibroblasts containing three X chromosomes were a gift of Catherine Brisken (Whitehead Institute).
Isolation of Mouse Embryos.
Cells from E7.5 embryos were isolated as described (32).
Indirect Immunofluorescence.
Human fibroblast cells were grown overnight on glass microscope slides and fixed with 2% formaldehyde for 15 min at room temperature. Fixed cells were treated with acetone at −20°C for 5 min, then at room temperature blocked with 3% BSA in PBS (2 mM KH2PO4/8 mM Na2HPO4/2.5 mM KCl/140 mM NaCl, pH 7.2) for 15 min, incubated with diluted autoantiserum for 1 h, washed with PBS, incubated with FITC-conjugated goat anti-human Ig for 1 h, washed again with PBS, counterstained with 1 μg/ml DAPI in PBS for 10 min, mounted in 1 mg/ml p-phenylenediamine/10 mM Tris⋅HCl, pH 8.5/90% glycerol, and examined with an Olympus fluorescence microscope. Images in Figs. 1 and 3 were collected with a Bio-Rad 1024-ES confocal microscope.
Figure 1.
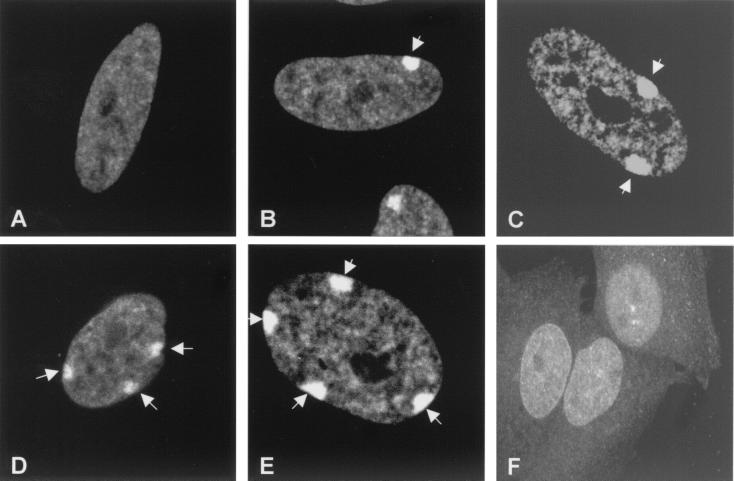
Detection of Barr bodies by autoimmune serum 154. Indirect immunofluorescence was performed on diploid human fibroblasts containing different numbers of X chromosomes using autoimmune serum 154 (1:200 dilution) and an FITC-conjugated secondary antibody against human immunoglobulins. Images were collected with a Bio-Rad 1024 ES confocal microscope. (A) GM00468 (46,XY). (B) GM06111 (46,XX). (C) GM00254 (47,XXX). (D) GM01415E (48,XXXX). (E) GM05009C (49,XXXXX). (F) Human-hamster hybrid cell line 8121 (containing an inactive human X chromosome). Arrows indicate Barr bodies.
Figure 3.
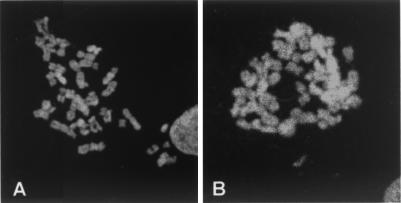
Immunostaining of mitotic chromosomes with autoimmune serum 154. Metaphase chromosomes were prepared from human fibroblast cells. (A) GM00254 (47,XXX). (B) GM00468 (46,XY). The chromosomal staining patterns shown are representative of multiple fields and preparations from each cell line.
Immunostaining Followed by RNA FISH.
ES cells and trypsinized embryos were affixed to glass slides by using a cytospin apparatus. Fibroblasts were grown directly on glass slides. Immunostaining was performed first as described above, except that yeast tRNA (0.5 mg/ml) and RNasin (0.4 units/μl, Promega) were included at every step to prevent RNA degradation. After immunostaining, the labeled secondary antibody was fixed with 2% formaldehyde in PBS, and the slides were dehydrated through an ethanol series from 70 to 100%. Double-stranded DNA probes were indirectly labeled by incorporation of biotin-conjugated dCTP by random priming (GIBCO/BRL) reactions using the plasmid pXist3K for mouse Xist (33) and pG1A (17) as templates. Labeled probes were precipitated with yeast tRNA, mouse COT-1 DNA, and sheared salmon sperm DNA, then dried and resuspended in hybridization solution (Hybridsol VII, Oncor). Hybridizations were carried out at 37°C overnight in a humidified chamber. Slides were washed, and probe hybridization was detected with Cy3 conjugated avidin (Amersham Pharmacia), stained with DAPI, and mounted in antifade mounting media (Vectashield, Vector Laboratories) as described (34).
