Abstract
MilB is a CMP hydrolase involved in the early steps of biosynthesis of the antifungal compound mildiomycin. An enzyme from the bacimethrin biosynthetic pathway, BcmB, is closely related in both sequence and function to MilB. These two enzymes belong to the nucleoside 2′-deoxyribosyltransferase (NDT) superfamily. NDTs catalyze N-glycosidic bond cleavage of 2′-deoxynucleosides via a covalent 2-deoxyribosyl-enzyme intermediate. Conservation of key active site residues suggests that members of the NDT superfamily share a common mechanism; however, the enzymes differ in their substrate preferences. Substrates vary in the type of nucleobase, the presence or absence of a 2′-hydroxyl group, and the presence or absence of a 5′-phosphate group. We have determined the structures of MilB and BcmB and compared them to previously determined structures of NDT superfamily members. The comparisons reveal how these enzymes differentiate between ribosyl and deoxyribosyl nucleotides or nucleosides, and among different nucleobases. The 1.6 Å structure of the MilB-CMP complex reveals an active site feature that is not obvious from sequence comparisons alone. MilB and BcmB that prefer substrates containing 2′-ribosyl groups have a phenylalanine positioned in the active site whereas NDT family members with preference for 2′-deoxyribosyl groups have a tyrosine residue. Further studies show that the phenylalanine is critical for MilB and BcmB specificity towards CMP, and mutation of this phenylalanine residue to tyrosine results in a 1000-fold reversal of substrate specificity from CMP to dCMP.
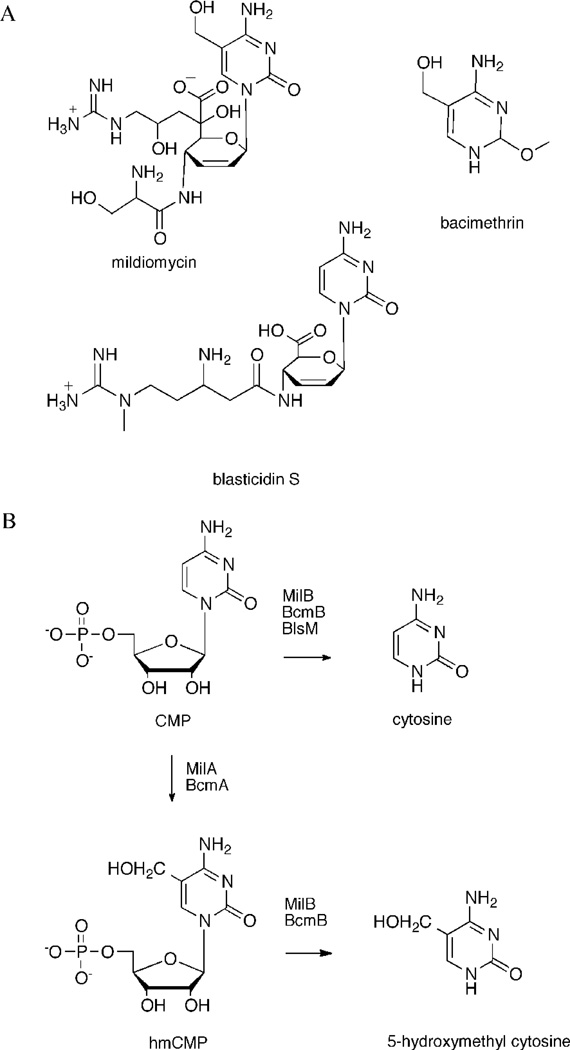
MilB is an enzyme involved in the biosynthesis of the natural product mildiomycin (Figure 1A), a commercially available antifungal agent isolated from treptomyces rimofaciens.1–3 MilB preferentially hydrolizes the N-glycosidic bond of 5-hydroxymethyl cytidine 5′-monophosphate (hmCMP) to release 5-hydroxymethyl cytosine, which is later incorporated into mildiomycin (Figure 1B).1 Previous studies found that MilB also hydrolyzes cytidine 5′-monophosphate (CMP) to its cytosine and ribose 5′-phosphate (Figure 1B).1 MilB is a member of the 2′-deoxynucleoside ribosyltransferase (NDT) superfamily. Typically, NDTs catalyze the reversible transfer of 2′-deoxyribose from a donor 2′-deoxynucleoside to an acceptor base. Their structural and functional properties have been investigated in detail.4–6 NDTs are found in a limited number of organisms and are thought to be important for nucleoside recycling when alternative nucleoside salvage pathways do not exist.5 In contrast to most NDTs, MilB participates in a hydrolysis reaction, in which the acceptor base is replace by a water molecule. While NDT exhibits hydrolysis activity in the absence of an acceptor base, the hydrolysis activity is about 100-fold lower than its transferase reaction.6
Figure 1.
Natural products with cytosine precursors. (A) Cytosine, or a cytosine derivative, is incorporated in the natural products mildiomycin, bacimethrin and blasticidin S. (B) The first steps in their biosyntheses involve the enzymatic release of cytosine or hydroxymethylcytosine from a CMP precursor by a CMP hydrolase enzyme.
BLAST analyses starting with the MilB sequence suggest homology to BcmB from Clostridium botulinum (47% sequence identity). Like MilB, BcmB also shows CMP hydrolase activity, but is utilized in a different biosynthetic pathway. BcmB is responsible for the production of the 5-hydroxymethylcytosine precursor for the biosynthesis of bacimethrin, a thiamin antimetabolite found in C. botulinum (Figure 1) (T.P. Begley, manuscript in preparation). While MilB and BcmB both belong to the NDT superfamily, they differ from other members of this family in their substrate preferences. Most enzymes containing the NDT-motif are specific for 2′-deoxynucleosides.4, 7 MilB and BcmB hydrolize hmCMP, a ribonucleotide containing 5′-monophosphate and 2′-hydroxy groups.
In addition to MilB and BcmB, several other enzymes containing the NDT motif function as nucleoside transferases. BlsM from Streptomyces griseochromogenes (52% sequence identity to MilB) is a nucleotide hydrolase, with CMP as the preferred substrate. The cytosine produced from the BlsM reaction is incorporated into blasticidin S, a commercially available antibiotic compound (Figure 1).8 Rcl, another member of the NDT superfamily, catalyzes the hydrolysis of purine 2′-deoxyribosylnucleotides and is inhibited by ribonucleotides 9. Its solution structure10, 11, and recent crystal structures with bound inhibitors12, provide highly detailed information about its substrate-enzyme interactions. Rcl is an unusual because it is the only NDT family member found in mammals. Rcl is upregulated in some cancers and contributes to tumorigenesis; however, the details of its cellular function are unclear.13,14
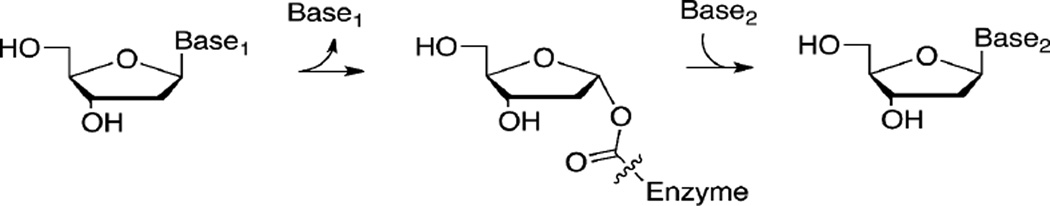
Because members of the NDT superfamily are not widely distributed and they show significant variation in substrate specificity, their evolution is not clearly understood. Conservation of key active site residues suggests a common catalytic mechanism even though the substrates for different family members vary in their preference for ribosyl or 2′-deoxyribosyl groups, 5′-hydroxyl or 5′-phosphate groups, and the type of nucleobase. Scheme 1 shows the reaction for NDT with a deoxyribosyl covalent intermediate, where Base1 and Base2 may be any purine or pyrimidine base. Each half reaction proceeds with inversion of stereochemistry at C1′, thus, the full reaction proceeds with retention of stereochemistry.
Scheme 1.
Here we report the X-ray structures of MilB and BcmB, and of MilB complexed with CMP. Comparisons with the structures of NDT6 and Rcl10–12 revealed a structural basis, which was not obvious from sequence comparisons alone, by which NDT family members differentiate between ribosyl and 2′-deoxyribosyl substrates. The selectivity is attributed to a phenylalanine residue found in MilB and BcmB that is replaced by tyrosine in NDT and Rcl. In support of this hypothesis, in vitro enzymatic activity assays showed that for both MilB and BcmB, mutation of the active site phenylalanine residue to tyrosine resulted in a reversal of substrate preference from CMP to dCMP.
