Abstract
Background
During implantation the blastocyst trophectoderm attaches to the endometrial epithelium and continues to differentiate into all trophoblast subtypes, which are the major components of a placenta. Aberrant trophoblast proliferation and differentiation are associated with placental diseases. However, due to ethical and practical issues, there is almost no available cell or tissue source to study the molecular mechanism of human trophoblast differentiation, which further becomes a barrier to the study of the pathogenesis of trophoblast-associated diseases of pregnancy. In this study, our goal was to generate a proof-of-concept model for deriving trophoblast lineage cells from induced pluripotency stem (iPS) cells from human fibroblasts. In future studies the generation of trophoblast lineage cells from iPS cells established from patient’s placenta will be extremely useful for studying the pathogenesis of individual trophoblast-associated diseases and for drug testing.
Methods and Results
Combining iPS cell technology with BMP4 induction, we derived trophoblast lineage cells from human iPS cells. The gene expression profile of these trophoblast lineage cells was distinct from fibroblasts and iPS cells. These cells expressed markers of human trophoblasts. Furthermore, when these cells were differentiated they exhibited invasive capacity and placental hormone secretive capacity, suggesting extravillous trophoblasts and syncytiotrophoblasts.
Conclusion
Trophoblast lineage cells can be successfully derived from human iPS cells, which provide a proof-of-concept tool to recapitulate pathogenesis of patient placental trophoblasts in vitro.
Introduction
The segregation of the blastocyst trophectoderm and inner cell mass denotes the first cell-fate decision in mammalian development. During human embryo implantation, the outer trophectoderm layer attaches to endometrial epithelial cells and continues to differentiate into lineage committed cytotrophoblasts and ultimately syncytio-, villous and extravillous trophoblasts, all required for the formation of a functional placenta. In human assisted reproductive technologies (ART) nearly 60% of embryos do not implant or are lost during the 1st trimester of pregnancy [1]. Identification of embryos prior to transfer that are capable of successful implantation and increased fecundity rates, would provide support for widespread acceptance of single embryo transfer in mitigating ART associated multiple pregnancies. The causes of implantation failure and/or 1st trimester loss are complex and may be related to an abnormal trophectoderm epithelium. It has been demonstrated that the morphological quality of trophectoderm in human blastocysts correlates with rates of ongoing pregnancy and miscarriage in fresh and frozen-thawed blastocysts [2]. Trophectoderm biopsy of day-5 blastocysts is an approach for preimplantation genetic diagnosis of single gene disorders to select unaffected embryos for uterine transfer. This supports the concept that the state of blastocyst trophectoderm dictates, to a significant extent, the success of ongoing pregnancy and can be sampled for testing.
It well accepted that defective human syncytiotrophoblasts and extravillous trophoblasts will result in implantation failure and abnormal placental formation [3]. Despite the important role that trophoblasts have in implantation and maintaining a successful pregnancy, the underlying molecular and cellular mechanisms responsible for trophoblast lineage establishment and multipotency in humans are largely unknown. The deficiency in this knowledge is due to a lack of available human blastocysts and 1st trimester placentas (<6 weeks gestation) as research specimens. Establishment of human trophoblast lineage cells (TCs) which show temporal and spatial fidelity to the developmental program of human trophoblast lineages will shed light on these cellular and molecular mechanisms.
Over the last decade several laboratories have demonstrated that human embryonic stem (ES) cells can be directed to differentiate into TCs when exposed to BMP4 under specific culture conditions [4; 5; 6; 7; 8; 9; 10; 11; 12; 13]. Two major limitations with using human ES cells for generating trophoblasts are the ethics associated with destroying human embryos for ES derivation and the inability to obtain human trophoblasts that are genetically related to the patient and/or patient’s placenta. Human induced pluripotent stem (iPS) cells which are derived from fibroblasts resemble human ES cells in terms of molecular signature and biofunction [14; 15] and are a genetic match to the human donor. Hence, human iPS cells derived from a patient’s fibroblasts are an ideal source of cells for differentiation into TCs. Furthermore, establishment of TCs using a patient’s placental trophoblasts will provide unique insight to the pathogenesis of placental trophoblasts of the patient, or individual drug testing in personalized medicine. Therefore, the objective of this study was to use iPS cells derived from human fetal fibroblasts to generate TCs in order to establish a proof-of-concept model to study the pathogenesis of trophoblast-associated diseases common during pregnancy.
