Abstract
Background
Penicillium marneffei is the most important thermal dimorphic fungus causing systemic mycosis in China and Southeast Asia. While miRNAs are increasingly recognized for their roles in post-transcriptional regulation of gene expression in animals and plants, miRNAs in fungi were less well studied and their potential roles in fungal dimorphism were largely unknown. Based on P. marneffei genome sequence, we hypothesize that miRNA-like RNAs (milRNAs) may be expressed in the dimorphic fungus.
Methodology/Principal Findings
We attempted to identify milRNAs in P. marneffei in both mycelial and yeast phase using high-throughput sequencing technology. Small RNAs were more abundantly expressed in mycelial than yeast phase. Sequence analysis revealed 24 potential milRNA candidates, including 17 candidates in mycelial and seven in yeast phase. Two genes, dcl-1 and dcl-2, encoding putative Dicer-like proteins and the gene, qde-2, encoding Argonaute-like protein, were identified in P. marneffei. Phylogenetic analysis showed that dcl-2 of P. marneffei was more closely related to the homologues in other thermal dimorphic pathogenic fungi than to Penicillium chrysogenum and Aspergillus spp., suggesting the co-evolution of dcl-2 among the thermal dimorphic fungi. Moreover, dcl-2 demonstrated higher mRNA expression levels in mycelial than yeast phase by 7 folds (P<0.001). Northern blot analysis confirmed the expression of two milRNAs, PM-milR-M1 and PM-milR-M2, only in mycelial phase. Using dcl-1KO, dcl-2KO, dclDKO and qde-2KO deletion mutants, we showed that the biogenesis of both milRNAs were dependent on dcl-2 but not dcl-1 or qde-2. The mRNA expression levels of three predicted targets of PM-milR-M1 were upregulated in knockdown strain PM-milR-M1 KD, supporting regulatory function of milRNAs.
Conclusions/Significance
Our findings provided the first evidence for differential expression of milRNAs in different growth phases of thermal dimorphic fungi and shed light on the evolution of fungal proteins involved in milRNA biogenesis and possible role of post-transcriptional control in governing thermal dimorphism.
Author Summary
Penicillium marneffei is the most important thermal dimorphic pathogenic fungus in Southeast Asia. Despite findings on diverse genes and mechanisms involved in dimorphic switching, the key to signally pathways governing the switch is still unknown. Since miRNAs are important regulatory molecules in eukaryotes, we attempt to define if miRNAs are expressed in different growth phases of P. marneffei. Using high-throughput sequencing, we identified 24 potential milRNA candidates in P. marneffei, which were more abundantly expressed in mycelial than yeast phase. Two genes, dcl-1 and dcl-2, encoding Dicer-like proteins and the gene, qde-2, encoding Argonaute-like protein, were also identified. Phylogenetic analysis showed that dcl-2 of P. marneffei was more closely related to the homologues in other thermal dimorphic pathogenic fungi than to Penicillium chrysogenum and Aspergillus spp.. dcl-2 demonstrated higher mRNA levels in mycelial than yeast phase. Northern blot analysis confirmed expression of two milRNAs, PM-milR-M1 and PM-milR-M2, only in mycelial phase, whose expression was dependent on dcl-2 but not dcl-1 or qde-2. The mRNA levels of three predicted targets of PM-milR-M1 were upregulated in knockdown strain PM-milR-M1 KD, supporting its regulatory function. This study represents the first discovery of milRNAs in thermal dimorphic fungi, with differential expression in different growth phases.
Introduction
Penicillium marneffei is the most important thermal dimorphic fungus causing respiratory, skin and systemic mycosis in Southeast Asia [1]–[4]. Recently, it has been renamed as Talaromyces based on phylogenetic analyses [5]. The fungus was first discovered in Chinese bamboo rats, Rhizomys sinensis, and subsequently isolated from other species of bamboo rats [6], [7]. While only 18 cases of human diseases were reported until 1985 [8], the emergence of the HIV pandemic in the 1980's has resulted in increasing reports of HIV-associated P. marneffei infections in Southeast Asia where the fungus is endemic. Penicilliosis is the third most common indicator disease of AIDS In northern Thailand [2]. In Hong Kong, about 10% of HIV patients are infected with P. marneffei, which represents the sixth leading cause of death [9], [10]. Cases of imported P. marneffei infections have also been reported from countries outside Southeast Asia [11], [12]. In addition, P. marneffei infections are increasingly reported in other immunocompromised patients, such as transplant recipients and others on immunosuppressant therapy [13]–[16]. Despite its medical importance, the mode of transmission, and dimorphic and pathogenic mechanisms of P. marneffei remain largely unknown. In particular, P. marneffei exhibits distinct cellular morphologies in different temperatures, in mycelial phase at 25°C and yeast phase at 37°C. During the mycelial phase, hyphae can differentiate to produce conidia which are believed to be the infectious form being inhaled to the lungs of infected hosts. When these conidia are phagocytosed by alveolar macrophages, they germinate into yeast cells as the tissue form. Despite the efforts of using various gene knockout experiments in identifying diverse genes and complex mechanisms involved in dimorphic switching in P. marneffei, the signals that trigger the switch in response to temperature and signaling pathways leading to the transition remain elusive [17].
MicroRNAs or miRNAs are small non-coding endogenous RNAs of approximately 22 nt, which play important roles in post-transcriptional regulation of gene expression in animals and plants [18]. They are now known to comprise one of the most abundant classes of gene regulatory molecules in multicellular organisms. The mature miRNAs negatively regulate gene expression by targeting mRNAs mediated through complementary binding to the open-reading frame or untranslated (UTR) regions of specific target genes. Interactions with targets can be through imprecise base pairing leading to translational inhibition in animals, or near-perfect complementarity leading to mRNA cleavage in plants [19], [20]. In animals, miRNAs have been shown to play various roles in ranging from cell development, proliferation and differentiation, apoptosis, carcinogenesis to immunity [21]–[23]. In plants, they are also involved in plant development, stress response and antibacterial resistance [24], [25], [26].
The first known miRNA lin-4 was discovered in Caenorhabditis elegans in 1993 [27]. However, it was only until 2000 that the second miRNA, let-7, also in C. elegans, was identified [28]. With the advent of molecular and bioinformatics tools, numerous miRNAs have now been identified in animals, plants, viruses and unicellular organisms, with >25,000 miRNAs being currently included in the miRNA database, miRBase release 19.0 [29]. Although small RNA pathways have been found in various fungi, the existence of miRNAs and their roles in fungi has been less well understood. Recently, miRNA-like small RNAs (milRNAs) have been identified in the red bread mold, Neurospora crassa, the plant pathogenic fungus, Sclerotinia sclerotiorum, the entomopathogenic fungus, Metarhizium anisopliae and the human pathogenic yeast, Cryptococcus neoformans [30]–[33]. However, their existence in thermal dimorphic fungi and potential roles in fungal dimorphism were largely unknown.
In 2002, we started the P. marneffei genome project in an attempt to expedite the study of biology, epidemiology and virulence factors of this dimorphic fungus [34]–[41]. Based on the available genome sequence data, potential genes encoding proteins important for miRNA biogenesis can be identified in P. marneffei. Since miRNAs are important gene regulatory molecules in multicellular organisms, we hypothesize that milRNAs may be expressed in P. marneffei and involved in the regulation of thermal dimorphism. We attempted to identify milRNAs in P. marneffei in both mycelial and yeast phase using high-throughput Illumina DNA sequencing. Sequence analysis revealed 24 potential milRNA candidates, which were more abundantly expressed in mycelial than yeast phase of P. marneffei. Two genes, dcl-1 and dcl-2, encoding putative Dicer-like proteins and the gene, qde-2, encoding quelling-deficient-2, an Argonaute-like protein, were also identified. Northern blot analysis confirmed the differential expression of two milRNAs, PM-milR-M1 and PM-milR-M2, in mycelial phase, which was dependent on dcl-2 but not dcl-1 or qde-2. Our findings provided evidence for the existence of milRNAs in thermal dimorphic fungi and differential expression of milRNAs during different growth phases, which may provide new insights into the mechanism governing thermal dimophism.
Materials and Methods
Ethics statement
P. marneffei strain PM1 was obtained from an already-existing collection from the clinical microbiology laboratory in Queen Mary Hospital and the strain was anonymized.
P. marneffei strains and growth conditions
P. marneffei strain PM1 was isolated from a patient with culture-documented penicilliosis in Hong Kong. Knock-out mutant strains including dcl-1KO, dcl-2KO, dcl-1 dcl-2 double mutant (dclDKO) and qde-2KO were generated as described below. All P. marneffei strains were grown on Sabouraud dextrose agar (SDA) (Oxoid, Cambridge, UK) at 25°C for 7 days for the collection of conidia as described previously [40]. Conidia were collected by scraping and resuspension in 0.1% Tween-20 with PBS followed by three washes in sterile PBS before subculturing into liquid cultures in BHI medium (Difco, NJ, USA) in a shaker at 37°C for yeasts or at 25°C for mycelia for 48 hours. Cells were enumerated using a hemocytometer.
