Abstract
All eukaryotic DNA transposons reported so far belong to a single category of elements transposed by the so-called “cut-and-paste” mechanism. Here, we report a previously unknown category of eukaryotic DNA transposons, Helitron, which transpose by rolling-circle replication. Autonomous Helitrons encode a 5′-to-3′ DNA helicase and nuclease/ligase similar to those encoded by known rolling-circle replicons. Helitron-like transposons have conservative 5′-TC and CTRR-3′ termini and do not have terminal inverted repeats. They contain 16- to 20-bp hairpins separated by 10–12 nucleotides from the 3′-end and transpose precisely between the 5′-A and T-3′, with no modifications of the AT target sites. Together with their multiple diverged nonautonomous descendants, Helitrons constitute ≈2% of both the Arabidopsis thaliana and Caenorhabditis elegans genomes and also colonize the Oriza sativa genome. Sequence conservation suggests that Helitrons continue to be transposed.
Eukaryotic and prokaryotic genomes are populated by transposable elements (TEs) that are capable of intragenomic multiplication by transferring a DNA segment from one genomic site to another (1–3). On the basis of mechanisms of their transposition, TEs can be divided into two classes: retrotransposons, which proliferate via reverse transcription, and DNA transposons, which are transposed without RNA intermediates. DNA transposons found so far in eukaryotic genomes have characteristic structural hallmarks, including terminal inverted repeats (TIRs) and 2- to 10-bp flanking direct repeats, generated by target site duplications (TSD) on insertion of the transposons in the genome. Transposition of known DNA transposons in eukaryotes is described by the “cut-and-paste” model (4), according to which transposases encoded by transposons perform both DNA cleavage and transfer reactions, which are necessary to cut a transposon at both its termini and insert it into a new position. The majority of eukaryotic transposases belong to the DDE class, named after the highly conserved Asp (D), Asp (D), and Glu (E) amino acid residues, which belong to the catalytic core (4, 5). It has been suggested (6–8) that the bacterial transposons IS91, IS801, and IS1294 form an exotic family of prokaryotic rolling-circle (RC) transposons that do not belong to the DDE class, as they transpose via RC replication (RCR). These transposons share common 5′-AY and GTTC-3′ termini, do not possess TIRs, and do not generate TSDs. They encode only one protein, similar to the replication initiator proteins (Rep) from known RC replicons (6–9). There are three groups of episomal replicons using RCR: circular single-stranded DNA (ssDNA) bacteriophages (10), plasmids of bacteria or archaea (11), and geminiviruses (circular ssDNA viruses replicating in plant cells) (12). Usually replication of RC replicons is catalyzed by the nuclease/ligase activity of Rep and is assisted by host DNA helicases and ssDNA-binding proteins (SSBs) (6–12). Here, we report a previously unknown category of eukaryotic DNA transposons, named Helitron, that transpose as RC replicons in the Arabidopsis thaliana, Oriza sativa, and Caenorhabditis elegans genomes. Most TEs reported in this manuscript were active in recent evolutionary history and were reconstructed from their inactive copies accumulated in the respective genomes. Typically, the reconstruction process produces a consensus sequence without insertions, deletions, and false stop codons accumulated in the inactive copies. This approach is well known and best illustrated by a recent study of Sleeping Beauty, a Tc1-like transposon from fish (13), reconstructed from its inactive copies and demonstrated to be transpositionally active in a test tube. Another much more ancient example is a PiggyBac-like DNA transposon, Looper, discovered in the human genome [V.V.K. and J.J., Repbase Update (1998) www.girinst.org/Repbase_Update.html], whose consensus sequence is based on a multiple alignment of the inactive copies, which are ≈100 million years old. All genomic copies of Looper are mutated to the extent that no traces of its transposase could be detected at the sequence level. However, the transposase re-emerged from the virtual background noise after reconstructing the consensus sequence.
Materials and Methods
Computational Analysis.
TEs reported in the manuscript were identified by running DNA sequences of prospective TEs against GenBank by using the National Center for Biotechnology Information blast server (14), followed by censor analysis at Genetic Information Research Institute (GIRI) (15). censor is much more sensitive than blast and was applied to determine the precise locations of sequences similar to the query and to identify distantly related DNA sequences (≈60% identical to each other).