Results
Generation and characterization of human iPS cell lines
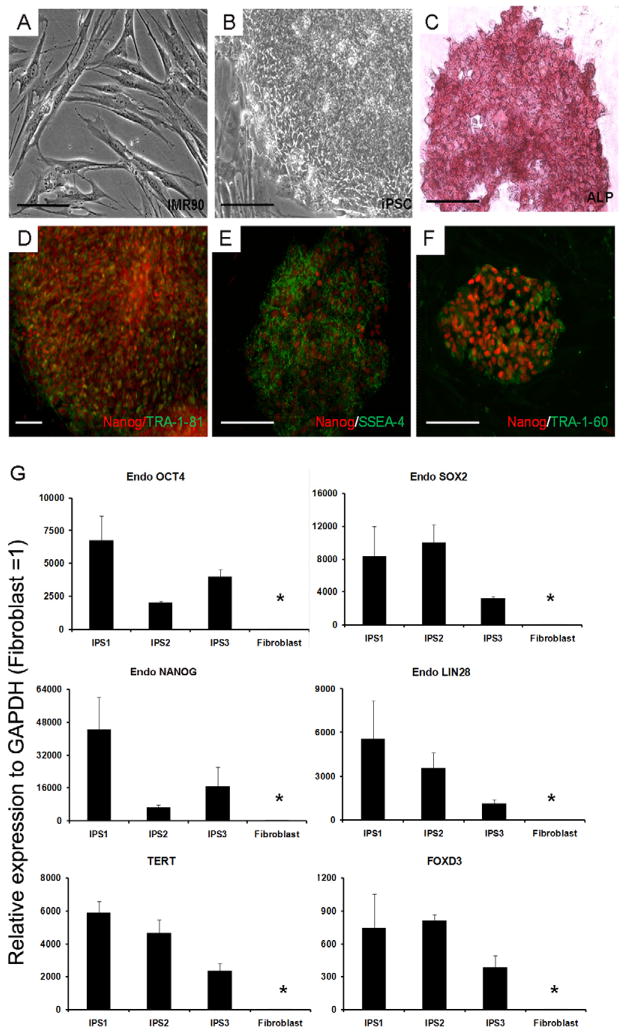
Human iPS cells, which form distinct colonies composed of small round cells (increased nuclear/cytoplasm ratio) were successfully established from human fetal fibroblasts (IMR-90) using a combination of six reprogramming factors as described previously [14; 15] [Fig. 1A–B]. All six of the iPS cell lines generated showed similar growth and morphology characteristics. Three of these iPS cell lines were characterized in detail (iPS1, iPS2, iPS3). These cells stained positive for alkaline phosphatase (ALP), [Fig. 1C] as well as pluripotency surface markers (TRA-1-81, SSEA-4, and TRA-1-60) [Fig. 1D–F]. The iPS cell lines maintained normal karyotype throughout the lentiviral transduction and the differentiation process [Fig. S1]. qPCR revealed that each of the three iPS cell lines highly expressed endogenous pluripotency genes (POU5F1, SOX2, NANOG and LIN28) as well as FOXD3 and TERT similar to ES cells [14; 15] and unlike the donor fibroblast [Fig. 1G]. The iPS cells also demonstrated pluripotent capability by forming embryoid bodies (EBs) [Fig. S2A]. EBs were plated for outgrowths and stained positive for beta-tubulin, alpha-SMA, and AFP; specific markers of ectoderm, mesoderm, and endoderm, respectively [Fig. S2B–D]. Furthermore, EBs embedded in Matrigel with BMP4 could secrete estradiol, progesterone and hCG [Fig. S3A] and form multi-nuclear cells [Fig. S3B].
Fig. 1.
Induced pluripotent stem cells (iPS cells) established from human fibroblast. (A) Human fetal fibroblast (IMR-90). (B) iPS cells generated from IMR-90. (C) Alkaline phosphatase (ALP) was expressed in iPS cells. (D, E and F) NANOG/TRA-1-81, NANOG/SSEA-4 and NANOG/TRA-1-60 were expressed in iPS cells. (Scale bars, D are 100 μm and the remaining is 200 μm.) (G). Endogenous 4 pluripotent factors POU5F1, SOX2, NANOG and LIN28 as well as FOXD3 and TERT were used to measure their respective expression levels. Expression was detected from all 6 loci in the iPS cells, compared to donor fibroblasts (P<0.05). The expression level relative to fibroblast=1.
