Abstract
We report a pioneering approach using Tetrahymena thermophila that permits rapid identification of genes based on their null or hypomorphic phenotypes. This technique involves cell transformation with a library of plasmids that encode 26S ribosomal subunits containing short insertions. The insertions correspond to antisense sequences for a large number of genes. The majority of cells each acquires a single antisense sequence, which silences a single genomic locus. Because the insertion site within the ribosomal sequence is known, the silenced gene is easily amplified. We demonstrate that this approach can be used to identify genes required for dense core granule exocytosis.
Rapid techniques for cloning genes based on mutant phenotypes, such as cloning by complementation, restriction endonuclease-mediated integration (REMI), and transposon-induced mutagenesis, are available in a limited number of eukaryotes including Saccharomyces cerevisiae and Dictyostelium discoideum (1–3), and have played key roles in illuminating many physiological pathways. The ciliate Tetrahymena thermophila has been a valuable experimental organism for discoveries including telomerase, enzymatic RNA, and histone acetylation (4–6). Easily transformed by homologous integration, Tetrahymena is established or emerging as a powerful system for studying histone and tubulin modifications, chromosome rearrangements, and regulated exocytosis (7–10). Extensive genetic and physical mapping is underway (11–13); in addition, classical genetic approaches have been used over four decades to isolate and characterize a large number of mutants (14). However, no general approaches have been available to identify the genetic bases of such mutant phenotypes.
A pathway that is studied easily in Tetrahymena involves the synthesis and exocytosis of dense-core granules (DCGs), vesicles that are used in eukaryotes for storage and regulated secretion of a wide variety of macromolecular cargoes (15). Tetrahymena cells contain several thousand DCGs, positioned at the cell periphery, that are capable of undergoing stimulus-triggered synchronous fusion with the plasma membrane. Stimulation results in massive exocytosis of DCG contents. Under controlled laboratory conditions, cells become trapped in a capsule of the released contents; such wild-type (WT) cells are designated caps(+) (16). Because entrapment prevents them from swimming freely, it is possible to enrich for cells defective in exocytosis based on their greater mobility. In addition, such mutants can be identified by visually screening for caps(−) cells. Such screens led to the isolation of a collection of nitrosoguanidine-induced mutants (17, 18), but uncovering the molecular bases for these defects is currently precluded by the absence of a method for cloning by complementation.
Here we describe a method based on an antisense library, whereby chimeric cells are created that express ribosomes carrying antisense sequences. This method relies on the previous discovery by Yao and colleagues (19) that the 26S subunit of Tetrahymena ribosomes could be engineered to contain a short additional loop that does not compromise ribosome function. Such recombinant ribosomes, expressed from an rDNA-based plasmid, were capable of rapidly replacing the endogenous ribosomes. If the short additional loop corresponded to a gene-specific antisense sequence, the recombinant “antisense ribosome” could specifically and reliably suppress the expression of the complementary gene, as long as the antisense sequences were less than ≈130 bp-long and corresponded to the 5′ untranslated region of the targeted gene (19).
Materials and Methods
Cells and Cell Culture.
T. thermophila strains CU428 and B2086 were kindly provided by Peter Bruns (Cornell University, Ithaca, NY), and are WT [caps (+)] with respect to exocytosis. Cells were cultured routinely as described in ref. 20. For drug selection of transformants, an alternative medium, SPP, was used (21).
Construction and Characterization of Antisense Ribosome Library.