Small RNA purification, library preparation and sequencing
Small RNA libraries were constructed for mycelial and yeast phases of P. marneffei. Total RNAs were extracted from cultures grown at the respective phase using Plant Isolation Aid (Ambion, Austin, TX, USA) and mirVana miRNA Isolation Kit (Ambion) after mechanical disruption with acid-washed glass beads (Sigma-Aldrich, Missouri, USA), and treated with DNA-free Kit (Ambion) to remove residual DNA. 10 µg of total RNAs from both mycelial and yeast phases was treated with RiboMinus Eukaryote Kit for RNA-Seq (Invitrogen, Carlsbad, CA) to remove ribosomal RNAs (rRNAs). The rRNA-depleted RNA was concentrated by ethanol precipitation in the presence of glycogen carrier (Ambion). RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). A strand-specific library construction protocol was used to generate template for Illumina DNA sequencing [42]. An adenylated 3′- adaptor (Integrated DNA Technologies, Coralville, IA) was first ligated to the 3′ ends of a small RNA (≤60 nt) fraction was extracted from 15% denaturing polyacrylamide gel. The 3′ adaptor-ligated small RNAs were then ligated with a 5′-adaptor (Integrated DNA Technologies). Adaptor-ligated small RNAs were reverse transcribed into first-strand cDNA using a primer hybridizing to the 3′-adaptor using SuperScript II reverse transcriptase (Invitrogen). First strand cDNA was amplified by polymerase chain reaction (PCR) from Illumina/Solexa PCR primer binding sites present on the 5′- and 3′-adaptors to generate templates for sequencing on the Illumina Genome Analyzer IIx (Illumina, San Diego, CA).
Small RNA analyses
Sequence reads were processed to remove low quality reads, adaptor and adaptor-dimer sequences, and nuclear and mitrochondrial rRNA sequences to yield 16,479,305 filtered reads for mycelial and 12,754,677 filtered reads for yeast phase respectively. Relative expression levels were estimated by normalizing read counts for each non-redundant small RNA species against RPM (number of reads per million mapped reads) as mapped to the draft P. marneffei PM1 genome sequence [39]. Small RNA sequences between 17–30 nt were selected to identify perfect matches to the genome using Bowtie (0.12.8) [43].
Identification of milRNAs and milRNA loci
To identify milRNA candidates, other non-coding RNAs including rRNAs and tRNAs were first excluded. Potential milRNA candidates were predicted with miRDeep [44] based on draft P. marneffei PM1 genome. Analysis was performed with the following adjustments: (1) Filtering ubiquitous alignments, keeping only reads that were perfectly mapped to no more than 5 different regions in the genome; (2) Potential precursor sequences were excised from the genome with the size of 250 nt flanking to the sequencing reads; and (3) Hybridization temperatures of 25°C and 37°C were used in the script regarding RNAfold for deep sequencing data from mycelial and yeast form of P. marneffei respectively. milRNA candidates were identified with the following criteria: small RNAs that formed a stem-loop structure (hairpin) with flanking sequences (up to 250 nt), as examined by RNAfold in miRDeep package.
Identification and sequencing analysis of dcl-1, dcl-2 and qde-2 genes in P. marneffei
Based on the predicted protein sequences of corresponding genes from N. crassa, putative dcl-1, dcl-2 and qde-2 genes in the P. marneffei strain PM1 draft genome sequence (GenBank accession no. AGCC00000000) were searched using BLASTP algorithm. Introns were predicted by performing pairwise alignment with the annotated Talaromyces stipitatus (teleomorph of Penicillium emmonsii) (GenBank accession no. ABAS00000000) and P. marneffei strain ATCC 18224 (GenBank accession no. ABAR00000000) genome sequences. The complete coding sequences of dcl-1, dcl-2 and qde-2 of P. marneffei were PCR amplified from cDNA using primers derived from P. marneffei genome sequence as described previously (Table 1) [36]. To perform phylogenetic analysis, putative dcl-1, dcl-2 and qde-2 homologues from representative fungal species were retrieved using BLASTP against the GenBank database. Nucleotide sequences of the internal transcribed spacer (ITS) regions were obtained from GenBank. Phylogenetic trees were constructed using the maximum-likelihood method with 1000 bootstrap replicates with Mega 5.1 [45]. WAG+F+G (for dcl-1 and dcl-2) and rtREV+G (for qde-2) amino acid substitution models, K2+G nucleotides substitution models (for ITS) with 5 gamma categories were used. Nine-hundred and fourteen, 764 and 525 amino acid positions of dcl-1, dcl-2 and qde-2 respectively, and 465 nucleotides positions of ITS, were used for analysis. Domains were predicted using the Conserved Domains Database of NCBI and PFAM (http://pfam.sanger.ac.uk/search?tab=searchSequenceBlock) and manual inspection of multiple alignments with homologous sequences.
Table 1. Primers used in this study.
| Gene Targets | Primers | Purpose |
| Upstream of dcl-1 | LPW10929 5′-GAAGATCTCCGTAGTGCTTCTGATTGGTCTGAG-3′ | pAN7-1 cloning |
| LPW10930 5′-GAAGATCTTCTTTTGCGGCCTTTGTAAGTCTG-3′ | (BglII and HindIII) | |
| Downstream of dcl-1 | LPW10931 5′-TGATTGAAGATCCTCCCAAGGTTG-3′ | |
| LPW10932 5′-CCCAAGCTTGGGTGGTTCGTGAGATAGGTGGTGGATA -3′ | ||
| Upstream of dcl-2 | LPW13339 5′-GAAGATCTCGCCGAACAAACGGAAGAAGGAGA-3′ | pAN7-1 cloning |
| LPW13340 5′-TGGCTTCTCCGAAGCTCTCTATGG-3′ | (BglII and SfoI) | |
| Downstream of dcl-2 | LPW13341 5′-ATAGGCGCCCCTAGTCGATTTTCATGAACGGACC-3′ | |
| LPW13342 5′-ATAGGCGCCGGATTACATAACATACCGTCGGCTG-3′ | ||
| Upstream of qde-2 | LPW12475 5′-ACCCAATAAGGATGAGGAAGTTCGG-3′ | pAN7-1 cloning |
| LPW12476 5′-GAAGATCT AAGTCAGTCGCAATCTCGTCCCG-3′ | (BglII and SbfI) | |
| Downstream of qde-2 | LPW12718 5′-GACCTGCAGGACACATCACCAAGTGAAGTGTCAC-3′ | |
| LPW12719 5′-GACCTGCAGGATCCGCTTGACTCCAGGTGGTA -3′ | ||
| Upstream of dcl-1 | LPW10929 5′-GAAGATCTCCGTAGTGCTTCTGATTGGTCTGAG-3′ | pAN8-1 cloning |