We built consensus sequences of the transposons on the basis of a simple majority rule applied to their multiply aligned copies. Additional copies of transposons obtained because of redundant sequencing or chromosomal duplications unrelated to transpositions were discarded on the basis of the identity of extended flanking regions.
Distantly related proteins were identified by using the position-specific iterated psi-blast search (16). Multiple alignments of protein sequences were produced by clustal w (17) and edited manually by using genedoc (18). Alignments of nucleotide sequences were performed by using VMALN2 and PALN2, programs developed at GIRI. Manual editing of nucleotide alignments was done by using mase, a UNIX-based sequence editor (19). We used the genscan (ref. 20; http://genes.mit.edu/GENSCAN.html) and fgenesh (ref. 21; http://genomic.sanger.ac.uk/gf/gf.html) programs to predict the exon/intron structures of genes encoded by Helitrons.
Monte Carlo Simulation.
We applied computer-assisted simulations to address whether conservation of the ssDNA-binding replication protein A (RPA)-like proteins in highly diverged families of Helitron indicates functional significance of those proteins for transposition. The premise behind the simulation is that a protein-coding sequence, free of functional constraints, will lose its coding capacity because of the accumulation of stop codons and breakup of splicing sites, leading to elimination of the protein-coding region. As a result, one could expect a little protein-coding capacity preserved in two sequences 40% diverged from each other, as in the case of the ATRPA1H1 and ATRPA2H genes (Fig. 1) encoding ≈500-aa proteins 44% identical to each other. To test the impact of unselected mutations, we used the 2,600-bp ATRPA2H DNA sequence, containing both exons and introns, as a query to generate 100 random sequences. Every random sequence was generated by random mutations at N random positions in the query, without insertions and deletions (the simulation program written in perl is available on request). N equals D⋅L/100, where D stands for the sequence divergence and L for length of the query. Using genscan, we were able to obtain a distribution of lengths for proteins predicted in the set of 100 random DNA sequences D% divergent from the query. Assuming no functional role of RPA-like proteins in Helitrons, one should expect that the lengths of these proteins in two D% diverged Helitrons follow the random distribution.
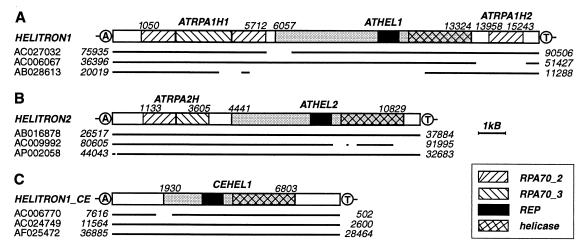
Figure 1.
Reconstruction of the Helitron1 (A), Helitron2 (B), and Helitron1_CE (C) consensus sequences. The consensus sequences are schematically depicted as rectangles. Contiguous copies of Helitrons that we used for reconstruction of the consensus sequences are shown as bold lines beneath the rectangles. Gaps in the lines mark deletions of corresponding regions of the consensus sequences. GenBank accession nos. and sequence coordinates are indicated. Genes and their coordinates in the consensus sequences are indicated above the rectangles. Genes coding for proteins composed of the Rep and helicase domains are shaded in gray. The AT target sites are encircled.
Databases.
Sequences of TEs reported in the manuscript are deposited in the A. thaliana and C. elegans sections of Repbase Update (ref. 22; www.girinst.org/Repbase_Update.html) and are also included in supplemental information, which is available at www.girinst.org/∼vladimir/RC/S.html.
Results and Discussion
Helitrons in the A. thaliana Genome.
