Abstract
Prepulse inhibition (PPI) of the startle response is a measure of sensorimotor gating, a process that filters out extraneous sensory, motor and cognitive information. Humans with neurological and psychiatric disorders, including schizophrenia, obsessive-compulsive disorder and Huntington’s disease, exhibit a reduction in PPI. Habituation of the startle response is also disrupted in schizophrenic patients. In order to elucidate the genes involved in sensorimotor gating, we phenotyped 472 mice from an F2 cross between LG/J × SM/J for PPI and genotyped these mice genome-wide using 162 single nucleotide polymorphism (SNP) markers. We used prepulse intensity levels that were 3, 6 and 12 dB above background (PPI3, PPI6 and PPI12, respectively). We identified a significant quantitative trait locus (QTL) on chromosome 12 for all three prepulse intensities as well as a significant QTL for both PPI6 and PPI12 on chromosome 11. We identified QTLs on chromosomes 7 and 17 for the startle response when sex was included as an interactive covariate and found a QTL for habituation of the startle response on chromosome 4. We also phenotyped 135 mice from an F34 advanced intercross line (AIL) between LG/J × SM/J for PPI and genotyped them at more than 3000 SNP markers. Inclusions of data from the AIL mice reduced the size of several of these QTLs to less than 5 cM. These results will be useful for identifying genes that influence sensorimotor gaiting and show the power of AIL for fine mapping of QTLs.
Keywords: Advanced intercross lines, habituation, prepulse inhibition, quantitative trait loci, schizophrenia, sensorimotor gating, startle
The startle response is an involuntary contraction of muscles caused by an intense, sudden onset stimulus. When a startling stimulus is preceded by a smaller ‘prepulse’ stimulus the startle response is inhibited; this process is known as prepulse inhibition (PPI). PPI is used as a measure of sensorimotor gating (Geyer et al. 2001), which is a process by which extraneous stimuli are filtered out to allow the brain to focus on more important stimuli and avoid sensory overload (McGhie & Chapman 1961).
Humans with certain psychiatric and neurological disorders exhibit a reduction in PPI. Decreased PPI is most often considered an endophenotype for schizophrenia (Braff et al. 1992, 2008), but it has also been associated with obsessive-compulsive disorder (Hoenig et al. 2005; Swerdlow et al. 1993), Huntington’s Disease (Swerdlow et al. 1995) and bipolar disorder (Giakoumaki et al. 2007; Perry et al. 2001). All of these disorders are associated with a loss of gating in sensory, motor and cognitive domains and involve an impairment of higher order cognition (Bolbecker et al. 2009). Deficits in habituation of the startle response over the course of a single testing session have also been associated with schizophrenia (Braff et al. 1992; Moriwaki et al. 2009).
There is strong evidence that deficits in PPI have a genetic basis. PPI deficits are more frequent among non-affected relatives of schizophrenia patients when compared with controls (Cadenhead et al. 2000). The heritability of PPI has been estimated to be between 32% and 58% in humans (Anokhin et al. 2003; Greenwood et al. 2007). Similarly, inbred strains of both mice and rats have been reported to show differences in PPI (Palmer et al. 2000; Willott et al. 2003). Heritability has been estimated to be 48%, ranging from 31% to 67% among different strain crosses (Willott et al. 2003); quantitative trait loci (QTLs) have also been reported in both species (Hitzemann et al. 2001, 2008; Joober et al. 2002; Leussis et al. 2009; Liu et al. 2003; Palmer et al. 2003; Petryshen et al. 2005; Watanabe et al. 2007).
In this study, we tested the F2 generation of an intercross between LG/J and SM/J mice at three prepulse intensities in order to map QTLs for PPI, startle and habituation of the startle response. These strains were selected both because they have a large difference in PPI and because of the availability of both recombinant inbred and advanced intercross lines (AILs) (Ehrich et al. 2005). We also used mice from the 34th generation of an AIL between LG/J and SM/J to fine map our QTLs. AILs are a powerful tool for fine mapping because of the accumulation of recombinations that occur over generations. Elucidating the genes involved in PPI, and thereby those important to sensorimotor gating, will lead to a greater understanding of how sensory information is processed and how defects in this process can affect neurological disorders.
Materials and methods
Animals
We tested 472 F2 mice (237 male, 235 female) derived from a cross between LG/J and SM/J. We obtained inbred male SM/J and female LG/J mice from Jackson Labs (Bar Harbor, ME, USA). Some of these mice were bred to produce F1 and subsequently F2 mice, while others were used to produce inbred SM/J (n = 20) and LG/J (n = 28) mice that were phenotyped. We also tested 135 F34 mice (56 male, 79 female) derived from a cross between LG/J and SM/J. These mice were the progeny of F33 breeders obtained from Dr. James Cheverud of Washington University in St. Louis. All mice were housed in cages of 2–5 same-sex individuals (most commonly 4–5), and were kept on a 12-h light–dark cycle (lights on at 0630 h) with food and water administered ad libitum, except during behavioral testing. Other behavioral tests were conducted on the mice prior to PPI testing; however, all mice in this study were exposed to identical prior testing experience. All testing was conducted in accordance with the National Institutes of Health guidelines for care and use of laboratory animals and with the approval of the University of Chicago’s Institutional Animal Care and Use Committee.