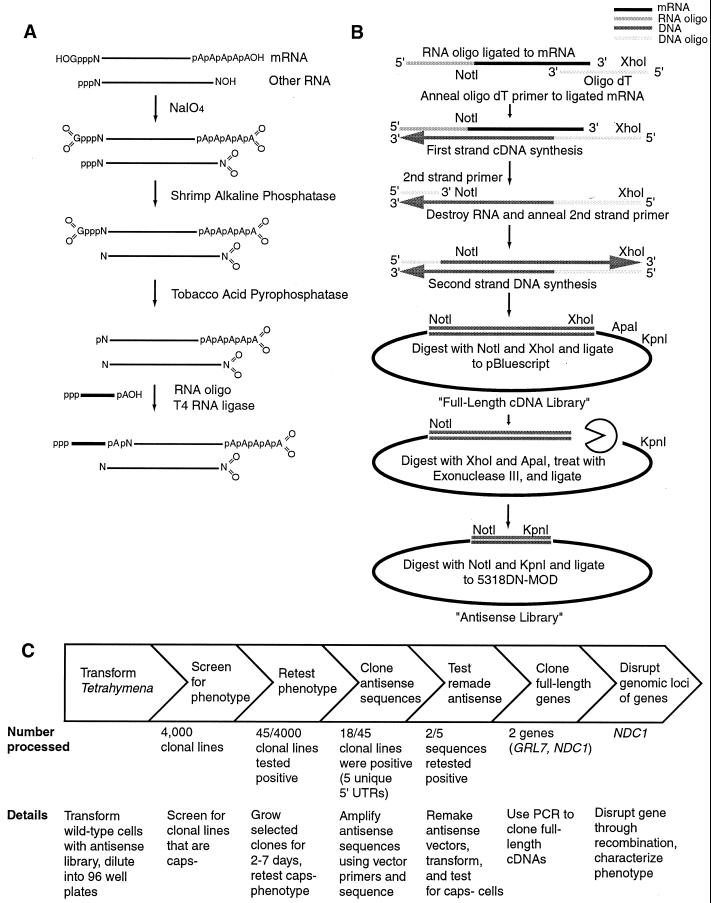
Because conventional cDNA libraries do not preserve information near the 5′ end of the mRNA (22, 23), the method used here is based on several described techniques that were designed to retain complete 5′ information (24–27). The 5′ ends of mRNAs were tagged selectively by using an oligo-capping protocol. Other RNAs (e.g., rRNA, degraded mRNA, etc.) were not tagged by this procedure, because the starting RNA [poly(A)+ RNA isolated from exponentially growing Tetrahymena strain CU428.1] was oxidized first with sodium periodate (NAIO4; Sigma), to prevent its later ligation, and then dephosphorylated with shrimp alkaline phosphatase (Roche Molecular Biochemicals), to prevent ligation of uncapped RNAs. Then mRNAs were decapped with tobacco acid pyrophosphatase (Epicentre Technologies, Madison, WI) to generate a 5′ monophosphate suitable for ligation, and the products were tagged by ligation to an RNA 42-mer (5′-pppGGGAACAAAAGCTGGAGCTCCACCGCGGTGGCGGCCGCTCTA-3′).
Tagged mRNA was used to generate a library of short 5′ sequences. The tagged mRNAs were converted first into a full-length double-stranded (ds) cDNA library. Avian myeloblastosis virus reverse transcriptase (Roche Molecular Biochemicals)-mediated reverse transcription was primed with an oligo(dT) (XhoI) primer (5′-GAGAGAGAGAGAGAGAGAGAACTAGTCTCGAGTTTTTTTTTTTTTTTTTT-3′). Then RNA was removed by treatment with a mixture of RNAses (Ambion, Austin, TX). Second-strand synthesis by Pfu polymerase (Stratagene) was primed by using a DNA oligo whose sequence overlapped the RNA oligo used for tagging (5′-AACAAAAGCTGGAGCTCCAC-3′). The resulting dsDNA molecules then were digested with NotI and XhoI, ligated into pBluescript II SK(+) (Stratagene), and transformed into Escherichia coli. Independent clones (3.3 × 105) were generated, with an average size of 1 kb. This library of mostly full-length cDNAs (>90% of sequenced inserts contain 5′ untranslated regions) was used to make the antisense ribosome library by treatment with exonuclease III and mung bean nuclease (New England Biolabs; ref. 28). Molecules 30–150 bp-long were isolated by agarose gel electrophoresis and ligated into ribosomal antisense plasmid 5318DN (19), which was generously provided by M.-C. Yao (Fred Hutchinson Cancer Research Center). The multiple-cloning site (MCS) of the antisense vector was shortened so that larger antisense inserts could be accommodated. The following sequence of the original MCS (5′-TCAGGTACCCGCAAAGCGGCCGCGTCGACGGGCCCCCCGGGGTAACCTTTGCGGGTACCCTGA-3′) was replaced with (5-TCACCTGAGGGTACCCCCGGCGGCCGCGTACCCTGA-3′). Emboldened portions of the above sequence remained the same. Restriction sites were also altered for convenience, leaving a NotI and an XhoI site. This new vector was named 5318DN.MOD. The 5318DN.MOD clones containing antisense molecules were transformed into E. coli to generate 1.5 × 106 independent clones, of which about 10% contained inserts of 30–160 bp and averaged 69 bp. Construction of the library is summarized in Fig. 1 A and B.
