Abstract
Background
Researchers have provided evidence that telomere dysfunction play an important role in cancer development. MNS16A is a polymorphic tandem repeats minisatellite of human telomerase (hTERT) gene that influences promoter activity of hTERT and thus implicates to relate with risk of several malignancies. However, results on association between MNS16A and cancer risk remain controversial. We therefore conduct a meta-analysis to derive a more precise estimation of association between MNS16A and cancer risk.
Methods
A systematic literature search was conducted by searching PubMed, ISI Web of Knowledge, Human Genome and Epidemiology Network Navigator and Google Scholar digital database for publications on associations between MNS16A and cancer risk. Variants with statistically significant associations by meta-analysis were assessed using Venice criteria.
Results
10 case-control articles enrolling 6101 cases and 10521 controls were brought into our meta-analysis. The relationships were strong epidemiological credibility in cerebral cancer and breast cancer population (P for heterogeneity > 0.1). The cumulative analysis in chronologic order suggested a clear tendency towards a significant association with additional study samples.
Conclusions
The results provided a more accurate depiction of the role of MNS16A in cerebral cancer and breast cancer susceptibility. Additional larger studies were warranted to validate our findings.
Introduction
Telomeres (a distinctive DNA-protein structure at the distal end of eukaryotic chromosomes) are crucial for genomic stability [1]–[5]. Somatic cells have a progressive shortening of telomeres after each cell division, however, telomeres reach a critical short length and lose capping function at the senescence stage in immortal tumor cells. Uncapped chromosomal ends will then trigger DNA-damage-like responses [6], [7]. The expressions of telomerase can prevent the loss of telomeres [8]–[10]. Human telomerase reverse transcriptase (hTERT) as the key constituent of telomerase, is highly expressed in essentially all immortal tumor cells, but is restricted in normal tissues, leading investigators to considerate hTERT as a critical role with cancer susceptibility [11]–[13]. MNS16A, a polymorphic tandem repeats minisatellite in downstream of hTERT gene, has been first reported to affect promoter activity in lung cancer cell lines [14]. The variants containing short tandem repeats (S allele) have stronger promoter activity than long repeats (L allele), indicating number of tandem repeats associated with lung cancer risk. Subsequently, several malignancies such as cerebral [15], [16], lung [17], [18], breast [19], [20], colorectal [21], nasopharyngeal [22], prostate cancer [23] and one meta-analysis [24] had investigated MNS16A in the etiology of cancer but with inconsistent results. Considering the important role of MNS16A in promoter activity of hTERT gene, we therefore conduct a meta-analysis on eligible articles to estimate association of MNS16A with cancer risk.
Materials and Methods
Search strategy, eligibility criteria and data extraction
All methodology was based on guidelines proposed by the Human Genome Epidemiology Network (HuGENet) [25] and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [26] for systematic review of genetic association studies. A systematic review of original publications analyzing the association between MNS16A and cancer risk was performed by searching PUBMED, ISI Web of knowledge and Google Scholar database on and before February 2013, without language restriction. The strategy of keywords were: ("Neoplasm" [Mesh] OR "Carcinoma"[Mesh]) AND ("Telomerase"[Mesh] OR hTERT) AND MNS16A. Furthermore, we screened the Human Genome and Epidemiology Network Navigator as well as the references lists of key studies and reviews for additional publications [27]. We then performed the following criteria for literature selection: (a) original relevant case-control articles were included in this paper; (b) articles dealing with association between MNS16A and cancers in humans were available; (c) articles providing sufficient data to calculate ORs and 95% confidence intervals (CIs) were considered eligible. Information was extracted independently by two investigators (Rui and Zou) to ensure homogeneity of data collection and to rule out subjectivity effect in data gathering and entry. The following data should be noted: first author’s name, published year, location where the study was conducted, ethnicity, study period, mean age of case and control, source population, cancer type, sample size, variant counts in both cases and controls. For studies investigating more than one type of cancer, data were extracted separately as independent study [15], [16].
Statistical analysis
Meta-analysis
For statistical analysis, number of tandem repeats was classified as either short (S) or long (L) alleles (LS classification system): S alleles, 213bp, 240bp, 243bp, 271bp, 272bp, 274bp; L alleles, 299bp, 302bp, 331bp, 333bp, 364bp, frequently applied in literature. On basis of classification, MNS16A genotypes were assigned to SS, LS or LL genotype groups. ORs and 95% CIs were recalculated and assessed in gene models based on MNS16A length comparisons (S allele versus L allele): a co dominant genetic model (SS versus LL; LS versus LL), a dominant genetic model (SS+LS versus LL) and a recessive model (SS versus LS + LL). To explore in depth of different lengths of MNS16A under S allele group, we classified the 271bp, 272bp and 274bp allele as middle alleles (M allele) and 213bp, 240bp and 243bp alleles still as S alleles (LMS classification system) described by Jin et al [18].