| LPW10930 5′-GAAGATCTTCTTTTGCGGCCTTTGTAAGTCTG-3′ | (BglII and SfoI) | |
| Downstream of dcl-1 | LPW12799 5′-ATAGGCGCCTGATTGAAGATCCTCCCAAGGTTG-3′ | |
| LPW12800 5′-ATAGGCGCCTGGTTCGTGAGATAGGTGGTGGATA -3′ | ||
| dcl-1 | LPW13343 5′-TTTACGGGACGTAAATGGCGGCCTA-3′ | qPCR |
| LPW13344 5′-AATTCTAGGCGCTGGTAAGTCGGC-3′ | ||
| LPW21945 5′-ATGTTGGAGACTCTTACCCTGG-3′ | cDNA amplification | |
| LPW22072 5′-CTCGGCATTCCATAGTTTGT-3′ | & sequencing | |
| LPW22073 5′-GCCGAGGTTTCATGGAAAGA-3′ | ||
| LPW22074 5′-CGGTTCAGCTGGAGAAAACA-3′ | ||
| LPW22075 5′-CGTCAAACCTTCATTCTGGA-3′ | ||
| LPW22076 5′-CGGTGTTGAATAAATTCCTG-3′ | ||
| LPW22077 5′-ATATCTTTATTCGTTGGAAGTCCG-3′ | ||
| LPW21946 5′-TACATCACGGGACTCGGGGA-3′ | ||
| LPW21943 5′-ATGGCCATAGAGAGCTTCGG-3′ | ||
| LPW22066 5′-TCGGCCAAAACGTCCCTTTG-3′ | ||
| LPW22067 5′-CTCATCTCTTGAGCGTGCAC-3′ | ||
| dcl-2 | LPW13347 5′-GTGTGAAGTGATATTGCCAAAGGG-3′ | qPCR |
| LPW13348 5′-CATTTTGTAACGGTTCAGCTGGAG-3′ | ||
| LPW22068 5′-CGATGATGAATGGTCGTGAA-3′ | cDNA amplification | |
| LPW22069 5′-GATGCAAAATCTTGGAATGG-3′ | & sequencing | |
| LPW22070 5′-GCATCAGGTGCATTTCTTGG-3′ | ||
| LPW22071 5′-CGATTCCTTGCTAGACCACTTCG-3′ | ||
| LPW21944 5′-CACTGCCGGCTTGACTGTCC-3′ | ||
| LPW21947 5′-ATGTCCAGCGGGTATAGACG-3′ | ||
| LPW22078 5′-TCTAGCCGAGCCTTGCCCTT-3′ | ||
| LPW22079 5′-CTAGTACAGCCTCGGTAGACA-3′ | ||
| LPW22080 5′-ACCTTCAGAGACTCCATCGC-3′ | ||
| qde-2 | LPW14804 5′-GCCTCATCAAAATCCCCGGT-3′ | qPCR |
| LPW14805 5′-GGAGAAACGACGACACCCAT-3′ | ||
| LPW22081 5′-CCGCCCTTCCTGAGAACATT-3′ | cDNA amplification | |
| LPW22082 5′-TTAGATATAGAACATCGTGT-3′ | & sequencing | |
| Actin | LPW20631 5′-GAACGTGAAATCGTCCGT-3′ | qPCR |
| LPW20160 5′-AGCAAGAATGGAACCACC-3′ | ||
| PM-milR-M1 gene | LPW20742 5′-CCGCTCGAGGACGCACAAAACAATGCAAA-3′ | pSilent-1 cloning |
| locus | LPW20743 5′-CCCAAGCTTGGGGTATTGCCGTTGATCCATCG-3′ | (XhoI-HindIII) |
| LPW20740 5′-GGGGTACCGACGCACAAAACAATGCAAA-3′ | pSilent-1 cloning | |
| LPW20741 5′-GAAGATCTGTATTGCCGTTGATCCATCG-3′ | (BglII-KpnI) | |
| LPW23656 5′- TTGCCAATAACAAAGACTCTTC-3′ | qPCR | |
| LPW23657 5′- TCTTAGTCATAGCATCTGCG-3′ | ||
| RanBP10 | LPW23241 5′- CAAGTGCTGGCACAGGTCTA-3′ | qPCR |
| LPW23242 5′- TATATCCCAGCTTCACGCGG-3′ | ||
| Cyctochrome P450 | LPW23438 5′- GACGGCTATCAGTCTCACGG-3′ | qPCR |
| LPW23439 5′- GAGGCCGAACGGCATATACA-3′ | ||
| Conserved | LPW23499 5′- TGTCGGCAACCATGTGCTAT-3′ | qPCR |
| hypothetical protein | LPW23500 5′- CATTTCTTCGATGCAGGCGG-3′ |
Construction of dcl-1KO, dcl-2KO, dclDKO and qde-2KO mutants of P. marneffei
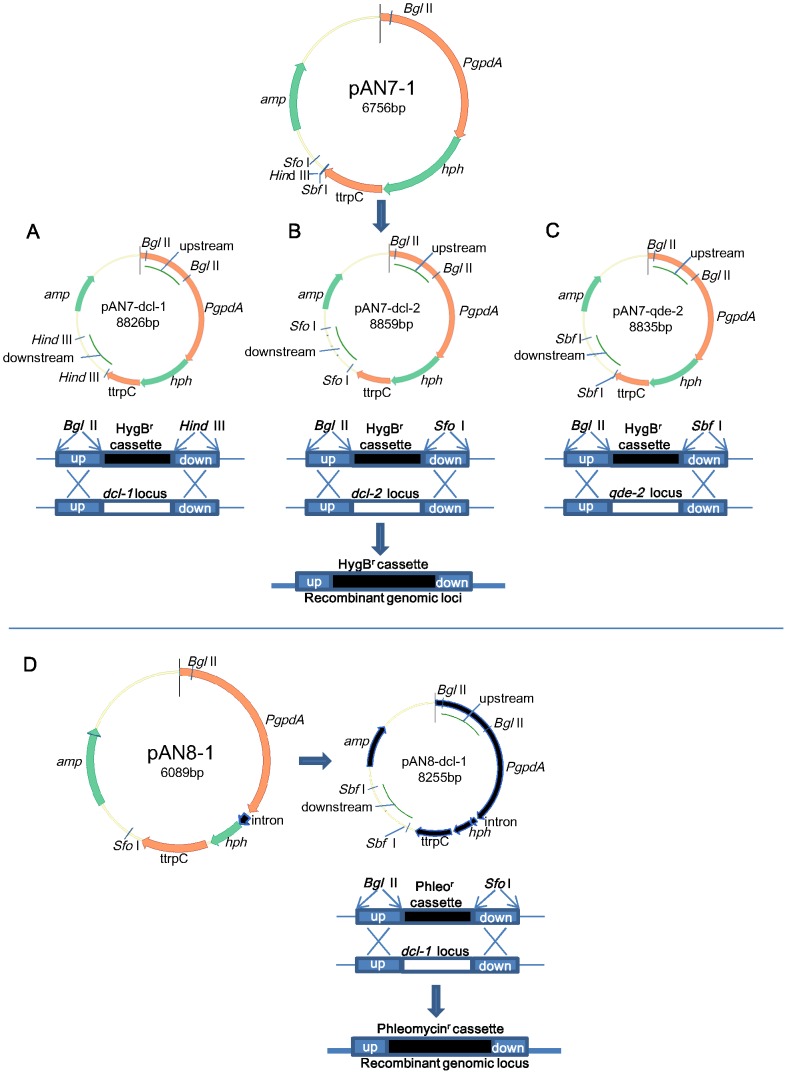
Deletion mutants were generated by homologous recombination (Fig. 1). Based on dcl-1, dcl-2 and qde-2 gene sequences from P. marneffei strain PM1, primers were designed to amplify upstream and downstream fragments of dcl-1, dcl-2 and qde-2 for the construction of the corresponding knockout constructs using the vector pAN7-1 (a gift from Dr. P. J. Punt) as described previously [46], [47]. The flanking sequences upstream and downstream of dcl-1, dcl-2 and qde-2 were amplified by PCR using DNA extracted from strain PM1 with primers shown in Table 1. PCR products of upstream and downstream flanking fragments were ligated into corresponding restriction sites of plasmid pAN7-1 to generate the knock-out plasmids pAN7-dcl-1, pAN7-dcl-2 and pAN7-qde-2 as shown in Fig. 1. The resultant plasmids were linearized with AhdI and transformed to strain PM1 according to previous publications [40], [48]. SDA supplemented with 150 µg/ml hygromycin B was used as selection medium. To construct dclDKO, PCR products of dcl-1 flanking fragments were ligated into vector pAN8-1 (a gift from Dr. P. J. Punt) as described previously [46], [49] to generate the knock-out plasmid pAN8-dcl-1 (Fig. 1). pAN8-dcl-1 was linearized with AhdI and transformed to dcl-2KO to generate the double mutant dclDKO, using SDA supplemented with 100 µg/ml phleomycin as selection medium.
Figure 1. Deletion of (A) dcl-1, (B) dcl-2, (C) qde-2, (D) dcl-1/dcl-2 in P. marneffei by homologous recombination.
Plasmids pAN7-1 and pAN8-1 were used to construct the knockout plasmids of pAN7-dcl-1, pAN7-dcl-2, pAN7-qde-2 and pAN8-dcl-1 respectively.
Northern blot analyses
Northern blot analysis was performed according to published protocols with modifications [30], [50]. Briefly, 10–20 µg of small RNAs was separated on 12% denaturing polyacrylamide gel and transferred onto a positively charged nylon membrane (Amersham Biosciences, United Kingdom) with NorthernMax Transfer buffer (Ambion) by means of capillary force for 1 h. Crosslinking of RNA to Hybond-NX was performed using a CL-1000 UV Cross-linker (UVP) according to the manufacturer's instructions, followed by baking at 80°C for 2 h. Hybridization was performed in ULTRAhyb-Oligo hybridization buffer (Ambion) for 3′ digoxigenin (DIG) labeled RNA probes (Sigma-Aldrich). Detection of the DIG-labeled probe on the blot was performed by using DIG Luminescent Detection kit (Roche).
Target prediction for milRNAs
The potential targets of milRNA candidates were predicted using the predicted gene sequences, including their 5′ and 3′ UTRs, of the P. marneffei strain PM1 and ATCC strain 18442 draft genomes by the RNAhybrid program [51] with or without mismatches or insertions at positions 9–11 of the milRNA and with parameters that encourage complete complementarity at the seed region (positions 2–7 of the milRNA) [52].
Construction of PM-milR-M1 gene knockdown plasmid of P. marneffei
To knockdown the PM-milR-M1 gene locus, plasmid pSilent-1 [53], obtained from the Fungal Genetics Stock Center, was used to construct the pSilent-M1 plasmid as previously described [40]. Briefly, the internal fragments (sense and antisense) were amplified with primers shown in Table 1 and cloned into the XhoI-HindIII and BglII-KpnI sites of pSilent-1 plasmid, resulting in pSilent-M1. The wild type P. marneffei strain PM1 was transformed with linearized pSilent-M1, using 200 µg/ml hygromycin for selection.