During computational identification of DNA repeats, we found that the A. thaliana genome harbors multiple dispersed ≈10-kb units that encode two proteins similar to the yeast Pif1p DNA helicase (23–24) and RPA (25–26). On the basis of pairwise nucleotide identity, these DNA units can be divided into several groups of 2–10 sequences each. Typically, sequences from any particular group are ≈90% identical to each other, whereas they are only 60–70% identical to sequences from separate groups. We have derived consensus sequences for four groups, called hereafter Helitron1-4, of which the first two are discussed in detail in this paper (Table 1). The 15,809-bp Helitron1 consensus sequence harbors three genes (Fig. 1A) composed of multiple exons (20–21), named ATRPA1H1, ATHEL1, and ATRPA1H2, respectively. ATRPA1H1 encodes an 842-aa protein, ATRPA1H1p, similar to RPA70, the largest subunit of RPA. RPA70 is conserved in human, fly, rice, frog, worm, and yeast and is composed of three domains, RPA70_1, RPA70_2, and RPA70_3 (25–26). ATRPA1H1p (Fig. 1A) is composed of two divergent RPA70_2-like domains separated by a RPA70_3-like domain (data not shown). ATRPA1H2 is another gene that encodes a RPA70_2-like protein. This gene resides in the 3′-terminal segment of Helitron1 (Fig. 1A) and encodes a 262-aa protein (ATRPA1H2p), 20 and 39% identical to the first and second RPA70_2-like domains of ATRPA1H1p, respectively. It is known that both RPA70_2 and RPA70_3 bind ssDNA (25–26). Therefore, both ATRPA1H1p and ATRPA1H2p are expected to be SSBs. ATHEL1 is the largest gene, residing in a central portion of Helitron1, between the ATRPA1H1 and ATRPA1H2 genes, and encodes a 1,697-aa protein (ATHEL1p). A ≈500-aa C-terminal portion of ATHEL1p is similar to numerous DNA helicases that unwind the DNA duplex in a 5′-to-3′ direction (10). These helicases belong to the SF1 superfamily, encompassing a broad spectrum of eukaryotic, prokaryotic, and viral proteins characterized by a specific set of seven conservative motifs (27). As shown in Fig. 2, all these motifs are present in ATHEL1p.
Table 1.
RC transposons in the A. thaliana, O. sativa, and C. elegans genomes
| Family | Length, bp | Rep-helicase | RPA | Nonautonomous families |
|---|---|---|---|---|
| A. thaliana | ||||
| Helitron1 (7) | 15,809 | ATHEL1p | ATRPA1H1p, ATRPA1H2p | Helitrony1A (1348, 10), Helitrony1B (1311, 10), Helitrony1C (3058, 10), Helitrony2 (11114, 10), Atrepx1 (2432, 20), Atrep1 (888, 100), Atrep2 (564, 150), Atrep3 (2097, 150), Atrep4 (2240, 50), Atrep5 (2386, 50), Atrep6 (1189, 30), Atrep7 (940, 50), Atrep8 (1077, 40), Atrep9 (899, 10), Atrep10 (899, 50), Atrep10A (1380, 20), Atrep10B (1821, 20), Atrep10C (653, 20), Atrep11 (1053, 50), Atrep12 (1342, 10), Atrep13 (648, 30) |
| Helitron2 (6) | 11,435 | ATHEL2p | ATRPA2Hp | |
| Helitron3 (2) | 15,333 | ATHEL3p | ATRPA3H1p, ATRPA3H2p, ATRPA3H3p | |
| Helitron4 (5) | 17,261 | ATHEL4p | ATRPA4H1p, ATRPA4H2p, ATRPA4Hp3 | |
| O. sativa | ||||
| Helitron1_OS (1) | 10,182 | OSHEL1p | OSRPA1Hp | |
| Helitron2_OS (2) | 15,167 | OSHEL2p | OSRPA2Hp | — |
| Helitron3_OS (1) | 12,693 | OSHEL3p | OSRPA3Hp | |
| C. elegans | ||||
| Helitron1_CE (7) | 8,484 | CEHEL1p | — | Helitrony1_CE (2593, 50), Helitrony1A_CE (3023, 50), Helitrony2_CE (245, 200), Helitrony3_CE (193, 100), Helitrony4_CE (1855, 100), NDNAX1 (2085, 100), NDNAX2 (2844, 100), NDNAX3 (1591, 100) |
| Helitron2_CE (1) | 5,514 | CEHEL2p | — |
Rep-helicase, proteins composed of the RCR Rep and DNA helicase domains. Numbers of copies per haploid genome are shown in parentheses. Length of the consensus sequence and numbers of copies per genome are shown together in parentheses for nonautonomous Helitrons.
Figure 2.