Differentiation of iPS cells into TCs
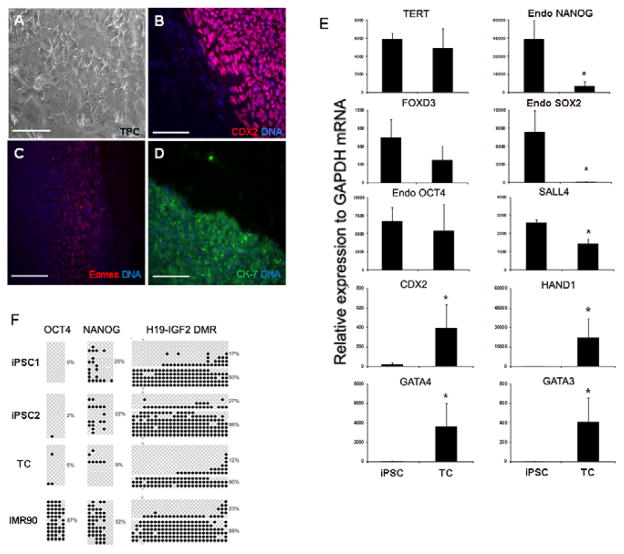
To determine whether human iPS cells could be differentiated into TCs. iPS cells were cultured in human ES cell medium in the presence of BMP4 and bFGF on feeders for approximately 14 days. By day 7–10 of culture, TCs derived from iPS cells stained positive for trophoblast lineage specific markers CDX2, CK-7, and EOMES which were detected in the cells along the perimeter of the colony [Fig. 2A–D]. These cells expressed trophoblast lineage specific genes including CDX2, GATA3, GATA4, and HAND1, while also expressing pluripotent genes POU5F1, TERT and FOXD3, but the expression of NANOG, SOX2 and SALL4 were significantly decreased (P<0.05) [Fig. 2E]. These results demonstrate that human iPS cells can differentiate into TCs in a manner that resembles human ES cells.
Fig. 2.
iPS cells (iPSC) generated from fibroblasts were differentiated into trophoblast lineage cells (TC) with BMP4. (A) TC derived from iPSC. (B–D) TC identity is confirmed with CDX2, EOMES and CK-7 protein expression in cultures. Scale bar, 200μM. (E) TC express pluripotent associated transcripts: FOXD3, TERT and POU5F1, as well as NANOG, SOX2 and SALL4 (P<0.05); and trophoblast specific transcripts: CDX2, HAND1, GATA3 and GATA4. (F) DNA methylation status of iPSC, TC and fibroblasts on POU5F1, NANOG, H19-IGF2.
Epigenetic signature of iPS cells and TCs
Given the tight epigenetic regulation of embryonic and trophoblast lineages, we sought to investigate CpG methylation of key genes in both iPS cells and TCs [Fig. 2F]. The differentiation of iPS cells to TCs showed that the CpG islands of the promoters of pluripotent genes POU5F1 and NANOG were hypomethylated in iPS cells and TCs, but were hypermethylated in IMR-90 donor fibroblasts consistent with the low levels of POU5F1 and NANOG expression in these cells (P<0.05). Additionally, the imprinted locus (H19-Igf2) exhibited a similar methylation pattern among IMR-90 donor fibroblasts, iPS cells and TCs, demonstrating that the pattern of CpG methylation was faithfully conserved during the reprogramming and differentiation process.
Transcriptome changes in fibroblasts, iPS cells, and TCs
To further define the differentiation continuum and differences between fibroblasts, iPS and TCs, we performed gene expression analysis by microarray. Cluster analysis revealed that iPS and TCs form a sub-cluster separately from fibroblasts [Fig. 3A]. Scatter plot comparison reveals that the correlation between iPS and TCs (coefficient r = 0.967) is much higher than the correlation between iPS cells and fibroblasts (r = 0.838) [Fig. 3B–D]. There were 3238 features common between the significantly altered 5120 features in iPS and 7714 features in TCs [Fig. 3E], indicating greater similarity between iPS and TCs expression profiles as compared to fibroblasts. From principal component analysis (PCA), the projection of two PCs covering 78% of the total variance reveals that both iPS and TCs are differentially expressed from fibroblasts along PC #1 (57% variance) [Fig. 3F]. The differences between iPS and TCs are smaller as depicted along PC #2 that covers only 21% of the total variance. The representative genes which show differences between iPS cells and TCs are listed in Table 2. Collectively, these results show that human TCs generated from iPS cells are more similar to iPS cells than fibroblasts, but exhibit a distinct gene expression signature that resembles trophoblasts.
Fig. 3.
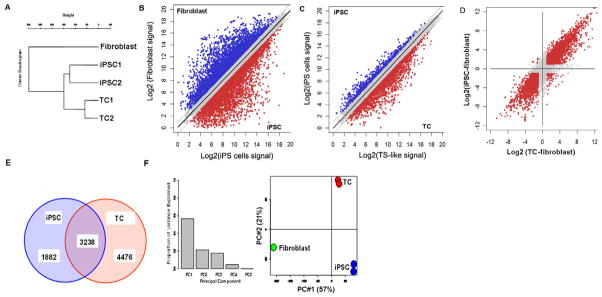
Transcriptome analysis iPS cells (iPSC), trophoblast lineage cells (TC), and fibroblasts. (A) Clustering analysis indicates that iPSC and TC form a sub-cluster separately from fibroblasts. (B–D) The scatter plots reveal that the correlation between iPSC and TC (coefficient r = 0.967) is much higher than the correlation between iPSC and fibroblast (r = 0.838). (E) There were 3238 features common between the significantly altered 5120 features in iPSC and 7714 features in TC. These observations indicate greater similarity between iPSC and TC expression profiles compared to fibroblasts. (F) From PCA, the projection on two principal components (PCs) covering 78% of the total variance reveals that both iPSC and TC are differentially expressed from fibroblasts along PC #1 (57% variance). The differences between iPSC and TC are smaller as depicted along PC #2 that covers only 21% of the total variance.
