Abstract
Cells continually assess their energy and nutrient state to maintain growth and survival and engage necessary homeostatic mechanisms. Cell autonomous responses to the fed state require the surveillance of the availability of amino acids and other nutrients. The mammalian target of rapamycin complex 1 (mTORC1) integrates information on nutrient and amino acid availability to support protein synthesis and cell growth. We identify the G protein-coupled receptor (GPCR) T1R1/T1R3 as a direct sensor of the fed state and amino acid availability. Knocking down this receptor, which is found in most tissues, reduces the ability of amino acids to signal to mTORC1. Interfering with this receptor alters localization of mTORC1, downregulates expression of pathway inhibitors, upregulates key amino acid transporters, blocks translation initiation, and induces autophagy. These findings reveal a mechanism for communicating amino acid availability through a GPCR to mTORC1 in mammals.
The mammalian target of rapamycin (mTOR) coordinates cell growth, protein translation, and autophagy with the availability of nutrients, growth factors, and energy (Dazert and Hall, 2011; Avruch et al., 2009; Ma and Blenis, 2009; Zoncu et al., 2011; Corradetti and Guan, 2006). At least two functionally distinct mTOR complexes have been identified, mTORC1 and mTORC2, that carry out different cellular functions.
mTORC1 is regulated by insulin and growth factors by mechanisms primarily dependent on phosphatidyl inositol-3 kinase (PI3K) and Ras. These pathways relieve mTORC1 repression by tuberous sclerosis 1 and 2 (TSC1/2). TSC1/2 are GTPase-activating proteins that inactivate RAS homolog enriched in brain (Rheb), which is required for the activation of mTORC1. Akt and ERK1/2, through the ERK1/2 substrate ribosomal protein S6 kinase (RSK), phosphorylate and inhibit the TSC1/2 complex (Roux et al., 2004; Carriere et al., 2008), releasing its negative control of mTOR and enhancing mTORC1 activity. ERK1/2 may also promote mTORC1 activation by directly phosphorylating Raptor, a subunit of mTOR complex 1 (Carriere et al., 2008; Carriere et al., 2011).
Among its most important functions, mTORC1 stimulates protein translation. Cap-dependent translation initiation involves the assembly of the small ribosomal subunit at the 7-methylGTP-capped 5′ end of mRNA, and its scanning to the start codon to form a complex with the large ribosomal subunit. In the translationally inactive state, the 7-methylGTP cap binds eIF4E and its inhibitor eIF4E binding protein 1 (4EBP-1) (Ma and Blenis, 2009; Sonenberg and Hinnebusch, 2009). Amino acid repletion and growth factors induce phosphorylation of 4EBP-1, releasing it from eIF4E and enhancing recruitment of eIF4G, other initiation factors, and the 40S ribosomal subunit to the mRNA cap. mTORC2 localizes to ribosomes to phosphorylate Akt and some of its relatives on hydrophobic motif sites required for high activity (Oh et al., 2010).
Nutrient starvation suppresses mTORC1 more than mTORC2, leading to increased rates of catabolic processes including autophagy (Dazert and Hall, 2011; Avruch et al., 2009; Zoncu et al., 2011; Ma and Blenis, 2009; Corradetti and Guan, 2006). Damaged organelles and long-lived proteins may be trapped in autophagosomes, which then fuse with lysosomes to degrade their contents. Under stress, autophagy can accelerate to provide nutrients for enhanced survival. With sufficient amino acids, mTORC1 phosphorylates and inhibits the autophagy kinase unc51-like protein kinase ULK1 slowing autophagy (Jung et al., 2009; Egan et al., 2011).
Several events have been implicated in mTORC1 activation by amino acids. The Rag GTPases associate with mTORC1 in an amino acid-dependent manner and localize it to a compartment that contains its activator Rheb. Also involved is the Ragulator complex which scaffolds mTORC1 to late endosomes and lysosomes, where it is activated (Sancak et al., 2010). The spatial organization of lysosomes in the cytoplasm is critical for mTORC1 activation.
Preceding these key steps, the initial amino acid sensing mechanisms are less clear. Amino acids are transported into cells where it is thought that they engage an unknown intracellular sensor that activates mTORC1. In addition, the transporters themselves are proposed to be components of the sensory apparatus. The bidirectional amino acid transporter SLC7A5/SLC3A2 is thought to activate mTOR by allowing for the influx of leucine in exchange for the efflux of glutamine (Nicklin et al., 2009). In yeast Ssy1, a member of the amino acid permease family, regulates gene transcription in response to extracellular amino acids, although it is not able to transport amino acids into the cell (Hundal and Taylor, 2009; Forsberg and Ljungdahl, 2001). Glutamatergic activation of mTORC1 has also been reported in neurons (Lenz and Avruch, 2005). Findings such as these suggest possible roles for membrane receptors in signaling to mTORC1.
The G protein-coupled receptor (GPCR) complex T1R1/T1R3 is an amino acid receptor that was discovered in gustatory neurons as a detector of the umami flavor (Matsunami et al., 2000; Nelson et al., 2002). The related receptor T1R2/T1R3, also discovered in gustatory neurons, is known as the sweet receptor and functions as a glucose sensor not only in gustatory neurons but also in the intestine and the hypothalamus (Ren et al., 2009; Jang et al., 2007; Mace et al., 2007; Nakagawa et al., 2009). Here we show that T1R1/T1R3 is an early sensor of amino acid availability. Reduced expression of this receptor impairs activation of mTORC1 by amino acids, results in mTORC1 mislocalization, and accelerates autophagy.
Results
The GPCR T1R1/T1R3 Is an Amino Acid Sensor in Pancreatic Beta Cells
Pancreatic β cells respond to increased nutrient concentrations by secreting insulin and inducing synthetic processes to replenish the insulin supply. Activation of ERK1/2 by glucose in β cells is necessary for glucose-stimulated insulin gene transcription (Khoo et al., 2003); yet, the mechanism(s) by which these cells sense and respond to amino acids is unclear. Because ERK1/2 are activated by glucose and hormones that promote insulin secretion, we hypothesized that ERK1/2 in β cells may also monitor extracellular amino acids. We noted that amino acids rapidly yet transiently activated ERK1/2 in MIN6 β cells. The kinetics of ERK1/2 activation were similar to those of carbachol, a ligand for muscarinic (M3) GPCRs on β cells (Figure 1A). Because of this shared kinetic profile, we determined if other aspects of the mechanism were similar. Muscarinic GPCR activation increases phospholipase C β (PLCβ) activity, which leads to an influx of extracellular calcium. We found that amino acids induced calcium entry into MIN6 cells with similar kinetics and amplitude as carbachol, and calcium entry was necessary for ERK1/2 activation by both ligands (Figures 1B and 1C). PLCβ activation was required for amino acid-induced calcium entry and ERK1/2 activation, as the PLCβ inhibitor U73122 blocked both of these events (Figure 1D, and Figure S1A), revealing common mechanistic features.