Multiple alignment of helicases encoded by the Helitron1 and Helitron1_CE transposons with a set of eukaryotic and prokaryotic DNA helicases. Domains I–VI that are conservative in DNA helicases from the SF1 superfamily (20) and distances between these domains are indicated. Domain IV/V has not been reported previously. Invariable positions are shaded in black, and those conserved in more than 60% of the sequences are shaded in gray. The following are names of helicases: PIF1 (GenBank protein identification no. 130196, yeast), BACULOVIRUS (7460536, the dsDNA Lymantia dispar nucleopolyhedrovirus), CHILO (5725645, the dsDNA chilo iridescent virus), TRAA_RHISN (2499024, a Ti-like plasmid from Rhizobium), TRAI_EC (136208, the F plasmid from Escherichia coli), EXOV_EC (2507018, the RecD subunit from the E. coli exodeoxyribonuclease V), TRWC (1084124, the R388 conjugative plasmid from E. coli), and HEL_T4 (416895, the dsDNA T4 bacteriophage).
The 11,435-bp Helitron2 consensus sequence is ≈96% identical to seven identified Helitron2 copies (Fig. 1B) and carries two genes called ATRPA2H and ATHEL2. ATRPA2H is composed of 10 exons (20–21) and encodes a 518-aa RPA70-like protein, 41% identical to ATRPA1H1p from Helitron1. There is only a 60% nucleotide identity between corresponding DNA segments that code for the two proteins. ATHEL2 is composed of 11 exons encoding a 1,743-aa protein (ATHEL2p), whose ≈500-aa C-terminal portion is similar to the SF1 helicases. ATHEL1p and ATHEL2p have different 300-aa N termini, and their remaining ≈1,400-aa portions are 55% identical. There is only a 64% nucleotide identity between DNA segments encoding these portions. Overall, about 10 families of 8- to 15-kb-long Helitrons are present in the A. thaliana genome (not shown). Despite a high nucleotide divergence between different Helitrons (≈40%), each has conserved structural hallmarks, which include 5′-TC and CTAR-3′ termini, the AT target sites, and an ≈18-bp hairpin separated by ≈11 nucleotides from the 3′ end (Fig. 3).
Figure 3.
Termini of Helitrons. Conserved 5′ and 3′ termini are in bold capital letters, 3′ terminal hairpins are shaded in gray, and inverted repeats are underlined.
Helitrons in the O. sativa Genome.
Arabidopsis is not the only plant species harboring Helitron-like transposons. We found three families of Helitrons, named Helitron1_OS, Helitron2_OS, and Helitron3_OS, in the rice genome (Table 1; also see supplemental information at www.girinst.org/∼vladimir/RC/S.html). They encode RPAs as well the Rep/DNA helicases and share the same structural hallmarks with the Helitrons from Arabidopsis (Fig. 2). Helitrons have been transposed recently in the rice genome, where they are represented by just a few copies (not shown). Their recent origin is indicated in the case of Helitron2_OS, represented by two ≈15-kb copies, which are 99% identical (GenBank accession nos. AP001278, positions 56948–41572, and AP001800, 115543–87503).
Helitrons in the C. elegans Genome.
The C. elegans genome also contains multiple copies of DNA helicases related to those encoded by the plant Helitrons. We extracted five DNA fragments coding for the Helitron-like helicase, which were ≈98% identical to each other. After iterative series of expansions of the original DNA fragments and multiple alignments of the expanded fragments, we derived an 8,484-bp consensus sequence (Fig. 1C). Analysis of the consensus sequence revealed a single gene, named CEHEL1, composed of nine exons (20–21), which encode a 1,466-aa protein (CEHEL1p), 33% identical with ATHEL1p (excluding their 125- and 308-aa N-terminal portions, correspondingly). This consensus sequence, named Helitron1_CE, shares common structural hallmarks with the plant Helitrons, including 5′-TC and CTGG-3′ termini and the 3′-hairpin (Fig. 3). On the basis of protein and nucleotide similarities to Helitron1_CE, we found another 5,514-bp element, Helitron2_CE, whose seven putative exons (20–21) encode a protein 60% identical to CEHEL1p (67% nucleotide identity). Again, despite the high divergence, Helitron2_CE shares structural hallmarks with the plant Helitrons. However, nematode Helitrons do not contain the RPA-like proteins.