Table 2.
| Gene name | Alternative Name/Function | |
|---|---|---|
| High in trophoblast Lineage cells vs. iPS cells | CDX2 | (caudal type homeobox 2) Important functions in early differentiation to trophoblast lineage |
| EGF | (epidermal growth factor) stimulates the growth of various epidermal and epithelial tissues in vivo and in vitro | |
| GATA2 | (GATA binding protein 2) Transcriptional activator which regulates endothelin-1 gene expression in endothelial cells. | |
| KRT8 | (keratin 8) together with KRT18, to be necessary for embryo survival, | |
| LIFR | (leukemia inhibitory factor receptor alpha) Signal-transducing molecule. May have a common pathway with IL6ST. | |
| ISL1 | (ISL LIM homeobox 1) Binds to one of the cis-acting domain of the insulin gene enhancer. | |
| IGF2 | (insulin-like growth factor 2) is influenced by placental lactogen and may play a role in fetal development | |
| COL3A1 | (collagen, type III, alpha 1) Collagen type III occurs in most soft connective tissues along with type I collagen | |
| HGF | (hepatocyte growth factor) acts as growth factor for a broad spectrum of tissues and cell types. | |
| GATA3 | (GATA binding protein 3) an important regulator of T-cell development and plays an important role in endothelial cell biology. | |
| GATA4 | (GATA binding protein 4) Acts as a transcriptional activator of ANF in cooperation with NKX2-5. | |
| High in iPS cells vs. Trophoblast lineage cells | SOX2 | (SRY (sex determining region Y)-box 2) Critical for early embryogenesis and for embryonic stem cell pluripotency |
| SLAIN1 | (SLAIN motif family, member 1) stem cell gene | |
| ZNF483 | (zinc finger protein 483) May be involved in transcriptional regulation | |
| EPB41 | (erythrocyte membrane protein band 4.1) tumor suppressor genes in meningioma pathogenesis. | |
| LRIG1 | (leucine-rich repeats and immunoglobulin-like domains) Act as a feedback negative regulator of signaling by receptor tyrosine kinases | |
| PRDM14 | (PR domain containing 14) may suppress expression of differentiation marker genes in human embryonic stem cells | |
| POLR3G | (polymerase (RNA) III (DNA directed) polypeptide G) human embryonic stem cell specific gene | |
| CAV1 | (caveolin 1, caveolae protein) CAV-1 is a transcriptional target of EWS/FLI-1 | |
| NANOG | (Nanog homeobox) Imposes pluripotency on ES cells and prevents their differentiation towards extraembryonic endoderm and trophectoderm lineages. | |
| SALL4 | (Sal –like 4) may be a zinc finger transcription factor |
Biofunction of TCs
To determine if TCs can be reliably differentiated into syncytiotrophoblasts, multi-nucleated cells expressing CK-7 and progesterone, estradiol and hCG were identified by day-7 differentiating culture of TCs, indicative of syncytiotrophoblast formation [Fig. 4A–C]. Furthermore, to determine whether TCs were functionally similar to human trophoblasts they were grown on Matrigel-coating plates to assess invasion. The invasion percentage of TCs versus fibroblasts was 58.4% versus 25.3% at 20 hours (n=3, p<0.01), indicative of invasive trophoblast formation [Fig. 4D].
Fig. 4.
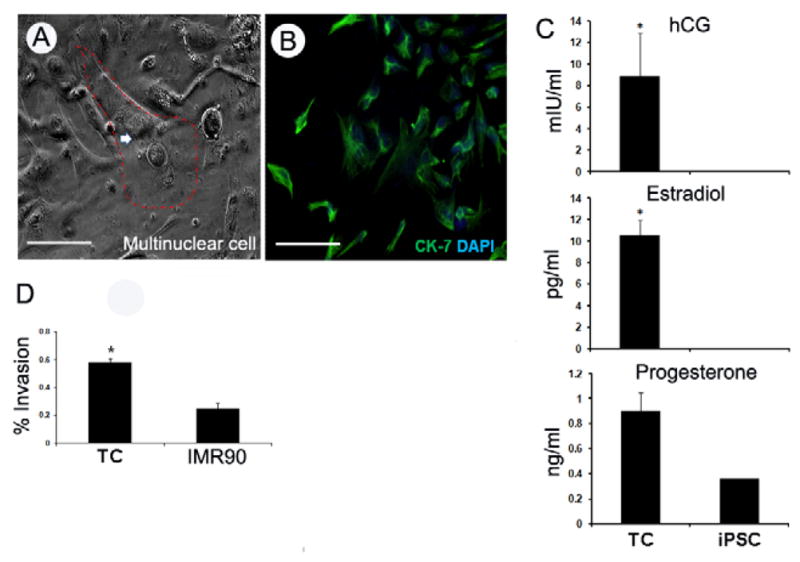
Trophoblast lineage cells (TC) derived from iPS cells (iPSC) differentiated into trophoblasts with 100ng/ml BMP4 for 7-day. (A) Multinuclear trophoblasts formed, nuclei marked by red. (B) CK-7 (green) positive cells. (C) Estradiol, hCG and Progesterone detected from D-7 culture medium. (n=3). (D) TC showed significant invasive capability compare to donor fibroblast in 20~22 hours invasion assay (n=3). Scale bar, 200μM.