Preparation of Mitotic Cells.
The method used was modified from Jeppesen et al. (35). Cells in log-phase were treated with colcemid (0.05 μg/ml) for 4 h, trypsinized, and washed twice with PBS. The cells were swollen in 0.075 M KCl supplemented with a protease inhibitor mixture (1 mM benzamide/0.5 μg/ml leupeptin/1.5 μM bestatin in H2O; 0.5 mM phenylmethylsulfonyl fluoride/1.5 μM pepstatin A/1 μg/ml chymostatin solublized in dimethyl sulfoxide) at room temperature for 15 min and spun onto a glass slide with a Shandon cytospin at 700 rpm at room temperature for 10 min. Slides were then incubated in KCM buffer (120 mM KCl/20 mM NaCl/10 mM Tris⋅HCl, pH 7.5/0.5 mM EDTA) supplemented with the protease inhibitor mixture at room temperature for 10 min, fixed with 2% formaldehyde at room temperature for 15 min, treated with acetone at −20°C for 3 min, and used for indirect immunofluorescence.
Western Analysis.
Total cellular proteins were prepared as described (36). Nuclear proteins were isolated according to Pederson (37) with modifications. Briefly, cells were incubated at room temperature in TM-2 buffer (0.01 M Tris, pH 7.4/0.002 M MgCl2) supplemented with the protease inhibitor mixture (see above) for 10 min, treated with 0.5% (vol/vol) Triton X-100 in TM-2 buffer at room temperature for 10 min, and lysed with a Dounce homogenizer. Nuclei were collected by spinning at 600 × g at 4°C for 10 min and washed twice with TM-2 buffer. Nuclear proteins were extracted by lysing the nuclei in sample buffer (4% SDS/83 mM Tris base/127 mM Tris⋅HCl, pH 8.0). The lysates were sonicated and fractionated by SDS/polyacrylamide gels.
Western blotting was performed according to standard methods (36). Proteins were fractionated on 10.5% SDS/polyacrylamide gels and transferred to a nitrocellulose membrane. After blocking with 5% nonfat milk plus 1% Tween-20, the membrane was incubated with serum 154 (diluted 1:4,000). Signals were detected with a horseradish peroxidase–anti-human IgG (1:35,000, Promega) and ECL reagents (Promega).
Results
Screening of Autoantisera.
Because antisera from patients with autoimmune diseases often contain antibodies against a variety of intracellular components (29, 30, 38), we examined the possibility that sera from certain autoimmune patients might contain antibodies against the Barr body. Antisera from 255 autoimmune patients with systemic lupus erythematosus, scleroderma, or mixed connective tissue disease were screened by indirect immunofluorescence for staining of the Barr body. Cultured male (GM00468; 46,XY) and female (GM00254; 47,XXX) fibroblasts were assayed pairwise with each autoimmune serum at 1:20 and 1:200 dilutions of serum and examined by fluorescence light microscopy. Over 100 cells from multiple fields and at different magnifications were examined for each serum dilution. Each serum sample was examined for a fluorescent staining pattern consistent with binding to the Barr body. Because all X chromosomes in excess of one per diploid genome are inactivated (the N-1 rule) (3, 4), 47,XXX female cells should contain two preferentially stained nuclear structures frequently associated with the nuclear membrane, and male cells should not show any staining of such structures if an antiserum contains antibodies against the Barr body. Initial screening of 255 autoimmune sera at two different dilutions showed that a majority of the sera contained autoantibodies against nuclear antigens, but staining of cytoplasmic antigens also was commonly observed (data not shown). One antiserum, no. 154, exhibited an immunofluorescence pattern by optical microscopy consistent with staining of the Barr body where 47,XXX female fibroblasts showed a distinctly more intense staining of two nuclear structures similar in size and location to Barr bodies, and 46,XY male fibroblasts lacked this staining pattern. Nearly every female nucleus that showed staining of Barr body-like structures by DAPI showed immunostaining of the same Barr body-like structures by serum 154.