MATERIALS AND METHODS
Cloning, Expression, and Purification of MilB and BcmB
All cloning procedures followed standard DNA manipulation methods. The S. rimofaciens ZJU5119 milB gene was commercially synthesized by DNA2.0 and bcmB was cloned using genomic DNA from C. botulinum A str. ATCC 19397. The milB and bcmB genes were inserted into pTHT, a modified pET-28 plasmid (Novagen) containing an N-terminal His6-tag and TEV protease recognition site. All mutant plasmids were prepared at the Cornell Protein Production and Purification Facility by site-directed mutagenesis of the native gene following standard, PCR-based mutagenesis.15
Escherichia coli B834(DE3), a methionine auxotrophic cell line, was transformed with the pTHT-milB wild-type (WT) plasmid. E. coli BL21(DE3) cells were used to express all other proteins. Starter cultures were grown from a single colony in 10 mL of sterile LB media containing 30 µg/mL kanamycin (LB-kan) at 37 °C overnight with shaking. Selenomethionine (SeMet)-containing MilB was prepared in 1.5 L- cultures in minimal media with 0.4% glucose, 1x MEM vitamin mix, 2 mM MgSO4, 0.1 mM CaCl2, 25 mg/L FeSO4, 20 mg/L all amino acids except methionine, and 50 mg/L of L-selenomethionine. To remove any LB media during SeMet protein preparation the overnight culture was centrifuged at 1000g for 20 min before the cell pellet was collected and resuspended in minimal media. The 1.5 L volume was then inoculated with 5 mL of starter culture and allowed to grow at 37 °C with shaking until reaching an OD600 of 0.6, at which point the incubator temperature was reduced. Once reaching an induction temperature of 15 °C and OD600 ∼0.8, protein expression was initiated by inoculating the cultures with 0.5 mM isopropyl β-D-thiogalactopyranoside. After 18 h, cells were centrifuged at 2000g for 20 min, collected, and frozen at −20 °C for storage. For native protein overexpression, 5 mL of overnight culture was added directly to 1.5 L of LB-kan media. The cells were then grown, collected and stored as described for the preparation of SeMet-MilB protein.
After thawing and resuspending the cell pellet in 45 mL of lysis buffer (50 mM Tris, 300 mM NaCl, and 20 mM imidazole at a pH of 8.0), the cells were lysed on ice with two rounds of sonication. The cell extract was centrifuged for 30 min at 40,000g at 4 °C after sonication. The supernatant was then collected and loaded onto a 2 mL Ni-NTA column (Qiagen) preequilibrated with the lysis buffer. The column was washed with 50 mL of lysis buffer to remove any nonspecifically bound contaminants. To elute the protein, the column was washed with elution buffer (50 mM TRIS, 300 mM NaCl, and 250 mM imidazole at a final pH of 8.0). The elution sample was collected and further purified using size exclusion chromatography (HiLoad Superdex 200 PG; GE Healthcare). The purification procedures for WT and mutant BcmB, WT and mutant MilB, and SeMet MilB all followed this protocol. The native MilbWT polyhistidine-tag was removed for crystallization by incubation with TEV protease for 12 h at 4 °C. The sample was passed over a Ni-NTA column preequilibrated with lysis buffer and the flow-through volume containing MilB was collected. The purity of the sample was analyzed by SDS-PAGE before concentration to ∼20 mg/mL. Protein was buffer exchanged by overnight dialysis into storage buffer (SeMet MilB: 50 mM Tris, pH 8.5, 500 mM NaCl; WT MilB: 10 mM Tris, pH 8.0, 100 mM NaCl; BcmB: 10 mM Tris pH 8, 50 mM NaCl) before flash freezing with liquid nitrogen for storage at −80 °C.
Crystallization of MilB and BcmB
The hanging-drop vapor diffusion method was used for crystallization of both MilB and BcmB. After mixing equal volumes of protein sample and reservoir solution, the samples were equilibrated overnight at 18 °C against a total reservoir volume of 500 µL. Protein concentrations for crystallization were 10 mg/mL for MilB and 15 mg/mL for BcmB. Initial crystallization conditions were determined using commercially available sparse matrix screens (Hampton Research, Emerald Biosystems).
SeMet-MilB crystals grew as hexagonal rods approximately 200 µm long and 40 µm wide under the optimized crystallization condition of 8% PEG 1000, 0.1 M phosphate-citrate pH 4.2, and 0.2 M Li2SO4 after two days. A second condition for MilB crystallization was found with the N-terminal polyhistidine tag removed. Square rods 400 µm long by 20 µm wide crystallized after two days in 0.1 M Tris pH 7.5, 0.2 M MgCl2, and 18% PEG 8000. The ligand soaked into the crystals added using a solution containing the components of the reservoir solution supplemented with 10 mM cytosine and 10 mM ribose 5-phosphate for 5 min before cryoprotection.
BcmB crystals formed 200 µm by 150 µm plates with 10 µm thickness in 18% PEG MME 2000, 0.1 M phosphate-citrate pH 4.4, 0.01 M spermine, and 0.2 M (NH4)2SO4 after five days. All crystals were cryoprotected with a solution consisting of their crystallization condition supplemented with 20% ethylene glycol before flash freezing with liquid nitrogen.
Data Collection and Processing
X-ray diffraction experiments were conducted at the Advanced Photon Source NE-CAT beamline 24-ID-C (Argonne National Lab) using a Quantum315 detector (Area Detector Systems Corp.) using vitrified crystals. A fluorescence scan was conducted to determine the data collection wavelength corresponding to the maximum f" for the SeMet-MilB crystals. All data collection runs used the oscillation method with 1.0° rotation per frame. HKL2000 was used for indexing, integrating and scaling the data.16 Data collection and processing statistics are listed in Table 1.
Table 1.
Summary of data collection and refinement statistics.
| SeMet MilB-SO4 | MilB-CMP | BcmB-PO4 | |
|---|---|---|---|
| beamline | APS 24-ID-C | APS 24-ID-C | APS 24-ID-C |
| resolution (Å) | 1.95 | 1.55 | 2.99 |
| wavelength (Å) | 0.97918 | 0.97918 | 0.97918 |
| space group | P6222 | C2 | C2 |
| a (Å) | 97.8 | 45.2 | 177.9 |
| b (Å) | 97.8 | 100.4 | 40.2 |
| c (Å) | 61.9 | 71.5 | 98 |
| β (°) | 90.0 | 99.6 | 98.0 |
| Matthews coefficient | 2.1 | 2.1 | 3.2 |
| % solvent | 41 | 41 | 61 |
| mol/asu | 1 | 2 | 3 |
| Measured reflections | 137124 | 162974 | 54150 |
| Unique reflections | 23697 | 44996 | 14604 |
| Average I/σ | 37.1 (5.3)a | 25.1(3.6) | 17.8 (4.7) |
| Redundancy | 5.7 (5.4) | 3.6(3.6) | 3.7(3.8) |
| Completeness (%) | 98 (97) | 99.8 (100) | 99.9(100) |
| Rsym (%)b | 5.4 (32.1) | 5.7 (35.2) | 10.0(34.2) |
| No. of protein atoms | 1164 | 2369 | 2700 |
| No. of ligand atoms | 10 | 41 | 15 |
| No. of water atoms | 96 | 196 | 10 |
| Reflections in working set | 23686 | 42663 | 14582 |
| Reflections in test set | 1168 | 2269 | 737 |
| R-factor/Rfree (%)c | 20.6/25.2 | 19.9/22.7 | 20.7/23.9 |
| rms deviation from ideals | |||
| bonds (Å) | 0.007 | 0.006 | 0.015 |
| angles (°) | 1.072 | 1.061 | 1.975 |
| average B factor for protein (Å2) | 28.1 | 18.0 | 49.1 |
| average B factor for water (Å2) | 34.1 | 25.6 | 54.5 |
| average B factor for ligand (Å2) | 31.3 | 17.0 | 84.2 |
| Ramachandram plot | |||
| most favored (%) | 97.3 | 98.0 | 96.3 |
| allowed (%) | 2.7 | 2.0 | 3.7 |
| disallowed (%) | 0.0 | 0.0 | 0.0 |
Values in parentheses are for the highest-resolution shell.
Rsym = ΣΣi | Ii – <I> | / Σ <I>, where <I> is the mean intensity of the N reflections with intensities Ii and common indices h,k,l.
Rwork = Σhkl| |Fobs| – k |Fcal| | / Σhkl |Fobs| where Fobs and Fcal are observed and calculated structure factors, respectively, calculated over all reflections used in the refinement. Rfree, is similar to Rwork but calculated over a subset of reflections (5%) excluded from all stages of refinement.
Structure Determination, Model Building, and Refinement
The crystal structure of MilB was solved by single-wavelength anomalous diffraction (SAD) using SeMet-MilB. The crystals belong to space group P6222 with a Matthews coefficient of 2.1 Å3 /Da and an estimated solvent content of 41%, assuming one protein chain per asymmetric unit. Initial phase calculation and model building were carried out using PHENIX AutoSol.18,19 Subsequent model improvement was performed using COOT for manual manipulations and PHENIX.refine with default parameters for structure refinement.19,20 Water molecules were added during later rounds of refinement in PHENIX and the final structure was analyzed using COOT and MolProbity.21 The structure of MilB complexed with CMP was determined in space group C2 with two protein chains per asymmetric unit. Under this new condition, MilB also crystallized with a Matthews coefficient of 2.1 Å3/Da corresponding to 41% solvent content. SKETCHER was used for ligand generation before placement into Fo – Fc electron density.22 Model improvement was performed as described above.
BcmB crystallized in space group C2 with three chains per asymmetric unit and a Matthews number of 3.2 Å3/Da, corresponding to a solvent content of approximately 61%. The structure of BcmB was determined using molecular replacement, with the newly solved MilB structure as the search model.23 MolRep placed three chains in the asymmetric unit and further model building and refinement was carried out using COOT and Refmac.20, 24 The three chains correspond to one dimer formed through noncrystallographic twofold symmetry and one dimer generated by crystallographic symmetry. Several rounds of refinement using Refmac and PHENIX.refine resulted in a BcmB model with R-factor and Rfree values of 21.7% and 24.2%, respectively. Refinement statistics for SeMet-MilB, MilB-CMP and BcmB are summarized in Table 1.