Figure 1.
Construction and use of antisense ribosome library. Details can be found in Materials and Methods.
Use of Antisense Ribosome Library.
Transformation of Tetrahymena mating pairs (CU428 × B2086) and screening for caps(−) transformants were done by using described protocols with several modifications (refs. 17 and 29; Fig. 1C). For transformations, five aliquots of 5318DN.MOD (50 μg) mixed with 250 μl of cells were each electroporated, pooled, and dispensed into 96-well plates at appropriate dilutions. A total of 4,000 clonal lines were screened for the caps(−) phenotype of which 45 initially tested positive. These clones were retested, and 18 regenerated the caps(−) phenotype. Ribosomal inserts of transformants showing caps(−) phenotypes were amplified by PCR, using primers on either side of the cloning site (forward primer, 5′-AAAAGGTCGATGAGTAAGGAAATG-3′; reverse primer, 5′-CAATCTCAGGGTACGCGG-3′). These 18 products ranged from 63 to 112 bp in length, with an average of 84 and a median value of 94. The two products that were later reconfirmed, corresponding to NDC1 and GRL7, were slightly larger than average (109 and 94 bp, respectively.) Fourteen of the 18 sequences, including those corresponding to NDC1 and GRL7, contained 5′ untranslated regions (UTRs) of which five were unique. Those sequences that arose more than once corresponded to clones that seem particularly abundant in the cDNA library (based on sequencing of ≈1,200 inserts from that library). For example, the 5′ UTR corresponding to a ribosomal protein gene, rpl17, is found in 7/18 clones. The GRL7 gene, also highly represented in the library, is found in 3/18 antisense inserts. In plasmids containing duplicated sequences, the inserts are not identical. For example, the 7 clones corresponding to rpl17 fall into three classes. The partial redundancy may be a result of the fact that these are particularly effective sequences for antisense suppression, or a result of the redundancy within the cDNA or antisense libraries.
The five unique amplified products were ligated into the antisense vector (PCR products and 5318DN.MOD were cut with NotI and KpnI and ligated), transformed into cells, and tested for the caps(−) phenotype. Two of these five clones retested positive for the caps(−) phenotype. One of the sequences that retested positive (NDC1) was (5′-AAAATACAAAGTACAAAAAAATAAATAAAAACGTAAAACGTAAAGGAAAATGAACAAGGCCTTAGTTTTCTTGGGGTGTAGGTGGGGGCCC-3′). Primers specific to this sequence, in combination with a vector-specific primer, were used to amplify the entire gene from the full-length library.
Disruption of NDC1 Through Homologous Recombination.