Sensitivity analyses and between-study heterogeneity
Between-study heterogeneity was assessed by the χ2-based Cochran’s Q statistic test and I2 metric [28]. Heterogeneity was considered significant at P<0.1 for the Q statistic (to assess whether observed variance exceeds expected variance). And for the I2 metric (I2 = 100% × (Q-df)/Q), the following cut-off points were used: I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity; I2 = 75–100%, extreme heterogeneity. The significance of the combined ORs was determined using the Z test (P<0.05 was considered statistically significant). The DerSimonian and Laird random effect model [29] was used to calculate pooled ORs and 95% CIs according to their heterogeneity, otherwise, a fixed effects model (the Mantel-Haenszel method) was applied. Stratified analysis was performed for two ethnicity groups in order to investigate the hypothesis of ethnicity-specific genetic mechanisms in the development of MNS16A. Summary ORs and 95% CI were also calculated after stratification for cancer type. Additionally, sensitivity analysis was performed consecutively by omitting every article from the meta-analysis in turn to determine the influence of each study on the overall estimate [30]. Cumulative meta-analysis was performed through an assortment of all eligible cancer studies within the publication years. Finally, publication bias was evaluated by Begg’s test and Egger’s test to detect the small study effect [31]. All statistical analyses were performed with STATA software (version 10.1), and a 2-sided P value of less than 0.05 was considered significant, except for Q test for heterogeneity, for which a less than 0.1 level of statistical significance was applied.
Estimating the credibility of statistically significant associations
Each variant with statistically significant associations by meta-analysis were assessed on the basis of the Human Genome Epidemiology Network Venice criteria. Credibility was defined as “strong,” “moderate,” or “weak” based on grades A, B, or C in three categories: 1) amount of evidence; 2) replication; and 3) protection from bias. Amount of evidence was assessed by size of test allele among case and controls in meta-analysis (nminor): grade A, B, C requires nminor > 1000, 100 ≤ nminor ≤ 1000, nminor<100. Replication was graded by the heterogeneity statistic: grades A, B, and C were assigned for I2 less than 25%, 25–50%, and greater than 50%, respectively. Assessment of protection from bias was graded as grade A if there was no observable bias, grade B if bias could be present, or grade C if bias was evidence ( the presence of a summary ORs less than 1.15 or loss of statistical significance after excluding the initial study) [32].
Results
Subjects characteristics
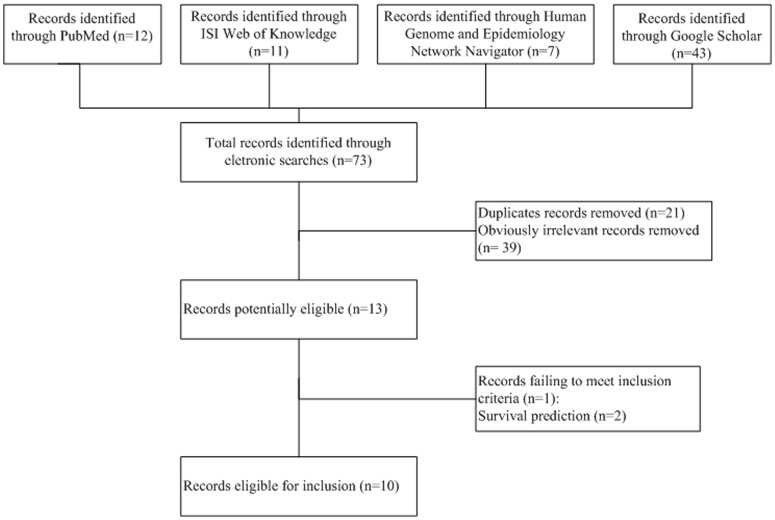
After comprehensive searching of 71 articles, we identified 10 relevant publications including 6101 cases and 10521 controls from 13 studies to assess the association between MNS16A and cancer risk (Figure 1): 2 studies focused on glioblastoma [15], [16], 2 studies focused on glioma [15], [16], 3 studies focused on non-small cell lung cancer [14], [17], [18], 2 studies focused on breast cancer[19], [20] and each was one for meningioma [15], colorectal carcinoma [21], nasopharyngeal carcinoma [22] and prostate cancer [23] (Table 1). All studies were case-control studies, of which the most frequently investigated was brain cancer (6451 subjects; 38.81%). Among these, 9 studies were conducted in Caucasians (10400 subjects; 62.57%) and 4 in Asians (6222 subjects; 37.43%).
Figure 1. Flow chart of study selection.