Quantitative real-time RT-PCR
Total RNA was extracted using RiboPure-Yeast (Ambion). Reverse transcription was performed using the SuperScript III kit (Invitrogen). Real-time RT-PCR assays were performed as described previously with modifications [36], using primers shown in Table 1. Results from actin were used for normalization. cDNA was amplified in a ABI 7900HT Fast Real-Time PCR System (Life Technologies) in 20-µL reaction mixtures containing FastStart DNA Master SYBR Green I Mix reagent kit (Roche, Basel, Switzerland), using the standard qPCR conditions (40 cycles of 95°C for 15 s, followed by 60°C for 1 min) and dissociation curve in the control software of SDS 2.4 (Life Technologies). Statistical analyses of the qRT-PCR data were performed using Student's t-test (SPSS version 19).
Nucleotide sequence accession number
The nucleotide sequences of the dcl-1, dcl-2 and qde-2 genes of P. marneffei have been deposited in GenBank under accession no. KC686608, KC686609 and KC686610 respectively. The Illumina small RNA sequences have been deposited in SRA NCBI database under accession no. SRX306604.
Results
Identification of P. marneffei small RNAs by deep sequencing
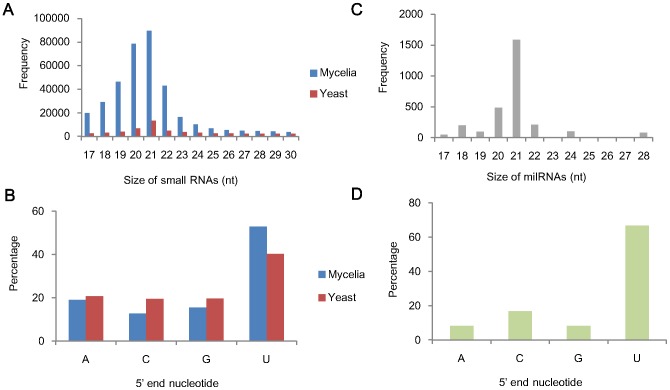
To examine small RNA species in the two growth phases of P. marneffei, cDNA libraries of small RNAs ≤60 nt extracted from mold and yeast cultures respectively were sequenced using the Illumina/Solexa Genome Analyzer IIx. The total number of both raw and filtered reads from mycelial and yeast phase was similar (Table 2). However, small RNAs were more abundant in mycelial than yeast phase of P. marneffei. We obtained a total of 3,155,063 and 270,782 high-quality, small RNA sequences of size 17–30 nt from mycelia and yeast phases respectively that perfectly match the P. marneffei genome. Among these, 362,805 and 56,543 unique small RNA sequences were identified from mycelial and yeast phases respectively (Fig. 2A). Most (89%) of the small RNAs identified from mycelial phase were 17–23 nt long, with the peak at 20–21 nt, and had a strong preference (52.82%) for 5′U (Fig. 2B), a known phenomenon in small RNAs of animals and plants [54], [55].
Table 2. Analysis of total and small RNA sequences in mycelial and yeast phase of P. marneffei.
| Total reads | Unique reads | |
| Mycelial | ||
| Raw reads | 39,809,400 | |
| Filtered reads | 27,914,677 | |
| Adaptors or rRNA reads | 11,435,372 | |
| Small RNA reads (17–30 nt) | 6,910,710 | 1,077,964 |
| Small RNA reads (17–30 nt) mapped to P. marneffei genome | 3,155,063 | 362,805 |
| Yeast | ||
| Raw reads | 36,999,600 | |
| Filtered reads | 28,424,899 | |
| Adaptors or rRNA reads | 15,670,222 | |
| Small RNA reads (17–30 nt) | 768,705 | 165,545 |
| Small RNA reads (17–30 nt) mapped to P. marneffei genome | 270,782 | 56,543 |
Figure 2. Characterization of small RNAs and milRNAs in P. marneffei.
(A) Size distribution and (B) nucleotide frequency of the 5′ end of small RNAs in mycelial and yeast phase. (C) Size distribution and (D) nucleotide frequency of the 5′ end of the 24 milRNA candidates.
Potential milRNAs in P. marneffei
Based on the distinguishing feature of known plant and animal miRNAs, 24 milRNA candidates, with flanking sequences forming hairpin secondary structures and at least five reads, were identified (Table 3). Their size distribution was shown in Fig. 2C, with a peak at 21 nt. There was also strong preference for U at their 5′ termini (67%, 16 of the 24 milRNA candidates) (Fig. 2D). These include 17 potential milRNAs (2,502 reads) in mycelial phase and seven potential milRNAs (232 reads) in yeast phase respectively (Table 3).
Table 3. Potential milRNA candidates in mycelial and yeast phase of P. marneffei.
| milRNA | Sequence (5′–3′) | Length (nt) | Reads |
| PM-milR-M1 | GAGAAACGCCUUAUGAUCGAC | 21 | 1482 |
| PM-milR-M1* | UGACUCGAAGAGCCUCUA | 18 | 1 |
| PM-milR-M2 | GUCCUAUAGUAAAGCCAGUC | 20 | 10 |
| PM-milR-M2* | AUUUCUAGGCUAUAAAAGCUU | 21 | 1 |
| PM-milR-MC3 | UGAUAUCAAAGUGGGCUAUC | 20 | 351 |
| PM-milR-MC4 | UCAAGUCAACCCUUACUC | 18 | 198 |
| PM-milR-MC5 | UUGCUAUGAUGAAAGCUGAGCA | 22 | 127 |
| PM-milR-MC6 | AACGUUUAAAUUUCCGAUACAAUU | 24 | 101 |
| PM-milR-MC7 | UAGGAUUAGGAUUAGGAUUA | 20 | 97 |
| PM-milR-MC8 | UUUCUACAGCUGCUGAACGUC | 21 | 44 |
| PM-milR-MC9 | UUGGCGUUGGGUGUAAUUG | 19 | 22 |
| PM-milR-MC10 | UCGACUGGCUCACCUGAUGCC | 21 | 14 |
| PM-milR-MC11 | UCGAUGUACUUCCUUGUGGA | 20 | 12 |
| PM-milR-MC12 | UGUUCAUCGAUCUGCUGUAGA | 21 | 9 |
| PM-milR-MC13 | UGCCACUCGAUCAUCUUGGG | 20 | 8 |
| PM-milR-MC14 | UAAGAGCUGUACAUAUGUAAG | 21 | 8 |
| PM-milR-MC15 | AUCCGGAUCGAGUUAUUCAC | 20 | 8 |
| PM-milR-MC16 | CAUAAGGUCGAGAGUCUCGCA | 21 | 6 |
| PM-milR-MC17 | UGGCGGACGCGAUGGUGGAGG | 21 | 5 |
| PM-milR-YC1 | UGCCAUUGCUAAGUCAAGG | 19 | 76 |
| PM-milR-YC2 | CAGCGGUGAUGACAACC | 17 | 47 |
| PM-milR-YC3 | CCGCUUCUAAAAUUGCUAGAGC | 22 | 44 |
| PM-milR-YC4 | UUGCUAUGAUGAAAGCUGAGCA | 22 | 30 |
| PM-milR-YC5 | UUUCUUGUCUACCUUUCGAGU | 21 | 19 |
| PM-milR-YC6 | UUCUCGGUGGCGAUGUCCAUU | 21 | 8 |
| PM-milR-YC7 | CCUUCAGAUCUGGGCUAUGCCC | 22 | 8 |
Identification and sequence analysis of dcl-1, dcl-2 and qde-2 genes
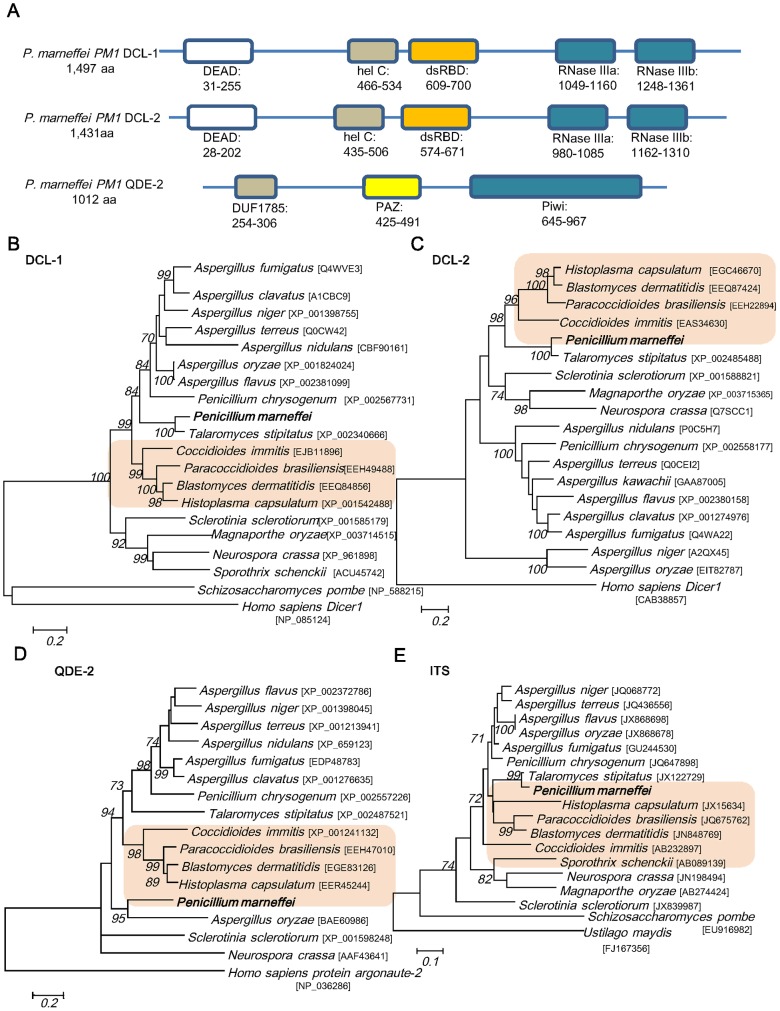
Using the respective homologues of N. crassa for BLAST search of P. marneffei strain PM1 draft genome sequence, two dcl genes, dcl-1 and dcl-2, encoding putative Dicer-like proteins and a gene, qde-2, encoding a putative Argonaute-like protein were identified (Fig. 3A). Dicer and Argonaute proteins are known to be involved in the biogenesis of miRNAs in animals and plants [56], [57], [58]. The dcl-1 gene is 5,383 bp in length, comprising 15 introns with total length of 889 bp. The resultant mRNA encodes 1,497 amino acid residues with a predicted molecular mass of 170.31 kDa. The dcl-2 gene is 4,636 bp in length, comprising six introns with total length of 340 bp. The resultant mRNA encodes 1,431 amino acids with a predicted mass of 161.15 kDa. These putative proteins possessed 42% and 32% amino acid identities to the DCL-1 and DCL-2 of N. crassa respectively. Both predicted proteins contain all four domains characteristic of the Dicer family. Two RNase III domains are present in the C-terminal region, and a DEAD-box ATP binding domain is present in the N-terminal region. In between there are RNA helicase and double stranded RNA binding domains. The qde-2 gene is 3,199 bp in length, comprising three introns with total length of 160 bp. The resultant mRNA encodes 1,012 amino acid residues with a predicted molecular mass of 111.75 kDa. The predicted QDE-2 protein possessed 35% amino acid identity to the QDE-2 of N. crassa. It contains two characteristic domains of the argonaute family, PAZ and Piwi domains, and the DUF1785 domain conserved in many argonaute proteins. The domain organization of DCL-1, DCL-2 and QDE-2 of P. marneffei is similar to that of the corresponding homologues in N. crassa [59].