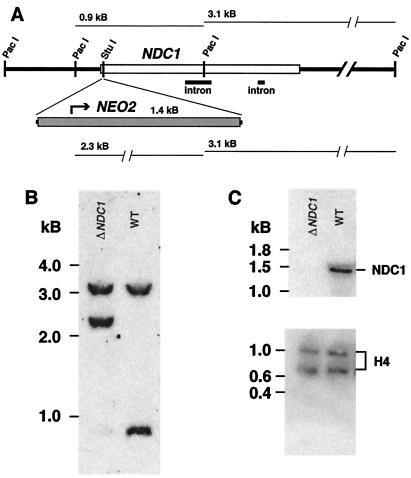
NDC1 was cloned and disrupted in the same manner as PGM1 (8). DNA sequences spanning all of NDC1, including upstream and downstream regions, were obtained by using PCR techniques. A knockout construct was generated with the NEO2 drug-resistance cassette (29) inserted at a StuI site just downstream of the predicted translational initiation site, thereby creating the null allele ndc1–1∷neo2 (30). Macronuclear knockouts were confirmed on the basis of the restriction maps, as detected by Southern blotting with a probe corresponding to the NDC1 gene. Cells in which all macronuclear copies of NDC1 are replaced with ndc1–1∷neo2 are referred to as ΔNDC1.
Northern Blotting.
Northern blotting of total RNA extracted from WT and ΔNDC1 cells was performed following standard techniques (28) by using an NDC1 riboprobe. Histone H4 mRNA served as a control; a plasmid containing the histone H4 gene was kindly provided by Martin Gorovsky, Univ. of Rochester.
Identification and Analysis of Caps(−) Cells.
Antisense transformants, selected after 4 days in 120 μg/ml of paromomycin sulfate (Sigma), were enriched for caps(−) cells, which then were identified by visual screening as described (17). Stimulation of exocytosis to induce capsule formation was performed by using the conditions described in ref. 17. Fixation conditions for transmission electron microscopy were identical to those in ref. 20.
Results
To assemble a plasmid library of antisense sequences, we first constructed a cDNA library from full-length mRNA to preserve complete 5′ untranslated regions. From this library, we generated a plasmid library of antisense ribosomes by truncating the full-length library from the 3′ end and directionally cloning the inserts into the antisense plasmid (Fig. 1 A and B). Like the initially described antisense ribosomes (19), our library is based on a vector encoding a variant of the Tetrahymena rDNA minichromosome, allowing efficient transformation and selection of drug-resistant transformants (31, 32). After transformation of Tetrahymena with this library, a brief period of selective growth is sufficient to permit the recombinant antisense ribosomes to replace the endogenous copies. Transformants with phenotypes of interest can then be isolated by appropriate selection or screening.
We used the antisense library to expand molecular studies of regulated exocytosis. From a collection of antisense ribosome transformants, we used a combination of enrichment and visual screening to isolate mutant clones that failed to show stimulus-dependent exocytosis of DCG contents. The rDNA inserts of such mutants were amplified and cloned, using primers based on the known flanking rDNA sequence. The ability of these sequences to interfere with exocytosis was confirmed by reinserting the antisense sequence into the rDNA backbone, retransformation, and confirming the caps(−) phenotype. Antisense sequences that reproduced the caps(−) phenotype were used to design primers, which then were used to amplify the gene by using the full-length cDNA library as template. Thus, cloning of the full-length gene required just a limited series of PCR reactions (Fig. 1C).
The first two genes identified by using this protocol serve to validate our approach. The first gene identified was GRL7 (Granule lattice protein 7), a gene we had characterized as one of a family of five genes encoding highly abundant calcium-binding proteins in the DCG core (33). Although the phenotype associated with a GRL7 gene knockout had not been examined, we previously found that disruption of the macronuclear (expressed) copies of encoding other granule lattice proteins genes (GRL1, GRL3, or GRL4) was sufficient to block normal DCG assembly and therefore exocytosis (ref. 20 and N.D.C., J. W. Verbsky, and A.P.T., unpublished data). These results strongly suggested that the suppression of GRL7 expression would have a similar effect. The isolation of GRL7 in this antisense-based screen fulfilled this prediction.