Table 1. Characteristics for case-control studies of MNS16A and risk of cancer included in a meta-analysis.
| First author | Year | Study | Ethnicity | Mean | age | Source | Cancer | No. of | No. of | No. of | No. of |
| location | case | control | population | type | case/ | LLa | LSb | SSc | |||
| control | case/ | case/ | case/ | ||||||||
| control | control | control | |||||||||
| Wang [14] | 2003 | USA | Caucasian | 65.5 | 54.9 | hospital | NSCLC | 53/72 | 30/33 | 17/29 | 6/10 |
| Carpentier[16] | 2007 | France | Caucasian | 56.3 | 49.0 | population | GBM | 205/305 | 69/133 | 111/144 | 25/28 |
| 46.25 | 49.0 | Glioma | 147/305 | 57/133 | 63/144 | 27/28 | |||||
| Wang [19] | 2008 | China | Asian | 51.71 | 51.77 | population | BC | 1006/1095 | 860/984 | 141/107 | 5/4 |
| Andersson[15] | 2009 | Europe | Caucasian | 47 | 51 | population | Glioma | 648/1359 | 282/650 | 277/560 | 89/149 |
| 52 | 51 | Meningioma | 473/1359 | 212/650 | 207/560 | 54/149 | |||||
| NA | NA | GBM | 291/1359 | 120/650 | 127/560 | 44/149 | |||||
| Jin [18] | 2010 | Korea | Asian | 61.7 | 61.5 | population | NSCLC | 937/943 | 820/840 | 110/101 | 7/2 |
| Hofer [21] | 2011 | Austria | Caucasian | 66.8 | 61.3 | population | CRC | 88/1712 | 36/770 | 44/747 | 8/195 |
| Zhang[22] | 2011 | China | Asian | NA | NA | population | NPC | 798/1019 | 725/891 | 71/121 | 2/7 |
| Chang[17] | 2011 | Taiwan | Asian | 67.58 | 67.09 | population | NSCLC | 205/219 | 181/197 | 24/21 | 0/1 |
| Zagouri[20] | 2012 | Greece | Caucasian | 55.1 | 55.7 | hospital | BC | 113/124 | 50/63 | 36/29 | 27/32 |
| Hofer [23] | 2013 | Austria | Caucasian | 63.8 | 67.4 | hospital | PC | 1137/650 | 501/308 | 499/277 | 137/65 |
The length of MNS16A were defined as L allele or S allele under LS classification system.
Abbreviation: NA, none anonymous; GBM, glioblastoma; BC, breast cancer; NSCLC, non-small cell lung cancer; CRC, colorectal cancer; NPC, nasopharyngeal cancer; PC, prostate cancer.
Results of the meta-analysis
As shown in Table 2, all studies were pooled into a meta-analysis, and the increased association between MNS16A and cancer risk were found for all genotypic models. Random-effect model pooling analyses yielded overall ORs of 1.15 (95% CI = 1.03–1.28; P for heterogeneity = 0.102, I2 = 35.0%) for LS genotype versus LL genotype, and 1.17 (95%CI = 1.05–1.31; P for heterogeneity = 0.064, I2 = 40.5%) for dominant model. In fixed-effects model, overall ORs were 1.32 (95%CI = 1.14–1.53; P for heterogeneity = 0.337, I2 = 10.8%) for SS genotype versus LL genotype, and 1.23 (95% CI = 1.07–1.41; P for heterogeneity = 0.307, I2 = 13.7%) for recessive model.
Table 2. Pooled ORs with 95% CIs for the association between MNS16A and cancer risk in meta-analysis.
| Category | Genetic model | ORs (95% CI) | P a | P for | I2 |
| Heterogeneity | |||||
| LS classification | S vs. L | 1.13 (1.03–1.25) | 0.013 | 0.012 | 53.3% |
| (No. of study = 13) | LS vs. LL | 1.15 (1.03–1.28) | 0.015 | 0.102 | 35.0% |
| SS vs. LL | 1.32 (1.14–1.53) | 0.000 | 0.337 | 10.8% | |
| Dominant | 1.17 (1.05–1.31) | 0.006 | 0.064 | 40.5% | |
| Recessive | 1.23 (1.07–1.41) | 0.003 | 0.307 | 13.7% | |
| LMS classification b | S vs. L | 1.21 (1.04–1.41) | 0.015 | 0.047 | 50.8% |
| (No. of study = 8) | M vs. L | 1.04 (0.75–1.42) | 0.830 | 0.041 | 52.1% |
| LM+MM vs. LL | 1.04 (0.73–1.50) | 0.823 | 0.003 | 54.8% | |
| LS+MS+SS vs. LL | 1.75 (1.02–1.73) | 0.041 | 0.000 | 93.0% | |
| LS+MS+SS vs. LL+LM+MM | 1.03 (0.73–1.45) | 0.862 | 0.000 | 79.3% |
P value was calculated by the Z test.
The length of MNS16A was defined as L, M or S allele under LMS classification system.
Subsequently we categorized the data in LMS classification described by Jin et al. to explore in depth the effect of MNS16A S allele (the short allele) and M allele (the middle allele) with cancer risk. As shown in Table 2, 8 studies were classified during LMS classification system. All genetic models revealed that S allele presented a great cancer risk than M allele and 95%CIs were nearby statistically significant.