Fig. 1. Amino acids activate ERK1/2 through the T1R1/T1R3 receptor.
(A) MIN6 pancreatic β cells in Kreb’s Ringer’s bicarbonate solution (KRBH) without amino acids (AA) for 2 hr were treated with AA as defined in Extended Experimental Procedures or 100 μM carbachol for the indicated times. ERK1/2 activation was analyzed by immunoblotting. (B) MIN6 cells treated as in (A) were loaded with Fura-2 and then stimulated with AA or carbachol as above. Data are the mean +/− SEM of the 340/380 values of three independent experiments. (C) MIN6 cells incubated in KRBH for 2 hr as above. 5 min prior to stimulation with AA, KCl, or carbachol (Carb), cells were placed in KRBH with our without CaCl2. Cells were stimulated for 2 min, harvested and lysates were immunoblotted as above. (D) Calcium was measured as above in MIN6 cells pretreated with vehicle (DMSO), 10 μM U73122, or 10 μM U73344 for 30 min prior to AA stimulation. Data are the mean +/− SEM of the 340/380 values of three independent experiments. (E–H) MIN6 cells stably expressing control shRNA1 and T1R3 shRNA1 (E, G) or T1R1 shRNA1 (F, H) in KRBH were stimulated with AA or 100 μM Carb for 2 min (E) or with AA for the indicated times (F). (I) RNA was isolated from tissues from C57BL/6J mice for qPCR analysis. Data are normalized to 18S RNA and are the means of triplicate measurements +/− SD. Data from all panels are representative 3 independent experiments except panels (E), one of 8 and (F) one of 2. See also Figure S1.
These data suggested that amino acids stimulated ERK1/2 through a cell surface GPCR. Because the GPCR complex T1R1/T1R3 is activated by all twenty amino acids, we tested the hypothesis that T1R1/T1R3 was the amino acid sensor in pancreatic β cells signaling to ERK1/2. We observed that amino acid-induced stimulation of ERK1/2 in MIN6 cells stably expressing either T1R3 or T1R1 shRNA was significantly reduced compared to cells expressing a control shRNA (Figures 1E–1H and S1E). Carbachol-induced ERK1/2 activation was not diminished with T1R3 knockdown, demonstrating that reduced expression of T1R3 leaves other signaling mechanisms to ERK1/2 intact (Figure 1E).
Expression of T1R1/T1R3 was initially thought to be restricted to gustatory neurons (Matsunami et al., 2000; Nelson et al., 2002). Because most cell types require amino acid sensing capabilities to coordinate cellular demands with nutrient availability, we asked if T1R1/T1R3 might serve as an amino acid sensor in other cell types as well. We found both T1R1 and T1R3 are present broadly in mouse tissues, human islets, and many types of cultured cells (Figures 1I and Figures S1B–S1D).
Amino Acids Signal to mTORC1 through T1R1/T1R3
mTORC1 coordinates cell growth with the availability of amino acids. That mTORC1 is activated by amino acids is well characterized, but less is understood about upstream mechanisms. In MIN6 cells in which either T1R3 or T1R1 was knocked down, there was a significant impairment of amino acid-induced phosphorylation of the mTORC1 substrate p70 ribosomal protein S6 kinase (S6K) (Figure 2A and Figure S2A). The phosphorylation of mTOR on S2488, which is positively correlated with increased mTORC1 activity (Nave et al., 1999), was also reduced in the T1R3 and T1R1 knockdown cells (Figure S2B).
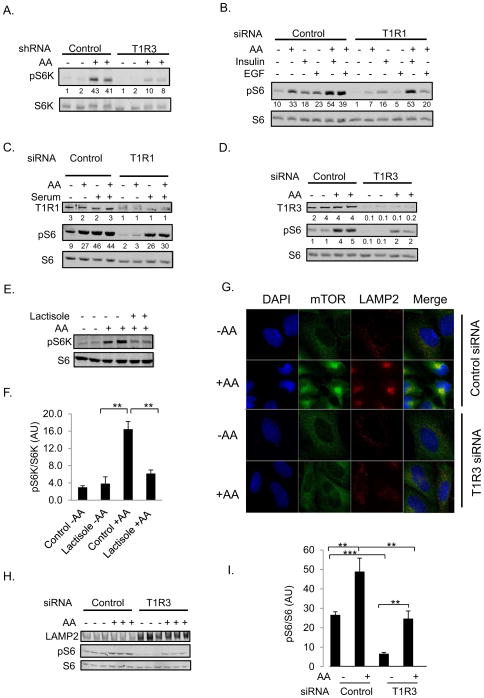
Fig. 2. The T1R1/T1R3 taste receptor is necessary for optimal amino acid induced-mTORC1 activation in numerous cell types.