Discussion
The goal of this study was to develop a proof-of-principle model to derive TCs from human fibroblasts in vitro by employing a combination of iPS cell technology and BMP4 induced differentiation. The findings reported here reveal that TCs derived from human iPS cells are similar to those derived from human ES cells in terms of morphology, functionality, gene expression, and epigenetic control [11].
Establishment of human iPS cells
The human iPS cells that were established from fibroblasts in this study are comparable to human ES cells in terms of pluripotency, gene expression, and epigenetic signature, which is consistent with others reports [14; 15]. EBs, which formed from iPS cells, secreted trophoblast hormones in different time courses in Matrigel with BMP4 consistent with findings in human ES cells [4; 5; 6; 7; 8; 10; 11; 12; 13].
BMP4-induced differentiation of human iPS cells into TCs
BMP4 can efficiently induce differentiation of human ES cells into trophoblasts [4; 5; 6; 7; 8; 10; 11; 12; 13]. In the present study, our TCs derived from iPS cells exhibited epithelial colony morphology in culture for 2 weeks. The TCs expressed human trophectoderm-specific genes and other pluripotent genes, which are consistent with the trophectoderm of human blastocysts [16; 17; 18; 19] [Table 2]. The complicated scenario of bFGF, BMP4, or both in maintaining and differentiation of human ES cells have been extensively studied. bFGF is found to help maintain pluripotent status of ES cells through MEK/ERK signaling [20], while BMP4 facilitates trophoblast lineage differentiation combined with TGF-beta signaling [13]. Recently, it has been challenged that human ES cells cultured in the presence of BMP4 alone or in combination with bFGF, do not give rise to “real” trophoblast lineage cells [21; 22]. For example, Bernado et al showed that human ES cells treated with BMP4 largely favored mesoderm lineage rather than trophoblast lineage [21]. However, the work of Roberts and colleagues demonstrated that human ES cells treated with BMP4, under specific culture conditions, predominantly form TCs [6; 9]. Our iPS cells based BMP4 model is in agreement with their findings. Nonetheless, additional research is necessary to fully understand the role of bFGF, BMP4 or both in the differentiation of human ES cells into TCs.
Human iPS cells differentiation into TCs
Due to limited information on trophectoderm formation in human blastocysts, the mouse system has been extensively characterized to better understand trophectoderm formation in mammals. Our trophoblast characterization showed some similarities as well as differences compared with mouse model. For example, similar to the trophectoderm of human and Rhesus blastocysts, our TCs exhibited high levels of CDX2 and POU5F1, but low levels of NANOG [16; 23]. Unlike the human and Rhesus counterparts, mouse trophectoderm and trophoblast stem (TS) cells highly express CDX2, but not POU5F1 [24]. It has been well documented that in mouse TS cells, POU5F1 and CDX2 exhibit reciprocal regulation [25]. Hence, combining others observations with ours, we postulate that in human trophectoderm, NANOG and CDX2 may exhibit reciprocal regulation instead of POU5F1 and CDX2 reciprocal regulation. Future studies are necessary to establish the exact relationship between CDX2 and NANOG in human trophectoderm.
To address whether or not our TCs exhibited trophoblast biofunction, we removed bFGF and feeder cells from the culture medium. We found multinuclear cells and detectable levels of progesterone, estradiol and hCG in our differentiating culture, indicating syncytio-like trophoblasts. Meanwhile, these cells showed significant invasion capability compared to donor fibroblasts, a feature of extravillous trophoblasts. These findings further suggest that these TCs have multiple differentiation potency in vitro, which is consistent with others [8; 11; 26]. Unfortunately, in this study we did not have access to primary cytotrophoblasts to serve as a side-by-side positive control for hormone and invasion assays when testing our TCs.
The epithelial-like phenotype of TCs we generated from iPS cells suggests that this cell model could be a useful tool to recapitulate human trophoblast development in vitro. Furthermore, the establishment of TCs from iPS cells provides a proof-of-concept model to recapitulate the pathogenesis of placental trophoblasts in vitro when using a patient’s cells as donor cells. Using a patient’s placental fibroblasts as donor cells will be the focus for a future study.