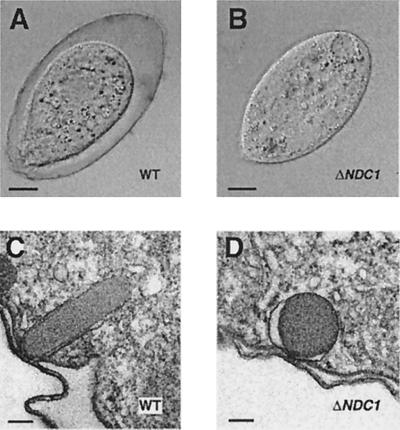
The second gene identified has been named NDC1 (Non-discharge of granule contents). To confirm that this gene is required for DCG function, we cloned the macronuclear copy of NDC1 and disrupted all expressed copies by gene replacement with an allele interrupted by the neomycin-resistance cassette (ndc1–1∷neo2; Fig. 2). Southern and northern analyses of transformants demonstrated that these cells have no intact copies of NDC1 and no visible NDC1 transcripts; such cells are referred to as ΔNDC1. More importantly, and as expected, NDC1 is essential for normal DCG biosynthesis (Fig. 3). Although ΔNDC1 cells synthesize DCGs that are docked correctly, the granules are morphologically aberrant and closely resemble those in ΔGRL1 cells. Like ΔGRL1cells, ΔNDC1 cells are caps(−), confirming a defect in exocytosis.
Figure 2.
Disruption of NDC1 through homologous recombination. (A) A physical map of the NDC1 locus, indicating the disruption site of the NEO2 cassette. (B) Southern blot of WT and ΔNDC1 cells, probed for the NDC1 locus. (C) Northern blot of WT and ΔNDC1 cells, demonstrating the absence of visible NDC1 transcript in the knockout cell line. Histone H4 serves as a control.
Figure 3.
Phenotype of ΔNDC1 cells. (A and B) Light micrographs of WT and ΔNDC1 cells under conditions that trigger synchronous fusion of DCGs and release of their contents. WT cells are surrounded by a capsule of secreted protein that is absent in ΔNDC1 cells. (Scale bar = 10 μm.) (C and D) Transmission electron microscopy of DCGs in WT and ΔNDC1 cells. The WT granule is roughly cylindrical, tethered to the plasma membrane, and contains a visible protein lattice. DCGs in ΔNDC1 also are tethered at the plasma membrane but are roughly spherical, and the contents are not visibly organized in a lattice. (Scale bar = 200 nm.)
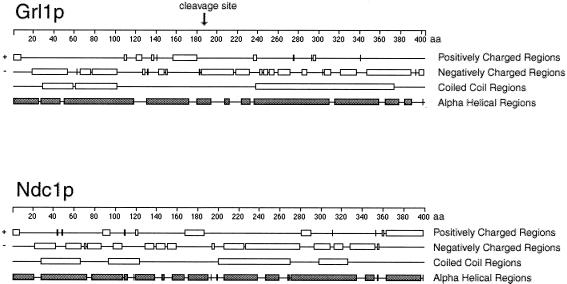
The similarities between the phenotypes of ΔNDC1 and ΔGRL1 suggested that the protein product of NDC1, Ndc1p, is part of the DCG cargo. Consistent with this, the predicted coding sequence begins with a canonical signal peptide that targets translocation into the endoplasmic reticulum and is expected on all secreted proteins. NDC1 is related to, but interestingly different from, all of the characterized GRLs. The secondary structure of GRL gene products is predicted to consist largely of coiled coils, and the proteins undergo proteolytic processing during granule synthesis (33). Although the GRL products share relatively little sequence identity with one another, the processing sites have conserved motifs and each is preceded by a short stretch of basic amino acids in otherwise highly acidic proteins (33). The same organization is seen in orthologous proteins in Paramecium tetraurelia DCGs (34), suggesting the possibility that these basic regions may be determinants of the downstream cleavage sites. NDC1 encodes a comparably acidic polypeptide of similar length to the GRL products, and also is predicted to assemble largely as coiled coils. Like the GRL products, Ndc1p contains short basic stretches that are likely to precede processing sites (Fig. 4). However, the single most basic stretch in the predicted polypeptide lies at its extreme C terminus, an arrangement not seen in any of the known GRL-encoded proteins. Because this stretch cannot precede a cleavage site, it suggests that basic regions in DCG proteins play a role distinct from determining downstream processing sites. This suggestion led us to test whether deletion of the basic region preceding the characterized cleavage site in Grl1p was required for cleavage site determination. By using site-directed mutagenesis, we replaced GRL1 with variants that lack the entire stretch of basic amino acids that lie immediately upstream of the normal processing site at Lys-188 (20), and found that cleavage-site specificity is unaltered in these mutants (N.D.C., J. W. Verbsky, and A.P.T., unpublished data).