(A) MIN6 cells stably expressing either control shRNA2 or T1R3 shRNA1 in KRBH for 2 hr were treated with AA or vehicle (H2O) for the indicated times. Protein expression and phosphorylation were analyzed by immunoblotting. Signals were quantitated with the Odyssey Licor infrared scanning system. Numbers under lanes are arbitrary units of pS6K/S6K. The means of pS6K/S6K +/− SEM from three independent experiments are the following: Control (2 +/− 0.42), Control + AA (33 +/− 4.2), T1R3 (2 +/− 0.44), and T1R3 + AA (8 +/− 0.98). The p values for Control - AA vs. Control + AA, T1R3 vs. T1R3 +AA, and Control + AA vs. T1R3 + AA are all < 0.01. (B) H9C2 cardiac myoblast cells transiently transfected with either control non target siRNA1 or T1R1 siRNA1 in KRBH were treated with vehicle, AA, 100 nM insulin, or 50 ng/ml EGF for 30 min. Protein expression and phosphorylation were analyzed by immunoblotting. The means of signal from pS6 relative to S6 or alpha tubulin +/− SEM from three samples from two independent experiments are: Control (12.5 +/− 3.3), Control + AA (33.9 +/− 3.9), Control + insulin (19.3 +/− 1.0), Control + EGF (26.9 +/− 3.2), Control + insulin and AA (43.2 +/− 6.8), Control + EGF and AA (43.9 +/− 3.7) T1R1 (1.9 +/− 0.3), T1R1 + AA (8.3 +/− 1.6), T1R1 + insulin (19.5 +/− 2.5), T1R1 + EGF (6.7 +/− 1.5), T1R1 + insulin and AA (48 +/− 6.0), and T1R1 + EGF and AA (22.8 +/− 4.6). The p values are the following: Control + AA vs. T1R3 + AA (< 0.01), Control + insulin vs. T1R3 + insulin (ns), Control + EGF vs. T1R3 + EGF (< 0.01), Control + insulin and AA vs. T1R3 + insulin and AA (ns), and Control + EGF and AA vs. T1R3 + EGF and AA (< 0.05). (C) H9C2 cells transfected with the indicated siRNAs as in (B) were stimulated with AA or 20 % fetal bovine serum (FBS). Quantitation was performed as above. The mean of pS6/S6 +/− SEM for three independent experiments are Control (10.3 +/− 1.3), Control + AA (30.7 +/− 1.9), Control + FBS (67.3 +/− 10.7), T1R1 (2.3 +/− 0.3), T1R1 + AA (8.7 +/− 2.9), and T1R1 + FBS (36.7 +/− 5.8). The p values are the following: Control + AA vs. T1R1 + AA (< 0.01) and Control + FBS vs. T1R1 + FBS (< 0.05). (D) H9C2 cardiac myoblasts transiently transfected with control siRNA1 or T1R3 siRNA1 in KRBH were treated with vehicle or AA for 30 min. Immunoblots were performed as above. The mean from three independent experiments +/− SEM are the following: control siRNA (1.7 +/− 0.2), control siRNA + AA (5.4 +/− 0.7), T1R3 siRNA (0.5 +/− 0.2), and T1R3 siRNA + AA (2 +/− 0.2). The p values of control siRNA vs T1R3 siRNA and control siRNA + AA vs T1R3 siRNA +AA are 0.019 and 0.018, respectively. (E and F) HeLa cells in KRBH for 2 hours were pretreated with 8 mM lactisole or vehicle (DMS0) for 15 min, and stimulated with AA for 30 min. (F) Graphed are means +/− SEM of the ratio of pS6K/S6K from three independent experiments. **, p < 0.01. (G–I) HeLa cells transiently transfected with either scrambled siRNA or T1R3 siRNA4 were incubated in EBSS for 90 min before stimulation with AA for 30 min. Cells were then either fixed and stained with DAPI (blue), anti-mTOR (green), or anti-LAMP2 (red) (G) or lysed and immunoblotted as above (H–I). (I), means of 3 independent experiments +/−SEM, **, *** p < 0.01 and 0.001, respectively. P values were determined using the two-tailed Student’s t-test. See also Figure S2.
Because we observed robust T1R1 and T1R3 expression in mouse heart (Figure S1D), we investigated the role of T1R1/T1R3 signaling to mTORC1 in H9C2 rat cardiac myoblasts. We depleted T1R1 and T1R3 transiently in H9C2 cells using siRNA and found that amino acid-stimulated and basal mTORC1 activity, assessed by phosphorylation of both S6K on S389 and ribosomal protein S6 on S235 and S236, were reduced (Figures 2B, 2C, 2D, and Figures S2D and S2G).
Activation of mTORC1 by growth factors requires the presence of amino acids (Avruch et al., 2009). Because we hypothesized that T1R1/T1R3 provides this amino acid signal, we determined if knockdown of this receptor would inhibit growth factor-induced mTORC1 activation. We observed that reduced T1R1 expression in H9C2 cells significantly reduced the ability of EGF and fetal bovine serum to induce phosphorylation of S6K and S6 even in the presence of amino acids (Figure 2B, 2C and Figure S2E). As additional support for the conclusion that T1R1/T1R3 is an amino acid sensor for mTORC1 activation, we acutely impaired human T1R3 signaling using small molecule inhibitors (Xu et al., 2004; Maillet et al., 2009). Pretreatment of cells with lactisole for 15 min reduced amino acid-stimulated phosphorylation of S6K and S6 in HeLa cells in a dose-dependent manner (Figures 2E and 2F and Figure S2E). Amino acid-induced phosphorylation of S6K in PANC-1, MCF7, and Jurkat human cell lines was also reduced by receptor inhibitors (Figure S2F). Thus, T1R1/T1R3 senses amino acids and communicates this information to mTORC1 in a wide variety of human and rodent cell lines.
Amino acids activate mTORC1 in part by localizing the kinase to lysosomal membranes where it interacts with Rag small GTPases and the Ragulator complex (Sancak et al., 2010). We tested the idea that T1R1/T1R3 may be necessary to trigger the movement of mTORC1 to lysosomes. In control cells, we observed an amino acid-induced redistribution of mTOR from a diffuse cytoplasmic pattern to a more concentrated localization, in proximity to the lysosomal marker LAMP2 (Figure 2G). However, amino acids did not cause redistribution of mTOR in cells with reduced T1R3 expression, nor did mTOR appear to co-localize to a substantial extent with LAMP2 in those cells. LAMP2 expression was greater, while S6 phosphorylation was decreased in the receptor knockdown cells (Figures 2H and 2I). The distribution of this lysosomal membrane protein also reflected a reorganization of lysosomes; they appeared more diffuse and peripheral in the cytoplasm, a pattern characteristic of starved cells. These results suggest that T1R1/T1R3 is involved in early events in amino acid sensing that lead to mTORC1 activation.
Loss of T1R1/T1R3 Reduces Translation Initiation and Insulin Content of β Cells
mTORC1 controls translation initiation (Ma and Blenis, 2009). In starved cells 4EBP-1 binds eIF4E, blocking initiation of translation from capped mRNAs. Phosphorylation of 4EBP-1 by mTORC1 frees eIF4E to recruit eIF4G, the small ribosomal subunit and other factors to these mRNAs to initiate their translation (Brunn et al., 1997). Stable knockdown of T1R3 in MIN6 cells or transient knockdown of T1R1 in H9C2 cells reduced phosphorylation of 4EBP1 and interfered with cap-dependent translation initiation (Figure 3A, and Figures S3D and S3E). Amino acids were much less effective in recruiting eIF4G to the mRNA cap from either T1R1 or T1R3 knockdown cells compared to control (Figures 3B, 3C and Figures S3A, S3B, S3C). We conclude that amino acid sensing through T1R1/T1R3 supports enhanced cap-dependent translational activity.
Fig. 3. Signaling through T1R1/T1R3 is necessary for translation initiation.