Materials and Methods
A. Cell Culture
iPS cells generation
Human iPS cells were generated using previously reported methods [14; 15]. In brief, Human lenti-virus constructs for POU5F1, SOX2, NANOG, LIN28, C-MYC, and KLF4 (OpenBiosystems) were used to produce iPS cells from human fetal fibroblasts (IMR-90, ATCC). 1.25X10^5 fibroblasts were transduced by a mixture of 6 viral genes in fibroblast medium (DMEM, high glucose + 10% FBS + 1% L-Glu + 1% NEAA + 1% penicillin/streptomycin(P/S)) and cells were incubated with virus cocktail for 24 hours. After 96 hours, cells were passaged onto in-activated CF-1 mouse embryonic fibroblast (Millipore) in fibroblast medium. The following day fibroblast medium was replaced with human ES medium (DMEME/F12+ 20% serum replacement + 1% L-Glu+ 1% NEAA + 0.5% P/S + 0.1mM 2-ME + 10ng/ml bFGF), and cells were subsequently cultured in human ES medium. Colonies were manually picked approximately day-25~30 after viral transduction. Colonies were passaged mechanically every 5~7 days. All of the reagents used in this study were from invitrogen unless otherwise noted.
iPS cells differentiation to TCs
Human iPS cells differentiating into TCs using previously reported methods [8] with some modifications. After 48 hours iPS cells were split onto a feeder and treated with 100ng/ml BMP4 (R&D) in human ES medium. The BMP4/ES medium was changed every other day. Cells were passaged every 5 days at a ratio of 1:3~4.
Biofunction assay of TCs
Biofunction assay using previously reported methods [8]. In brief, day-15 TCs were split onto a 4-well plate and cultured with 100ng/ml BMP4 in ES medium (−bFGF). The medium was changed every other day. The cells and supernatant were analyzed on day-7 after the onset of treatment.
ELISA assay
Supernatant of day-7 trophoblasts, day-15 TCs and iPS cells were collected to detect the hCG, progesterone, and estradiol by ELISA kit (R&D), following the manufacture’s instruction. The readout in spectrum was between 490nm~560nm.
Invasion assay
Invasion assays were carried out using previously described methods [27]. In brief, TCs and fibroblast (IMR-90) were seeded 2×10^4 per upper cells (BD cop.) with/without matrigel in the upper wells respectively. The medium used in upper well is: DMEM/F12 + 20% serum replacement + 1% NEAA + 1% Glu + 0.1mM 2-ME. The medium used in bottom well is: RPMI1640 + 20% FBS + 1% sodium pyruvate + 1% L-Glu + 0.1mM 2-ME. Around 20~22 hours, the cells were collected and stained by 0.09% Crystal violet solution. The upper cells were carefully removed by cotton swab, and then 5 fields per backside upper cell were photographed and counted (Nikon software).
B. qRT-PCR
Total RNA was isolated with PicoPure RNA isolation kit (Moleculardevices), and first strand cDNA synthesis was performed by Superscript II (invitrogen). qPCR was conducted using SYBR green PCR master Mix (Applied Biosystems) and Primers used were summarized in Table 1.teepOne Plus.)
Table 1.
Primers used in this study
| Gene names | Forward primer | Reverse primer |
|---|---|---|
| POU5F1 | GCCTGCCCTTCTAGGAATGG | AGTGAATGAAGAACTTAATCCCA |
| LIN28 | AGTGGAGAAAGTGGGAATAGG | TCCTTTGACTCACCCTTTCC |
| KLF4 | GAGGGAAGGAGCCCAGCCAG | TTTGTCTTTTGGATTCCTCAT |
| CMYC | CTAACAGAAATGTCCTGAGCAATC | CCCAAAGTCCAATTTGAGGC |
| SOX2 | AGCGAACTGGAGGGGGGAGA | TTCTTTTTGAGCGTACCGGG |
| NANOG | ACTTGGAGGCTGCCTTGGAA | AGGTTGCATGTTCATGGAGTAG |
| viral POU5F1 | GAAGCCTTTCCCCCTGTCTC | CACAAATTTTGTAATCCAGAGG |
| Viral NANOG | GGGTGGAGTATGGTTGGAGC | CACAAATTTTGTAATCCAGAGG |