PPI testing
The mice were tested over the course of 5 months between the hours of 1000 and 1730. The age of F2 mice at testing was 89 ± 0.25 days (mean ± standard error; range was 75–99 days); the age of F34 mice was 92 ± 0.29 days (range 83–97 days). PPI was measured using a protocol that has been described previously (Palmer et al. 2003; Shanahan et al. 2009). Briefly, mice were moved from the vivarium to a sound-attenuated pre-test room at least 30 min prior to the beginning of the test to allow them to acclimate to the testing room. At the beginning of the test, mice were placed into holding cages and moved into the testing room. Each mouse was placed into a cylindrical Plexiglas container, 5 cm in diameter, which rested on a platform within a lighted and ventilated chamber (San Diego Instruments, San Diego, CA, USA). The mouse’s movements were measured by a piezoelectric accelerometer, converted to digital data and recorded on a computer. This apparatus was calibrated in accordance with the manufacturer’s instructions before the start of each test day.
Once in the test chamber, mice were presented with 5 min of 70 dB white noise which persisted throughout the remainder of the test. The test consisted of the presentation of 62 trials that were a mixture of the following five types: a ‘pulse alone trial’, which consisted of a 40-millisecond 120-dB burst, a ‘no stimulus’ trial where no stimulus was presented, and three prepulse trials containing a 20-millisecond prepulse that was either 3, 6 or 12 dB above the 70-dB background noise level followed 100 milliseconds later (onset to onset) by a 40-millisecond 120-dB pulse. Trials were arranged into four consecutive blocks. The first and fourth blocks consisted of six pulse-alone trials. Blocks two and three consisted of a mixture of 25 of the following five trial types – six pulse-alone trials, four no stimulus trials, and five of each prepulse trial – in a pseudorandom order. The response to each trial was recorded for 65 milliseconds after the beginning of the 120-dB stimulus, or at the beginning of the ‘no stimulus’ trail. The intertrial interval was 9–20 seconds (average 15 seconds) throughout all 62 trials.
The startle response measure (‘startle’) was the average startle amplitude for all of the pulse-alone trials and is expressed in arbitrary units. PPI at each intensity was calculated using the following formula:
| (1) |
where SRprepulse is the average startle amplitude for prepulse trials and SRpulse is the average startle amplitude for pulse-alone trials in the second and third testing blocks. Habituation of the startle response (‘habituation’) was the average startle amplitude of Block 4 (six pulse-alone trials) minus the average startle amplitude of Block 1 (six pulse-alone trials). The ‘no stimulus’ trials were used to identify technical problems but were not used to calculate any of the phenotypes assessed in this study.
Genotyping
F2 genotyping was performed by KBioscience Ltd. (Hoddesdon, Hertfordshire, UK) using KASPar, which is a fluorescence-based PCR assay. One hundred and sixty-two single nucleotide polymorphisms (SNPs), selected from Petkov et al. (2004), were used as markers. They were spaced evenly across all autosomes and the X chromosome thus establishing a spacing of about 10 cM per marker. Genotyping of the F34 mice used a custom-designed Illumina array to obtain more than 3000 SNPs, as described previously (Cheng et al. in press).
Analysis
Analyses of variance (ANOVAs) to compare the phenotypes of inbred strains and correlations among phenotypes in the F2 mice were performed using the freely available statistical package R (www.r-project.org). The QTL analysis of the F2 mice was analyzed using R/qtl (Broman et al. 2003), which is a package that can be installed in R. The ‘scanone’ command was used to identify QTLs for the three prepulse trials using the expectation maximization (EM) algorithm. A similar procedure was used to evaluate startle and habituation. For each analysis, we estimated genome-wide significance levels by using 1000 permutations. We experimented with using sex and age as either additive or interactive covariates. For startle, data from the inbred strains indicated an interaction between sex and strain which led us to examine sex as an interactive covariate. Sex and age had little impact on the results of the analyses of the other traits and were therefore not included as covariates. We also tried to use the ‘scantwo’ command to identify epistatic relationships between loci.
The F34 data were analyzed as previously described by Cheng et al. (in press). Because not many AIL mice were phenotyped and genotyped in this study, we combined the F2 and AIL data rather than analyzing the AIL data separately. Prior to combining the two datasets all trait values were adjusted to have a mean of 0 and standard deviation of 1. This allows combination of datasets without using an indicator variable for population (F2 or AIL; Cheng et al. in press). Our approach uses a mixed effect model with a random term accounting for polygenic effects and fixed terms representing QTL effects at both markers and scanning loci between markers. A likelihood ratio test statistic was calculated at more than 5000 loci, which densely and evenly covered the whole genome using the Haley–Knott method (Haley & Knott 1992). Genome-wide significance thresholds were estimated by gene dropping, which is a procedure that simulates genotypes consistent with the pedigree to determine the distribution of the test statistic when the null hypothesis is true. Using the results from the gene dropping procedure, we were able to define a significance threshold that should only be exceeded 5% of the time in the absence of a true association.
Results
Inbred LG/J and SM/J mice
We assessed the effect of sex and strain (LG/J and SM/J) on startle magnitude, PPI and habituation. First, we used a two-way ANOVA to examine the effect of these factors on the startle response. This test showed a significant main effect of strain (F1,44 = 6.10; P < 0.001), sex (F1,44 = 18.72; P < 0.05) and a significant interaction between the two (F1,44 = 8.91; P < 0.01; Fig. 1a). We used a three-way ANOVA to assess the effect of strain, sex and prepulse intensity (3, 6 and 12 dB) on PPI. We observed significant interactions between sex and strain (F1,132 = 4.16; P < 0.05) and between prepulse intensity and strain (F2,132 = 8.75; P < 0.001). Based on these interactions, we chose to examine the factors sex and strain separately for each prepulse intensity. Strain (but not sex) was significant for all prepulse intensities (PPI3: (F1,44 = 6.39; P < 0.05); PPI6: (F1,44 = 21.21; P < 0.001); PPI12: (F1,44 = 71.39; P < 0.001); Fig. 1b. Finally, we used a two-way ANOVA to assess the effects of sex and strain on habituation; this test did not yield any significant results (Fig. 1c).