Stratified analysis
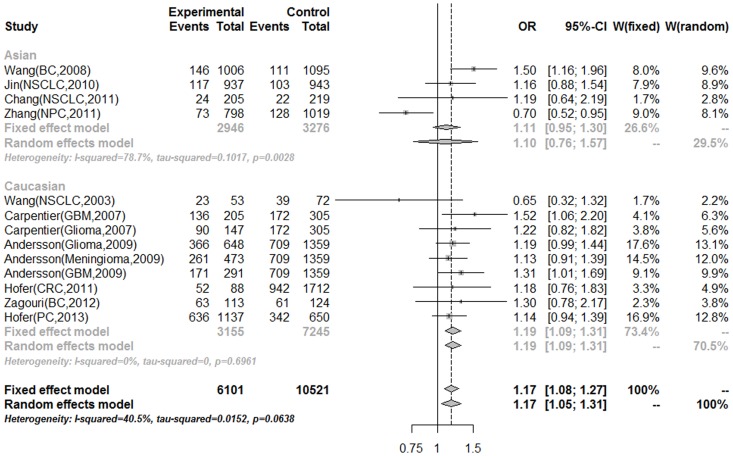
Stratified analysis was performed for two ethnicity groups in order to investigate the hypothesis of Asian and Caucasian genetic mechanisms in the development of MNS16A. (Table 3). No evidence of heterogeneity was revealed in Caucasian population (P for heterogeneity > 0.1), and all genetic models presented a significantly increased cancer risk, with ORs of 1.16 (95%CI = 1.05–1.28), 1.33 (95%CI = 1.15–1.54), 1.19 (95%CI = 1.09–1.31), and 1.23 (95%CI = 1.07–1.42) for LS versus LL genotype, SS versus LL genotype, dominant model, and recessive model, respectively. However, all genetic models presented no statistical differences of cancer risk among Asian population (Figure 2).
Table 3. Pooled ORs with 95% CIs for the association between MNS16A and cancer risk by stratified analysis.
| Category | Genetic model | ORs (95%CI) | P a | P for heterogeneity | I2 | |
| Ethnicity | Caucasian | S vs. L | 1.16 (1.06–1.26) | 0.001 | 0.235 | 23.4% |
| (No. of study = 9) | LS vs. LL | 1.16 (1.05–1.28) | 0.003 | 0.689 | 0.00% | |
| SS vs. LL | 1.33 (1.15–1.54) | 0.000 | 0.383 | 6.20% | ||
| Dominant | 1.19 (1.09–1.31) | 0.000 | 0.696 | 0.00% | ||
| Recessive | 1.23 (1.07–1.42) | 0.003 | 0.322 | 13.5% | ||
| Asian | S vs. L | 1.08 (0.76–1.55) | 0.658 | 0.002 | 80.0% | |
| (No. of study = 4) | LS vs. LL | 1.10 (0.78–1.56) | 0.591 | 0.005 | 76.3% | |
| SS vs. LL | 1.13 (0.54–2.35) | 0.747 | 0.188 | 37.3% | ||
| Dominant | 1.10 (0.76–1.57) | 0.621 | 0.003 | 78.7% | ||
| Recessive | 1.12 (0.53–2.33) | 0.768 | 0.204 | 34.8% | ||
| Cancer type | Cerebral Cancer | S vs. L | 1.19 (1.10–1.30) | 0.000 | 0.503 | 0.00% |
| (No. of study = 5) | LS vs. LL | 1.17 (1.04–1.32) | 0.008 | 0.708 | 0.00% | |
| SS vs. LL | 1.42 (1.19–1.70) | 0.000 | 0.303 | 17.6% | ||
| Dominant | 1.22 (1.09–1.37) | 0.000 | 0.686 | 0.00% | ||
| Recessive | 1.32 (1.11–1.56) | 0.001 | 0.248 | 26.0% | ||
| Lung Cancer | S vs. L | 0.96 (0.62–1.49) | 0.842 | 0.066 | 63.3% | |
| (No. of study = 3) | LS vs. LL | 1.07 (0.84–1.37) | 0.571 | 0.379 | 0.00% | |
| SS vs. LL | 1.14 (0.51–2.57) | 0.744 | 0.180 | 41.7% | ||
| Dominant | 1.07 (0.81–1.42) | 0.627 | 0.315 | 13.4% | ||
| Recessive | 1.23 (0.56–2.73) | 0.609 | 0.229 | 32.2% | ||
| Breast Cancer | S vs. L | 1.31 (1.00–1.72) | 0.046 | 0.214 | 35.2% | |
| (No. of study = 2) | LS vs. LL | 1.52 (1.19–1.94) | 0.001 | 0.914 | 0.00% | |
| SS vs. LL | 1.12 (0.64–1.99) | 0.687 | 0.691 | 0.00% | ||
| Dominant | 1.46 (1.16–1.84) | 0.002 | 0.620 | 0.00% | ||
| Recessive | 0.97 (0.57–1.66) | 0.904 | 0.576 | 0.00% |
P value was calculated by the Z test.
Figure 2. Forest plot of MNS16A association with cancer risk under dominant model stratified by ethnicity.