Interestingly, five previously reported families of 200- to 400-bp minisatellite-like nematode repetitive DNA (RcA1, RcC9, RcD1, Rc35, and Rc123) have been found frequently adjacent to one another in the same order and orientation (28). It has been suggested that gene conversion and molecular drive are responsible for the conservation of these repetitive elements in different clusters located far apart in the genome (28). We found that these repeats are different fragments of Helitrons, and the “clusters” observed earlier are just different copies of Helitrons. Overall, Helitrons carry and propagate multiple minisatellites in the C. elegans genome. For example, the internal portion of the ≈3-kb nonautonomous element Helitrony1_CE is built predominantly of different 15-, 35-, and 40-bp minisatellites. The number of minisatellite units present in some copies of the nematode Helitrons is close to 300 (not shown).
Nonautonomous Helitrons.
Most Helitrons are “nonautonomous” elements. They share common termini and other structural hallmarks with “autonomous” Helitrons, but they do not encode any complete set of proteins encoded by the autonomous elements. This phenomenon is common for other known DNA transposons and indicates that structural hallmarks, present in both 5′ and 3′ termini, ensure a transposition of the nonautonomous elements because of the interaction between the termini and transposase expressed by the autonomous elements. It has been reported recently (29–30) that the A. thaliana genome harbors multiple families of 1- to 3-kb-long repetitive elements reported under the names ATREP (29) and AthE1 (30), which constitute more than 1% of the haploid genome. We found that these elements share the same structural hallmarks with Helitrons and are 60–80% similar to other nonautonomous Helitrons, like Helitrony1A-Helitrony1C (not shown); therefore, they are also classified here as nonautonomous Helitrons.
Multiple nonautonomous derivatives of Helitrons are present in the C. elegans genome (Table 1). For example, we found two families of short nonautonomous transposons, called Helitrony2_CE and Helitrony3_CE, which share similar terminal regions with Helitron1_CE and Helitron2_CE. The ≈200 copies of Helitrony2_CE in the genome are ≈5% divergent from their 249-bp consensus sequence and ≈10% divergent from each other. Helitrony3_CE copies are only 1% divergent from their 195-bp consensus sequence. Moreover, several copies of Helitrony3_CE, inserted in different places, are identical (GenBank accession nos. AF106583, positions 13953–13789, AL023831, 213–407, and U97016, 7488–17294). The last observation suggests that Helitrons are transpositionally active in C. elegans.
Approximately 2% of both the A. thaliana and C. elegans DNA sequences deposited in GenBank (over 90% of both genomes) are composed of repetitive elements significantly similar to Helitrons present currently in Repbase Update. These elements were identified by censor, and their coordinates are listed in maps of TEs from the A. thaliana and C. elegans (www.girinst.org/Repbase_Update.html). The proportion of Helitrons in these genomes is slightly underestimated, because most Helitrons reside in heterochromatin regions, underrepresented in the available sequence data. Moreover, multiple highly divergent families of nonautonomous Helitrons are represented by one or two copies per genome, and only some of them can be detected on the basis of distant similarity to known TEs.
Target Sites.
Numerous nonautonomous Helitrons were found inserted into copies of other well-known mobile elements in the A. thaliana genome (29–30), and it has been suggested that those elements do not produce target site duplications on their integration in the genome. On the basis of our analysis of Helitron insertions into other well-defined TEs in both the A. thaliana and C. elegans (Fig. 4), we also conclude that Helitrons transpose specifically into host AT target sites. The integration occurs precisely between the host A and T nucleotides, without duplications or deletions of the target sites, consistent with the RC mechanism discussed below.
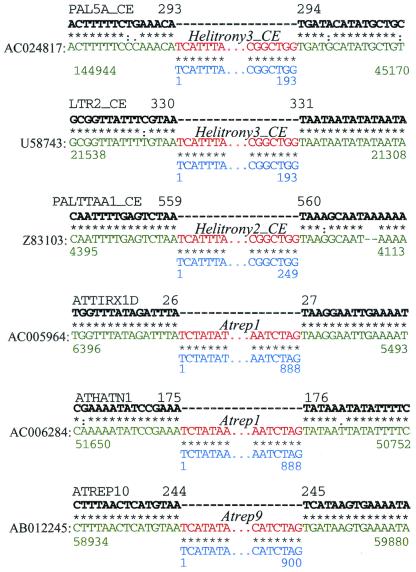
Figure 4.