Figure 1. Sex and strain differences in startle, PPI and habituation.
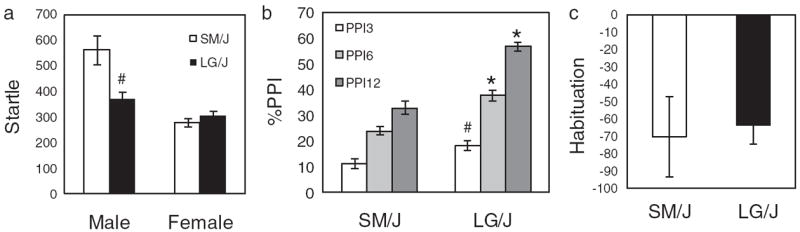
We observed significant differences between the strains for startle and PPI; however, there was no significant difference between the two strains for habituation. There was a significant interaction between strain and sex for startle but not for any of the other traits. Startle and habituation are both expressed in arbitrary units. # and * indicate differences between the two strains with P < 0.05 and P < 0.001, respectively.
LG/J x SM/J F2 analysis
We used genotype and phenotype data from the F2 mice to identify QTLs for startle, PPI and habituation.
Because there was a significant interaction between sex and strain in the inbred mice for startle (Fig. 1a), we used a model in which sex was an interactive covariate. Using this approach, we identified a significant peak on the proximal end of chromosome 17 [Logarithm of odds (LOD) = 8.19; Fig. 2] near marker rs29530504 that accounts for 7.68% of the variance. The 1.5 LOD support interval for this QTL was from 7 to 15 cM. There was also a significant peak on chromosome 7 (LOD = 7.02; Fig. 2) near marker rs31102161. This QTL accounts for 6.62% of the variance and has a 1.5 LOD support interval from 56 to 72 cM (Table 1).
Figure 2. Genome-wide LOD scores for startle show a significant peak on chromosomes 7 and 17 in the F2 generation when sex was treated as an interactive covariate.
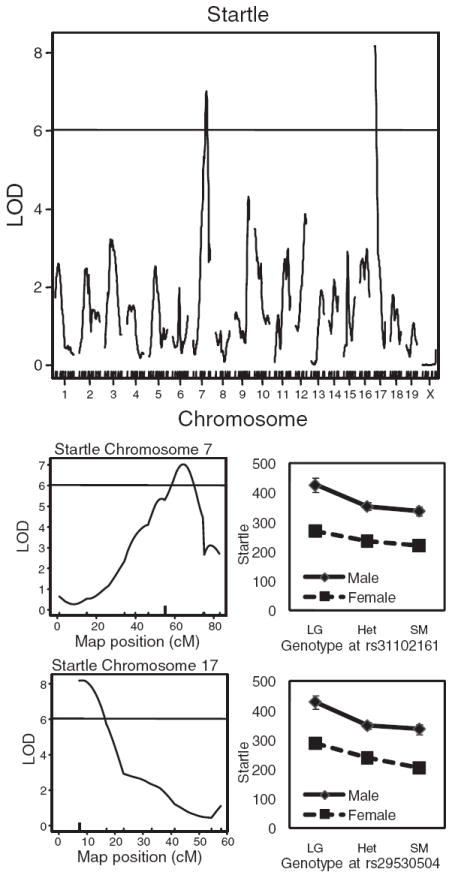
The effect plot of each genotype at the marker nearest to the peak LOD score (indicated with a heavy tick mark) is shown for markers rs31102161 and rs29530504. The horizontal line indicates the genome-wide significance threshold (P < 0.05). Startle response is in arbitrary units.
Table 1.
Peak LOD scores and 1.5 LOD support intervals (SI) for significant QTLs for startle, PPI and habituation in both the F2 and the integrated analysis (F2 + AIL)
| Trait | population | chromosome | Peak (cM) | LOD | 1.5 LOD SI (% reduced) |
|---|---|---|---|---|---|
| Startle | F2 | 7 | 64 | 7.02 | 56–72 |
| F2 + AIL | 7 | 60 | 6.05 | 54–65 (31%) | |
| Startle | F2 | 17 | 8 | 8.19 | 7–15 |
| F2 + AIL | 17 | 13 | 7.25 | 8–14 (25%) | |
| PPI6 | F2 | 11 | 26 | 4.25 | 14–40 |
| PPI12 | F2 | 11 | 22 | 4.70 | 9–35 |
| F2 + AIL | 11 | 40 | 5.31 | 39–42 (88%) | |
| PPI3 | F2 | 12 | 45 | 6.70 | 34–52 |
| F2+AIL | 12 | 46 | 6.78 | 45–50 (72%) | |
| PPI6 | F2 | 12 | 45 | 7.65 | 11–52 |
| F2 + AIL | 12 | 45 | 6.62 | 41–46 (88%) | |
| PPI12 | F2 | 12 | 47 | 6.73 | 13–56 |
| F2 + AIL | 12 | 44 | 7.82 | 42–46 (91%) | |
| Habituation | F2 | 4 | 33 | 4.40 | 15–43 |
| F2 + AIL | 4 | 30 | 4.77 | 29–35 (79%) |
SI range and peak LOD values (Peak) are expressed in centiMorgans.