(A) MIN6 cells with T1R3 silenced, as indicated, were placed in AA-free medium as in Figures 1 and 2. Cells were treated with or without AA for 30 min. Representative immunoblots of total 4EBP/1 and p4EBP from three independent experiments are displayed. (B) To investigate the ability of cells to initiate translation with T1R3 silenced, cleared lysates from MIN6 cells treated as described in (A) were incubated with Sepharose beads conjugated to 7-methylGTP. Proteins in lysates and bound to 7-methylGTP were detected by immunoblotting. (C) Amounts of eIF4G and eIF4E in lysates and bound to the 7-methylGTP beads in (B) were quantified using a Licor Odyssey imaging system and graphed as the ratio of bound eIF4G to total eIF4G in the lysate. Data are as mean +/− SEM of the 4 samples (C). (D–E) Insulin was measured in the medium (D) or lysates (E) of stable MIN6 cell lines by ELISA and normalized to soluble protein. Mean +/− SEM of three samples from two independent experiments (D). Insulin measurements in (E) are mean +/− SEM of 4 independent experiments. (F) Insulin mRNA was measured in MIN6 cells via qPCR. Data are normalized to actin RNA and are mean +SEM of triplicate measurements. *, **, p < 0.05, and 0.01, respectively (two-tailed Student’s t-test). See also Figure S3.
In pancreatic β cells, translation of preproinsulin mRNA is stimulated by insulin demand and is dependent on mTORC1 activity (Welsh et al., 1986; Mori et al., 2009). Basal and amino acid-stimulated insulin secretion was reduced in T1R3 knock down cells (Figure 3D). To determine the basis for decreased secretion, we examined insulin protein content in control and T1R3 knockdown MIN6 cells (Figure 3E). Immunoreactive insulin was decreased by approximately 50 % relative to control cells. A smaller but significant loss was also noted in T1R1 knockdown cells. Quantitation of preproinsulin mRNA indicated no reduction in message (Figure 3F), suggesting that loss of insulin protein is not due to loss of mRNA but rather due to insulin degradation or insufficient translation or processing resulting from reduced mTORC1 activity.
Activation of mTORC1 Is Reduced in T1R3−/− Mice
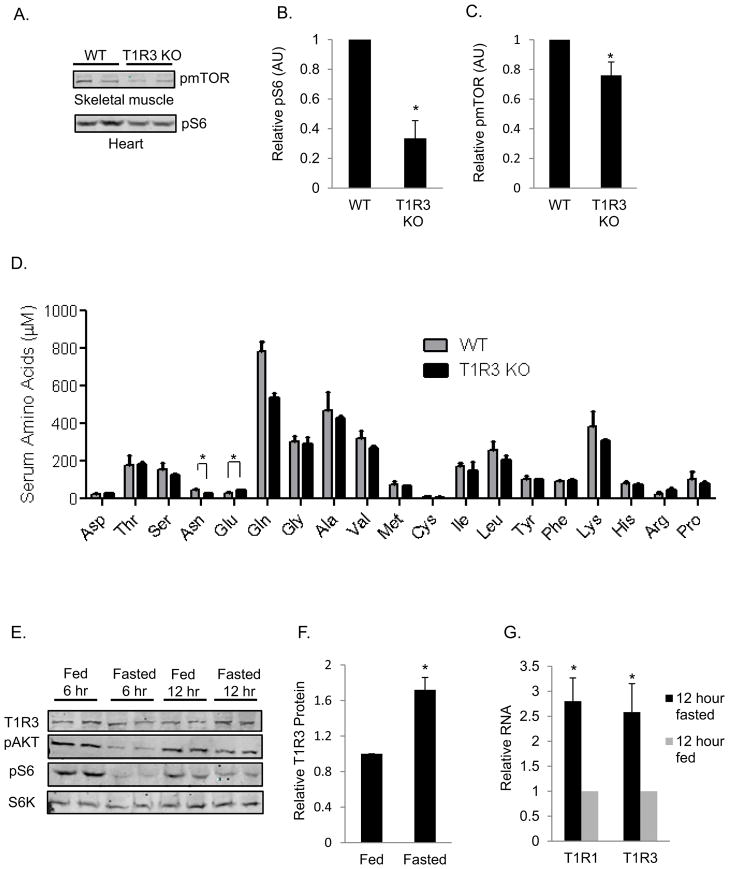
To determine the impact of T1R3 deficiency in mice, we measured mTORC1 activity in the skeletal muscle and heart from wild-type and T1R3 knock out mice fasted overnight. There was a ~25% reduction in S2448 phosphorylation of mTOR in the skeletal muscle and a 60% reduction in S6 phosphorylation in hearts of T1R3 knock out mice compared to wild-type mice (Figures 4A, 4B, and 4C).
Fig. 4. mTORC1 activity is lower in tissues from T1R3−/− mice.
(A–D) C57BL/6J wild-type mice or T1R3 knockout mice were fasted 12–15 hr before sacrifice. (A) Immunoblots to detect pmTOR or pS6 from the indicated organs are shown. Refer to the S6K immunoblot in Figure 7I for protein normalization for the corresponding tissues. (B and C) Means of abitrary units of either pmTOR/S6K or pS6/S6K +/− SEM from 6 wild type and 6 T1R3 knockout mice. (D) AA concentrations were determined in serum isolated from mouse blood collected by cardiac puncture. Data are mean +/− SEM from 3 wild-type mice and 3 T1R3 knockout mice. (E–G) C57BL/6J mice were either fed or fasted for 12 hours before sacrifice. Protein and RNA were isolated from leg muscles for immunoblotting (E–F) and qPCR (G). Data are normalized to S6 protein (E and F) or 18S RNA (G), and means +/− SEM of a total of 6 fed and 6 fasted mice from three independent experiments. *, **, p < 0.05, and 0.01, respectively (two-tailed Student’s t-test). See also Figure S4.
Because serum amino acid concentrations fluctuate as a consequence of food intake, we asked if mTORC1 activity was lower in the T1R3 knockout mice because of an overall decrease in serum amino acid concentrations. We found that most amino acid concentrations were unchanged between control and T1R3 knockout mice. Exceptions include a slight, but statistically significant, decrease in asparagine and increase in glutamate in the T1R3 knockout mice (Figure 4D).
A previous study reported that the concentrations of a number of amino acids in plasma, skeletal muscle, and liver decreased in C57BL/6J mice after 6–12 hours of fasting, with a transient increase after 24 hours of fasting (Ezaki et al., 2011). Because agonist availability can induce compensatory changes in the expression of receptors, we tested if T1R3 receptor expression was affected in the skeletal muscle of C57BL/6J mice that were fasted for 6 and 12 hours. Immunoreactive T1R3 in skeletal muscle was highest following a 12 hour fast (Figures 4E and 4F) and receptor mRNA was also increased 3.5-fold (Figure 4G). Similar changes were also observed in cultured cells. As expected, the phosphorylation of both AKT and S6 were reduced in the muscle of mice after 6 hours and 12 hours of fasting when compared to fed mice.