Precise integration of Helitrons into the host AT target sites. Six insertion cases of Helitrons (red) into different transposons (green) are shown separately. Two copies of ATREP1, two copies of Helitrony3_CE, and single copies of Helitrony2_CE and ATREP9 are inserted into copies of the ATTIRX1D, ATHATN1, PAL5A_CE, LTR2_CE, PALTTAA1_CE, and ATREP10 transposons, respectively. The consensus sequences of the elements harboring Helitrons are marked by the bold black letters and are described in the A. thaliana and C. elegans sections of Repbase Update at www.girinst.org/Repbase_Update.html. Consensus sequences of the corresponding Helitrons are marked in blue. Asterisks, semicolons, and dots indicate identical nucleotide positions, transitions, and transversions, respectively. Only termini of Helitrons are shown. Black, green, and blue numbers show positions in the consensus sequences of the harboring transposons, GenBank, and Helitron consensus sequences, respectively.
RPA-Like Proteins Are Functional Components of Helitrons.
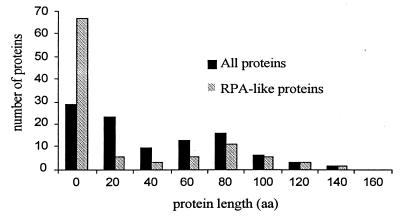
We generated a set of 100 random DNA sequences 61% identical to the 2,600-bp ATRPA2H gene from Helitron2, encoding the 518-aa RPA-like protein ATRPA2Hp. These random sequences imitated a ≈2,767-bp portion of ATRPA1H1 from Helitron1 encoding a 475-aa portion of ATRPA1H1p 44% identical to ATRPA2Hp at the protein level and 60% at the DNA level. After alignment by VMALN2, the average identity between the random sequences and ATRPA2H was even higher, 63%, because of gaps introduced by the alignment. Fig. 5 shows the distribution of lengths of proteins predicted by genscan in the randomly mutated sequences (proteins encoded by the minus strand have been discarded). The average length was 46 aa with a standard deviation of 40 aa. No proteins longer than 152 aa were found in the random set of DNA sequences. Overall, only 71 random sequences encoded proteins. Moreover, only 31 sequences encoded proteins similar to ATRPA2Hp (blastp, E < 0.05). The average length of the “random” proteins similar to ATRPA2Hp was only 15 aa, with a standard deviation of 25 aa (Fig. 5). These random proteins contrast strongly with ATRPA1H1p, whose 475-aa length differs from the average “random” length by the 20 standard deviations. Therefore, we conclude that the RPA protein is a functional component of Helitrons, evolving under selective pressures. The same conclusion is well supported by the observation of RPA-like proteins encoded by the rice Helitrons, which do not show any significant nucleotide identity to Helitrons from A. thaliana.
Figure 5.
Distributions of protein lengths predicted in 100 random DNA sequences. Every random sequence was 61% identical to ATRPA2H, without insertions or deletions. Black marks all proteins predicted by genscan; gray marks proteins similar to ATRPA2Hp.
Replication Initiator Motifs in Helitrons.
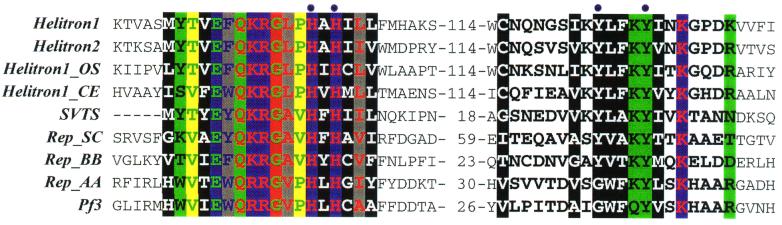
Despite their DNA transposon-like characteristics, Helitrons do not code for any proteins similar to known transposases. However, iterative screening by psi-blast shows that ATHEL1p contains an 11-aa motif (Fig. 6) similar to the “two-His” motif conserved in the Reps encoded by a diverse set of plasmids and ssDNA viruses that use RC DNA replication (9). Most importantly, Reps perform both cleavage and ligation of DNA, reactions that initiate and terminate RCR, and contain three conserved motifs (9). The first, most variable motif is of unknown function and lies 30–80 aa upstream of a “two-His” motif, which functions putatively as a ligand to Mg2+ and Mn2+, which are required for RCR. Another Rep motif lies 30–90 aa downstream of “two His” and catalyzes DNA cleavage and ligation, two key reactions of RCR. The last motif is also conserved in Helitrons and is separated by the invariant number of 114 aa following the “two-His” motif (Fig. 6). It includes two conserved tyrosine residues, which are central to DNA nick formation and ligation by the Rep proteins during RCR (9, 31–33).