To further examine the possibility of Barr body staining, serum 154 was used in indirect immunofluorescence assays of human fibroblasts carrying one to five X chromosomes and examined by confocal microscopy (Fig. 1). The number of intensely fluorescent nuclear structures stained by serum 154 was equal to that expected from the N-1 rule for Barr body formation (Fig. 1 A–E). Male fibroblasts exhibited no staining of a Barr body-like nuclear structure (Fig. 1A), XX female fibroblasts showed immunofluorescent staining of a single Barr body-like nuclear structure (Fig. 1B), XXX female fibroblasts showed staining of two Barr body-like structures (Fig. 1C), etc. The absence of a highly localized Barr body-like staining pattern in cell line 8121 (Fig. 1F), a rodent–human hybrid cell line containing an inactive human X chromosome that does not form a Barr body (5), further supports the notion that serum 154 contains antibodies against the Barr body.
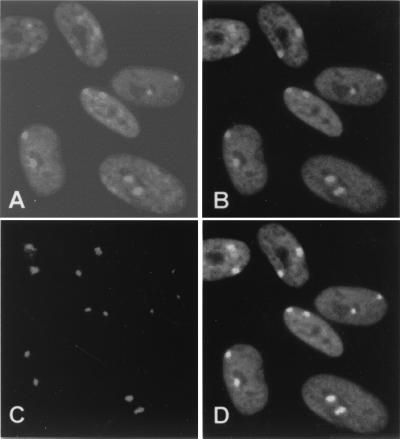
To confirm that serum 154 stains the Barr body, we performed colocalization studies with XIST RNA by FISH. Previous studies have shown that XIST RNA accumulates over the inactive X chromosome and “paints” the inactive X by FISH (17). Therefore, human fibroblasts (47,XXX) were first stained by indirect immunofluorescence with serum 154, then subjected to FISH with an XIST DNA probe. As shown in Fig. 2, fluorescent signals for serum 154 (Fig. 2B) and the XIST probe (Fig. 2C) colocalized (Fig. 2D) to nuclear structures consistent in size, location, and number with Barr bodies, as confirmed with DAPI staining (Fig. 2A). These studies confirm that our screening of 255 human autoimmune sera identified one sample that contains antibodies recognizing one or more components of the Barr body.
Figure 2.
Colocalization of XIST RNA and major sites of immunostaining by autoimmune serum 154. Indirect immunofluorescence with autoimmune serum 154 (1:200) followed by RNA FISH with a probe against the XIST RNA were performed on human female fibroblast cells GM00254 (47,XXX). (A) DAPI staining. (B) Immunostaining with serum 154. (C) FISH with probe for XIST RNA. (D) The merged images of B and C.
Analysis of Metaphase Chromosomes.
To determine whether the Barr body-associated antigen(s) recognized by serum 154 remains associated specifically with the inactive X chromosome at metaphase, metaphase chromosome spreads of 47,XXX female fibroblasts were analyzed by indirect immunofluorescence. As shown by representative metaphase spreads in Fig. 3, the autoantiserum showed strong immunostaining of all chromosomes in both female (Fig. 3A) and male (Fig. 3B) cells, with some chromosomal regions showing slightly more intense staining than others. There does not appear to be preferential staining of centromeric heterochromatin in metaphase chromosomes. A similar pattern of immunostaining also was observed with metaphase chromosomes from female mouse embryo fibroblasts (data not shown). These data suggest that the antigen(s) recognized by serum 154 is not preferentially localized only to the inactive X chromosome at metaphase but is present on all metaphase chromosomes. This further suggests that the antigen(s) is unlikely to play a role unique to X inactivation. Nonetheless, at interphase, the antigen(s) is preferentially concentrated over the inactive X (see Fig. 1). The staining pattern of metaphase chromosomes with serum 154 is in contrast to the pattern seen with antibodies to macroH2A1.2, which preferentially stained only the inactive X chromosome in metaphase spreads (23).
Analysis of Mouse Fibroblasts, ES Cells, and Embryos.