Signaling Mechanisms that Contribute to mTORC1 Activity
Amino acid regulation of mTORC1 may occur in part through ERK1/2 directly and through the ERK1/2 substrate kinase Rsk (Roux et al., 2004; Carriere et al., 2008). Although it has been reported that ERK1/2 are not sensitive to amino acids (Gulati et al., 2008), we found that amino acids rapidly but transiently stimulated ERK1/2 activity at concentrations as little as 1/4 of those in growth medium, comparable to the sensitivity of mTORC1 to amino acids (Figure 5A). Because we observed that T1R1/T1R3 signals to ERK1/2 (Figures 1E and 1F), we determined if ERK1/2 contribute to mTORC1 activation by amino acids. U0126, which prevents ERK1/2 activation by blocking MEK1/2, inhibited not only amino acid ERK1/2 stimulation but also phosphorylation of S6K, implicating ERK1/2 in mTORC1 regulation in response to amino acids in multiple cells types (Figures 5B, 5C, S5A, S5B). These results suggest that ERK1/2 is a significant amino acid-dependent input to mTORC1. In contrast, AKT, another activator of mTORC1, was not activated to a detectable extent by amino acids. Depletion of T1R1 had little if any effect on serum stimulation of Akt (Figure 5D).
Figure 5. T1R1/T1R3-induced signaling to mTORC1.
Following the indicated treatments, cell lysates were immunoblotted with designated antibodies. (A) MIN6 cells in KRBH were stimulated with the indicated concentrations of AA (0.25X - 1X) for 2 min or 30 min. (B) MIN6 cells in KRBH were pretreated with 10 μM U0126 and then stimulated with 1X AA for the indicated times. (C) H9C2 pretreated as in (B) were stimulated with AA for 15 min. (D) H9C2 cells transiently expressing control siRNA1 or T1R1 siRNA1 were incubated in KRBH prior to stimulation with 25 mM glucose and/or AA, or with 20% serum for 30 min. (E) The PLC inhibitor U73122 (10 mM), the inactive analog U73343, or vehicle were added to MIN6 cells in KRBH 15 min prior to treatment with AA for the indicated times. (F) MIN6 cells stably expressing either control shRNA2 or T1R3 shRNA1 were loaded with Fura-2 in KRBH before AA stimulation for 30 min. Data are means +/− SEM of the 340/380 values of three independent experiments. (G) MIN6 cells in KRBH were pretreated with vehicle, or 10 μM nifedipine for 15 min prior to addition of vehicle or AA for the indicated times. Data from all panels are representative of 3 independent experiments, except panel (D), one of 2. See also Figure S5.
In gustatory neurons, amino acid binding to T1R1/T1R3 activates the G protein gustducin to stimulate PLCβ2, which leads to a release of calcium from intracellular stores. The role of PLCβ activity in mTORC1 activation was tested by comparing the effects of the PLCβ inhibitor U73122 to its inactive analog U73343. The PLCβ inhibitor, but not U73343, inhibited calcium influx induced by amino acids (Figure 1D), and reduced amino acid-induced mTORC1 activity (Figure 5E), suggesting that PLCβ is required for maximum amino acid-induced activity. Increased intracellular calcium is necessary for amino acid-induced mTOR activation in HeLa cells (Gulati et al., 2008). We found that T1R1/T1R3 was necessary for amino acid-induced calcium influx, and that this calcium influx was necessary for optimal mTORC1 activation (Figure 5F). Consistent with a role for calcium, blockade of L-type calcium channels interfered with amino acid stimulation of mTORC1 in β cells (Figure 5G). Our findings suggest that amino acids bind T1R1/T1R3 leading to activation of PLCβ, calcium entry, and ERK1/2, thus stimulating mTORC1 (Figure 7J).
Figure 7. T1R3 depletion induces autophagy.
(A) H9C2 cells in complete growth medium or in KRBH were permeabilized and LC3 was detected by immunofluorescence. (C) H9C2 cells transfected with either control non target siRNA1 or T1R3 siRNA1 were incubated in complete growth medium and LC3 was detected as above. (B and D) LC3 punctae in Z-stack images from (A and C, respectively) were quantitated as described in Extended Experimental Procedures and are the mean +/− SEM of LC3 punctae/cell. (E) Lysates of H9C2 cells that had been transfected with the indicated siRNA were incubated in KRBH for 2 hrs before lysis. Lysates were immunoblotted to detect the indicated proteins. (F) Quantitation of immunoblots is graphed as ratios of the means +/− SEM of LC3-II/GAPDH of a total of 4 samples from three independent experiments. (G) H9C2 cells were transfected with the indicated siRNA and cultured in complete growth medium before harvest. (H) Quantitation was as in (F) and is graphed as the ratio +/− SEM of the amounts of indicated proteins divided by the amount of S6. (I) C57BL/6 wild-type or T1R3 knockout mice were fasted for 12 hrs before sacrifice. The indicated proteins were immunoblotted. (J) Model. Amino acids are sensed directly via T1R1/T1R3. PLCβ is then activated and calcium influx increases, activating ERK1/2 and Rsk. These kinases aid in activation of mTORC1 by phosphorylating Raptor and TSC2. Growth factors feed into this pathway at multiple levels. T1R1/T1R3 may also positively regulate mTORC1 by affecting the localization of Rag proteins.
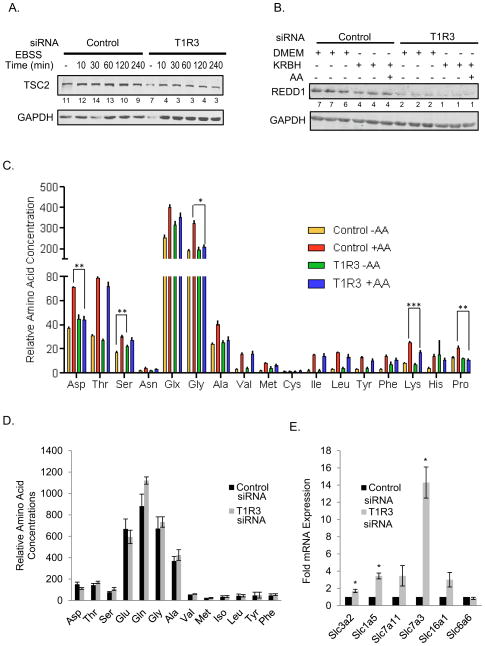
TSC2 and REDD1 are among negative regulators of mTORC1 activity (Brugarolas et al., 2004). We tested the idea that knockdown of T1R3 might decrease mTORC1 activation by increasing the expression of these negative regulators. In HeLa cells we found instead that the opposite occurred; silencing T1R3 resulted in a decrease in TSC1 and REDD1 (Figures 6A and 6B). These findings suggest that silencing T1R3 causes changes that would be expected if cells were engaging mechanisms to compensate for loss of mTORC1 activity. Because loss of TSC2 increases mTORC1 activity, mTORC1 is less readily inhibited by amino acid withdrawal in TSC2 null MEFs (Figure S5C). However, knockdown of TSC2 in HeLa cells does not rescue amino acid activation of mTORC1 if T1R3 is also knocked down (Figures S5D and S5E). The ability of constitutively active Rag GTPases to keep mTORC1 active in the absence of amino acids was also tested. However, we observed a similar decrease in mTORC1 activity upon amino acid deprivation in both the control cells and cells transfected with active RagB (Figure S5G). On the other hand, mTORC1 was less inhibited by amino acid removal in HeLa cells highly overexpressing active RasV12 than control cells (Figure S5F).