| viral LIN28 | AGTGCACAGGGAAAGCCAAC | CACAAATTTTGTAATCCAGAGG |
| viral KLF4 | CCCGTTCCAGTGCCAAAAAT | CACAAATTTTGTAATCCAGAGG |
| viral CMYC | GGACTTGTTGCGGAAACGAC | CACAAATTTTGTAATCCAGAGG |
| viralSOX2 | CTTCACATGTCCCAGCACTA | CACAAATTTTGTAATCCAGAGG |
| GATA 4 | AGTCTCCACAGACCAGCTCC | AGGTCCGTGCAGGAATTTGAG |
| GATA 3 | ACATGCTGACCACGCCCACG | GCAGGGCTCTAACCCATGGC |
| FOXD3 | GGGCAAGGGCAACTACTGGA | GTAAGCGCCGAAGCTCTGCAT |
| EOMES | AGCTCTCCAAGGAGAAAGTG | GCCTTCGCTTACAAGCACTG |
| HAND1 | TCAAGGCTGAACTCAAGAAGG | TGCGTCCTTTAATCCTCTTC |
| CDX2 | CCCTCGGCAGCCAAGTGAAA | TCCTCCGGATGGTGATGTAG |
| G6PDH | TGGCCTCCAAGGAGTAAGAC | CTCTTCAAGGGGTCTACATGG |
| TERT | TGATGAGTGTGTACGTCGTCGA | AAGTGCTGTCTGATTCCAATGC |
C. Immunocytochemistry
Cells were fixed in 4.0% paraformaldehyde for 10 minutes, and then penetrated with 0.3% triton X-100 in PBS for 15 minutes. Cells were blocked with 5% BSA for 30 minutes. Cells were incubated in primary antibody for 4°C overnight, and then incubated with secondary antibody (Alexa Flours, Invitrogen) for 45 minutes. The cells were finally stained for the nuclei by Hochest33342 (sigma). Imaging used a Nikon 2000 inverted microscope and was analyzed by Openlab system (Apple). Primary antibodies used in this study were SSEA-4 (1:100, Millipore), TRA-1-60 (1:100, Millipore), TRA-1-81 (1:100, Millipore), NANOG (1:100, Abcam), CDX2 (1:100, Abcam), EOMES (2μg/ml, Abcam), CK-7 (1:100, Dako), ALP activity was detected by ALP substrate kit (Vector) according to manufacturer’s protocol.
D. DNA Methylation
The method as previously described [28]. Briefly, 3×10^4 iPS cells, fibroblasts, or TCs were collected. Extraction of genomic DNA and bisulfate mutagenesis were performed using the ReadyAmp Genomic Kit (Promega, Madison, WI) and the EZ DNA Methylation Kit (Zymo Research, Orange, CA), respectively. PCR products were verified by running on a 2% agarose gel, and then purified and ligated into the pTOPO 10 vector system (invitrogen) and more than 10 clones per cell sample were randomly picked up for sequencing.
E. Gene Array and Analysis
We analyzed RNA with human genome 4×44K expression chip (Agilent Technologies, Santa Clara, CA) using the manufacture’s Reader and collected primary data using the supplied Scanner software. Background subtracted signals were extracted using Agilent Feature Extraction Software (v9.5). The signals of each array were normalized by median centering of ratios considering fibroblast as reference RNA. The features flagged as detected in all of iPS cells or TCs replicates or fibroblast were included in the analysis [29]. All the statistical calculations were performed on logarithmic values of signals to the base 2. The statistical significance of iPS cells or TCs versus fibroblast differential expression was evaluated by two-tailed one-sample T-tests. The features which differentially expressed in iPS cells and TCs respectively that altered by 2-fold at p < 0.05 and having at least one geometric average signal > 100 will be analyzed. The global expression profiles of fibroblast, iPS cells and TCs were examined by hierarchical clustering analysis, principal component analysis (PCA), and scatter plots using R-statistical package version 2.10.1 (http://www.R-project.org). About 25,700 probes which were flagged as detected in all of the arrays were included for these analyses.
F. Statistical analysis
Student’s t-test (paired or unpaired) was used for calculation of statistical significance. Error bars on the graphs show standard deviation of three or more replicates (n = 3) or samples.