For PPI3, we identified a significant peak on chromosome 12 (LOD = 6.70) near marker rs29172545 that accounts for 6.33% of the phenotypic variance. For PPI6 we identified significant peaks on chromosomes 11 (LOD = 4.25, near marker rs26967743) and 12 (LOD = 7.65) near the QTL for PPI3. These peaks account for 4.06% and 7.19% of the phenotypic variance, respectively. For PPI12 we identified a significant QTL on chromosome 11 (LOD = 4.70) near the QTL for PPI6 that explained 4.48% of the variance. There was also a significant peak on chromosome 12 (LOD = 6.73) near the peak marker for the other PPI traits, which explained 6.36% of the variance. For all three prepulse intensities we also observed suggestive QTLs on several other chromosomes; these results are shown in Fig. 3 and Table 1. For the QTLs on chromosome 12, the LOD curves for all three prepulse intensities showed one peak at about 45 cM; however, the LOD curves for PPI6 and PPI12 also showed what appeared to be a second peak towards the proximal end of chromosome 12.
Figure 3. LOD scores for all three PPI intensities on chromosomes 11 and 12 based on analysis of the F2 mice.
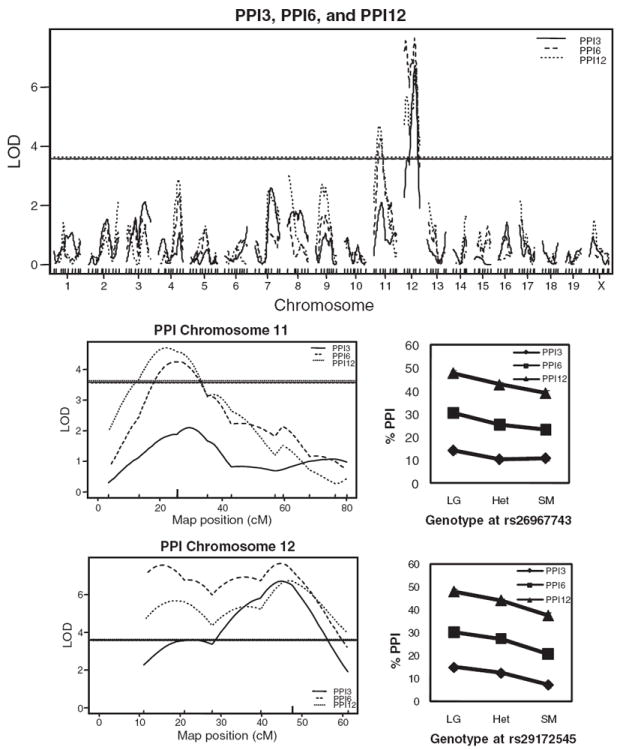
The mean phenotype for mice with the indicated genotypes at the marker closest to the peak LOD score for chromosomes 11 and 12 are shown in the lower right. Horizontal lines indicate the genome-wide significance thresholds (P < 0.05).
The similarities between the results for all three prepulse intensities are not surprising in light of the high correlations between these traits, as shown in Table 2. Because we were not specifically interested in the individual prepulse intensities, we used the principal components function in R (princomp) to identify principal components for these three variables (Table 3). The results were very similar for both the F2 and AIL mice, with the first principal component (PC1) accounting for about 80% of the total variance.
Table 2.
Correlation matrix between startle, PPI3, PPI6 and PPI12
| Startle | PPI3 | PPI6 | PPI12 | |
|---|---|---|---|---|
| Startle | — | — | — | — |
| PPI3 | −0.0633 | — | — | — |
| PPI6 | −0.1400 | 0.6960* | — | — |
| PPI12 | −0.1468 | 0.6059* | 0.7831* | — |
The correlations noted with an asterisk have P values less than 1 × 10−5.
Table 3.
Principal components analysis of PPI3, PPI6 and PPI12
| F2
|
AIL
|
|||||
|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | |
| Proportion of Var | 0.800 | 0.131 | 0.0690 | 0.809 | 0.124 | 0.0672 |
| PPI3 | −0.452 | 0.721 | 0.525 | −0.391 | 0.839 | 0.379 |
| PPI6 | −0.61 | 0.18 | −0.772 | −0.577 | 0.0976 | −0.811 |
| PPI12 | −0.651 | −0.669 | 0.358 | −0.717 | −0.536 | 0.445 |
Our analysis identified three principal components that were similar in the F2 and AIL in terms of the proportion of variance and the loadings of the prepulse variables. The first principal component (PC1) accounted for more then 80% of the total trait variance and was used for QTL analysis (see Fig. S1).
We also examined the PPI data from the F2 mice for two-way epistatic interactions between loci (Broman & Sen 2009). There was evidence of an additive interaction between the QTL on chromosome 11 and the proximal and more distal QTLs on chromosome 12 for PPI6 and PPI12, respectively, but no significant epistatic interactions were found (data not shown). This analysis was not able to determine whether or not the two different LOD peaks on chromosome 12 were truly independent.
For habituation, we identified a significant QTL on chromosome 4 close to marker rs27895401 (LOD score = 4.40; Fig. 4), which accounts for 4.20% of the phenotypic variance. The 1.5 LOD support interval for this QTL was from 15 to 43 cM (Table 1).
Figure 4. LOD scores for habituation show a significant peak on chromosome 4 in the F2 generation.