Figure 3. Sequence analysis of dcl-1, dcl-2 and qde-2 genes in P. marneffei.
(A) Predicted domains of Dicer and QDE-2 proteins in P. marneffei strain PM1. Black bars represent the full protein sequence. The boxes represent the identified domains, each with its starting and stopping amino acid. Both DCL-1 and DCL-2 of P. marneffei contain a DEAD box, a helicase C domain (hel C), a double stranded RNA binding domain (dsRBD), and two RNase III domains (RNase IIIa and RNase IIIb). QDE-2 contains a PAZ domain ,a Piwi domain and a DUF1785 domains. Phylogenetic tree showing the relationship of predicted protein sequences of (B) dcl-1, (C) dcl-2, (D) qde-2 and (E) ITS of P. marneffei to homologues in other fungi constructed by maximum-likelihood method with Homo sapiens (DCL-1,DCL-2 and QDE-2) and Ustilago maydis (ITS) as the root. The thermal dimorphic pathogenic fungi are highlighted. A total of 914, 764 and 525 amino acid positions for dcl-1, dcl-2 and qde-2 and 465 nucleotide positions for ITS were included in the analysis respectively. Bootstrap values were calculated as percentages from 1000 replicates and only values ≥70% were shown. The scale bars indicate the estimated number of substitutions per 5, 5, 5 amino acids and 10 bases respectively. Names and accession numbers are given as cited in GenBank database.
Our previous study based on mitochondrial genome sequence has shown that P. marneffei is phylogenetically more closely related to those of filamentous fungi, including Aspergillus species, than yeasts [34]. Phylogenetic analysis of both ITS, another important marker for fungal identification and phylogeny, and dcl-1 gene showed that the corresponding sequences in P. marneffei were most closely related to Talaromyces stipitatus (a teleomorph of Penicillium emmonsii), Penicillium chrysogenum and Aspergillus spp. (Fig. 3). In contrast, phylogenetic analysis of dcl-2 and qde-2 genes showed a different evolutionary topology. The dcl-2 of P. marneffei and its homologue in T. stipitatus are more closely related to those of the thermal dimorphic pathogenic fungi, Histoplasma capsulatum, Blastomyces dermatitidis, Paracoccidioides brasiliensis and Coccidioides immitis than to P. chrysogenum and Aspergillus spp., suggesting the co-evolution of dcl-2 among the thermal dimorphic fungi. On the other hand, qde-2 of P. marneffei is most closely related to its homologues in other thermal dimorphic fungi than to that in T. stipitatus, P. chrysogenum and Aspergillus spp.
Differential mRNA expression of dcl-1, dcl-2 and qde-2 in mycelial and yeast phase
The mRNA expression level of dcl-1 in yeast phase was significantly higher than mycelial phase by 25 folds (P<0.001 by student t test). In contrast, the mRNA expression levels of dcl-2 and qde-2 were higher in mycelial phase than in yeast phase by 7 folds and 2 folds respectively (P<0.001 by Student's t-test) (Fig. 4).
Figure 4. Relative mRNA expression of (A) dcl-1, (B) dcl-2 and (C) qde-2 genes in mycelial and yeast phase of P. marneffei by qRT-PCR.
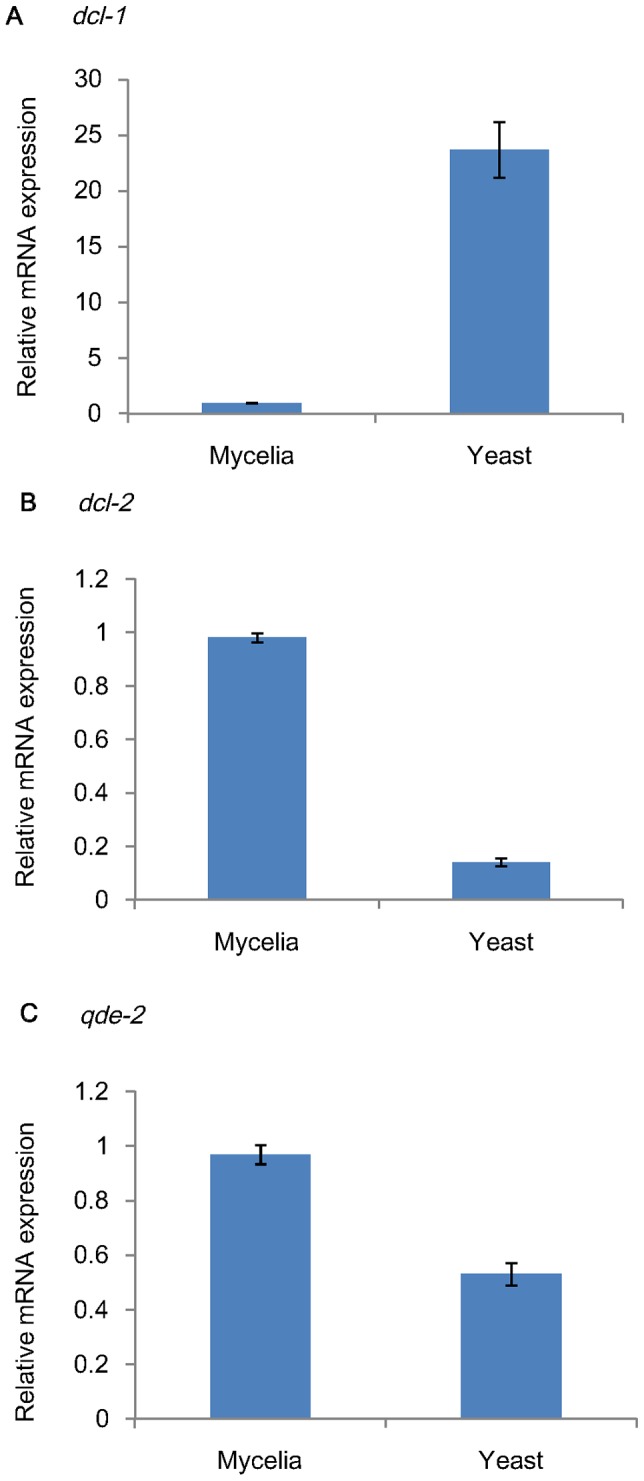
Results were obtained from five independent experimental replicates.