Analytical HPLC
Reaction mixtures were analyzed by reverse-phase HPLC on an Agilent 1200 HPLC system equipped with a quaternary pump and thermostatted autosampler (4 °C). The stationary phase was a Supelcosil LC-18-T column (15 cm × 4.6 mm, 3 µm particles). The LC eluent (1 mL/min flow rate) consisted of a gradient of methanol and 6 mM N,N-dimethylhexylamine (DMHA) in water that was adjusted to pH 6.5 by addition of acetic acid. In the analytical method the percentages of DMHA (D) and methanol (M) balanced with water at time t varied according to the following scheme: (t,M,D): (0,0,100), (2,10,80), (12,20,60), (14,65,10), (16,0,100), (21,0,100). Chromatograms were detected using the absorbance at 270 nm. UV-Vis spectra of substrate and product peaks were also collected and assessed.
Substrate Specificity for MilB and BcmB
Substrate specificity studies for MilB and MilB F17Y were carried out with adenosine 5′-monophosphate (AMP), CMP, 2′-deoxycytidine 5′-monophosphate (dCMP), guanosine 5′-monophosphate (GMP), and inosine 5′-monophosphate (IMP) (Figure S1). Experiments were designed based on the previously described assay conditions used for MilA/MilB 25. In vitro enzymatic assays were prepared with a final volume of 50 µL. AMP, CMP, dCMP, GMP, or IMP (1 mM) were added to MilB WT or MilB F17Y (10 µM) in 50 mM Tris buffer, pH 7.5 and then the samples were incubated at 37 °C for 3 h. Control samples omitting enzyme were also prepared. Reactions were quenched by boiling at 100 °C for 5 min. Samples were diluted with DMHA, filtered using a 3 kDa MWCO membrane, and analyzed by HPLC as described above. Reaction products were compared to commercially available standards that were analyzed under identical conditions. Substrate specificity studies for BcmB and BcmB F6Y were carried out with CMP and dCMP as described above.
Kinetics for MilB and BcmB
Kinetic parameters were monitored based on cytosine production from CMP or dCMP for MilB WT, MilB F17Y, BcmB WT, and BcmB F6Y. Each enzyme was incubated with varying substrate concentrations (see Table S1) in 50 mM Tris, pH 7.5 for 30 min at 37 °C and then the reactions were quenched by boiling at 100 °C for 5 min. Samples were diluted with DMHA, filtered using a 3 kDa MWCO membrane, and analyzed by HPLC as described above. Reaction products were compared to commercially available standards that were analyzed under identical conditions. The product absorbance signal was measured by integration (ChemStation, Agilent Technologies) and compared to cytosine standards of known concentration. Kinetic parameters were calculated using the Michaelis-Menten function in KaleidaGraph 4.0 (Synergy Software).
Figure Preparation
All figures were prepared using PyMOL 26, ChemDraw (Cambridge Biosoft), KaleidaGraph 4.0 (Synergy Software), or Excel (Microsoft), and compiled in Photoshop (Adobe).
RESULTS
Structure of MilB
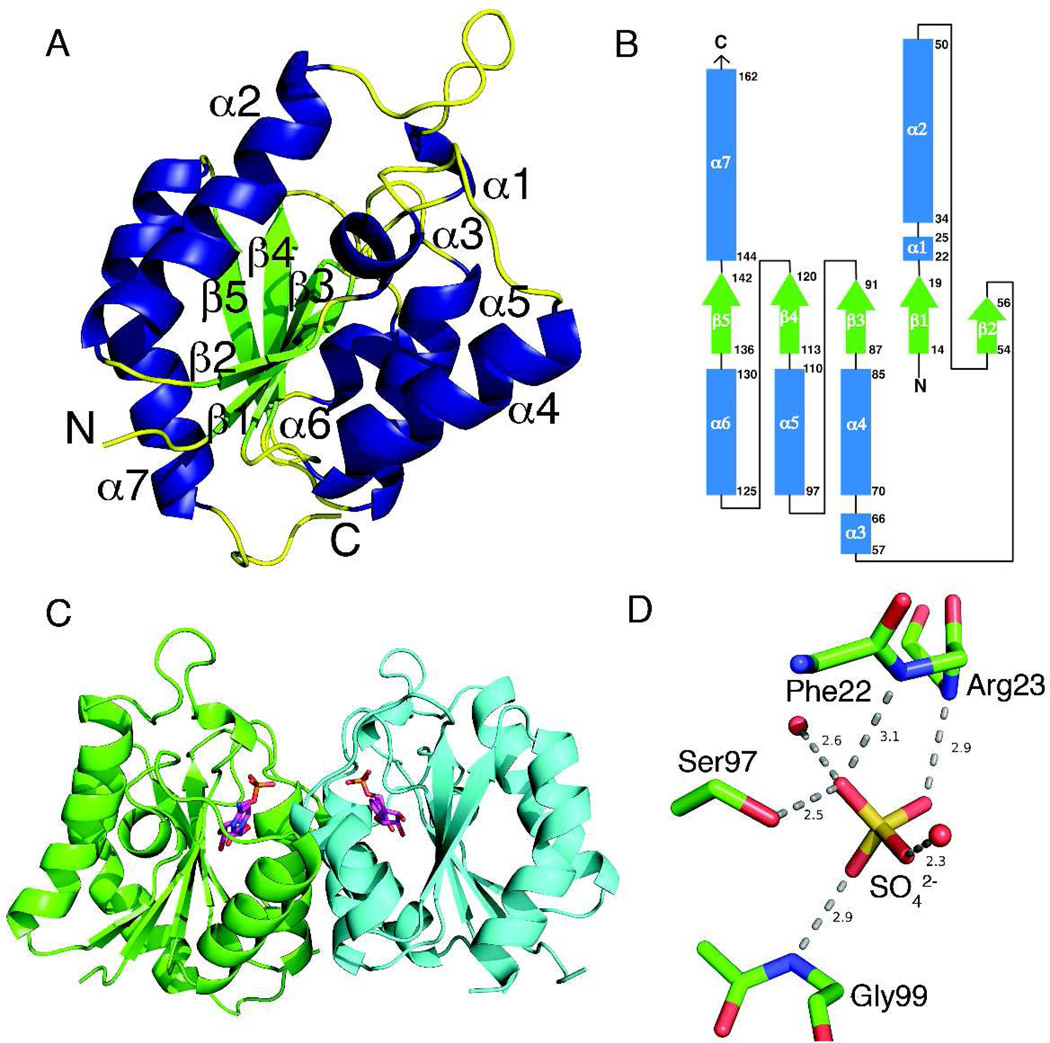
The MilB with bound SO4 contains one protomer per asymmetric unit; however, a dimer is formed by crystallographic twofold symmetry. The final model at 1.95 Å resolution contains residues 11–162 of the possible 170 amino acid residues. The protomer of MilB contains an α/β fold with a five-stranded parallel β-sheet with a strand order of 21345 (flavodoxin-like). The β-sheet is flanked on both sides by α-helices in an asymmetric manner (Figure 2A and 2B). Dimer formation occurs through interactions between helices α4, α5, and α6 of the two protomers with an interface area of 1170 Å2. Size exclusion chromatography confirmed the dimeric state of MilB (data not shown), which is consistent with the quaternary structures of both NDT (trimer of dimers) and Rcl.
Figure 2.
Structure of MilB. (A) The MilB protomer structure adopts an overall α/β-twist fold, depicted with β-strands in green and α-helices in blue. (B) The topology diagram of MilB with β-strands in light green and α-helices in light blue. (C) The dimeric structure of MilB with chain A in green and chain B in cyan and ligands bound in the active sites. (D) The active site phosphate-binding pocket is occupied by a sulfate ion in the MilB/SO4 complex. The sulfate binding site corresponds to the phosphate binding site in the CMP complex.
The MilB/CMP complex also crystallizes with one dimer per asymmetric unit. The final model at 1.55 Å resolution contains residues 12–168 of chain A and residues 10–162 of chain B. Each protomer is occupied by one CMP molecule and the overall structure is essentially the same as that of the MilB/SO4 complex. Each dimer features two equivalent active sites.
Mi lB Active Site
The MilB active site is located at the C-terminal edge of the β-sheet, between helices α4 and α5 (Figure 2C). All of the interactions necessary for substrate binding and catalysis are contained within a single MilB protomer, with no direct participation from the adjacent protomer. A phosphate/sulfate binding pocket contains hydrogen bonding interactions with the Ser97 sidechain, the amide groups of Gly99, Arg23, and Phe22, and two well ordered water molecules (Figure 2D). This binding site corresponds to the phosphate binding site in the CMP complex. Ordered water molecules occupy the rest of the active site in the MilB/SO4 structure.