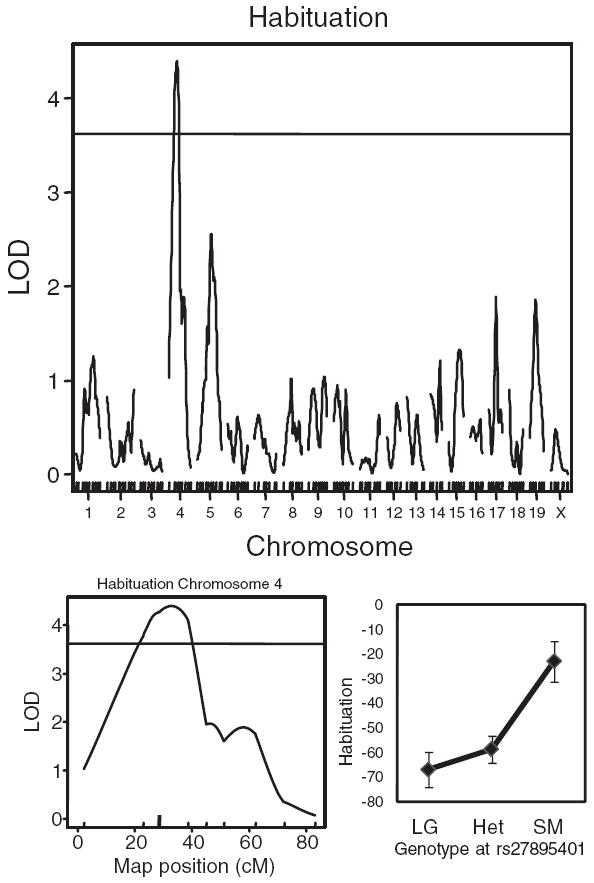
The plot in the lower right shows the mean phenotype associated with each genotype at the marker nearest to the peak LOD score. The horizontal line indicates the genome-wide significance threshold (P < 0.05). Habituation is expressed as the difference between the first six and the last six startle alone trials, in arbitrary units.
LG/J × SM/J AIL analysis
The analysis of data from the F2 mice identified several significant QTLs; however, the 1.5 LOD support intervals of these QTLs were generally quite large, often encompassing tens of centiMorgans. This is a common observation for QTLs obtained using F2 crosses, but presents a serious impediment to the identification of the underlying genes. AILs provide a means of fine mapping QTLs and thus partially address this limitation (Darvasi & Soller 1995). When we incorporated phenotype and genotype data from 135 AIL mice into our analysis, we were able to significantly narrow the QTLs identified using the F2 mice; these results are shown in Fig. 5 and Table 1. Our integrated analysis took advantage of the fact that both the F2 and the AIL mice are derived from the same two inbred strains. This analysis was performed on a genome-wide basis; however, because the AIL mice contributed little power, the significant QTLs closely mirrored those obtained by analyzing the F2 mice, and so only the results from chromosomes implicated in the F2 study are presented in Fig. 5.
Figure 5. Integrated analysis of F2 and AIL for startle, PPI and habituation.
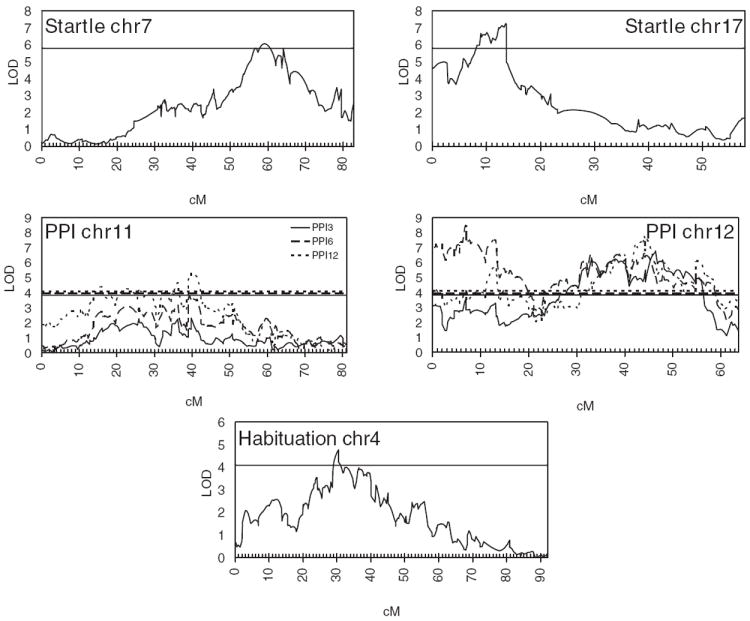
LOD scores are shown on the y-axis and genetic distance in cM is plotted on the x-axis. Horizontal lines indicate the genome-wide significance thresholds (P < 0.05).
For startle, the integrated analysis of the F2 and AIL mice identified a peak on chromosome 7, which was located near SNP rs31816605 and had an LOD score of 6.05; the 1.5 LOD support interval spanned from 54 to 65 cM. We also identified a QTL on chromosome 17 that had an LOD score of 7.25 and was located near rs29515258; this QTL had a 1.5 LOD support interval from 8 to 14 cM.
The integrated analysis of PPI significantly narrowed the width of several QTLs. For PPI3, we identified a significant peak on chromosome 12 (LOD = 6.78) near marker rs29219198 that had a 1.5 LOD support interval from 45 to 50 cM. Whereas the F2 analysis identified a significant QTL for PPI6 on chromosome 11, the integrated analysis fell just short of significance when examining PPI6 on chromosome 11. We identified two significant QTLs for PPI6 on chromosome 12; the more proximal one had an LOD of 8.48 (near marker rs29132070) and had a 1.5 LOD support interval from 6 to 10 cM. In addition, we also identified a more distal QTL for PPI6 on chromosome 12 with an LOD of 6.62 and a 1.5 LOD support interval of 41–46 cM. We identified a significant peak on chromosome 11 for PPI12 with an LOD of 5.31 that was near marker rs26922491 and had a 1.5 LOD support interval of 39–42 cM. A QTL for PPI12 on chromosome 12 with an LOD of 7.82 that was near rs6349472 had a 1.5 LOD interval that spanned from 42 to 46 cM. Finally, we performed QTL mapping of PC1 using data from both the F2 and AIL mice (Fig. S1); the results were very similar to those obtained by analyzing the individual PPI variables.