Figure 6.
Alignment of the RC motifs in the Helitrons. Following is a list of the RCR initiator-like proteins: SVTS2 (GenBank accession no. AAF18310, the SVTS2 ssDNA spiroplasma plectrovirus); Rep_SC (BAA34784, the pSA1.1 conjugative plasmid from Streptomyces cyaneus); Rep_BB (BAA07788, the pHT926 Bacillus borstelensis cryptic plasmid); Rep_AA (AAC37125, the pVT736–1 RCR plasmid from Actinobacillus actinomycetemcomitans); and Pf3 (AAA88392, the Pf3 ssDNA bacteriophage from Pseudomonas aeruginosa). Color shading shows different physicochemical properties of conserved amino acids (17). Conserved tyrosines, corresponding to the RCR nicking/ligation catalytic center, are marked by dots.
Helitrons Are RC DNA Transposons.
The current model for RCR involves several basic stages (11). Replication starts from a site-specific nicking of the replicon plus strand by the Rep protein. A free 3′-OH end of the nicked plus strand serves as a primer for leading-strand DNA synthesis and is elongated by several host replication proteins, such as DNA helicase, DNA Pol, and SSB. The newly synthesized leading plus strand remains covalently linked to the 3′-OH end of the parent plus strand during the continuous displacement of its 5′-OH end. When the leading strand makes a complete turn, Rep catalyzes a strand transfer reaction followed by release of an ssDNA intermediate, the parent minus strand, and a double-stranded DNA (dsDNA) replicon composed of both the parental plus and a newly synthesized strand. The RCR model (6–8) explains why there are no target site duplications during integration of Helitrons in the genome. Presumably, the 3′ terminal hairpin, conserved in Helitrons, serves as a terminator of RCR.
Evolutionary Implications.
It has been suggested that geminiviruses have evolved from prokaryotic circular ssDNA replicons (9, 12). If this scenario is correct, one would expect to observe geminiviruses in different eukaryotic kingdoms. However, no geminiviruses are found outside plant species. Our finding of eukaryotic RC transposons suggests that geminiviruses might have evolved from plant RC transposons rather than from prokaryotic RC replicons.
Given characteristics of prokaryotic and eukaryotic RC transposons, it seems that the hypothesis of their evolution from the same common ancestral elements would be the most parsimonious. The prokaryotic RC transposons encode the Rep proteins only, and their transposition depends presumably on the host DNA helicase and SSB (8). As shown here, in addition to Rep, the eukaryotic RC transposons encode their own helicase and SSB (the latter in plants only). Given their exon–intron structure and similarities to known proteins, Helitron's SSB and helicase are both likely to have evolved from former host proteins recruited by ancestral eukaryotic RC transposons. Apparently, RC transposons form the only class of eukaryotic mobile elements that depend so critically on proteins related to those directly involved in a host DNA replication.
Two alternative scenarios describe the most likely fate of a host gene captured by a transposon: (i) The captured gene would be destroyed by multiple mutations if it did not provide any selective advantage to the transposon; (ii) it would be kept as a gene related to the original host gene if its capture is beneficial for the transposon, which is tolerated by the host. Given the conservation of the RPA and helicase proteins in different families of Helitron, DNA sequences of which are sometimes as much as 40–50% divergent, one has to presume that both proteins are functional parts of the transposition machinery. Spectacularly, Helitrons, as most of other mobile elements in the A. thaliana and C. elegans genomes (ref. 29; V.V.K. and J.J., www.girinst.org/Repbase_Update.html), are represented in the genomes by multiple highly diverged families. Given the young age of these families and the extent of protein conservation, it is highly unlikely that the divergence observed is a result of mutations accumulated by the transposons integrated in the host genome. In that case, Helitron transposons work as a powerful tool of evolution. They have recruited host genes, modified them to an extent that is unreachable by the Mendelian process, and multiplied them in the host genomes.