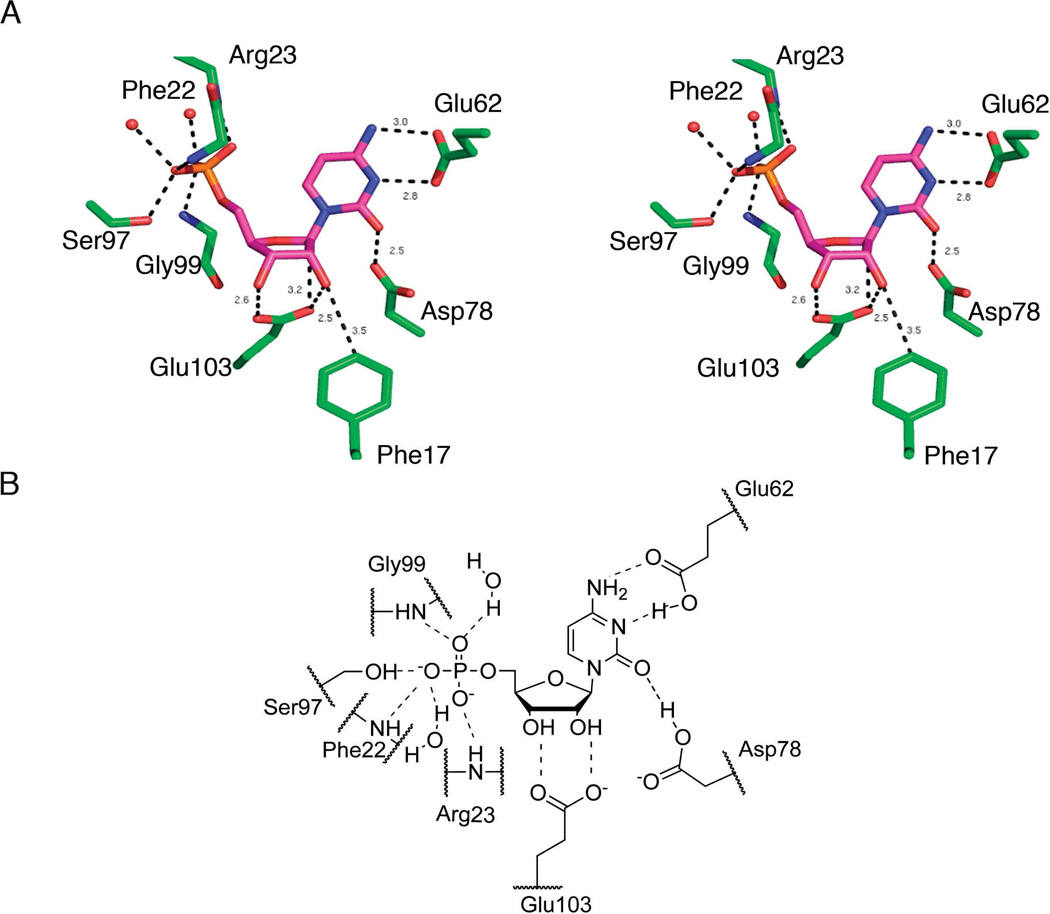
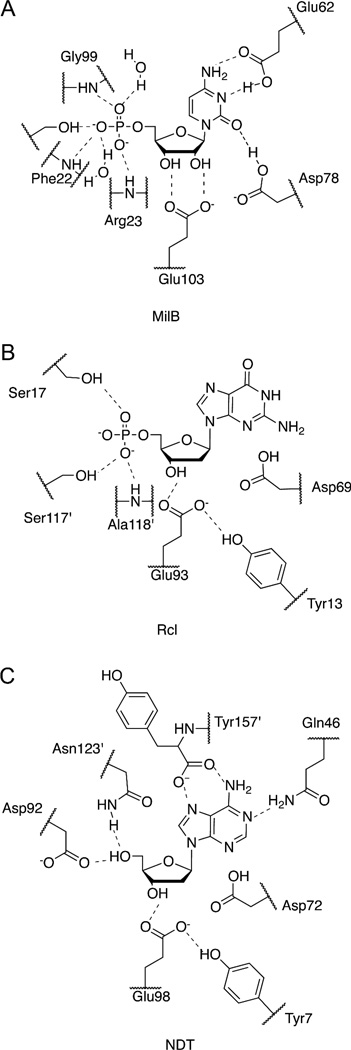
The structure of the MilB/CMP complex provides the molecular details of the nucleobase and ribosyl binding sites (Figure 3). The products of the CMP hydrolysis reaction, cytosine and ribose 5-phosphate, were soaked into the MilB crystals. Continuous electron density is observed in the active site representing CMP, the product of the reverse reaction, yet our in vitro studies did not show significant formation of CMP upon incubation of MilB, cytosine, and ribose 5-phosphate (data not shown). Thus, the presence of CMP in the MilB/CMP active site is a result of enzyme activity despite the fact that we could not detect activity in vitro. Glu62 accepts a hydrogen bond from the 4-amino group of the cytosine base and, if protonated, would donate a hydrogen bond to CMP N5. Glu103 accepts hydrogen bonds from the CMP 2'- and 3'-hydroxyl groups. Additionally, the side chain of Asp78 is within hydrogen bonding distance of the cytosine O2. Phe17 C4 is positioned 3.5 Å from the CMP 2'-oxygen in the MilB structure.
Figure 3.
Substrate binding in MilB. (A) MilB-CMP binding interactions represented in stick view. (B) Schematic diagram of the MilB active site with CMP bound.
Structure of BcmB
BcmB crystallized in space group C2, with three protein chains in the asymmetric unit. The structure of BcmB was determined to 3.0 Å resolution using the MilB structure (47% sequence identity) described above as the molecular replacement search model. The protomeric structure of BcmB displays the same α/β-twist fold as MilB, with five parallel β-strands forming a parallel β-sheet flanked on either side by α-helices (Figures S2A and S2B). The final BcmB model contains 120 amino acids in chain A, 123 in chain B, and 110 in chain C. The missing regions include residues 11–20 and 110–113 in chain A, 11–21 and 112 in B, and 10–24, and 109–112, which correspond to loop regions. Missing residues 47–55, 48–56, and 46– 58 in protomers A, B and C, respectively, correspond to a short α-helix and loop when aligned with MilB.
Chains A and B form a dimer by twofold noncrystallographic symmetry. The buried surface area at the interface is 1320 Å2. Chain C forms a dimer with an equivalent chain C using crystallographic twofold symmetry. The buried surface area of this interaction is 1210 Å2. This small discrepancy between the A/B and C/C interface surfaces areas can be attributed to additional disordered regions in chain C. Residues 58 and 59 in A and B and residue 114 in chain A are present at the A/B dimer interface, but are disordered at the C/C dimer interface. The oligomeric state of BcmB observed in the crystal structure is supported by size exclusion chromatography results (data not shown) and closely resembles the dimer formed by MilB (Figure S2C).
BcmB Active Site
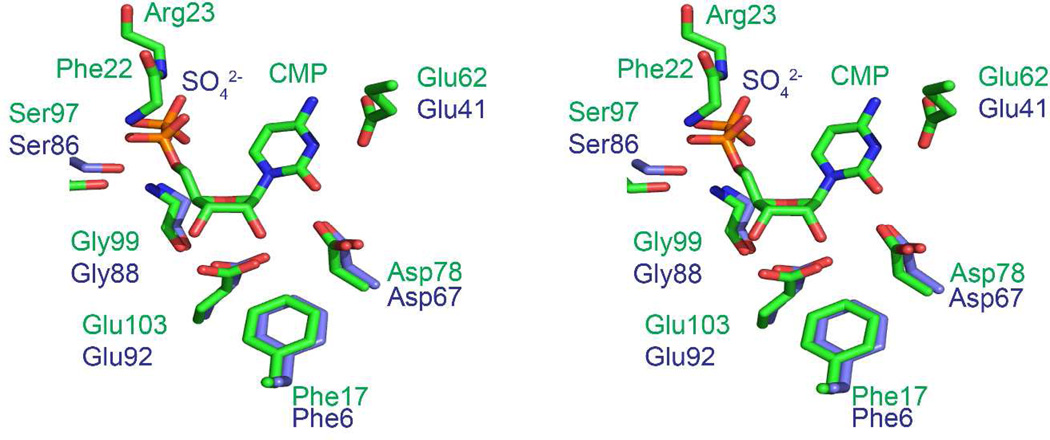
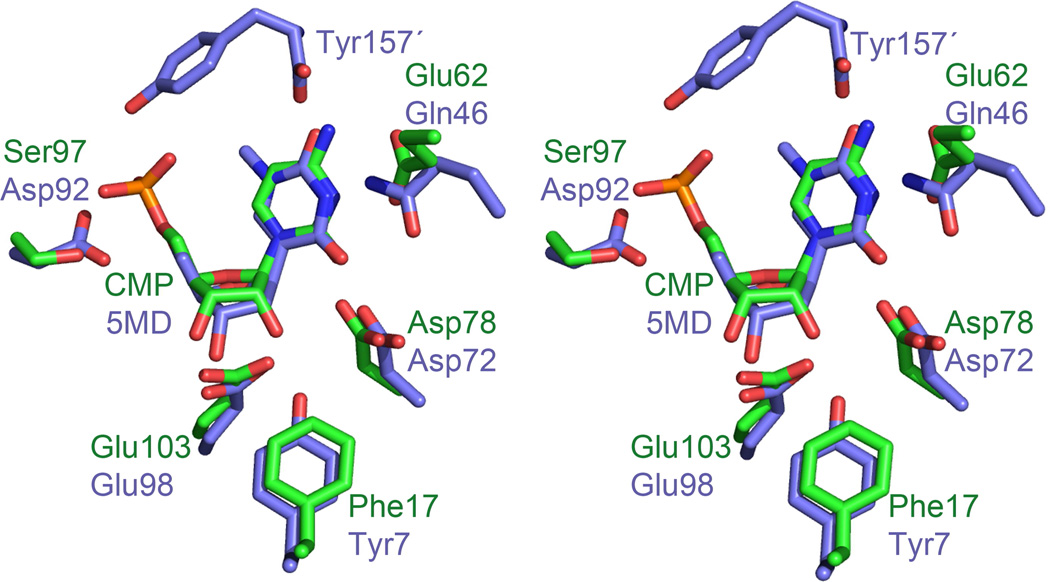
The active site of BcmB is similar to that of MilB. Each active site contains a bound phosphate ion from the crystallization solution. The phosphate makes hydrogen bonds with the side chain of Ser86 and the amide backbone of Gly88 in the phosphate-binding pocket (Figure S2D). Based on structural comparison to MilB, the BcmB active site residues that participate in substrate binding include Asp67, Glu92, and Phe6 (Figure 4). Glu41 falls in a disordered region in the BcmB crystal structure, but is expected to be equivalent to Glu62 in MilB on the basis of sequence alignments.