Supplementary Material
Epithelial-like phenotype of trophoblast lineage cells derived from human iPS cells
Trophoblast lineage cells derived from human iPS cells exhibit trophoblast function
Trophoblasts from iPS cells provides a proof-of-concept in regenerative medicine
Acknowledgments
The authors would like to thank Dr. Trixie Smith and Ms Susan Ferguson for critically reading the manuscript and providing constructive criticism. This research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institute of Health cooperative agreement (U54 HD 40093 to R.E.L) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research, and in part by a grant from the National Institute of General Medical Sciences to J.G.K. (R01GM09534).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luke B, Brown MB, Wantman E, Lederman A, Gibbons W, Schattman GL, Lobo RA, Leach RE, Stern JE. Cumulative birth rates with linked assisted reproductive technology cycles. N Engl J Med. 366:2483–91. doi: 10.1056/NEJMoa1110238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honnma H, Baba T, Sasaki M, Hashiba Y, Ohno H, Fukunaga T, Endo T, Saito T, Asada Y. Trophectoderm morphology significantly affects the rates of ongoing pregnancy and miscarriage in frozen-thawed single-blastocyst transfer cycle in vitro fertilization. Fertil Steril. 98:361–7. doi: 10.1016/j.fertnstert.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Hemberger M. Health during pregnancy and beyond: Fetal trophoblast cells as chief co-ordinators of intrauterine growth and reproductive success. Ann Med. 44:325–37. doi: 10.3109/07853890.2012.663930. [DOI] [PubMed] [Google Scholar]
- 4.Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM. Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res. 2007;1:61–74. doi: 10.1016/j.scr.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erb TM, Schneider C, Mucko SE, Sanfilippo JS, Lowry NC, Desai MN, Mangoubi RS, Leuba SH, Sammak PJ. Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev. 20:1601–14. doi: 10.1089/scd.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezashi T, Telugu BP, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. 349:809–24. doi: 10.1007/s00441-012-1371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golos TG, Pollastrini LM, Gerami-Naini B. Human embryonic stem cells as a model for trophoblast differentiation. Semin Reprod Med. 2006;24:314–21. doi: 10.1055/s-2006-952154. [DOI] [PubMed] [Google Scholar]
- 8.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–4. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 9.Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci U S A. 110:E1212–21. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerami-Naini B, Dovzhenko OV, Durning M, Wegner FH, Thomson JA, Golos TG. Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology. 2004;145:1517–24. doi: 10.1210/en.2003-1241. [DOI] [PubMed] [Google Scholar]
- 11.Harun R, Ruban L, Matin M, Draper J, Jenkins NM, Liew GC, Andrews PW, Li TC, Laird SM, Moore HD. Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Hum Reprod. 2006;21:1349–58. doi: 10.1093/humrep/del017. [DOI] [PubMed] [Google Scholar]
- 12.Udayashankar R, Baker D, Tuckerman E, Laird S, Li TC, Moore HD. Characterization of invasive trophoblasts generated from human embryonic stem cells. Hum Reprod. 26:398–406. doi: 10.1093/humrep/deq350. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, Gao Y, Dai H, Yuan J, Cheng L, Xiao L. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem. 2008;283:24991–5002. doi: 10.1074/jbc.M803893200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Cauffman G, De Rycke M, Sermon K, Liebaers I, Van de Velde H. Markers that define stemness in ESC are unable to identify the totipotent cells in human preimplantation embryos. Hum Reprod. 2009;24:63–70. doi: 10.1093/humrep/den351. [DOI] [PubMed] [Google Scholar]
- 17.Bai Q, Assou S, Haouzi D, Ramirez JM, Monzo C, Becker F, Gerbal-Chaloin S, Hamamah S, De Vos J. Dissecting the first transcriptional divergence during human embryonic development. Stem Cell Rev. 8:150–62. doi: 10.1007/s12015-011-9301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, Gosden RG, Lehrach H. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005;23:1514–25. doi: 10.1634/stemcells.2005-0113. [DOI] [PubMed] [Google Scholar]
- 19.Chen AE, Egli D, Niakan K, Deng J, Akutsu H, Yamaki M, Cowan C, Fitz-Gerald C, Zhang K, Melton DA, Eggan K. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell. 2009;4:103–6. doi: 10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–46. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 21.Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, Bardwell L, Moffett A, Pedersen RA. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 9:144–55. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 8:326–34. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harvey AJ, Armant DR, Bavister BD, Nichols SM, Brenner CA. Inner cell mass localization of NANOG precedes OCT3/4 in rhesus monkey blastocysts. Stem Cells Dev. 2009;18:1451–8. doi: 10.1089/scd.2009.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 25.Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev. 2007;19:111–8. doi: 10.1071/rd06125. [DOI] [PubMed] [Google Scholar]
- 26.Douglas GC, VandeVoort CA, Kumar P, Chang TC, Golos TG. Trophoblast stem cells: models for investigating trophectoderm differentiation and placental development. Endocr Rev. 2009;30:228–40. doi: 10.1210/er.2009-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leach RE, Kilburn B, Wang J, Liu Z, Romero R, Armant DR. Heparin-binding EGF-like growth factor regulates human extravillous cytotrophoblast development during conversion to the invasive phenotype. Dev Biol. 2004;266:223–37. doi: 10.1016/j.ydbio.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Wang K, Chen Y, Chang EA, Knott JG, Cibelli JB. Dynamic epigenetic regulation of the Oct 4 and Nanog regulatory regions during neural differentiation in rhesus nuclear transfer embryonic stem cells. Cloning Stem Cells. 2009;11:483–96. doi: 10.1089/clo.2009.0019. [DOI] [PubMed] [Google Scholar]
- 29.Mold JE, Venkatasubrahmanyam S, Burt TD, Michaelsson J, Rivera JM, Galkina SA, Weinberg K, Stoddart CA, McCune JM. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science. 330:1695–9. doi: 10.1126/science.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.