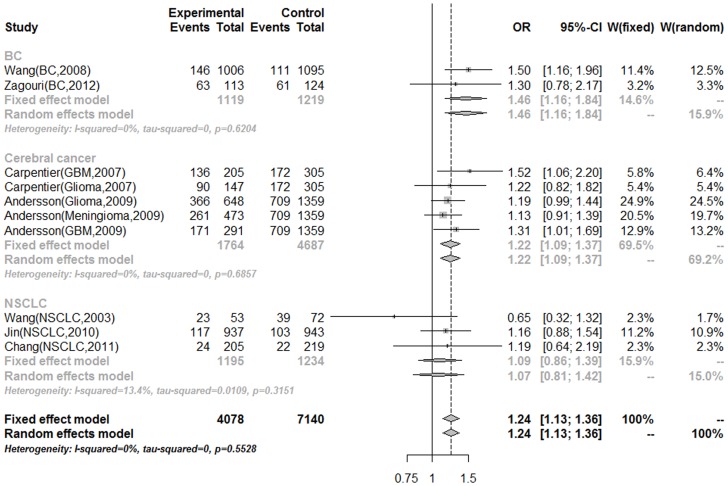
Then, we assessed the source of heterogeneity by cancer type (Table 3). On the basis of five cerebral cancer studies, there was no heterogeneity for all genetic models (P for heterogeneity > 0.1). Patients with MNS16A-S allele had a significant statistically association with cerebral cancer risk: with ORs of 1.42 (95%CI = 1.19–1.70), 1.22 (95%CI = 1.09–1.37), 1.32 (95%CI = 1.11–1.56) for SS versus LL genotype, dominant and recessive model (P for heterogeneity > 0.1). For breast cancer, patients carried with LS genotype had higher risk than SS genotype, which ORs and 95%CI were 1.52 (1.19–1.94) and 1.46 (1.16–1.84) for LS versus LL genotype and dominant models. However, no statistically significant associations were observed with lung cancer patients (Figure 3).
Figure 3. Forest plot of MNS16A association with cancer risk under dominant model stratified by cancer type.
Cumulative meta-analysis
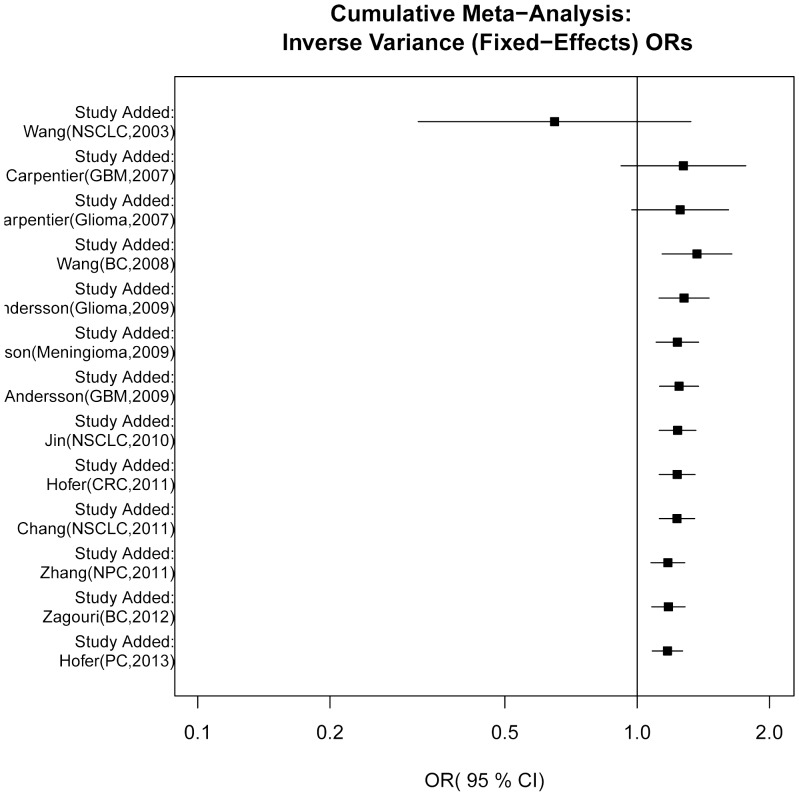
Cumulative meta-analyses of MNS16A were conducted via an assortment of studies in chronologic order. Figure 4 shows the results from the cumulative meta-analyses in fixed-effects model. The effect of MNS16A tended to show a significant association over time in all genetic models. Moreover, the 95% CIs became increasingly narrow with increasing data, suggesting that the precision of the estimates was progressively boosted by continually adding more studies.
Figure 4. Cumulative meta-analysis of association MNS16A with risk of cancer under dominant model.
Sensitivity analysis
Since moderate heterogeneity was observed under the genotypic model of LS versus LL and dominant models, we conducted a sensitivity meta-analysis to assess effects of each study on the combined ORs and 95% CIs. A random-effect model was employed since heterogeneity was indicated. Sensitivity analysis indicated the independent study contributing the most heterogeneity was conducted by Zhang et al. The heterogeneity was completely reduced by exclusion of that study: under the genotypic model of LS versus LL, ORs = 1.15 (95%CI = 1.03–1.28, P for heterogeneity = 0.102, I2 = 35.0%) and ORs = 1.20 (95%CI = 1.10–1.31, P for heterogeneity = 0.656, I2 = 0.00%) before and after removal, respectively. Omission of studies by Andersson et al. changed the pooled ORs fractionally (Table 4).