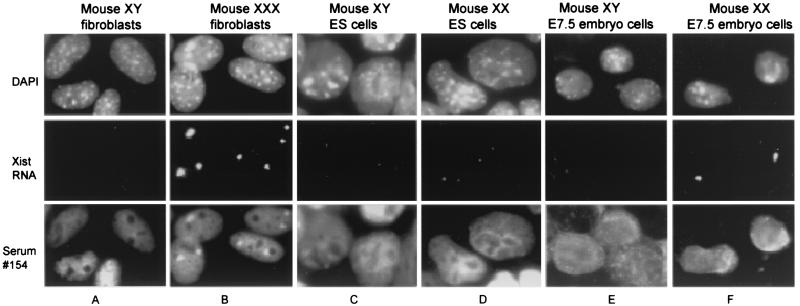
As shown in Fig. 4, we also examined the immunostaining pattern of serum 154 in female mouse fibroblasts, ES cells, and E7.5 embryos. Fig. 4 A and B show indirect immunofluorescence of XY male and XXX female mouse fibroblasts, respectively, stained with serum 154, then subjected to FISH with a probe for Xist RNA. The XXX female nuclei showed an accumulation of immunofluorescence at two sites in three of four nuclei, sites that also colocalized with Xist RNA. These data are consistent with immunostaining of the mouse inactive X chromosome. Examination of multiple preparations and fields (≥100 nuclei) indicated ≈75% of XXX mouse fibroblasts showed intense staining of two nuclear bodies. Similar indirect immunofluorescence experiments on normal XX female mouse fibroblasts showed a single intensely stained nuclear structure with a size and location consistent with staining of the inactive X chromosome (data not shown). XY male fibroblast nuclei (Fig. 4A) showed no evidence of a localized accumulation of antibody similar to that seen in XX or XXX female nuclei. These results strongly suggest serum 154 crossreacts with the inactive X chromosome in mouse cells and that the inactive X chromosome-associated epitope(s) recognized by the autoimmune serum appears to be evolutionarily conserved in humans and mice. This is consistent with the observation that both human and mouse metaphase chromosomes are also immunostained by the autoimmune serum 154 (see above).
Figure 4.
Analysis of mouse cells with autoimmune serum 154. Immunostaining with autoimmune serum 154 and Xist FISH were performed on the following. (A) XY male fibroblasts. (B) XXX female fibroblasts. (C) XY male ES cells. (D) XX female ES cells. (E) Cells from XY male E7.5 embryos. (F) Cells from XX female E7.5 embryos. The staining patterns shown are representative of multiple preparations and fields for each cell type.
Undifferentiated male and female mouse ES cells were also subjected to staining by indirect immunofluorescence with serum 154 to examine the immunostaining pattern before the onset of X inactivation. Undifferentiated female ES cells retain two active X chromosomes and, therefore, do not form a Barr body in interphase cells. Male and female ES cells were first immunostained with serum 154, then subjected to FISH with a probe for Xist RNA. As shown in Fig. 4 C and D, both male and female ES cells exhibited a very diffuse immunostaining pattern in the nucleus with no highly localized accumulation of fluorescence at one or more discrete sites. There appeared to be no preferential colocalization of the antigen(s) with Xist RNA or accumulation of the antigen(s) over either of the X chromosomes in female ES cells before the initiation of X inactivation (Fig. 4D). These results are consistent with the notion that the major antigen(s) recognized by serum 154 is associated with formation of a Barr body. There also appeared to be no preferential staining of a structure suggestive of a macrochromatin body (more recently identified as the centrosome), an intracellular structure stained in both undifferentiated male and female ES cells by antibodies against histone macroH2A1.2 (24, 25). This further indicates that the Barr body-associated antigen recognized by serum 154 is unlikely to be macroH2A1.2.