Figure 4.
Secondary structure modeling of Ndc1p and Grl1p. Both Grl1p and Ndc1p are highly acidic and are predicted to form predominantly α-helical structures with large regions of coiled coils. Both have a basic region near the center of the primary sequence (which directly precedes a known processing site in Grl1p), but Ndc1p also contains a C-terminal basic region.
Discussion
We have developed an approach for isolating genes based on their null and/or hypomorphic phenotypes in T. thermophila. Culpable genes can be cloned within several weeks of isolating transformed cells with a desired phenotype. Confirming the usefulness of this approach, we have used it to identify two genes that are involved in regulated secretion. Comparison of the organization of the gene product, Ndc1p, with those of previously identified DCG cargo proteins suggests that the characteristic basic regions in these proteins serve a function distinct from processing site determination, as had been hypothesized previously.
Before this work, no Tetrahymena gene had been cloned based on an associated phenotype, although a collection of interesting mutant lines has been well studied. The antisense method for gene identification can be applied to any somatic process that depends on transcribed genes and whose null/hypomorphic phenotype can be detected. Colleagues with whom we have shared this library have succeeded with a screen designed to identify genes associated with defective cell division. A gene in this pathway, identified by using the antisense library, has been confirmed already (E. Cole, personal communication). Nonetheless, that this method relies on screening for null or hypomorphic phenotypes may limit its application for identifying genes whose functions are essential for cell viability (35).
Because the antisense library described was derived from total mRNA, genes will be represented according to the abundance of their transcripts. This biased abundance is likely to explain why the first antisense plasmids coming out of our screen correspond to abundant DCG cargo proteins. The current library, as well as libraries derived from normalized or stage-specific mRNA, should facilitate the isolation of novel genes in a variety of biological pathways.
Acknowledgments
We thank A. Haddad, E. Zweifel, and M. Hund for valuable discussion. We thank Yimei Chen for assistance with transmission electron microscopy. This work was supported by National Institutes of Health Grants GM50946 and ACS IRG-41-30 (to A.T.P.). N.D.C. and N.C.E. were supported in part by National Institutes of Health Predoctoral Training Grants GM07197 and GM07197-26, respectively. Electron microscopy was carried out with support of the University of Chicago Digestive Diseases Research Center (DK42086).
Abbreviations
- DCG
dense-core granule
- WT
wild type
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF331706).