Dicer-dependent biogenesis of milRNA in P. marneffei
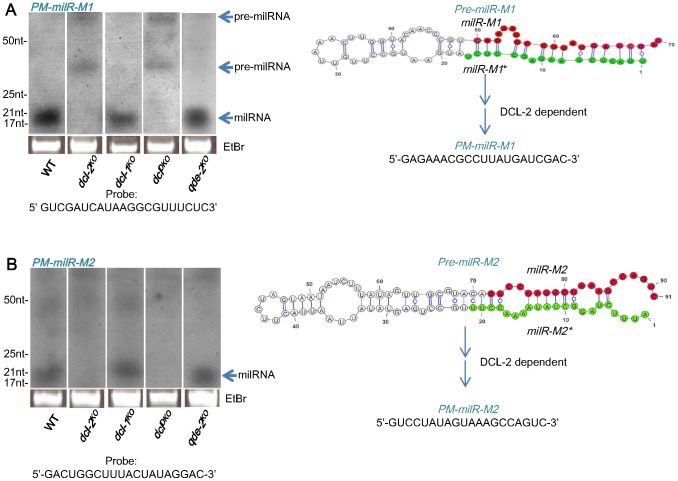
Northern blot analyses showed the production of milRNAs from two of the predicted milRNA loci, PM-milR-M1 and PM-milR-M2, both from mycelial phase of P. marneffei, with their predicted milRNA precursor (pre-milRNA) structures shown in Fig. 5. Their predicted precursors were approximately 70-nt and 91-nt in size and had negative folding free energies of −17.86 kcal mol−1 and -23.88 kcal mol−1 according to RNAfold (http://www.tbi.univie.ac.at/~ivo/RNA/RNAfold.html) for PM-milR-M1 and PM-milR-M2 respectively, comparable to those of known miRNA or milRNA precursors [31], [60], [61]. The majority of small RNA sequences of PM-milR-M1 and PM-milR-M2 correspond to one arm of the hairpin (the milRNA arm), with a total of 1482 small RNAs sequenced from PM-milR-M1 and 10 small RNAs sequenced from PM-milR-M2 (Table 3). In addition, small RNAs (milRNA*) matched to the complementary arm of the hairpin of PM-milR-M1 and PM-milR-M2 were also sequenced, but at much lower frequencies. In contrast to many small RNAs in which the miRNA arm possesses a 5′U position, the milRNA of both PM-milR-M1 and PM-milR-M2 have a 5′G position (Fig. 4). The existence of milRNA* and the presence of a 2 nt 3′ overhang in these milRNA/milRNA* pairs are strong evidence that they are produced from a Dicer-like enzyme (Fig. 5) [62]. Since loci which produce mature miRNAs and miRNA* sequences are considered miRNA loci, the two loci are tentatively named as P. marneffei milR-1 (PM-milR-1) and PM-milR-2. The locus, PM-milR-1, was situated within the coding region of a hypothetical protein, whereas PM-milR-2 was situated in the opposite strand of a pogo transposable element within a repeat region in the P. marneffei genome. The other 22 loci were considered milRNA candidates (named PM-milR-MC3, MC4…MC17 for milRNA candidates in mycelial phase and PM-milR-YC1…YC7 for those in yeast phase). These novel milRNAs or milRNA candidates showed no sequence similarity to known miRNAs miRBase as of March 2013.
Figure 5. milRNA biogenesis mechanism for PM-milR-M1 and PM-milR-M2 in P. marneffei.
Northern blot analyses of small RNA samples in wild-type (WT), dcl-1KO, dcl-2KO, dclDKO and qde-2KO strains of P. marneffei showing that the production of milRNA of (A) PM-milR-M1 and (B) PM-milR-M2 requires DCL-2 but not DCL-1 or QDE-2. The ethidium bromide-stained denaturing gel in the bottom panel showed equal loading of RNA. Predicted structures of pre-milRNA of PM-milR-M1 and PM-milR-M2, with their milRNA and paired milRNA* sequences as labeled in red and green respectively, are shown next to the northern blot analyses. The probe sequences used for northern blot analyses are marked.
To study the expression profile of PM-milR-M1 and PM-milR-M2 in mycelial and yeast phases, Northern blot analyses of small RNAs were performed, which cross-validated the Illumina sequencing results indicating mycelial-specific expression (Fig. 5A) with little or no expression in yeast phase (data not shown) of wild-type strain PM1. To assess the role of dcl-1, dcl-2 and qde-2 in the biogenesis of PM-milR-M1 and PM-milR-M2, dcl-1KO, dcl-2KO, dclDKO and qde-2KO mutants were generated using homologous recombination. All deletion mutants exhibited similar growth rates and phenotypic characteristics to wild-type strain in both mycelial and yeast phase cultures, although the dclDKO mutant exhibited poor sporulation and reduced red pigment production compared to wild-type strain upon transition from yeast to mycelial phase on sabouraud agar (data not shown).
Northern blot analysis of PM-milR-M1 in wild-type and deletion mutants showed that a band corresponding to the mature milRNA product with approximate size of 21 nt was present in wild-type strain, dcl-1KO and qde-2KO mutants, but absent in dcl-2KO and dclDKO mutants (Fig. 5A). Moreover, a band with approximate size of 70 nt, which matches the size of the predicted precursor of milRNA (pre-milRNA) of PM-milR-M1, was present in dcl-2KO and dclDKO mutants but not in wild-type strain, dcl-1KO or qde-2KO mutants. In addition, a band of approximately 30 nt is also seen in dcl-2KO and dclDKO mutants but not in wild-type strain, dcl-1KO or qde-2KO mutants, which may represent an intermediate product of the precursor. This suggested that DCL-2 protein is required for the biogenesis of mature milRNA from PM-milR-M1 and that the band at about 70 nt is likely the pre-milRNA. In the dcl-1KO and qde-2KO mutants, the levels of mature milRNA were similar to that of wild-type, indicating that DCL-1 and QDE-2 are not required for milRNA production from PM-milR-M1. As for PM-milR-M2, the band corresponding to its mature milRNA product, with approximate size of 20 nt, was also present in wild-type strain, dcl-1KO and qde-2KO mutants, but was absent in dcl-2KO and dclDKO mutants (Fig. 5A). This suggested that DCL-2 protein is also required for the biogenesis of mature milRNA from PM-milR-M2. In the dcl-1KO and qde-2KO mutants, the levels of mature milRNA were similar to that of wild-type, indicating that DCL-1 and QDE-2 are not required for milRNA production from PM-milR-M2.
Predicted milRNA targets in P. marneffei
Among the 24 potential milRNA candidates identified in the present study, 21 were predicted to have potential targets while three have no predicted targets (Supplementary Table S1). One of the candidates, PM-milR-MC17, was predicted to have up to 353 potential targets. These milRNAs candidates with predicted targets bind either perfectly or imperfectly complementary sequences. However, both PM-milR-M1 and PM-milR-M2 were predicted to bind complementary sequences of their targets imperfectly, similar to miRNAs in animals and the filamentous fungus, N. crassa [30]. The predicted targets of PM-milR-M1 include a putative Ran-binding protein RanBP10, a putative benzoate 4-monooxygenase cytochrome P450 and a conserved hypothetical protein. RanBP10 is a cytoplasmic guanine nucleotide exchange factor that modulates noncentrosomal microtubules involved in mitosis, while cytochrome P450 catalyses diverse reactions in fungal primary and secondary metabolism, and xenobiotic detoxification. As for PM-milR-M2, 20 potential targets were predicted, which include 13 transposon or transposable elements and seven conserved hypothetical proteins.
Regulation of target gene expression by PM-milR-M1
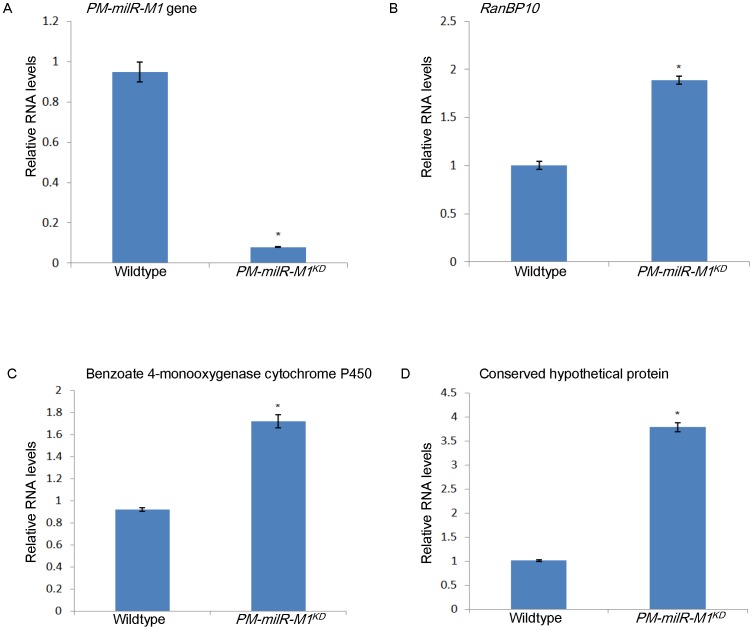
To test for potential regulation of target gene expression by these milRNAs, we generated a knockdown strain of PM-milR-M1 gene and measured the mRNA expression levels of the three predicted target genes. The knockdown strain, PM-milR-M1KD, only exhibited 8% transcription level of PM-milR-M1 gene in mycelial phase compared to wild type strain PM1 (Fig. 6A). The mRNA expression levels of the three predicted targets, putative RanBP10, putative benzoate 4-monooxygenase cytochrome P450 and a conserved hypothetical protein, were upregulated in PM-milR-M1KD by 1.9 (Fig. 6B) , 1.7 (Fig. 6C) and 3.8 folds (Fig. 6D) respectively compared to wild type strain PM1 (P<0.05 by student t test).