We also examined the staining pattern of serum 154 in cells from male and female mouse E7.5 embryos. At this stage, most cells of female embryos have just undergone initiation of X inactivation as suggested by the large proportion (80–90%) of cells that exhibit high-level monoallelic association of Xist RNA with the inactive X. The remaining cells exhibit a differential biallelic pattern of Xist RNA association (i.e., a site of low-level Xist RNA accumulation associated with the eventual active X chromosome, and a site of high level Xist RNA accumulation associated with the eventual inactive X chromosome) believed to indicate cells that are undergoing initiation of X inactivation (32, 39). Cells from trypsinized male and female E7.5 embryos were stained by indirect immunofluorescence with serum 154 followed by FISH with a probe for Xist RNA. As shown in Fig. 4 E and F, nuclei from female (and male) E7.5 embryos demonstrated a highly diffuse immunostaining pattern over the entire nucleus with serum 154 and did not exhibit an immunostaining pattern suggestive of localization to the Barr body (or to a macrochromatin body). There was clearly no evidence for colocalization of discrete sites of antibody accumulation with sites of Xist RNA accumulation. Furthermore, close examination of the panel in Fig. 4F of XX E7.5 embryo cells showed one nucleus (on the right) with a differential biallelic pattern of Xist RNA localization (both a weak and a strong site of Xist accumulation) suggestive of a cell in the process of X inactivation. This nucleus did not demonstrate discrete localization of autoantibody staining with either site of Xist RNA expression and accumulation (i.e., the active or inactive X chromosomes), indicating that the autoimmune serum does not recognize an epitope that accumulates on the inactive X chromosome at the same time as Xist RNA. Taken together, these data would suggest that the antigen(s) recognized by serum 154 is unlikely to be involved in the initiation of X inactivation.
Western Blot Analysis.
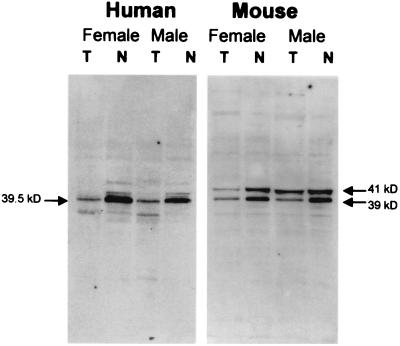
To determine the range of cellular proteins that react with serum 154 and detect the major proteins recognized by the autoimmune serum, Western blot analysis was performed on total cellular and nuclear proteins from male and female fibroblasts (see Fig. 5). The autoimmune serum recognized a single major 39-kDa polypeptide in human extracts, which was significantly enriched in the nuclear fraction. This 39-kDa band was present at apparently similar levels in both male and female extracts, indicating this major antigen is not female-specific. Two polypeptides at 39 and 41 kDa in mouse extracts also were recognized by serum 154. These murine polypeptides were again enriched in the nuclear fraction and present in both female and male cells at similar levels. If these major polypeptides represent the Barr body-associated antigens recognized by serum 154, they are constituents of the nucleus in both male and female cells and unlikely to function only in X inactivation. This is supported by the observation that the antigen(s) recognized by the autoimmune serum is also present on both male and female metaphase chromosomes (see Fig. 3). Nonetheless, these polypeptides appear to be specifically recruited to the Barr body in female cells at interphase (given the intense immunofluorescence signal at the Barr body relative to background staining) and are likely to be integral components of the interphase Barr body, possibly functioning in the condensation of chromatin of the inactive X (see Discussion). The size of the major polypeptides recognized by the autoimmune serum argues against the possibility that the Barr body-associated immunofluorescence signal from serum 154 could be because of core histones (27).
Figure 5.
Western analysis of human and mouse fibroblasts with autoimmune serum 154. Nuclear (N) or whole cell (T) extracts were prepared from male or female fibroblasts and subjected to Western blot analysis by using autoimmune serum 154. Molecular weights were estimated from molecular weight standards run in adjacent lanes (not shown) and are indicated as labeled.