Figure 4.
BcmB/PO4 and MilB/CMP active site comparison. A stereoview of BcmB/PO4 and MilB/CMP active sites superimposed. The active site residues of BcmB and MilB display a conserved architecture. Glu41 is disordered in the BcmB structure but conserved in sequence with MilB Glu62.
Kinetics and Substrate Specificity for MilB and BcmB
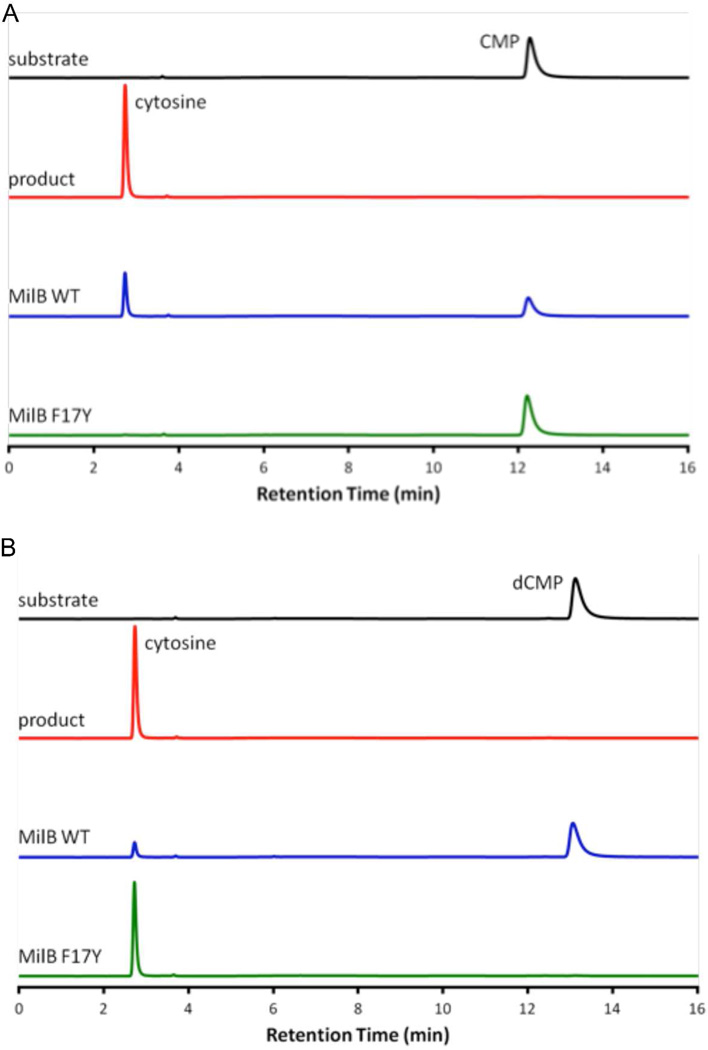
Initial studies determined the substrate specificity of MilB for cytosine containing nucleotides.1 MilB preferentially hydrolyzed CMP, but cytosine formation was also observed, to a lesser extent, from dCMP (Figure 5). MilB showed little to no hydrolysis of purine containing substrates (Figure S1). CMP or dCMP was incubated with MilB F17Y and the production of cytosine was monitored to measure N-glycosidase activity. MilB F17Y successfully cleaved dCMP but was no longer able to effectively hydrolyze CMP (Figure 5). Similar studies were conducted for both BcmB wild type (WT) and the corresponding active site Phe mutant (F6Y). As with MilB WT, BcmB WT can hydrolyze CMP and, to a lesser extent, dCMP (Table 2). Conversely, BcmB F6Y hydrolyzed dCMP with a 2'-deoxyribosyl group, while showing only minor activity with CMP (Figure S3).
Figure 5.
Substrate specificity of MilB WT and MilB F17Y. (A) When MilB WT is incubated with CMP a large peak representing free cytosine is observed. For MilB F17Y incubated with CMP, little hydrolysis is observed. (B) The opposite effect is observed with dCMP as substrate. MilB F17Y hydrolyzes dCMP producing free cytosine. There is comparably less dCMP hydrolysis by MilB WT.
Table 2.
Summary of kinetic data
| Protein (Substrate) | kcat (min−1) | Km (mM) | kcat/Km (M−1s−1) |
|---|---|---|---|
| MilB WT (CMP) | 1.19 (0.06) | 3.8 (0.5) | 5.2 (0.7) |
| MilB WT (dCMP) | 0.36 (0.02) | 5.4 (0.8) | 1.1 (0.2) |
| MilB F17Y (CMP) | 0.0126 (0.0003) | 4.5 (0.4) | 0.046 (0.004) |
| MilB F17Y (dCMP) | 7.6 (0.3) | 1.2 (0.2) | 100 (10) |
| BcmB WT (CMP) | 0.090 ( 0.003) | 0.19 (0.02) | 7.8 (0.9) |
| BcmB WT (dCMP) | 0.055 (0.001) | 0.57 (0.05) | 1.6 (0.1) |
| BcmB F6Y (CMP) | 0.0048 (0.0004) | 0.9 (0.2) | 0.09 (0.02) |
| BcmB F6Y (dCMP) | 0.081 (0.001) | 0.042 (0.004) | 32 (3) |
Steady-state kinetic parameters were determined for MilB WT, MilB F17Y, BcmB WT, and BcmB F6Y using either CMP or dCMP as substrates (Table 2). The kcat/Km value for MilB WT with hmCMP was previously reported as 22.1 M−1s−1.1 The kcat/Km values observed for MilB WT hydrolysis of CMP or dCMP were of 5.2 M−1 s−1 and 1.1 M−1 s−1, respectively.