Figure 6. Silencing T1R3 induces compensatory changes.
(A) HeLa cells transiently transfected with scrambled siRNA, or T1R3 siRNA4 were either left in complete growth medium or placed in EBSS for the indicated times before harvest. (B) HeLa with or without T1R3 knocked down as in (A), were in complete growth medium or KRBH for 1 h and 45 min before harvest; one sample was stimulated for 1 h with AA as indicated. Proteins were detected by immunoblotting. Numbers under lanes are ratios of TSC2/GAPDH (A), or REDD1/GAPDH (B). (A) The means of TSC2/GAPDH from control siRNA or T1R3 siRNA transfected cells either in DMEM, KRBH, or EBSS for various times from a total of 10 separate samples from two independent experiments are: Control (11.0 +/− 0.7) and T1R3 siRNA (5.6 +/− 0.8). P < 0.01. (B) The means +/− SEM of REDD1/GAPDH are: Control (5.7 +/− 0.39) and T1R3 siRNA (1.9 +/− 0.22). P < 0.01. (C) H9C2 cells with or without T1R3 silenced as above in KRBH were stimulated with AA for 30 min. Intracellular AA concentrations were determined. Data are area under the curve and are the means of three replicates +/− SEM. *, **, and *** p < 0.05, 0.01, and 0.001, respectively. (D) Relative intracellular amino acid concentrations were determined from H9C2 cells transiently transfected with either control siRNA1 or T1R3 siRNA1 and grown in complete growth medium. Data are graphed as above and are the means +/− SEM of three samples. Asn and Cys were below detection limits. His, Arg, and Lys were detected in only one of two experiments. Their concentrations in T1R3 knockdown cells either did not change (His), increased 1.3-fold (Arg), or increased 2-fold (Lys) when compared to cells transfected with control siRNA. (E) RNA from H9C2 cells with T1R3 knocked-down was analyzed by qPCR. Data are means +SEM of three independent experiments. * P < 0.05 control siRNA versus the indicated transcripts.
Reduced T1R1/T1R3 Expression Does Not Deplete Intracellular Amino Acids
To determine if impairing T1R3 affected intracellular amino acid concentrations, we compared amino acid concentrations in lysates from control and T1R3 knockdown cells (Figure 6D). The amounts of most amino acids including glutamine and the nonessential amino acids, in particular β-branched amino acids, were similar in control and knockdown cells. The relative amounts of amino acids were also comparable to those reported in muscle and other tissues (Kalhan et al., 2011). To determine if a defect could be observed as a result of amino acid depletion, we also examined amino acids in cells that had been deprived of and then refed with amino acids (Figure 6C). The amounts of most amino acids were again similar in knockdown and control cells. Among differences, concentrations of aspartic acid, serine, glycine, lysine, and proline after refeeding were significantly higher in the control cells; however, there were no differences in the concentrations of branched chain amino acids between the refed control and T1R3 knockdown cells.
Glutamine taken up by solute carrier family 1 member 5 (SLC1A5) is exchanged for leucine and other essential amino acids through a SLC3A2/SLC7A5 heterodimer. This exchange is rate limiting for activation of mTORC1 by essential amino acids and growth factors (Nicklin et al., 2009). To determine if knockdown of the receptor also affected mTORC1 in glutamine-loaded cells, cells were deprived of amino acids for one hour and then pretreated with insulin and glutamine for an additional hour prior to addition of leucine (Figure S5H). Under these conditions, loss of T1R3 impaired activation of mTORC1 by leucine. Based on these analyses, reduced mTORC1 activity is unlikely to be due to insufficient intracellular glutamine available for exchange.
In a microarray experiment (data not shown), we noted that SLC1A5, the glutamine transporter, appeared to be upregulated several-fold by loss of T1R3, as did SLC3A2, a subunit of the transporter that exchanges glutamine for essential amino acids. Quantitative PCR (qPCR) analysis confirmed that mRNAs encoding both transporters were increased by T1R3 knockdown in H9C2 cells (Figure 6E), further suggesting that a decrease in neither the expression nor functionality of these transporters contributed to reduced amino acid activation of mTORC1 in the knockdown cells. Other transporters were also upregulated including SLC7A11, a glutamate-cystine antiporter, which is induced by amino acid starvation (Sato et al., 2004), and SLC16A1, a lactate/pyruvate monocarboxylic acid transporter. SLC7A3, a cationic amino acid transporter, was upregulated 15-fold in the knockdown cells.
Reduced T1R3 Activated Autophagy
Amino acid deprivation increases the rate of autophagy (Yang and Klionsky, 2010). Conversion of the ubiquitin-like protein LC3-1 to its lipidated form (LC3-II) and accumulation of LC3 in autophagosomes are measures of autophagy that are visibly increased by amino acid deprivation (Figures 7A and 7B). Because mTORC1 suppresses autophagy, we probed the impact of receptor loss on this process. Knockdown of T1R3 in H9C2 or HeLa cells cultured in either complete growth medium or in the absence of amino acids induced the accumulation of LC3-II and the translocation of LC3 to autophagosomes (Figures 7C, 7D, 7E, and 7F, S6A). mTORC1 negatively regulates autophagy by phosphorylating ULK1 S757 (Kim et al., 2011). We observed that ULK1 S757 phosphorylation was significantly reduced in T1R3 knockdown cells in nutrient replete conditions. AMP-activated protein kinase (AMPK) positively regulates autophagy by phosphorylating ULK1 S555 (Egan et al., 2011). We found a decrease in AMPK activity in T1R3 knockdown cells as measured by decreased phosphorylation of AMPK substrates ULK1 S555 and acetyl-CoA carboxylase (ACC) (Figures 7G and 7H). Thus, autophagy induced by T1R3 knockdown is apparently not due to increased AMPK activity, but rather decreased mTORC1 activity.
Autophagy is induced in mouse after 12–24 hours of fasting (Ezaki et al., 2011). To determine if T1R3 loss exacerbates autophagy caused by caloric restriction, we measured p62 levels in tissues from wild-type or T1R3−/− mice after a 12–15 hour fast. Decreased p62, which is sequestered on autophagosomes and degraded in lysosomes, is an indicator of increased autophagic flux. We found decreased p62 in tissues of T1R3−/− mice compared to wild-type (Figures 7I, S6B), suggesting that loss of T1R3 increases autophagic flux.