Table 4. Pooled ORs with 95% CIs for the association between MNS16A and cancer risk by omitting each article in sensitivity analysis.
| First author | Year | Genetic model | ORs (95%CI) | P a | P for | I2 |
| heterogeneity | ||||||
| Wang [14] | 2003 | LS vs. LL | 1.16 (1.07–1.27) | 0.000 | 0.136 | 31.8% |
| Dominant | 1.19 (1.07–1.33) | 0.001 | 0.097 | 36.7% | ||
| Carpentier [16] | 2007 | LS vs. LL | 1.14 (1.01–1.29) | 0.028 | 0.091 | 38.8% |
| Dominant | 1.15 (1.02–1.30) | 0.023 | 0.053 | 44.9% | ||
| Wang [19] | 2008 | LS vs. LL | 1.12 (1.03–1.23) | 0.009 | 0.224 | 22.3% |
| Dominant | 1.15 (1.06–1.25) | 0.001 | 0.122 | 33.5% | ||
| Andersson [15] | 2009 | LS vs. LL | 1.15 (0.97–1.36) | 0.103 | 0.034 | 50.3% |
| Dominant | 1.15 (0.98–1.37) | 0.187 | 0.023 | 53.4% | ||
| Jin [18] | 2010 | LS vs. LL | 1.16 (1.03–1.31) | 0.017 | 0.076 | 39.8% |
| Dominant | 1.18 (1.04–1.33) | 0.009 | 0.044 | 45.3% | ||
| Hofer [21] | 2011 | LS vs. LL | 1.15 (1.02–1.29) | 0.019 | 0.077 | 39.5% |
| Dominant | 1.18 (1.04–1.32) | 0.007 | 0.044 | 45.3% | ||
| Zhang [22] | 2011 | LS vs. LL | 1.20 (1.10–1.31) | 0.000 | 0.656 | 0.00% |
| Dominant | 1.23 (1.13–1.33) | 0.000 | 0.719 | 0.00% | ||
| Chang [17] | 2011 | LS vs. LL | 1.15 (1.03–1.29) | 0.016 | 0.075 | 39.8% |
| Dominant | 1.18 (1.05–1.32) | 0.006 | 0.044 | 45.3% | ||
| Zagouri [20] | 2012 | LS vs. LL | 1.15 (1.02–1.28) | 0.018 | 0.097 | 36.7% |
| Dominant | 1.17 (1.04–1.31) | 0.008 | 0.046 | 45.0% | ||
| Hofer [23] | 2013 | LS vs. LL | 1.15 (1.02–1.31) | 0.027 | 0.074 | 40.0% |
| Dominant | 1.17 (1.03–1.33) | 0.015 | 0.044 | 45.3% |
P value was calculated by the Z test.
Publication bias
As reflected by either visualization of funnel plot or Egger’s and Begg’s test, there was no indication of publication bias in the genotypic models of LS versus LL, SS versus LL, dominant, and recessive model (P = 0.482, P = 0.537, P = 0.551, and P = 0.745, respectively), indicating the results were statistically robust.
Grading of associations
Based on the previously proposed guidelines and applying the Venice criteria, the amount of evidence was categorized as A, since its nminor is above 1,000 (nminor = 2558); replication was assigned to category B, because the amount of between-study heterogeneity (I2 = 40.5%); and protection from bias was graded as category B, due to the presence of summary ORs less than 1.15, which can easily be dissipated even by relatively small biases in a meta-analysis of published data. The overall assessment of association between MNS16A and cancer risk were moderate cumulative evidence (ABB). After stratification by ethnicity, the meta-analysis consistently showed a significant association cancer risk in Caucasian population and were assigned an overall strong epidemiological credibility (AAA). Asian population lacked of statistically significant findings and was placed to weak evidence. In addition, strong epidemiological credibility (AAA) was also observed for association between MNS16A with cerebral cancer and breast cancer (Table 5).
Table 5. Assessment of cumulative evidence for the association of MNS16A dominant model and risk of cancer.
| No. of | No. of | ORs | P for | P for | I2 | Level of evidence | |
| Case | Control | 95% CI | Z test | Heterogeneity | |||
| Overall | 6101 | 10521 | 1.17 (1.05–1.31) | 0.006 | 0.064 | 40.5% | ABB/Moderate |
| Caucasian | 3155 | 7245 | 1.19 (1.09–1.31) | 0.000 | 0.696 | 0.00% | AAA/Strong |
| Asian | 2946 | 3276 | 1.10 (0.76–1.57) | 0.621 | 0.003 | 78.7% | Weak |
| Cerebral Cancer | 1263 | 2974 | 1.22 (1.09–1.37) | 0.000 | 0.686 | 0.00% | AAA/Strong |
| Lung Cancer | 177 | 157 | 1.07 (0.81–1.42) | 0.627 | 0.229 | 32.2% | Weak |
| Breast Cancer | 1119 | 1219 | 1.46 (1.16–1.84) | 0.002 | 0.620 | 0.00% | AAA/Strong |
Abbreviation: NSCLC, non-small cell lung cancer; A, B, and C represent the Venice criteria grades for amount of evidence, replication of association and protection from bias, which ultimately define the level of cumulative evidence (strong, moderate, weak).