Discussion
We have identified a human autoimmune serum that immunostains a nuclear structure in female human and mouse fibroblasts that is consistent in size, location, and expected number with the Barr body. Furthermore, the immunostained structure colocalizes with XIST/Xist RNA in both human and mouse fibroblasts, demonstrating its association with the inactive X chromosome. The antigen(s) recognized by this serum shows features different from the known Barr body-associated factors, perichromin, XIST/Xist RNA, and histone macroH2A1.2. The staining pattern of serum 154 does not show strong immunofluorescence at the nuclear periphery, where perichromin is preferentially localized (14), suggesting the antigen to serum 154 is not perichromin. The autoimmune serum also does not colocalize with Xist RNA in cells from female E7.5 embryos, suggesting that the antigen(s) recognized by the autoimmune serum is not associated with Xist RNA at, or shortly after, the time X inactivation is initiated. Although macro H2A1.2 and the major antigen recognized by serum 154 have similar molecular weights (as suggested by the Western blot in Fig. 5), their immunolocalization on metaphase chromosomes and in mouse ES cells exhibit distinctly different patterns. Antibodies against macro H2A1.2 show preferential immunostaining of the inactive X chromosome in metaphase chromosomes, whereas serum 154 strongly stains all metaphase chromosomes (see Fig. 3). In undifferentiated ES cells, antibodies against macro H2A1.2 show localization to a distinct macrochromatin body in both male and female ES cells, whereas serum 154 shows no highly localized staining suggestive of a macrochromatin body in these cells (see Fig. 4). Serum 154 also does not show staining of a macrochromatin body in cells from E7.5 embryos (Fig. 4). Furthermore, we also have expressed a macroH2A1.2-GFP (green fluorescent protein) fusion protein in 293 cells, and Western blot analysis of these recombinant cells using serum 154 does not show a notable band at the expected size of the fusion protein (B.H., B.P., and T.P.Y., unpublished data). These results indicate the Barr body-associated antigen recognized by serum 154 is unlikely to be macroH2A1.2. Furthermore, the molecular weights of the major bands seen in Western blots of human and mouse cell extracts by using serum 154 (Fig. 5) are significantly larger than each of the core histones, arguing against the possibility that serum 154 recognizes any of the core histones that are reported to be concentrated at the Barr body (27). In addition, the antigen(s) recognized by serum 154 is not acid extractable (data not shown), further evidence that serum 154 is unlikely to recognize a histone family member.
Because serum 154 shows colocalization with XIST/Xist RNA in female fibroblast cells (Figs. 2 and 4B), but not in cells from female E7.5 embryos (Fig. 4F) when cells are undergoing or just completing initiation of X inactivation (32), the Barr body-associated antigen(s) recognized by the antiserum is more likely to be involved in the process of X inactivation, if at all, at a stage subsequent to the initiation of inactivation. This might include potential roles in the global stabilization of the inactive state after X inactivation has been established and/or maturation and condensation of the inactive X into the heterochromatic Barr body during female development. A role for the antigen(s) in chromatin/chromosome condensation is suggested by the strong staining of metaphase chromosomes by the autoantiserum (see Fig. 3). However, because the autoimmune serum does not exhibit preferential staining of centromeric heterochromatin in metaphase chromosomes (see Fig. 3), it is unlikely that the corresponding antigen(s) plays a role unique to formation of heterochromatin.
At present, we do not know the identity, age, sex, or disease state of the patient who provided serum 154. However, we have just completed a screening of another independent collection of 185 autoantisera and identified a second serum sample that immunostains the Barr body in human female fibroblast cells at very high frequency with strong signal intensity (W. H. Brooks, M. Sato, W. H. Reeves, and T.P.Y., unpublished data). This newly identified serum sample is derived from a female patient with systemic lupus erythematosus.
Our identification of autoimmune sera with antibodies against the Barr body indicates that antigens associated with the Barr body are capable of eliciting an autoimmune response. This leads to the possibility that sera from a distinct subpopulation of autoimmune patients (≈0.5%) may contain antibodies against a variety of Barr body-associated antigens. Thus, these autoantisera may offer an experimental approach for examining the molecular basis for Barr body formation and/or establishing and maintaining X chromosome inactivation by providing tools for the identification and characterization of molecular components of the Barr body. We currently are screening phage expression libraries with serum 154 to identify the Barr body-associated antigen.
Acknowledgments
We thank Ralph Williams and Robert Eisenberg for kindly providing autoimmune sera. We also thank Daniel Driscoll for use of his fluorescence microscope, and Wesley Brooks for assistance in preparation of figures. This work was supported by National Institutes of Health Grant RO1 GM44286 (to T.P.Y.) and the Sandler Family Foundation (to B.P.).