The integrated analysis of F2 and AIL mice identified a significant QTL for habituation of the startle response on chromosome 4 with an LOD of 4.77 that was near marker rs32621408 and had a 1.5 LOD support interval that spanned from 29 to 35 cM.
Discussion
In the present study we report a highly significant strain difference in PPI among the inbred LG/J and SM/J mice. This is the first study to our knowledge that has conducted a QTL screen for PPI with LG/J and SM/J mice. We used 472 mice from the F2 generation of a cross between LG/J and SM/J and 135 LG/J and SM/J AIL mice to identify significant QTLs for startle, PPI and habituation. We identified QTLs for startle on chromosomes 7 and 17, QTLs for PPI on both chromosomes 11 and 12 and QTLs for habituation of the startle response on chromosome 4.
We found significant QTLs on chromosomes 7 and 17 for the acoustic startle response when sex was treated as an interactive covariate (Fig. 2). We considered the possibility that the interaction with sex reflected a correlation between body weight and sex; however, this explanation is unlikely to be correct because we observed only very minor (r < 0.05) positive correlations between body weight and startle within each sex and because the female SM/J and LG/J have very different weights but similar startle responses (Fig. 1a). Several other studies have found QTLs for startle (Fernandez-Teruel et al. 2002; Joober et al. 2002; Palmer et al. 2003), but only Watanabe et al. (2007) and Leussis et al. (2009) identified QTLs for startle on mouse chromosome 7. Watanabe et al. (2007) had a peak around 65.9 megabases (Mb), which is likely to be different from our QTL that was located between 103 and 115 Mb. The study by Leussis et al. (2009) found that CSS-7 had an elevated startle response and CSS-17 had a lower startle response in comparison to B6 mice; however, their study did not attempt to further localize these peaks.
The QTLs for PPI6 and PPI12 on chromosome 11 (Figs. 3 and 5) appear to reflect the same locus despite the fact that the QTL for PPI6 fell just short of obtaining significance in the integrated analysis. The SM/J allele was associated with lower PPI (Fig. 3c), consistent with the lower PPI observed in inbred SM/J mice (Fig. 1). The integrated analysis narrowed the QTL for PPI12 from 26 to 3 cM (88% reduction; Table 1), showing the utility of this approach. Previous QTL studies using different mouse strains have identified a QTL on chromosome 11 for PPI (Hitzemann et al. 2008; Joober et al. 2002; Leussis et al. 2009; Watanabe et al. 2007); however, most of those QTLs were located more distally than ours. The QTL we identified on chromosome 11 may be caused by the same alleles that cause QTLs previously reported by Hitzemann et al. (2008) and Watanabe et al. (2007). The study by Leussis et al. (2009) used a panel of C57BL/6J (B6) × A/J chromosome substitution strains (CSS). The other three studies used F2 crosses between B6 and A/J (Joober et al. 2002), B6 and C3H/He (Watanabe et al. 2007) and B6 and DBA/2J (Hitzemann et al. 2008). The A/J allele on chromosome 11 was associated with elevated levels of PPI compared with B6 (Joober et al. 2002; Leussis et al. 2009), whereas Hitzemann et al. (2008) found that the B6 was associated with elevated PPI over DBA/2J. Taken together, these data suggest that the QTL we observed on chromosome 11 may or may not be because of the same alleles as previously reported QTLs for PPI on chromosome 11.
The QTLs on chromosome 12 suggest the presence of more than one QTL for PPI (Figs. 3 and 5). All three prepulse intensities showed distal peaks that reflected lower PPI among mice carrying the SM/J allele. In addition, a second more proximal QTL was observed for PPI6 and PPI12. While the F2 data were unable to statistically dissociate the distal and proximal peaks, the results from the integrated analysis provided strong support for the existence of two separate QTLs for PPI on chromosome 12. The integrated analysis significantly narrowed the location of the QTLs on chromosome 12, as shown in Table 1; all analyses showed remarkable consistency in terms of the peak of the distal QTL, which was estimated from 44 to 47 cM. The analysis of the first principal component obtained from all three prepulse intensities was also consistent with this pattern (Fig. S1). We are not aware of any previous studies reporting QTLs for PPI on mouse chromosome 12 (Hitzemann et al. 2008; Joober et al. 2002; Leussis et al. 2009; Palmer et al. 2003; Petryshen et al. 2005; Watanabe et al. 2007), suggesting that this QTL is a novel finding.
We identified a QTL for habituation on chromosome 4. The SM/J allele at QTL was associated with less habituation and showed an apparently recessive pattern of inheritance (Fig. 4). A recent study by Leussis et al. (2009) found QTLs for habituation on chromosomes 7 and 8, but not chromosome 4. Their measure of habituation was percent habituation and they used different inbred strains. These differences, along with incomplete power for smaller effect QTLs, likely explain the differences in the QTLs identified. Interestingly, we did not identify any QTLs influencing both PPI and habituation, suggesting that these two putative endophenotypes are genetically dissociable.