Figure 6. Regulation of target gene expression by PM-milR-M1.
Relative mRNA expression of (A) PM-milR-M1 gene, (B) RanBP10, (C) benzoate 4-monooxygenase cytochrome P450 and (D) a conserved hypothetical protein in mycelial phase of wild type strain PM1 and knockdown strain PM-milR-M1KD by qRT-PCR. Results were obtained from three independent experimental replicates.
Discussion
This is the first report of milRNAs in a human thermal dimorphic pathogenic fungus and their differential expression in mycelial and yeast phases. RNAi proteins such as Dicer and Argonaute have been identified in many fungi, such as the model filamentous fungus N. crassa [63] and fission yeast Schizosaccharomyces pombe [64]. Although RNAi proteins were lost in the famous budding yeast Saccharomyces cerevisiae, the closely related species Saccharomyces castelli encoded a defected but functional Dicer-like homolog [65]. However, till 2005, no endogenous miRNAs have been reported in fungi but only reports of antisense transcripts encoded in the genome of C. neoformans [66]. No plant or animal-like miRNAs was found in Aspergillus species by computational analysis of six Aspergillus genomes (Aspergillus nidulans, Aspergillus oryzae, Aspergillus fumigatus, Aspergillus terreus, Aspergillus clavatus, and Neosartorya fischeri) [67]. It was therefore uncertain whether fungi have microRNAs until the recent discovery of milRNAs in the filamentous fungi, N. crassa, S. sclerotiorum and M. anisopliae, as well as the human pathogenic yeast, C. neoformans [30]–[33]. Nevertheless, the presence of milRNAs in human pathogenic filamentous and dimorphic fungi was largely unknown. We have previously shown that target gene expression can be specifically knocked down by an RNAi-based method in P. marneffei [36], [40]. Moreover, we found that two dcl genes encoding putative dicer-like proteins and a qde-2 gene encoding a putative Argonaute-like protein, QDE-2, can be identified in P. marneffei strain PM1 draft genome, which are known to play key roles in the biogenesis of miRNAs and siRNAs [68]. Since miRNAs are important gene regulatory molecules in multicellular organisms, we hypothesized that P. marneffei possesses functional RNAi machinery and may encode miRNAs, which may be involved in the regulation of thermal dimorphism. In this study, using high throughput sequencing of small RNAs extracted from mycelial and yeast cultures of P. marneffei, we showed that small RNAs are more abundantly expressed in mycelial than yeast phase by >10 folds. The sequencing result is also in line with the more abundant small RNAs (approximately 20–24 nt) observed in mycelial than yeast phase upon Sybr Gold stained 12% denaturing polyacylamide gel electrophoresis (data not shown). After exclusion of other non-coding RNAs, a total of 2,734 reads were identified as potential milRNA candidates including 17 candidates in mycelial phase and seven in yeast phase, suggesting that milRNAs are differentially expressed in the two growth phases and may be more abundant in mycelial than yeast phase of P. marneffei. Two milRNAs, PM-milR-M1 and PM-milR-M2, both expressed in mycelial phase, were confirmed by Northern blot analyses. They share similar characteristics to miRNAs in animals and plants, being dependent on a Dicer-like protein for production and arisen from highly specific stem-loop RNA precursors. PM-milR-M1 was also shown to regulate the mRNA expression of its predicted target genes. The present results supported that dimorphic fungi may encode milRNAs which are likely conserved regulators of gene expression in diverse eukaryotes including fungi [18].
DCL-2 is likely a conserved protein involved in milRNA biogenesis among thermal dimorphic fungi. Dicer is a member of RNAse III family of nucleases and is responsible for miRNA processing in animals and plants [18]. While dicer-like proteins are known to be important for RNAi silencing in various fungi [36], [40], [69], [70], its role in milRNAs in fungi has been less well studied. A recent study on N. crassa has revealed diverse pathways in the generation of milRNAs and Dicer-independent small interfering RNAs (disiRNAs) [30]. In this study, the production of PM-milR-M1 and PM-milR-M2, as well as the pre-milRNA of PM-milR-M1, was dependent on the presence of DCL-2 but not DCL-1 or QDE-2 in P. marneffei. The pre-milRNA of PM-milR-M2 was not obvious upon Northern blot analyses, which may be due to degradation into small RNAs because of instability. No identifiable homologues of PM-milR-M1 and PM–milR-M2 could be in animals and plants, which supported the independent evolution of milRNAs in fungi [30], [33]. On the other hand, homologues of their precursors can be identified in T. stipitatus (data not shown). Nevertheless, it remains to be determined if such milRNA homologues are also expressed and processed in the same way. Interestingly, in contrast to ITS and dcl-1 sequences which were both phylogenetically most closely related to the homologues in T. stipitatus, P. chrysogenum and Aspergillus spp., the dcl-2 gene of P. marneffei is more closely related to the homologues in other geographically restricted thermal dimorphic fungi than to P. chrysogenum and Aspergillus spp.. This suggested that the dcl-2 gene may have co-evolved among the thermal dimorphic fungi and serve similar function. Since these thermal dimorphic fungi are different from other fungi by their ability to cause systemic mycosis as intracellular yeasts and survive in natural environments as molds, it would be interesting to explore the potential role of DCL-2 in fungal dimorphism as well as virulence. In N. crassa, at least four different mechanisms that involved a combination of factors were identified for the production of milRNAs. In fact, apart from dicers and QDE-2, homologues of QDE-2 interacting protein (QIP) and mitochondrial ribosomal protein L3 (MRPL3), which were also involved in biogenesis of some milRNAs in N. crassa [30], can also be found in the P. marneffei genome, with 27–49% amino acid identities (data not shown). Further studies are required to explore for possible role of these proteins in milRNA biogenesis in P. marneffei.
In contrast to miRNAs from animals and plants which are known to play different functions from multicellular development to stress response, the potential function(s) of milRNAs in fungi remain to be determined. Some miRNAs in plants and animals are known to exhibit temporal or tissue-specific expression patterns [18], [71], [72]. As for fungi, a recent study showed that some milRNAs are differentially expressed in sclerotial development of S. sclerotiorum [31]. In C. neoformans, milRNAs were shown to cause transgene silencing via the canonical RNAi pathway and proposed to be play a role in regulating transposons and pseudogene expression [33]. In this study, we showed the mRNA expression level of dcl-2 was higher in mycelial than yeast phase, suggesting that DCL-2 may function predominantly in the mycelial phase. This, in turn, may explain why PM-milR-M1 and PM–milR-M2 were only expressed in mycelial but not yeast form of P. marneffei. Therefore, it is likely that PM-milR-M1 and PM-milR-M2 are only produced from DCL-2 and serve important function during mycelial phase. A number of potential targets were predicted for both PM-milR-M1 and PM-milR-M2. For example, the predicted targets of PM-milR-M1 include RanBP10 and cytochrome P450, while transposon or transposable elements were the predominant predicted targets of PM-milR-M2. The targets of PM-milR-M1 were also confirmed to be upregulated at the RNA level in the knockdown strain, PM-milR-M1KD, supporting the mRNA cleavage function of milRNAs. These results suggested that milRNAs in P. marneffei may regulate cell division, metabolism as well as transposons, although further studies are required to investigate their biological function. Nevertheless, the present study demonstrated the potential role of differential post-transcriptional control in different growth phases of thermal dimorphic fungi, which may provide new insights into the mechanism governing thermal dimorphism.
Supporting Information
Predicted targets of milRNAs in P. marneffei.
(DOCX)
Acknowledgments
We thank Dr P.J. Punt for providing us with the pAN7-1 and pAN8-1 plasmids.