DISCUSSION
Structure Comparison
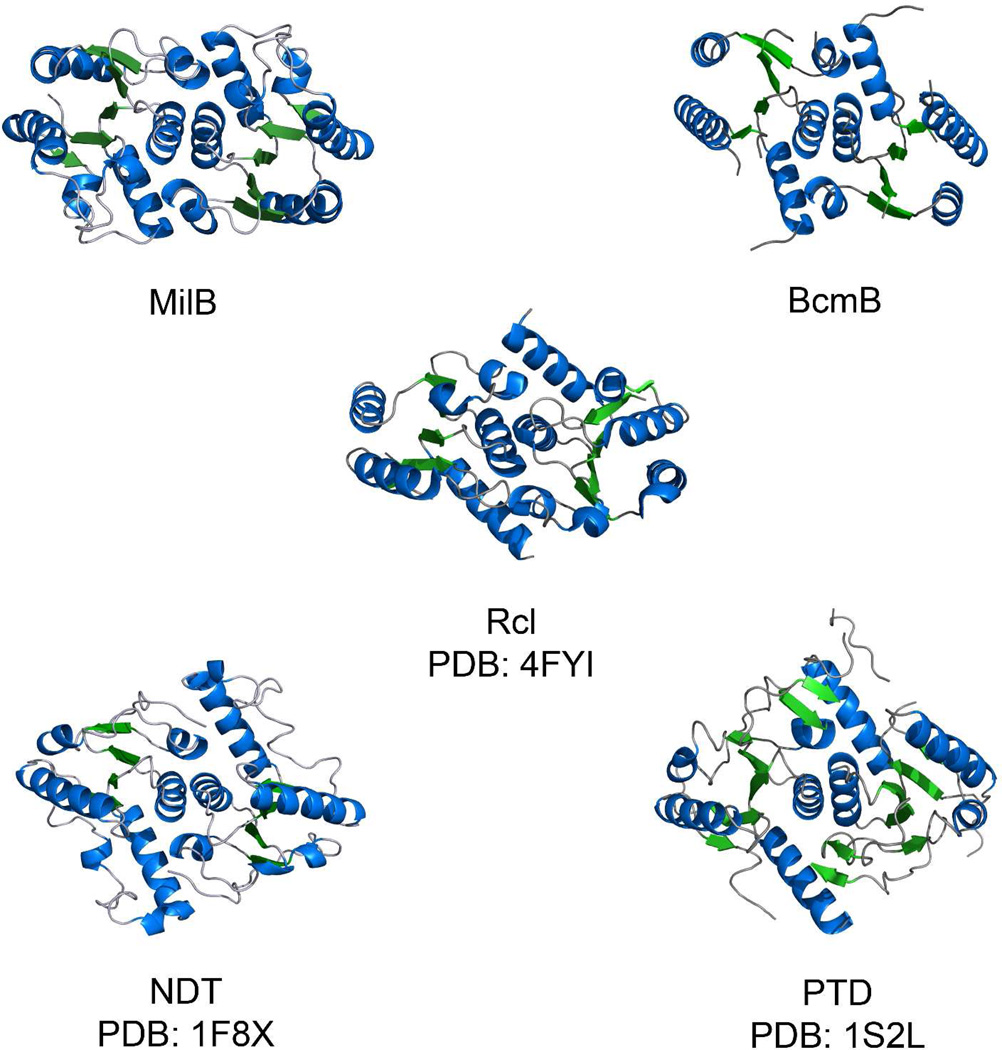
MilB and BcmB both adopt an α/β-fold composed of five parallel β-strands forming a core β-sheet flanked on either side by several α-helices. This fold is highly conserved among the NDT superfamily enzymes. A pairwise DALI comparison between the MilB and BcmB structures results in a Z-score of 16.9 and RMSD of 1.7, confirming their highly conserved tertiary structure (Table 3).27 A DALI structure search against the PDB starting with MilB reveals structural homology to the known 2′-deoxyribosyltransferases and Rcl (Table 3). Though their sequence identities are relatively low (11–22%), the enzymes retain similar folds. Based on a Z-score of 15.5 with a RMSD value of 2.3Å, MilB has the highest structural similarity to R. norvegicus Rcl (PDB ID 4FYI)12. Similar RMSD values of 2.5 Å and 2.9 Å are observed upon comparison of MilB to the Rcl solution structures with PDB IDs 2KHZ11 and 2KLH10, 11, respectively. Several 2′-deoxyribosyltransferases, such as the Trypanosoma brucei30 and L. leichmannii 6, 28, 29 NDTs (PDB ID 2F2T and 1F8X) and L. helveticus purine nucleoside 2′-deoxyribosyltransferase (PTD) (PDB ID 1S2L)29 are also among the most homologous structures.
Table 3.
Enzymes structurally similar to MilB
| Protein | PDB ID | Z score | rmsd | %identity | No. aligned residues |
|---|---|---|---|---|---|
|
C. botulinum BcmB |
4JEL | 16.9 | 1.7 | 46 | 123 |
|
R. norvegicus Rcl |
4FYI | 15.5 | 2.3 | 21 | 126 |
|
R. norvegicus Rcl (NMR) |
2KHZ | 14 | 2.5 | 22 | 134 |
|
R. norvegicus Rcl (NMR) |
2KLH | 13.7 | 2.9 | 21 | 131 |
| T. brucei NDT | 2F2T | 12 | 2.8 | 13 | 126 |
|
L. helveticus PTD |
1S2L | 11.9 | 2.4 | 19 | 122 |
|
L. leichmannii NDT |
1F8X | 10.6 | 2.8 | 11 | 123 |
While the enzymes in the NDT superfamily maintain a similar tertiary structure, their oligomeric assembly is not fully conserved. Both NDTs and PTD form a hexamer composed of a trimer of homodimers, while Rcl, MilB, and BcmB are observed to form only dimers.6, 11, 28 Comparison of the dimer interfaces reveals a conserved mode of interaction (Figure 6). Like many members of the NDT superfamily10, the interactions observed at the dimer interfaces in MilB and BcmB are mainly hydrophobic (>97%).
Figure 6.
Comparison of enzyme dimerization. MilB and BcmB are observed to be dimers. Like MilB, BcmB and Rcl have a dimeric tertiary structure. In their functional forms, NDT and PTD are hexamers composed of a trimer of dimers, in which the dimer (shown) is similar to that of MilB.
Base Specificity
Structural analysis of MilB, BcmB, and related enzymes reveals the basis of their substrate specificities. MilB shows specificity for the pyrimidine containing hmCMP or CMP substrates. It does not hydrolyze the purine containing nucleotides, demonstrated by an inability to hydrolyze AMP, GMP, or IMP (Figure S1). While highly specific for hmCMP or CMP, its relatives display variable nucleobase specificity (Table 4). NDT accepts all naturally occurring and some synthetic nucleosides, while PTD30 and Rcl7 are specific for purine containing nucleosides or nucleotides, respectively. In the structure of MilB/CMP, the Glu62 carboxylic group is positioned approximately 3 Å from the cytosine, forming hydrogen bonds with the cytosine N3 and 4-amine group (Figure 3). Helix α3 (on which Glu62 is located), helix α4, and the loop region formed by residues 67–70 shield the MilB active site. As a result, MilB has a relatively compact substrate-binding site that cannot accommodate the bulkier purine containing nucleotides. BcmB specificity is predicted to function similarly to MilB based on sequence and structural homology.
Table 4.
Summary of NDT-superfamily enzymes
| Enzyme | Preferred Base | 5'- specificity |
2'- specificity |
Acceptor Nucleophile |
|---|---|---|---|---|
| MilB | hydroxymethylcytosine/cytosine | phosphate | OH | water |
| BcmB | hydroxymethylcytosine/cytosine | phosphate | OH | water |
| BlsM | cytosine | phosphate | OH | water |
| Rcl | purine | phosphate | H | water |
| NDT | purine/pyrimidine | OH | H | purine/pyrimidine |
| PTD | purine | OH | H | purine |
The carboxylic acid from the NDT C-terminal Tyr157′ and Gln46 amine interact with its substrate nucleobase. NDT can accommodate a variety of nucleobases because of its flexibility in both accepting and donating hydrogen bonds.6 Like NDT, PTD interacts with nucleobases via hydrogen bonds from the carboxylic tail of the C-terminal tyrosine located in the active site. PTD diverges from NDT, as there are no equivalent interactions to those provided by Gln46 in NDT.28 The complexed Rcl structures 10–12 reveal a large nucleobase binding area with no specific enzyme-nucleobase interactions, allowing Rcl to accommodate a variety of nucleobases in its active site. Because the Rcl active site is rather open, it is solvent accessible and favors the larger purine containing substrates that can aid in shielding the active site from the solvent. As evidenced by the structures of MilB and other NDT superfamily members, their nucleobase specificities are derived from a combination of favorable enzyme-substrate hydrogen bonding interactions and active site accessibility.
Specificty at the 5′-position
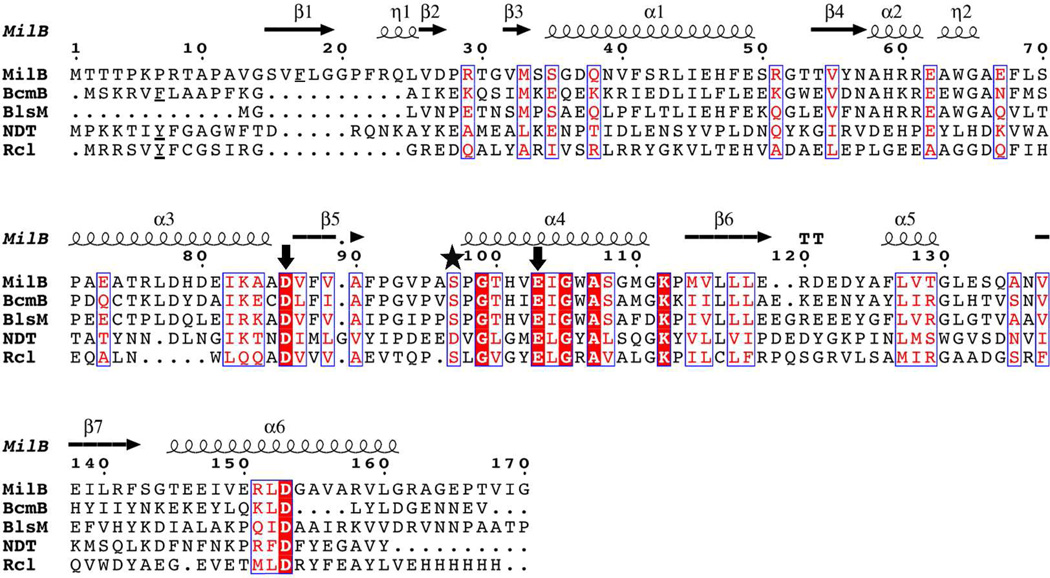
The NDT family members differ in their substrate preference for either a hydroxl or phosphate group at the 5′-position (Table 4). The high-resolution MilB/CMP structure reveals key information about the enzyme-substrate binding interactions at the CMP 5′-position, including seven hydrogen bonds. Two ordered water molecules each contribute one hydrogen bond, while Ser97 is hydrogen bonding distance from either of the two 5′-phosphate oxygen atoms. While the active site serine residue observed in MilB is conserved in both BcmB and BlsM (Figure 7), the remaining interactions in the MilB phosphate-binding pocket come from amide backbone groups and ordered water molecules. The amide backbones of Phe22, Arg23, and Gly99 act as hydrogen bond donors, completing the phosphate-binding pocket (Figure 8A). Although the identities of the backbone residues are not conserved in sequence, the phosphate ion in BcmB/PO4 is observed in an analogous binding pocket to MilB (Figure 8B). This pocket is also rich with hydrogen bond donors resulting is preference for phosphate groups at the 5′-position.