This study has several limitations. One limitation common to virtually all conventional QTL studies is that power to detect smaller effect QTLs is limited. For example, in this study we observed suggestive but not significant QTLs for PPI on chromosomes 7, 8 and 9 (Fig. 3); a larger sample size might have identified additional QTLs that have smaller impacts on the phenotype. Another common limitation of conventional QTL studies is the inability to narrow the confidence intervals of QTL to a manageable size. We were able to significantly improve our mapping resolution by using AIL mice, which is a notable strength of this study; however, we were still unable to identify specific genes. Another limitation is that we used three different prepulse intensities, but we did not vary the time between the prepulse and the pulse stimuli; evidence from rats indicates that different strains may have different optimal times between these two stimuli (Swerdlow et al. 2004). Therefore, the QTLs that we identified may reflect differences in the optimal times between prepulse and pulse stimuli.
In summary, we identified several QTLs for startle response, PPI and habituation of the startle response and used AIL mice to significantly narrow the 1.5-LOD support intervals for these QTLs. These results show the potential of using even a modest number of AIL mice in conjunction with conventional F2 studies and will facilitate the identification of the underlying genes. Elucidation of genes that influence PPI may stimulate a deeper understanding of the molecular mechanisms that underlie PPI and may thus enhance our understanding of neurological and psychiatric disorders that are characterized by deficits in sensorimotor gating.
Supplementary Material
Integrated analysis of F2 and AIL for the first principal component (PC1) derived from the three prepulse variables (PPI3, PPI6 and PPI12).
Results are generally quite similar to those obtained from the individual Prepulse variables; LOD scores are shown on the y-axis and centiMorgans are plotted on the x-axis. The horizontal line indicates the genome-wide significance threshold (P < 0.05).
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Acknowledgments
The authors would like to thank Dr. James Cheverud for kindly providing us with F33 LG/J × SM/J AIL mice and accompanying pedigree information about these mice, Drs. Mark Abney and Andrew Skol for help with the development of the software used in this analysis, Mr. Hsien Chang and Mr. Andrew Chen for their help with PPI testing and Dr. Ney Alliey for his organization of the phenotype data. This work was supported by T32MH020065 (JEL), R21DA024845 (AAP) and R01DA021336 (AAP).
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
References
- Anokhin AP, Heath AC, Myers E, Ralano A, Wood S. Genetic influences on prepulse inhibition of startle reflex in humans. Neurosci Lett. 2003;1:45–48. doi: 10.1016/j.neulet.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Bolbecker AR, Mehta CS, Edwards CR, Steinmetz JE, O’Donnell BF, Hetrick WP. Eye-blink conditioning deficits indicate temporal processing abnormalities in schizophrenia. Schizophr Res. 2009;1–3:182–191. doi: 10.1016/j.schres.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL, Grillon C, Geyer MA. Gating and habituation of the startle reflex in schizophrenic patients. Arch Gen Psychiatry. 1992;3:206–215. doi: 10.1001/archpsyc.1992.01820030038005. [DOI] [PubMed] [Google Scholar]
- Braff DL, Greenwood TA, Swerdlow NR, Light GA, Schork NJ The Investigators of the Consortium on the Genetics of Schizophrenia. Advances in endophenotyping schizophrenia. World Psychiatry. 2008;1:11–18. doi: 10.1002/j.2051-5545.2008.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. Springer; New York: 2009. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;7:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Cadenhead KS, Swerdlow NR, Shafer KM, Diaz M, Braff DL. Modulation of the startle response and startle laterality in relatives of schizophrenic patients and in subjects with schizotypal personality disorder: evidence of inhibitory deficits. Am J Psychiatry. 2000;10:1660–1668. doi: 10.1176/appi.ajp.157.10.1660. [DOI] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. [12 July 2010];Genetics. doi: 10.1534/genetics.110.116863. in press http://www.genetics.org/cgi/rapidpdf/genetics.110.116863v1. [DOI] [PMC free article] [PubMed]
- Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;3:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich TH, Kenney-Hunt JP, Pletscher LS, Cheverud JM. Genetic variation and correlation of dietary response in an advanced intercross mouse line produced from two divergent growth lines. Genet Res. 2005;3:211–222. doi: 10.1017/S0016672305007603. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teruel A, Escorihuela RM, Gray JA, Aguilar R, Gil L, Gimenez-Llort L, Tobena A, Bhomra A, Nicod A, Mott R, Driscoll P, Dawson GR, Flint J. A quantitative trait locus influencing anxiety in the laboratory rat. Genome Res. 2002;4:618–626. doi: 10.1101/gr.203402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer MA, Krebs-Thomson K, Braff DL, Swerdlow NR. Pharmacological studies of prepulse inhibition models of sensorimotor gating deficits in schizophrenia: a decade in review. Psychopharmacology (Berl) 2001;2–3:117–154. doi: 10.1007/s002130100811. [DOI] [PubMed] [Google Scholar]