Comparison with previously published meta-analyses
In a meta-analysis that regarding all hTERT locus polymorphisms with cancer susceptibility, Simone et al. investigated that MNS16A S allele was statistically associated with increased risk of central nervous system tumors (CNS). In comparison, our meta-analysis added more publications to consider association of MNS16A with all available type of cancer; analyzed data in different MNS16A classification system (LS and LMS classification system); stratified ethnicity and cancer types for further research.
Discussion
A number of well designed genome wide association studies (GWAS) had implicated variants at hTERT locus to be significantly associated with almost all malignant tumors [33]. MNS16A, a 23 bp (or 26 bp) tandem repeat sequence (TCCTCTTAT (cat) CTCCCAGTCTC) in putative promoter region of the antisense RNA transcript, was first reported to increase expression of hTERT mRNA in lung cancer tissues. In the current study, we conducted a meta-analysis of 10 previously published articles comprising 6101 cases and 10521 controls concerning association of MNS16A with cancer risk. Although all genetic models of MNS16A showed a moderate association with cancer risk, the effect could very well be driven by the effect on cerebral cancer. Thereupon, we stratified cancer types and found cerebral cancer and breast cancer patients showed strong cumulative evidence for associations, but lung cancer was not. Apart from this, ethnicity was also stratified in this work. Caucasian population presented a significantly increased relationship with cancer risk, whereas Asians not. The variance of effect between Caucasians and Asians might be contribute to that approximate 70% Caucasians were cerebral cancer, while similarly the absence of effect in Asians might well be due to the fact that only non-cerebral cancer were carried in this population. In addition, there was almost no obviously heterogeneity by stratified for cancer type, which suggested differential effects of MNS16A in diverse kinds of cancer. However, functional importance of the antisense transcript activity and exact molecular mechanisms of MNS16A with different cancer types were still unclear.
In this work, we analyzed data in different classification system: LS and LMS (described by Hofer et al. [21]) classification system for further excavation. The results figured that S allele had higher relationship than M allele with MNS16A. The cause might due to length of MNS16A: M allele contains three 26 bp repeats; whereas S alleles contain two 26 bp repeats. Hence we could see that, 26 bp sequence may influence as a repressor for promoter of antisense TERT mRNA [18]. It is more reasonable to analysis MNS16A S allele and M allele separately in future research to find accurate genotype with cancer risk.
Through sensitivity analysis, omission of one article by Zhang et al. eliminated heterogeneity of LS versus LL genotype and dominant models (P for heterogeneity > 0.1). The reason might due to lower frequencies of S allele in Asians. Additionally, omission article by Carpentier, the ORs were still presented increased risk, and 95%CI were nearby statistically significant (OR = 1.15, 95%CI = 1.03–1.28; OR = 1.14, 95%CI = 1.01–1.29, before and after removal), which not meaningfully changed the pooled ORs, as well as the article by Andersson.
Some limitations needed serious consideration. First, our result was based on unadjusted estimates. Individual data were not available for an adjusted estimate by age and sex, which might potentially lead to false positive results. Another limitation was lacking original data to limit our further evaluation of gene-environment interaction such as smoking, alcohol use and other clinical characteristics. Finally, lacking of sufficient original studies limited our further evaluation of colorectal cancer, breast cancer and nasopharyngeal carcinoma risk with MNS16A.
Conclusion
This work verified the important role of MNS16A minisatellites in cerebral and breast cancer predisposition. Additional larger studies were warranted to validate our findings.
Supporting Information
(DOC)
Acknowledgments
This work was copyedited by Helen Neumann from Cell Stress & Chaperones Editorial Office and Cell Stress Society International Dept. of Molecular & Cell Biology, University of Connecticut.
Funding Statement
The authors have no support or funding to report.