Figure 7.
Sequence alignment of enzymes containing the nucleoside 2′-deoxyribosyltransferase motif. Sequence comparison between MilB/BcmB and other family members reveals conservation of the key catalytic residues (arrows) but not of those conferring substrate-binding specificity. The serine residue involved in phosphate binding is indicated with a star. Phenylalanine and tyrosine residues known to be positioned in the active site are underlined. Representative sequences for NDT from L. leichmannii and Rcl from R. norvegicus were used.
Figure 8.
Active site comparison. (A) Observed MilB interactions with CMP. (B) Rcl active site with dGMP. (C) NDT active site with deoxyadenosine.
A 5′-phosphate binding pocket utilized for nucleotide binding was also observed in Rcl.10–12 The structure of Rcl shows hydrogen bonding contacts between the GMP phosphate group and Ser17, Arg19, Ser87, and Ser117′ (Figure 8B). This motif is highly conserved in the sequences of 13 different species in Rcl.10 While MilB does not share this motif, the positively charged pocket is consistent with preference for nucleotide monophosphate binding. In NDTs and PTD, an aspartate residue plus either an asparagine or glutamine residue stabilizes nucleoside 5′-hydroxyl groups, favoring nucleoside binding (Figure 8C)6, 28 and preventing the binding of 5′-phosphate containing nucleotides preferred by MilB and BcmB.4
Specificity at the 2′-position
While MilB and BcmB contain the conserved NDT motif, their ability to hydrolyze the N-glycosidic bonds of ribosylnucleotides makes them unique from other enzymes in this class. NDTs, PTD, and Rcl show no glycosidase activity towards ribosyl-containing substrates, which in some cases are inhibitory.9 Conversely, MilB and BcmB show higher activity towards substrates containing 2′-hydroxy groups (Table 2). In the related Rcl and NDT structures, a tyrosine residue is observed near the substrate 2′-binding position (Figure 8). Not obvious from sequence analysis alone, the structures of MilB and BcmB reveal a phenylalanine in the position equivalent to this tyrosine near the conserved catalytic glutamate (Figure 7). Compared to Rcl or NDTs, this phenylalanine residue is the only residue near the 2′-binding position that differs in either MilB or BcmB.
The identity of the active site phenylalanine or tyrosine residue proves to be important in differentiating between ribosyl or deoxyribosyl containing substrates by MilB and BcmB. Structural studies of PTD28, which has a tyrosine residue near the 2′-binding position, suggested that ribosylated substrates form hydrogen bonds with the catalytic glutamate, rendering it unreactive. This rational contradicts our findings in which MilB and BcmB react with ribosylated substrates, despite also showing interactions between their catalytic glutamate and the substrate 2′-hydroxy group. When both a hydroxy containing substrate and active site tyrosine are present, the glutamate may be too constricted by hydrogen bonds to position itself for catalysis. In the absence of an active site tyrosine, the catalytic glutamate is not restricted by the ribosylated substrate. Rather, the ribosylated substrate may help orient the glutamate for catalysis.
Acceptor/Nucleophile Binding Site
Previous studies of NDT-superfamily members have explored the structural basis of nucleophile binding at the active site. In NDT, the interaction between Gln46 and the nucleobase positions the loop region, on which Gln46 is located, shielding the active site from solvent.28 Along with a hydrophobic binding pocket, this excludes water as a nucleophile. After initial N-glycosidic bond cleavage and release of the base, NDT binds the incoming acceptor base, positioning it for the second half of the NDT transferase reaction. Due to more limited substrate-enzyme interactions in PTD, the corresponding loop region does not shield its active site28 and bulkier purine bases that shield the active site are favored over smaller pyrimidines, both as substrate and acceptor nucleobases. Compared to NDT, Rcl has a longer loop region that does not shield the active site from solvent. It also does not have any residues that specifically interact with and bind acceptor nucleobases.10–12 These structural features allow a water molecule to act as the final acceptor. Because the MilB active site is shielded by α-helices, it is less solvent accessible as compared to PTD or Rcl. Compared to NDT, the MilB active site is similarly shielded, but is not as hydrophobic. As a result, an active site water molecule is available to act as the final acceptor. Accordingly, the MilB/CMP structure reveals a water molecule in the active site positioned 4.9 Å from the 1′-ribosyl position.
Mechanism of Hydrolysis
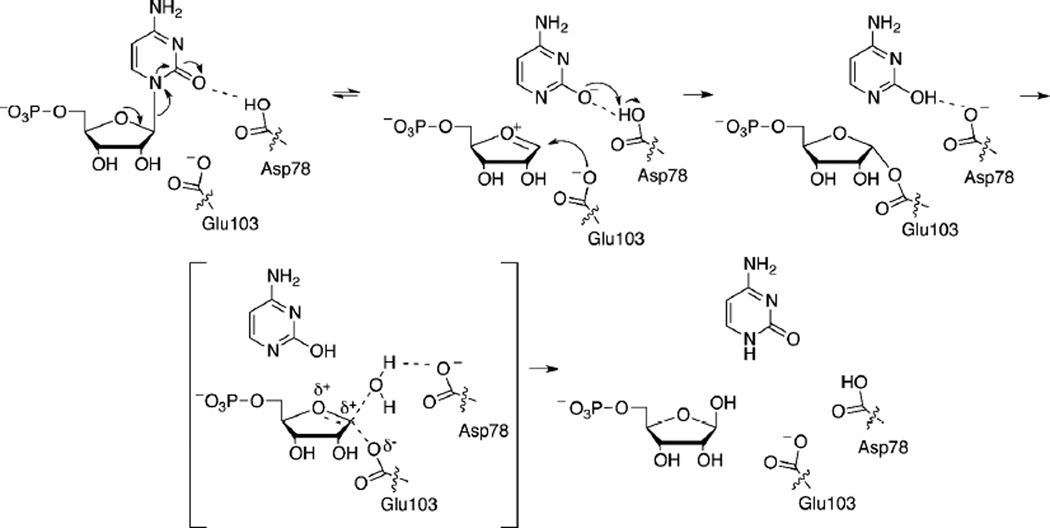
Previous studies showed that NDT family members catalyze transferase reactions utilizing a covalent enzyme-(deoxy)ribosyl intermediate.6, 9, 31 In the first half reaction the covalent intermediate forms between a glutamate side chain and C1′ of the (deoxy)ribosyl group (Scheme 1). The second half reaction takes place after the purine/pyrimidine base is released and a nucleophile (either a different base or a water molecule) displaces the glutamate side chain. Each half reaction is inverting at C1′ such that the stereochemistry at C1′ is retained for the full reaction. The catalytic glutamate residue is absolutely conserved among the NDTs, Rcl, MilB, BcmB, and BlsM (Figure 8). Based on this mechanism, Glu103 of MilB forms a covalent attachment to CMP at the ribosyl C1′-position in the first half of the hydrolysis reaction (Scheme 2). Previous studies on xylanases, which utilize a glutmate/glycoside covalent intermediate in a hydrolysis reaction similar to that of MilB, showed that each half reaction proceeds through an oxocarbenium-like transition state.32 In the crystal structure of the MilB/CMP complex the glutamate oxygen atom is not correctly oriented for a strictly SN2 mechanism suggesting the formation of an oxocarbenium ion intermediate followed by trapping of the intermediate by Glu103. The second half reaction, in which water displaces the glutamate side chain, likely proceeds through an oxocarbenium-like transition state. In MilB, Asp78 is positioned to protonate the cytosine base leaving group and to activate a water molecule for the second half of the reaction. This aspartate residue is conserved in MilB, BcmB, BlsM, and Rcl, also suggesting a conserved mechanism of hydrolysis.
Scheme 2.
Reversal of Substrate Specificity
Based on the structural data, we hypothesized that the active site phenylalanine is the key residue that confers specificity for either 2′-hydroxyl or 2′-deoxy groups. In MilB, Phe17 C4 is 3.6 Å from the catalytic glutamate residue and 3.5 Å from the CMP 2′-hydroxyl group. A tyrosine is at a position equivalent to MilB Phe17 in all previously solved structures of enzymes in the NDT superfamily, which prefer 2′-deoxy substrates (Figure 9). MilB F17Y was constructed using site-directed mutagenesis and used to test if the phenylalanine to tyrosine change is sufficient to increase the efficiency of dCMP hydrolysis. While MilB WT preferentially hydrolyzes CMP, with a five-fold lower efficiency towards dCMP, mutation of F17Y MilB resulted a 100-fold decrease in the hydrolysis of CMP and a 100-fold increase in the hydrolysis of dCMP, corresponding to a >10,000-fold inversion of substrate specificity (Table 2). A similar, but slightly smaller affect in substrate specificity was observed for BcmB.
Figure 9.
Active site comparison of MilB and NDT. A stereoview active site comparison between MilB with CMP bound (green) and NDT with 5-methyl-2’-deoxypseudouridine (5MD) (blue) reveals conserved positioning of the catalytic glutamate and aspartate residues. Other key residues for substrate binding deviate between the two structures.