Discussion
We provide evidence that the GPCR T1R1/T1R3 is a paramount mammalian amino acid detector that provides the earliest notice of amino acid availability from the extracellular milieu to essential intracellular energy-sensing mechanisms. Although not previously linked to mTORC1, amino acid-sensitive GPCRs are well known in mammals; yet, the majority of evidence has implicated amino acid uptake, metabolism and other unspecified intracellular events in activation of mTORC1. GPCRs participate in sensing nutrients including glucose and amino acids in fungi (Xue et al., 2008). In yeast amino acid detection by the TOR system involves nutrient transport proteins most of which have little or no ability to transport amino acids to the cell interior, and instead function as membrane receptors (Forsberg and Ljungdahl, 2001; Hundal and Taylor, 2009). Our conclusion that T1R1/T1R3 detects extracellular amino acids prior to their uptake and transmits that information to mTORC1 suggests that sensing amino acids at the plasma membrane by TOR is evolutionarily conserved in mammals.
Several observations suggest that cells attempt to compensate for the perceived deficiency in amino acids resulting from taste receptor knockdown. Receptor knockdown, even under nutrient-replete conditions, increased autophagy, consistent with perceived starvation. Although intracellular concentrations of amino acids in knockdown and control cells were comparable, cells also upregulated mRNAs encoding several transporters following receptor knockdown, to increase amino acid and monocarboxylic acid uptake and flux, as if these were impaired. Upregulated mRNAs included those encoding the transporters, SLC1A5 and SLC3A2, shown to be rate-limiting for mTORC1 activation, and SLC7A11 and SLC16A1, both of which are induced by starvation (Sato et al., 2004; Konig et al., 2008). Upregulation of these transporters upon depletion of T1R3 further supports the idea that this GPCR is a key amino acid sensor. In HeLa cells knockdown of this GPCR resulted in downregulation of two negative regulators of mTORC1, consistent with efforts by the cell to relieve repression and reactivate mTORC1. In β cells with stable T1R3 knockdown, insulin content was the lowest immediately following antibiotic selection, but increased over the subsequent months. These findings suggest that mechanisms instigated to offset the translational blockade due to mTORC1 inhibition facilitate increased protein synthesis and processing. How cells overcome the loss of this receptor is not yet defined. Compensation could be due to upregulation of related class C GPCRs that may be able to transmit amino acid signals to mTORC1, suppression of autophagy, even though it remains elevated over the basal rate in T1R3 knockdown cells, or through intracellular actions of leucine, arginine, or β-branched amino acids, for example.
Amino acids induce association of mTORC1 with intracellular membrane compartments. With signaling from T1R1/T1R3 interrupted, mTORC1 displays neither the characteristic lysosomal immunostaining pattern nor strong localization with lysosomal markers. Thus, this receptor is required to direct the amino acid-dependent intracellular trafficking of mTORC1. Activation of mTORC1 by high concentrations of serum or strong overexpression of V12 Ras was detected in receptor knockdown cells, suggesting that mTORC1 activity can be stimulated. Whether these effects actually reflect mechanistic bypass of actions of this receptor or are due largely to the pathophysiological stimuli remains a question. On the other hand, mTORC1 activity was poorly stimulated by EGF in receptor knockdown cells, consistent with the requirement for amino acids to support growth factor signaling to mTORC1. It remains possible that T1R1/T1R3 communicates a signal to mTORC1 that may be redundant with certain serum factors but that is not provided well by individual growth factors. These results conform to the expectation that maximal mTORC1 activation requires a convergence of pressure from multiple extracellular molecules.
Fasted T1R3 knockout mice have significantly less mTORC1 activity in both heart and skeletal muscle than wild-type mice. We were unable to consistently observe a significant decrease in mTORC1 activity between the livers of fasted T1R3 knockout mice and wild-type mice. Liver mTORC1 activity may be more resistant to fasting than in other tissues due to increased intra-organ nutrient availability caused by higher amounts of liver autophagy. In fact, mTORC1 activity was higher in livers from rats fasted for 48 hours than in fed controls (Anand and Gruppuso, 2005).
Our findings reveal that T1R1/T1R3 conveys information on amino acid availability to mTORC1. The fact that other GPCRs such as GPR40 mediate some actions of free fatty acids has demonstrated that nutrients have the potential to regulate cell signaling and metabolism at extracellular as well as intracellular levels in mammals (Itoh et al., 2003). Because membrane receptors are readily accessible to drugs, T1R1/T1R3 may be a valuable therapeutic target not only for cancer but for protein-wasting and degenerative diseases including diabetes, cachexia and Alzheimer’s disease.
Experimental Procedures
Sources of materials and antibodies are in Extended Experimental Procedures.
Transfection of shRNA and siRNA
MIN6 cells were electroporated with short-hairpin constructs (Extended Experimental Procedures) using the AMAXA electroporation system with solution V and program G-016. Stable cell lines were created by incubating cells in 2.5 μg/ml puromycin for ~one month and were subsequently maintained in puromycin. H9C2 rat cardiac myocytes were transiently transfected with either control non-target siRNA (Sigma, # SIC001) or MISSION® siRNA directed against T1R1 (#1), T1R3 (#1, or #2) designed and made by Sigma. HeLa cells were transfected with either siGENOME Non-Targeting siRNA pool #1 (Thermo Scientific) or T1R3 (#3) MISSION® siRNA. The siRNA sequences are in Extended Experimental Procedures. H9C2 cells were transfected with siRNA using 5–20 nM siRNA with Lipofectamine™ RNAiMAX (Invitrogen) following the manufacturer’s protocol and used 48–72 hours later.
Cell Culture, Immunofluorescence and Image Analysis, Cap Binding Assays, Immunoblotting and Quantitation
These procedures are described in Extended Experimental Procedures.
Tissue Harvest, RNA Isolation, and cDNA Synthesis
The experimental animal protocols used in this study were approved by the Institutional Animal Care and Use Committee (IACUC) and are listed in Extended Experimental Procedures.
Amino Acid Analysis
70–90% confluent H9C2 cells in 150 mm plates were washed 2-3X with 10 ml PBS. Cells lysed in 50% methanol were snap-frozen in N2 (liquid) and thawed at 37 °C. Freeze/thaw was repeated 4X. Insoluble material was sedimented at 16,000 X g for 10 min at 4 °C. Amino acids were measured in the supernatants. Amino acid amounts were normalized to protein concentration, determined in pellets by the BCA method (Pierce), to compare samples. Aliquots (50 μL) of 50% methanol cell extracts were deproteinized with 50 μL 3% (w/v) sulfosalicyclic acid (Sigma) in 0.02 N HCl. Supernatants were analyzed on an L-8900 Amino Acid Analyzer (Hitachi, Tokyo, Japan) equipped with a 6.0 mm ID × 40.0 mm PF-High Speed-packed ion-exchange column, fronted by a packed ion-exchange guard column, and using manufacturer reagents. Detection was by ninhydrin post-column derivatization.