References
- 1.Burns N, Grimwade B, Ross-Macdonald P B, Choy EY, Finberg K, Roeder G S, Snyder M. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 2.Kuspa A, Loomis W F. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beggs J D. Nature (London) 1978;275:104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- 4.Greider C W, Blackburn E H. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 5.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth SY, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 6.Kruger K, Grabowski P J, Zaug A J, Sands J, Gottschling D E, Cech T R. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 7.Coyne R S, Chalker D L, Yao M-C. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- 8.Chilcoat N D, Turkewitz A P. J Cell Biol. 1997;139:1197–1207. doi: 10.1083/jcb.139.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaertig J, Cruz M A, Bowen J, Gu L, Pennock D G, Gorovsky M A. J Cell Biol. 1995;129:1301–1310. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia L, Hai B, Gao Y, Burnette D, Thazhath R, Duan J, Bre M H, Levilliers N, Gorovsky M A, Gaertig J. J Cell Biol. 2000;149:1097–1106. doi: 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong L, Klionsky L, Wickert S, Merriam V, Orias E, Hamilton E P. Genetics. 2000;155:1119–1125. doi: 10.1093/genetics/155.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickert S, Nangle L, Shevel S, Orias E. Genetics. 2000;154:1155–1167. doi: 10.1093/genetics/154.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickert S, Orias E. Genetics. 2000;154:1141–1153. doi: 10.1093/genetics/154.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleyman L K. In: Ciliate Genetics in Ciliates: Cells as Organisms. Hausmann K, Bradbury P C, editors. Stuttgart: Fischer; 1996. pp. 291–324. [Google Scholar]
- 15.Arvan P, Castle D. Biochem J. 1998;332:593–610. doi: 10.1042/bj3320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiedtke A. Naturwissenschaften. 1976;63:93. doi: 10.1007/BF00622415. [DOI] [PubMed] [Google Scholar]
- 17.Melia S M, Cole E S, Turkewitz A P. J Cell Sci. 1998;111:131–140. doi: 10.1242/jcs.111.1.131. [DOI] [PubMed] [Google Scholar]
- 18.Orias E, Flacks M, Satir B H. J Cell Sci. 1983;64:49–67. doi: 10.1242/jcs.64.1.49. [DOI] [PubMed] [Google Scholar]
- 19.Sweeney R, Fan Q, Yao M-C. Proc Natl Acad Sci USA. 1996;93:8518–8523. doi: 10.1073/pnas.93.16.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chilcoat N D, Melia S M, Haddad A, Turkewitz A P. J Cell Biol. 1996;135:1775–1787. doi: 10.1083/jcb.135.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaertig J, Kapler G. Methods Cell Biol. 2000;62:485–500. doi: 10.1016/s0091-679x(08)61552-6. [DOI] [PubMed] [Google Scholar]
- 22.Gubler U, Hoffman B J. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 23.D'Alessio J M, Gerard G F. Nucleic Acids Res. 1988;16:1999–2014. doi: 10.1093/nar/16.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruyama K, Sugano S. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S. Gene. 1997;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- 26.Efstratiadis A, Vournakis J N, Donis-Keller H, Chaconas G, Dougall D K, Kafatos F C. Nucleic Acids Res. 1977;4:4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Gorovsky M A. Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E J, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Gaertig J, Gu L, Hai B, Gorovsky M A. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen S L, Altschuler M I, Bruns P J, Cohen J, Doerder F P, Gaertig J, Gorovsky M A, Orias E, Turkewitz A. Genetics. 1998;149:459–462. [Google Scholar]
- 31.Yu G L, Hasson M, Blackburn E H. Proc Natl Acad Sci USA. 1988;85:5151–5155. doi: 10.1073/pnas.85.14.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tondravi M, Yao M-C. Proc Natl Acad Sci USA. 1986;83:4369–4373. doi: 10.1073/pnas.83.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verbsky J W, Turkewitz A P. Mol Biol Cell. 1998;9:497–511. doi: 10.1091/mbc.9.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautier M-C, Sperling L, Madeddu L. J Biol Chem. 1996;271:10247–10255. doi: 10.1074/jbc.271.17.10247. [DOI] [PubMed] [Google Scholar]
- 35.Fan Q, Sweeney R, Yao M-C. Methods Cell Biol. 2000;62:533–545. doi: 10.1016/s0091-679x(08)61555-1. [DOI] [PubMed] [Google Scholar]