The largest change is seen in the preference of F17Y MilB for dCMP compared to CMP. The value of kcat for WT MilB with CMP is about 3-fold larger than for that of dCMP. For F17Y MilB, the value of kcat with dCMP is about 600-fold larger than for CMP. For both WT and F17Y MilB, the KM values for both substrates are in a similar range with the value for F17Y MilB with dCMP being the lowest. The value of kcat for WT BcmB with CMP is about 20-fold larger than for that of dCMP. For F6Y BcmB, the kcat with dCMP is about 20-fold larger than for CMP. The KM values for WT and mutant BcmB show a larger range compared to those of MilB. The lowest value is 0.042 mM for F6Y BcmB with dCMP and the highest value is 0.9 mM for WT BcmB with dCMP.
In the structure of MilB/CMP, the CMP 2′-hydroxyl group interacts with the catalytic Glu103, orienting the molecule in the active site. Based on the location of the Phe17 in MilB/CMP, the MilB F17Y Tyr17 hydroxyl group would be 3.1 Å from the 2′-hydroxyl group of bound CMP. This may cause unfavorable interactions for CMP binding or cause electronic repulsions effecting hydrolysis by the catalytic glutamate residue. The latter is consistent with the observed values of kcat, which is significantly enhanced while the KM values are in a similar range. Additionally, WT MilB less efficiently hydrolyzes dCMP possibly because the 2′-group of dCMP cannot form any such hydrogen bonding interaction to orient it for catalysis. Thus, WT MilB catalytic residues are best positioned for hydrolysis of CMP with the aid of the 2′-hydroxyl group orienting CMP in the binding pocket. These observations would also apply to WT and F6Y BcmB, which show patterns similar to WT and F17Y MilB in efficiency for hydrolysis of CMP and dCMP.
Supplementary Material
Acknowledgements
We thank Dr. Cynthia Kinsland for cloning MilB and BcmB, Leslie Kinsland for assistance in preparing the manuscript. We thank the staff of the NE-CAT at the APS for assistance with the data collection.
ABBREVIATIONS
- NDT
nucleoside 2′deoxyribosyltransferase
- hmCMP
5-hydroxymethyl cytidine 5′-monophosphate
- CMP
cytidine 5′-monophosphate
- SeMet
selenomethionyl
- AMP
adenosine 5′-monophosphate
- IMP
inosine 5′-monophosphate
- UMP
uridine 5′-monophosphate
- dCMP
2′-deoxycytidine 5′-monophosphate
- GMP
guanosine 5′-monophosphate
- SAD
single-wavelength anomalous diffraction
- PTD
purine deoxyribosyltransferase
- DMHA
N,N-dimethylhexylamine
- 5MD
5-methyl-2’-deoxypseudouridine;
Footnotes
This work was supported by NIH grants GM73220 and T32GM008500, and by the Robert A. Welch Foundation (A-0034 to TPB). This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award GM103403 from the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
The coordinates of MilB/SO4, MilB/CMP, and, BcmB/PO4 have been deposited in the Protein Data Bank under accession codes 4JEN, 4JEM, 4JEL, respectively.
Supplemental Information
Table S1 and Figures S1 and S2. This material is available free of charge at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Li L, Xu Z, Xu X, Wu J, Zhang Y, He X, Zabriskie TM, Deng Z. The mildiomycin biosynthesis: initial steps for sequential generation of 5-hydroxymethylcytidine 5'-monophosphate and 5-hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. Chembiochem. 2008;9:1286–1294. doi: 10.1002/cbic.200800008. [DOI] [PubMed] [Google Scholar]
- 2.Feduchi E, Cosin M, Carrasco L. Mildiomycin: a nucleoside antibiotic that inhibits protein synthesis. J Antibiot (Tokyo) 1985;38:415–419. doi: 10.7164/antibiotics.38.415. [DOI] [PubMed] [Google Scholar]
- 3.Harada S, Kishi T. Isolation and characterization of mildiomycin, a new nucleoside antibiotic. J Antibiot (Tokyo) 1978;31:519–524. doi: 10.7164/antibiotics.31.519. [DOI] [PubMed] [Google Scholar]
- 4.Macnutt WS. The enzymically catalysed transfer of the deoxyribosyl group from one purine or pyrimidine to another. Biochem J. 1952;50:384–397. doi: 10.1042/bj0500384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawdhri RF, Hutchinson DW, Richards AO. Nucleoside Deoxyribosyltransferase and Inosine Phosphorylase-Activity in Lactic-Acid Bacteria. Archives of Microbiology. 1991;155:409–411. [Google Scholar]
- 6.Armstrong SR, Cook WJ, Short SA, Ealick SE. Crystal structures of nucleoside 2-deoxyribosyltransferase in native and ligand-bound forms reveal architecture of the active site. Structure. 1996;4:97–107. doi: 10.1016/s0969-2126(96)00013-5. [DOI] [PubMed] [Google Scholar]
- 7.Ghiorghi YK, Zeller KI, Dang CV, Kaminski PA. The c-Myc target gene Rcl (C6orf108) encodes a novel enzyme, deoxynucleoside 5'-monophosphate N-glycosidase. J Biol Chem. 2007;282:8150–8156. doi: 10.1074/jbc.M610648200. [DOI] [PubMed] [Google Scholar]
- 8.Grochowski LL, Zabriskie TM. Characterization of BlsM, a nucleotide hydrolase involved in cytosine production for the biosynthesis of blasticidin S. Chembiochem. 2006;7:957–964. doi: 10.1002/cbic.200600026. [DOI] [PubMed] [Google Scholar]
- 9.Dupouy C, Zhang C, Padilla A, Pochet S, Kaminski PA. Probing the active site of the deoxynucleotide N-hydrolase Rcl encoded by the rat gene c6orf108. J Biol Chem. 2010;285:41806–41814. doi: 10.1074/jbc.M110.181594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doddapaneni K, Mahler B, Pavlovicz R, Haushalter A, Yuan C, Wu Z. Solution structure of RCL, a novel 2'-deoxyribonucleoside 5'-monophosphate N-glycosidase. J Mol Biol. 2009;394:423–434. doi: 10.1016/j.jmb.2009.08.054. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Padilla A, Zhang C, Labesse G, Kaminski PA. Structural characterization of the mammalian deoxynucleotide N-hydrolase Rcl and its stabilizing interactions with two inhibitors. J Mol Biol. 2009;394:435–447. doi: 10.1016/j.jmb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Padilla A, Amiable C, Pochet S, Kaminski PA, Labesse G. X-ray structure of the oncoprotein Rcl bound to three nucleotide analogs. Acta Crystallographia Section D. 2013;D69:247–255. doi: 10.1107/S0907444912045039. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BC, Shim H, Li Q, Wu CS, Lee LA, Maity A, Dang CV. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin S, Bosc DG, Ingle JN, Spelsberg TC, Janknecht R. Rcl is a novel ETV1/ER81 target gene upregulated in breast tumors. J Cell Biochem. 2008;105:866–874. doi: 10.1002/jcb.21884. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Vol. 3. Plainview, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CWJ, Sweet RM, editors. Methods Enzymol. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 17.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 18.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 19.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen VB, Arendall WB, 3rd., Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Xu Z, Xu X, Wu J, Zhang Y, He X, Zabriskie TM, Deng Z. The mildiomycin biosynthesis: initial steps for sequential generation of 5-hydroxymethylcytidine 5'-monophosphate and 5-hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. ChemBioChem. 2008;9:1286–1294. doi: 10.1002/cbic.200800008. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. CA: DeLano Scientific, San Carlos; 2002. [Google Scholar]
- 27.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand R, Kaminski PA, Ealick SE. Structures of purine 2’-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution. Biochemistry. 2004;43:2384–2393. doi: 10.1021/bi035723k. [DOI] [PubMed] [Google Scholar]
- 29.Bosch J, Robien MA, Mehlin C, Boni E, Riechers A, Buckner FS, Van Voorhis WC, Myler PJ, Worthey EA, DeTitta G, Luft JR, Lauricella A, Gulde S, Anderson LA, Kalyuzhniy O, Neely HM, Ross J, Earnest TN, Soltis M, Schoenfeld L, Zucker F, Merritt EA, Fan E, Verlinde CL, Hol WG. Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase. J Med Chem. 2006;49:5939–5946. doi: 10.1021/jm060429m. [DOI] [PubMed] [Google Scholar]
- 30.Kaminski PA. Functional cloning, heterologous expression, and purification of two different N-deoxyribosyltransferases from Lactobacillus helveticus. J Biol Chem. 2002;277:14400–14407. doi: 10.1074/jbc.M111995200. [DOI] [PubMed] [Google Scholar]
- 31.Porter DJ, Merrill BM, Short SA. Identification of the active site nucleophile in nucleoside 2-deoxyribosyltransferase as glutamic acid 98. J Biol Chem. 1995;270:15551–15556. doi: 10.1074/jbc.270.26.15551. [DOI] [PubMed] [Google Scholar]
- 32.Tull D, Withers SG. Mechanisms of cellulases and xylanases: a detailed kinetic study of the exo-beta-1,4-glycanase from Cellulomonas fimi. Biochemistry. 1994;33:6363–6370. doi: 10.1021/bi00186a041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.