- Giakoumaki SG, Roussos P, Rogdaki M, Karli C, Bitsios P, Frangou S. Evidence of disrupted prepulse inhibition in unaffected siblings of bipolar disorder patients. Biol Psychiatry. 2007;12:1418–1422. doi: 10.1016/j.biopsych.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Braff DL, Light GA, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the consortium on the genetics of schizophrenia. Arch Gen Psychiatry. 2007;11:1242–1250. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;4:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Bell J, Rasmussen E, McCaughran J. Mapping the genes for the acoustic startle response (ASR) and prepulse inhibition of the ASR in the BXD recombinant inbred series: effect of high-frequency hearing loss and cochlear pathology. In: Willott F, editor. Handbook of Mouse Auditory Research: From Behavior to Molecular Biology. CRC Press; Boca Raton: 2001. pp. 441–455. [Google Scholar]
- Hitzemann R, Malmanger B, Belknap J, Darakjian P, McWeeney S. Short-term selective breeding for high and low prepulse inhibition of the acoustic startle response; pharmacological characterization and QTL mapping in the selected lines. Pharmacol Biochem Behav. 2008;4:525–533. doi: 10.1016/j.pbb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenig K, Hochrein A, Quednow BB, Maier W, Wagner M. Impaired prepulse inhibition of acoustic startle in obsessive-compulsive disorder. Biol Psychiatry. 2005;10:1153–1158. doi: 10.1016/j.biopsych.2005.01.040. [DOI] [PubMed] [Google Scholar]
- Joober R, Zarate JM, Rouleau GA, Skamene E, Boksa P. Provisional mapping of quantitative trait loci modulating the acoustic startle response and prepulse inhibition of acoustic startle. Neuropsychopharmacology. 2002;5:765–781. doi: 10.1016/S0893-133X(02)00333-0. [DOI] [PubMed] [Google Scholar]
- Leussis MP, Frayne ML, Saito M, Berry EM, Aldinger KA, Rockwell GN, Hammer RP, Jr, Baskin-Hill AE, Singer JB, Nadeau JH, Sklar P, Petryshen TL. Genomic survey of prepulse inhibition in mouse chromosome substitution strains. Genes Brain Behav. 2009;8:806–816. doi: 10.1111/j.1601-183X.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Singh RP, Khan AH, Bhavsar K, Lusis AJ, Davis RC, Smith DJ. Identifying loci for behavioral traits using genome-tagged mice. J Neurosci Res. 2003;4:562–569. doi: 10.1002/jnr.10765. [DOI] [PubMed] [Google Scholar]
- McGhie A, Chapman J. Disorders of attention and perception in early schizophrenia. Br J Med Psychol. 1961;34:103–116. doi: 10.1111/j.2044-8341.1961.tb00936.x. [DOI] [PubMed] [Google Scholar]
- Moriwaki M, Kishi T, Takahashi H, Hashimoto R, Kawashima K, Okochi T, Kitajima T, Furukawa O, Fujita K, Takeda M, Iwata N. Prepulse inhibition of the startle response with chronic schizophrenia: a replication study. Neurosci Res. 2009;3:259–262. doi: 10.1016/j.neures.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Dulawa SC, Mottiwala AA, Conti LH, Geyer MA, Printz MP. Prepulse startle deficit in the Brown Norway rat: a potential genetic model. Behav Neurosci. 2000;2:374–388. doi: 10.1037//0735-7044.114.2.374. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Breen LL, Flodman P, Conti LH, Spence MA, Printz MP. Identification of quantitative trait loci for prepulse inhibition in rats. Psychopharmacology (Berl) 2003;3:270–279. doi: 10.1007/s00213-002-1258-0. [DOI] [PubMed] [Google Scholar]
- Perry W, Minassian A, Feifel D, Braff DL. Sensorimotor gating deficits in bipolar disorder patients with acute psychotic mania. Biol Psychiatry. 2001;6:418–424. doi: 10.1016/s0006-3223(01)01184-2. [DOI] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;9:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryshen TL, Kirby A, Hammer RP, Jr, Purcell S, O’Leary SB, Singer JB, Hill AE, Nadeau JH, Daly MJ, Sklar P. Two quantitative trait loci for prepulse inhibition of startle identified on mouse chromosome 16 using chromosome substitution strains. Genetics. 2005;4:1895–1904. doi: 10.1534/genetics.105.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan NA, Holick Pierz KA, Masten VL, Waeber C, Ansorge M, Gingrich JA, Geyer MA, Hen R, Dulawa SC. Chronic reductions in serotonin transporter function prevent 5-HT1B-induced behavioral effects in mice. Biol Psychiatry. 2009;5:401–408. doi: 10.1016/j.biopsych.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Benbow CH, Zisook S, Geyer MA, Braff DL. A preliminary assessment of sensorimotor gating in patients with obsessive compulsive disorder. Biol Psychiatry. 1993;4:298–301. doi: 10.1016/0006-3223(93)90300-3. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Paulsen J, Braff DL, Butters N, Geyer MA, Swenson MR. Impaired prepulse inhibition of acoustic and tactile startle response in patients with Huntington’s disease. J Neurol NeurosurgPsychiatry. 1995;2:192–200. doi: 10.1136/jnnp.58.2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Shoemaker JM, Auerbach PP, Pitcher L, Goins J, Platten A. Heritable differences in the dopaminergic regulation of sensorimotor gating. II. Temporal, pharmacologic and generational analyses of apomorphine effects on prepulse inhibition. Psychopharmacology (Berl) 2004;4:452–462. doi: 10.1007/s00213-003-1480-4. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Toyota T, Owada Y, Hayashi T, Iwayama Y, Matsumata M, Ishitsuka Y, Nakaya A, Maekawa M, Ohnishi T, Arai R, Sakurai K, Yamada K, Kondo H, Hashimoto K, Osumi N, Yoshikawa T. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 2007;11:e297. doi: 10.1371/journal.pbio.0050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willott JF, Tanner L, O’Steen J, Johnson KR, Bogue MA, Gagnon L. Acoustic startle and prepulse inhibition in 40 inbred strains of mice. Behav Neurosci. 2003;4:716–727. doi: 10.1037/0735-7044.117.4.716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Integrated analysis of F2 and AIL for the first principal component (PC1) derived from the three prepulse variables (PPI3, PPI6 and PPI12).
Results are generally quite similar to those obtained from the individual Prepulse variables; LOD scores are shown on the y-axis and centiMorgans are plotted on the x-axis. The horizontal line indicates the genome-wide significance threshold (P < 0.05).
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.