Supplementary Material
Acknowledgments
We thank David Mangelsdorf, Steven Kliewer, Joseph Albanesi, Elliott Ross, Gray Pearson (Dept of Pharmacology), Michael White (Dept of Cell Biology), Ondine Cleaver (Dept of Molecular Biology), James Brugarolas (Dept of Internal Medicine) and Sylvia Vega-Rubin-de-Celis from the Brugarolas lab, Natalie Ahn (U. Colo, Boulder) and members of the Cobb lab for comments and suggestions, Nizar Ghneim for early analysis of amino acid starvation, Svetlana Earnest for technical assistance, and Dionne Ware for administrative assistance. The islets and non-endocrine pancreatic tissue were provided by the Islet Resource Facility supported by the University of Alabama, Birmingham, Comprehensive Diabetes Center. This work was supported by grants from the National Institutes of Health (R01 DK55310 and R37 DK34128 to MHC and R01 CA157996 to RJD) and the Robert A. Welch Foundation (I1243 to MHC). During the majority of this work EMW was supported by a mentor-based postdoctoral fellowship from the American Diabetes Association, AYL was supported by a training grant from the Cancer Prevention and Research Institute of Texas (CPRIT), and ALB was supported by NIGMS Pharmacological Sciences training grant 5-T32 GM007062.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand P, Gruppuso PA. The regulation of hepatic protein synthesis during fasting in the rat. J Biol Chem. 2005;280:16427–16436. doi: 10.1074/jbc.M410576200. [DOI] [PubMed] [Google Scholar]
- Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JCJ, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Carriere A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, Roux PP. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1) J Biol Chem. 2011;286:567–577. doi: 10.1074/jbc.M110.159046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Guan KL. Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene. 2006;25:6347–6360. doi: 10.1038/sj.onc.1209885. [DOI] [PubMed] [Google Scholar]
- Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23:744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, Iwata J, Tanida I, Furuya N, Zheng DM, Tada N, Tanaka K, Kominami E, Ueno T. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H, Ljungdahl PO. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr Genet. 2001;40:91–109. doi: 10.1007/s002940100244. [DOI] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundal HS, Taylor PM. Amino acid transceptors: gate keepers of nutrient exchange and regulators of nutrient signaling. Am J Physiol Endocrinol Metab. 2009;296:E603–E613. doi: 10.1152/ajpendo.91002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Kawamata Y, Harada M, Kobayashi M, Fujii R, Fukusumi S, Ogi K, Hosoya M, Tanaka Y, Uejima H, Tanaka H, Maruyama M, Satoh R, Okubo S, Kizawa H, Komatsu H, Matsumura F, Noguchi Y, Shinohara T, Hinuma S, Fujisawa Y, Fujino M. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422:173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- Jang HJ, Kokrashvili Z, Theodorakis MJ, Carlson OD, Kim BJ, Zhou J, Kim HH, Xu X, Chan SL, Juhaszova M, Bernier M, Mosinger B, Margolskee RF, Egan JM. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci U S A. 2007;104:15069–15074. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan SC, Uppal SO, Moorman JL, Bennett C, Gruca LL, Parimi PS, Dasarathy S, Serre D, Hanson RW. Metabolic and genomic response to dietary isocaloric protein restriction in the rat. J Biol Chem. 2011;286:5266–5277. doi: 10.1074/jbc.M110.185991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo S, Griffen SC, Xia Y, Baer R, German MS, Cobb MH. Regulation of insulin gene transcription by extracellular-signal regulated protein kinases (ERK) 1 and 2 in pancreatic beta cells. J Biol Chem. 2003;278:32969–32977. doi: 10.1074/jbc.M301198200. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig B, Koch A, Giggel K, Dordschbal B, Eder K, Stangl GI. Monocarboxylate transporter (MCT)-1 is up-regulated by PPARalpha. Biochim Biophys Acta. 2008;1780:899–904. doi: 10.1016/j.bbagen.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Lenz G, Avruch J. Glutamatergic regulation of the p70S6 kinase in primary mouse neurons. J Biol Chem. 2005;280:38121–38124. doi: 10.1074/jbc.C500363200. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Mace OJ, Affleck J, Patel N, Kellett GL. Sweet taste receptors in rat small intestine stimulate glucose absorption through apical GLUT2. J Physiol. 2007;582:379–392. doi: 10.1113/jphysiol.2007.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet EL, Margolskee RF, Mosinger B. Phenoxy herbicides and fibrates potently inhibit the human chemosensory receptor subunit T1R3. J Med Chem. 2009;52:6931–6935. doi: 10.1021/jm900823s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- Mori H, Inoki K, Opland D, Muenzberg H, Villanueva EC, Faouzi M, Ikenoue T, Kwiatkowski D, MacDougald OA, Myers MG, Jr, Guan KL. Critical roles for the TSC-mTOR pathway in {beta}-cell function. Am J Physiol Endocrinol Metab. 2009;297:E1013–E1022. doi: 10.1152/ajpendo.00262.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Nagasawa M, Yamada S, Hara A, Mogami H, Nikolaev VO, Lohse MJ, Shigemura N, Ninomiya Y, Kojima I. Sweet taste receptor expressed in pancreatic beta-cells activates the calcium and cyclic AMP signaling systems and stimulates insulin secretion. PLoS ONE. 2009;4:e5106. doi: 10.1371/journal.pone.0005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999;344(Pt 2):427–431. [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An aminoacid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Zhou L, Terwilliger R, Newton SS, de Araujo IE. Sweet taste signaling functions as a hypothalamic glucose sensor. Front Integr Neurosci. 2009;3:12. doi: 10.3389/neuro.07.012.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Nomura S, Maebara K, Sato K, Tamba M, Bannai S. Transcriptional control of cystine/glutamate transporter gene by amino acid deprivation. Biochem Biophys Res Commun. 2004;325:109–116. doi: 10.1016/j.bbrc.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M, Scherberg N, Gilmore R, Steiner DF. Translational control of insulin biosynthesis. Evidence for regulation of elongation, initiation and signal-recognition-particle-mediated translational arrest by glucose. Biochem J. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci U S A. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Hsueh YP, Heitman J. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev. 2008;32:1010–1032. doi: 10.1111/j.1574-6976.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.