Acknowledgments
We thank Dr. Gabor Toth for helpful discussions and Jolanta Walichiewicz, Dr. Zelek Herman, Alison McCormack, and Michael Jurka for help with editing the manuscript. This work was supported by National Institutes of Health Grant 2 P41 LM06252–04A1 (to J.J.).
Abbreviations
- RC
rolling-circle
- RCR
RC replication
- TE
transposable element
- RPA
replication protein A
- RPA70
the largest subunit of RPA
- Rep
replication initiator protein
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- SSB
ssDNA-binding protein
- GIRI
Genetic Information Research Institute
References
- 1.Berg D E, Howe M M, editors. Mobile DNA. Washington, DC: Am. Soc. Microbiol.; 1989. [Google Scholar]
- 2.Kidwell M G, Lish D. Proc Natl Acad Sci USA. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fedoroff N V. Ann NY Acad Sci. 1999;870:251–264. doi: 10.1111/j.1749-6632.1999.tb08886.x. [DOI] [PubMed] [Google Scholar]
- 4.Craig N L. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 5.Turlan C, Chandler M. Trends Microbiol. 2000;8:268–274. doi: 10.1016/s0966-842x(00)01757-1. [DOI] [PubMed] [Google Scholar]
- 6.Mendiola M V, Bernales I, de la Cruz F. Proc Natl Acad Sci USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter G Y, Björklöf F, Romantschuk M, Mills D. Mol Gen Genet. 1998;5:381–387. doi: 10.1007/s004380050907. [DOI] [PubMed] [Google Scholar]
- 8.Tavakoli N, Comanducci A, Dodd H M, Lett M-C, Albiger B, Bennett P. Plasmid. 2000;44:66–84. doi: 10.1006/plas.1999.1460. [DOI] [PubMed] [Google Scholar]
- 9.Koonin E V, Ilyina T V. BioSystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 11.Khan S A. Mol Microbiol. 2000;37:477–484. doi: 10.1046/j.1365-2958.2000.02001.x. [DOI] [PubMed] [Google Scholar]
- 12.Rigden J E, Dry I B, Krake L R, Rezaian M A. Proc Natl Acad Sci USA. 1996;93:10280–10284. doi: 10.1073/pnas.93.19.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivics Z, Hackett P B, Plasterk R H, Izsvak Z. Cell. 1997;14:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Jurka J, Klonowski P, Dagman V, Pelton P. Comput Chem. 1996;20:119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholas K B, Nicholas H B, Jr, Deerfield D W, II. EMBNETNEWS. 1997;4:1–4. [Google Scholar]
- 19.Faulkner D, Jurka J. Trends Biochem Sci. 1988;13:321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- 20.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 21.Salamov A A, Solovyev V V. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurka J. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 23.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J-Q, Monson E K, Teng S-C, Schultz V P, Zakian V A. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 25.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 26.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Nature (London) 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 27.Gorbalenya A E, Koonin E V. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 28.Naclerio G, Cangiano G, Coulason A, Levitt A, Ruvolo V, La Volpe A. J Mol Biol. 1992;226:159–168. doi: 10.1016/0022-2836(92)90131-3. [DOI] [PubMed] [Google Scholar]
- 29.Kapitonov V V, Jurka J. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- 30.Surzycki S A, Belknap W R. J Mol Evol. 1999;48:684–691. doi: 10.1007/pl00006512. [DOI] [PubMed] [Google Scholar]
- 31.Noirot-Gros M-F, Erlich S D. Science. 1996;274:777–780. doi: 10.1126/science.274.5288.777. [DOI] [PubMed] [Google Scholar]
- 32.Novick R P. Trends Biochem Sci. 1998;23:434–438. doi: 10.1016/s0968-0004(98)01302-4. [DOI] [PubMed] [Google Scholar]
- 33.Sherman J A, Matson S W. J Biol Chem. 1994;269:26220–26226. [PubMed] [Google Scholar]