Abbreviations
- FISH
fluorescence in situ hybridization
- ES
embryonic stem
References
- 1.Heard E, Clerc P, Avner P. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 2.Barr M L, Bertram E G. Nature (London) 1949;163:676–677. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 3.Lyon M F. Nature (London) 1996;379:116–117. doi: 10.1038/379116a0. [DOI] [PubMed] [Google Scholar]
- 4.Lee J T, Jaenisch R. Nature (London) 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 5.Hansen R S, Canfield T K, Stanek A M, Keitges E A, Gartler S M. Proc Natl Acad Sci USA. 1998;95:5133–5138. doi: 10.1073/pnas.95.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyer K A, Riley D, Gartler S M. Chromosoma. 1985;92:209–213. doi: 10.1007/BF00348695. [DOI] [PubMed] [Google Scholar]
- 7.Wolstenholme D R. Chromosoma. 1965;16:453–462. doi: 10.1007/BF00343173. [DOI] [PubMed] [Google Scholar]
- 8.Kerem B S, Goitein R, Richler C, Marcus M, Cedar H. Nature (London) 1983;304:88–90. doi: 10.1038/304088a0. [DOI] [PubMed] [Google Scholar]
- 9.Yang T P, Caskey C T. Mol Cell Biol. 1987;7:2994–2998. doi: 10.1128/mcb.7.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley D E, Goldman M A, Gartler S M. Somatic Cell Mol Genet. 1986;12:73–80. doi: 10.1007/BF01560729. [DOI] [PubMed] [Google Scholar]
- 11.Eils R, Dietzel S, Bertin E, Schrock E, Speicher M R, Ried T, Robert-Nicoud M, Cremer C, Cremer T. J Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker C L, Cargile C B, Floy K M, Delannoy M, Migeon B R. Proc Natl Acad Sci USA. 1991;88:6191–6195. doi: 10.1073/pnas.88.14.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietzel S, Eils R, Satzler K, Bornfleth H, Jauch A, Cremer C, Cremer T. Exp Cell Res. 1998;240:187–196. doi: 10.1006/excr.1998.3934. [DOI] [PubMed] [Google Scholar]
- 14.McKeon F D, Tuffanelli D L, Kobayashi S, Kirschner M W. Cell. 1984;36:83–92. doi: 10.1016/0092-8674(84)90076-x. [DOI] [PubMed] [Google Scholar]
- 15.Gartler S M, Dyer K A, Goldman M A. Mol Genet Med. 1992;2:121–160. doi: 10.1016/b978-0-12-462002-5.50010-8. [DOI] [PubMed] [Google Scholar]
- 16.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 17.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 19.Brockdorff N, Ashworth A, Kay G F, Cooper P, Smith S, McCabe V M, Norris D P, Penny G D, Patel D, Rastan S. Nature (London) 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 20.Brown C J, Ballabio A, Rupert J L, Lafreniere R G, Grompe M, Tonlorenzi R, Willard H F. Nature (London) 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 21.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Nature (London) 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 22.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 23.Costanzi C, Pehrson J R. Nature (London) 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen T P, Mastrangelo M, Eden A, Pehrson J R, Jaenisch R. J Cell Biol. 2000;150:1189–1198. doi: 10.1083/jcb.150.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mermoud J E, Costanzi C, Pehrson J R, Brockdorff N. J Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csankovszki G, Panning B, Bates B, Pehrson J R, Jaenisch R. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 27.Perche P, Vourc'h C, Konecny L, Souchier C, Robert-Nicoud M, Dimitrov S, Khochbin S. Curr Biol. 2000;10:1531–1534. doi: 10.1016/s0960-9822(00)00832-0. [DOI] [PubMed] [Google Scholar]
- 28.Chadwick B P, Willard H F. J Cell Biol. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan E M. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- 30.Lerner M R, Steitz J A. Proc Natl Acad Sci USA. 1979;76:5495–5497. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ledbetter S A, Schwartz C E, Davies K E, Ledbetter D H. Am J Med Genet. 1991;38:418–420. doi: 10.1002/ajmg.1320380254. [DOI] [PubMed] [Google Scholar]
- 32.Panning B, Dausman J, Jaenisch R. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 33.Panning B, Jaenisch R. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 34.Johnson C V, Singer R H, Lawrence J B. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- 35.Jeppesen P, Mitchell A, Turner B, Perry P. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- 36.Gallagher S R. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 1999. pp. 10.2A.1–10.2A.34. [Google Scholar]
- 37.Pederson T. In: Cell: A Laboratory Manual. Spector D L, Goldman R D, Leinwand L A, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. p. 43.1. [Google Scholar]
- 38.Hollingsworth P N, Pummer S C, Dawkins R L. In: Autoantibodies. Peter J B, Shoenfeld Y, editors. New York: Elsevier; 1996. pp. 74–90. [Google Scholar]
- 39.Sheardown S A, Duthie S M, Johnston C M, Newall A E, Formstone E J, Arkell R M, Nesterova T B, Alghisi G C, Rastan S, Brockdorff N. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]