Funding Statement
This work was partly supported by the Strategic Research Theme Fund, Research Grant Council Grant, University Grant Council; Committee for Research and Conference Grant, and University Development Fund, The University of Hong Kong; Shaw Foundation; and Providence Foundation Limited in memory of the late Dr. Lui Hac Minh. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hsueh PR, Teng LJ, Hung CC, Hsu JH, Yang PC, et al. (2000) Molecular evidence for strain dissemination of Penicillium marneffei: an emerging pathogen in Taiwan. J Infect Dis 181: 1706–1712. [DOI] [PubMed] [Google Scholar]
- 2. Supparatpinyo K, Khamwan C, Baosoung V, Nelson KE, Sirisanthana T (1994) Disseminated Penicillium marneffei infection in southeast Asia. Lancet 344: 110–113. [DOI] [PubMed] [Google Scholar]
- 3. Huang YT, Hung CC, Liao CH, Sun HY, Chang SC, et al. (2007) Detection of circulating galactomannan in serum samples for diagnosis of Penicillium marneffei infection and cryptococcosis among patients infected with human immunodeficiency virus. J Clin Microbiol 45: 2858–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yuen KY, Wong SS, Tsang DN, Chau PY (1994) Serodiagnosis of Penicillium marneffei infection. Lancet 344: 444–445. [DOI] [PubMed] [Google Scholar]
- 5. Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, et al. (2011) Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium . Stud Mycol 70: 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chariyalertsak S, Vanittanakom P, Nelson KE, Sirisanthana T, Vanittanakom N (1996) Rhizomys sumatrensis and Cannomys badius, new natural animal hosts of Penicillium marneffei . J Med Vet Mycol 34: 105–110. [PubMed] [Google Scholar]
- 7. Deng ZL, Yun M, Ajello L (1986) Human penicilliosis marneffei and its relation to the bamboo rat (Rhizomys pruinosus). J Med Vet Mycol 24: 383–389. [DOI] [PubMed] [Google Scholar]
- 8. Deng ZL, Connor DH (1985) Progressive disseminated penicilliosis caused by Penicillium marneffei. Report of eight cases and differentiation of the causative organism from Histoplasma capsulatum . Am J Clin Pathol 84: 323–327. [DOI] [PubMed] [Google Scholar]
- 9. Low K, Lee SS (2002) The pattern of AIDS Reporting and the implications on HIV surveillance. Public Health Epidemiol Bull 11: 41–49. [Google Scholar]
- 10. Wong KH, Lee SS (1998) Comparing the first and second hundred AIDS cases in Hong Kong. Singapore Med J 39: 236–240. [PubMed] [Google Scholar]
- 11. Sekhon AS, Stein L, Garg AK, Black WA, Glezos JD, et al. (1994) Pulmonary penicillosis marneffei: report of the first imported case in Canada. Mycopathologia 128: 3–7. [DOI] [PubMed] [Google Scholar]
- 12. Vanittanakom N, Jr Cooper CR, Fisher MC, Sirisanthana T (2006) Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin Microbiol Rev 19: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lo CY, Chan DT, Yuen KY, Li FK, Cheng KP (1995) Penicillium marneffei infection in a patient with SLE. Lupus 4: 229–231. [DOI] [PubMed] [Google Scholar]
- 14. Wang JL, Hung CC, Chang SC, Chueh SC, La MK (2003) Disseminated Penicillium marneffei infection in a renal-transplant recipient successfully treated with liposomal amphotericin B. Transplantation 76: 1136–1137. [DOI] [PubMed] [Google Scholar]
- 15. Wong SSY, Woo PCY, Yuen KY (2001) Candida tropicalis and Penicillium marneffei mixed fungaemia in a patient with Waldenstrom's macroglobulinaemia. Eur J Clin Microbiol Infect Dis 20: 132–135. [DOI] [PubMed] [Google Scholar]
- 16. Woo PC, Lau SK, Lau CC, Chong KT, Hui WT, et al. (2005) Penicillium marneffei fungaemia in an allogeneic bone marrow transplant recipient. Bone Marrow Transplant 35: 831–833. [DOI] [PubMed] [Google Scholar]
- 17. Boyce KJ, Andrianopoulos A (2013) Morphogenetic circuitry regulating growth and development in the dimorphic pathogen Penicillium marneffei . Eukaryot Cell 12: 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 19. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- 20. Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, et al. (2002) Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- 21. Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X, Carthew RW (2005) A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123: 1267–1277. [DOI] [PubMed] [Google Scholar]
- 23. Xiao C, Rajewsky K (2009) MicroRNA control in the immune system: basic principles. Cell 136: 26–36. [DOI] [PubMed] [Google Scholar]
- 24. Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53. [DOI] [PubMed] [Google Scholar]
- 26. Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- 27. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 28. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- 29. Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34 Database issue: D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, et al. (2010) Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell 38: 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou J, Fu Y, Xie J, Li B, Jiang D, et al. (2012) Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing. Mol Genet Genomics 287: 275–282. [DOI] [PubMed] [Google Scholar]
- 32. Zhou Q, Wang Z, Zhang J, Meng H, Huang B (2012) Genome-wide identification and profiling of microRNA-like RNAs from Metarhizium anisopliae during development. Fungal Biol 116: 1156–1162. [DOI] [PubMed] [Google Scholar]
- 33. Jiang N, Yang Y, Janbon G, Pan J, Zhu X (2012) Identification and functional demonstration of miRNAs in the fungus Cryptococcus neoformans . PLoS One 7: e52734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Woo PC, Zhen H, Cai JJ, Yu J, Lau SK, et al. (2003) The mitochondrial genome of the thermal dimorphic fungus Penicillium marneffei is more closely related to those of molds than yeasts. FEBS Lett 555: 469–477. [DOI] [PubMed] [Google Scholar]
- 35. Woo PC, Chong KT, Tse H, Cai JJ, Lau CC, et al. (2006) Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei . FEBS Lett 580: 3409–3416. [DOI] [PubMed] [Google Scholar]
- 36. Woo PC, Tam EW, Chong KT, Cai JJ, Tung ET, et al. (2010) High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei . FEBS J 277: 3750–3758. [DOI] [PubMed] [Google Scholar]
- 37. Yuen KY, Pascal G, Wong SS, Glaser P, Woo PC, et al. (2003) Exploring the Penicillium marneffei genome. Arch Microbiol 179: 339–353. [DOI] [PubMed] [Google Scholar]
- 38. Woo PC, Lau CC, Chong KT, Tse H, Tsang DN, et al. (2007) MP1 homologue-based multilocus sequence system for typing the pathogenic fungus Penicillium marneffei: a novel approach using lineage-specific genes. J Clin Microbiol 45: 3647–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woo PC, Lau SK, Liu B, Cai JJ, Chong KT, et al. (2011) Draft genome sequence of Penicillium marneffei strain PM1. Eukaryot Cell 10: 1740–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woo PC, Lam CW, Tam EW, Leung CK, Wong SS, et al. (2012) First discovery of two polyketide synthase genes for mitorubrinic Acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei . PLoS Negl Trop Dis 6: e1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Henk DA, Shahar-Golan R, Devi KR, Boyce KJ, Zhan N, et al. (2012) Clonality despite sex: the evolution of host-associated sexual neighborhoods in the pathogenic fungus Penicillium marneffei . PLoS Pathog 8: e1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, et al. (2008) Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, et al. (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415. [DOI] [PubMed] [Google Scholar]
- 45. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woo PC, Chong KT, Lau CC, Wong SS, Lau SK, et al. (2006) A novel approach for screening immunogenic proteins in Penicillium marneffei using the DeltaAFMP1DeltaAFMP2 deletion mutant of Aspergillus fumigatus . FEMS Microbiol Lett 262: 138–147. [DOI] [PubMed] [Google Scholar]
- 47. Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli . Gene 56: 117–124. [DOI] [PubMed] [Google Scholar]
- 48. Sanglard D, Ischer F, Monod M, Bille J (1996) Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother 40: 2300–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mattern I, Punt P, Van den Hondel C (1988) A vector of Aspergillus transformation conferring phleomycin resistance. Fungal Genet Newsl 35: 25. [Google Scholar]
- 50. Lau SK, Woo PC, Yip CC, Fan RY, Huang Y, et al. (2012) Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol 86: 5481–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 53. Nakayashiki H, Hanada S, Nguyen BQ, Kadotani N, Tosa Y, et al. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42: 275–283. [DOI] [PubMed] [Google Scholar]
- 54. Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, et al. (2008) High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rathjen T, Pais H, Sweetman D, Moulton V, Munsterberg A, et al. (2009) High throughput sequencing of microRNAs in chicken somites. FEBS Lett 583: 1422–1426. [DOI] [PubMed] [Google Scholar]
- 56. Mukherjee K, Campos H, Kolaczkowski B (2013) Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol 30: 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Mierlo JT, Bronkhorst AW, Overheul GJ, Sadanandan SA, Ekström JO, et al. (2012) Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog 8: e1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, et al. (2004) Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa . Mol Cell Biol 24: 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bonnet E, Wuyts J, Rouzé P, Van de Peer Y (2004) Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20: 2911–2917. [DOI] [PubMed] [Google Scholar]
- 61. Liu N, Yang J, Guo S, Xu Y, Zhang M (2013) Genome-Wide Identification and Comparative Analysis of Conserved and Novel MicroRNAs in Grafted Watermelon by High-Throughput Sequencing. PLoS One 8 2: e57359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes Dev 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fulci V, Macino G (2007) Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa . Curr Opin Microbiol 10: 199–203. [DOI] [PubMed] [Google Scholar]
- 64. Sigova A, Rhind N, Zamore PD (2004) A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe . Genes Dev 18: 2359–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, et al. (2009) RNAi in budding yeast. Science 326 5952: 544–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, et al. (2005) The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans . Science 307: 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McGuire AM, Galagan JE (2008) Conserved Secondary Structures in Aspergillus . PLoS ONE 3 7: e2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Carthew RW, Sontheimer EJ (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Segers GC, Zhang X, Deng F, Sun Q, Nuss DL (2007) Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci U S A 104: 12902–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kadotani N, Nakayashiki H, Tosa Y, Mayama S (2004) One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation. J Biol Chem 279: 44467–44474. [DOI] [PubMed] [Google Scholar]
- 71. Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, et al. (2003) The microRNAs of Caenorhabditis elegans . Genes Dev 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted targets of milRNAs in P. marneffei.
(DOCX)