References
- 1. Greider CW (1991) Chromosome first aid. Cell 67: 645–647. [DOI] [PubMed] [Google Scholar]
- 2. Murnane JP (2006) Telomeres and chromosome instability. DNA Repair (Amst) 5: 1082–1092. [DOI] [PubMed] [Google Scholar]
- 3. Ju Z, Rudolph KL (2006) Telomeres and telomerase in cancer stem cells. Eur J Cancer 42: 1197–1203. [DOI] [PubMed] [Google Scholar]
- 4.Cong YS, Wright WE, Shay JW (2002) Human telomerase and its regulation. Microbiol Mol Biol Rev 66: : 407–425, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blackburn EH (2001) Switching and signaling at the telomere. Cell 106: 661–673. [DOI] [PubMed] [Google Scholar]
- 6. Satyanarayana A, Manns MP, Rudolph KL (2004) Telomeres and telomerase: a dual role in hepatocarcinogenesis. Hepatology 40: 276–283. [DOI] [PubMed] [Google Scholar]
- 7. Vaziri H, Benchimol S (1996) From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: the telomere loss/DNA damage model of cell aging. Exp Gerontol 31: 295–301. [DOI] [PubMed] [Google Scholar]
- 8. Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, et al. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. [DOI] [PubMed] [Google Scholar]
- 9. Broccoli D, Young JW, de Lange T (1995) Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci U S A 92: 9082–9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lantuejoul S, Salon C, Soria JC, Brambilla E (2007) Telomerase expression in lung preneoplasia and neoplasia. Int J Cancer 120: 1835–1841. [DOI] [PubMed] [Google Scholar]
- 11. Wick M, Zubov D, Hagen G (1999) Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 232: 97–106. [DOI] [PubMed] [Google Scholar]
- 12. Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, et al. (1995) The RNA component of human telomerase. Science 269: 1236–1241. [DOI] [PubMed] [Google Scholar]
- 13. Stanta G, Bonin S, Niccolini B, Raccanelli A, Baralle F (1999) Catalytic subunit of telomerase expression is related to RNA component expression. FEBS Lett 460: 285–288. [DOI] [PubMed] [Google Scholar]
- 14. Wang L, Soria JC, Chang YS, Lee HY, Wei Q, et al. (2003) Association of a functional tandem repeats in the downstream of human telomerase gene and lung cancer. Oncogene 22: 7123–7129. [DOI] [PubMed] [Google Scholar]
- 15. Andersson U, Osterman P, Sjostrom S, Johansen C, Henriksson R, et al. (2009) MNS16A minisatellite genotypes in relation to risk of glioma and meningioma and to glioblastoma outcome. Int J Cancer 125: 968–972. [DOI] [PubMed] [Google Scholar]
- 16. Carpentier C, Lejeune J, Gros F, Everhard S, Marie Y, et al. (2007) Association of telomerase gene hTERT polymorphism and malignant gliomas. J Neurooncol 84: 249–253. [DOI] [PubMed] [Google Scholar]
- 17. Chang CC, Yu MC, Bai KJ, Chang JH, Lee CN, et al. (2011) The Analysis Between Functional Human Telomerase Reverse Transcriptase MNS16A Polymorphisms and the Risk of Developing Non-Small Cell Lung Cancer in the Taiwanese Population. Journal of Experimental and Clinica l Medicine 3: 3. [Google Scholar]
- 18. Jin G, Yoo SS, Cho S, Jeon HS, Lee WK, et al. (2011) Dual roles of a variable number of tandem repeat polymorphism in the TERT gene in lung cancer. Cancer Sci 102: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Hu Z, Liang J, Wang Z, Tang J, et al. (2008) A tandem repeat of human telomerase reverse transcriptase (hTERT) and risk of breast cancer development and metastasis in Chinese women. Carcinogenesis 29: 1197–1201. [DOI] [PubMed] [Google Scholar]
- 20. Zagouri F, Sergentanis TN, Gazouli M, Tsigginou A, Dimitrakakis C, et al. (2012) HTERT MNS16A polymorphism in breast cancer: a case-control study. Mol Biol Rep 39: 10859–10863. [DOI] [PubMed] [Google Scholar]
- 21. Hofer P, Baierl A, Feik E, Fuhrlinger G, Leeb G, et al. (2011) MNS16A tandem repeats minisatellite of human telomerase gene: a risk factor for colorectal cancer. Carcinogenesis 32: 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Zhang H, Zhai Y, Wang Z, Ma F, et al. (2011) A functional tandem-repeats polymorphism in the downstream of TERT is associated with the risk of nasopharyngeal carcinoma in Chinese population. BMC Med 9: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofer P, Zerelles J, Baierl A, Madersbacher S, Schatzl G, et al.. (2013) MNS16A tandem repeats minisatellite of human telomerase gene and prostate cancer susceptibility. Mutagenesis. [DOI] [PubMed]
- 24. Mocellin S, Verdi D, Pooley KA, Landi MT, Egan KM, et al. (2012) Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst 104: 840–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sagoo GS, Little J, Higgins JP (2009) Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med 6: e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Swartz MK (2011) The PRISMA statement: a guideline for systematic reviews and meta-analyses. J Pediatr Health Care 25: 1–2. [DOI] [PubMed] [Google Scholar]
- 27. Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ (2008) A navigator for human genome epidemiology. Nat Genet 40: 124–125. [DOI] [PubMed] [Google Scholar]
- 28. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 29. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 30. Tobias A (1999) Assessing the influence of a single study in meta-analysis. Stata Technical Bulletin 8: 7. [Google Scholar]
- 31. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langevin SM, Ioannidis JP, Vineis P, Taioli E (2010) Assessment of cumulative evidence for the association between glutathione S-transferase polymorphisms and lung cancer: application of the Venice interim guidelines. Pharmacogenet Genomics 20: 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baird DM (2010) Variation at the TERT locus and predisposition for cancer. Expert Rev Mol Med 12: e16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)