Background: Transcriptional elongation is a rate-limiting step in activation of stress response genes.
Results: Optimal expression of stress response regulator ATF3 requires the elongation activity but not the ubiquitination activity of Elongin A.
Conclusion: Elongin A plays a key role for the adequate expression of ATF3 in vivo.
Significance: RNAPII ubiquitination and transcriptional elongation are independent activities of Elongin A.
Keywords: Gene Regulation, Mammal, RNA Polymerase II, Stress Response, Transcription Elongation Factors, ATF3, Elongin A
Abstract
Elongin A was shown previously to be capable of potently activating the rate of RNA polymerase II (RNAPII) transcription elongation in vitro by suppressing transient pausing by the enzyme at many sites along DNA templates. The role of Elongin A in RNAPII transcription in mammalian cells, however, has not been clearly established. In this report, we investigate the function of Elongin A in RNAPII transcription. We present evidence that Elongin A associates with the IIO form of RNAPII at sites of newly transcribed RNA and is relocated to dotlike domains distinct from those containing RNAPII when cells are treated with the kinase inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole. Significantly, Elongin A is required for maximal induction of transcription of the stress response genes ATF3 and p21 in response to several stimuli. Evidence from structure-function studies argues that Elongin A transcription elongation activity, but not its ubiquitination activity, is most important for its function in induction of transcription of ATF3 and p21. Taken together, our data provide new insights into the function of Elongin A in RNAPII transcription and bring to light a previously unrecognized role for Elongin A in the regulation of stress response genes.
Introduction
Transcription initiation by mammalian RNA polymerase II (RNAPII)3 requires a set of general initiation factors TFIID (or TBP), -IIB, -IIF, -IIE and -IIH, which all assemble with RNAPII to form a preinitiation complex (PIC). After initiation, RNAPII completes transcribing successfully at a fairly constant rate of 3–4 kb/min even in the case of an extraordinarily long (e.g. megabase) transcript. However, in some cases, transcribing RNAPII is subject to promoter-proximal pausing. In addition, subsequent to release from the promoter-proximal pause site, RNAPII is subject to frequent pausing, resulting in premature termination or inefficient elongation of nascent transcripts.
Transcript elongation by RNAPII can be regulated by a collection of elongation factors of which there are at least 20 in mammals (1, 2). Negative elongation factors, such as DSIF and NELF, are required for promoter-proximal pausing. Reactivation of paused RNAPII depends on a multiprotein complex called the super elongation complex, which contains the positive transcription elongation factor P-TEFb (Cdk9/CyclinT), elongation factor ELL/EAF (eleven-nineteen lysine-rich in leukemia/ELL-associated factor), and a collection of additional proteins (3–6). Some factors, such as FACT (7) and Elongator (8), function in a chromatin-dependent manner. ELL/EAF (9, 10), TFIIF (11–13), Cockayne syndrome protein B (14), and Elongin (15, 16) are all capable of activating the overall rate of elongation by suppressing transient pausing or by arrest of RNAPII, whereas SII family members reactivate arrested RNAPII after partial cleavage of the 3′-end of the transcript (17, 18). Although the biochemical activities of these factors have been well documented, their contributions to gene regulation in cells are only now beginning to emerge. For example, an RNAPII elongation factor, Elongin A, plays an essential role in mouse development especially in neuronal differentiation (19).
Elongin is a multimeric elongation factor comprising three subunits, Elongins A, B, and C (15, 16, 20, 21). Elongin A interacts directly with RNAPII in vitro (22) and carries the elongation stimulatory activity of Elongin (21); at least three isoforms of Elongin A (A, A2, and A3) are present in human (23, 24). Elongin A contains an N-terminal region (residues 1–120) similar to the N terminus of SII that is dispensable for its elongation activity and a C-terminal elongation stimulatory domain that falls between residues 400 and 773 (25). Within this C-terminal domain is a SOCS box containing the Elongin B and C binding site at residues 550–588 that together with Elongins B and C is required for maximal transcriptional activity in vitro (25). Recently, we showed that the C-terminal elongation activation domain contains a binding site for RNAPII between residues 590 and 690; this region of Elongin A is crucial for in vitro elongation activity (22).
Elongin A acts not only as a component of a transcript elongation factor but also as the substrate recognition subunit of an Elongin BC-containing ubiquitin ligase complex in which Elongins B and C link Elongin A to Cullin 5 and the RING finger protein Rbx2. Several recent studies including our own have suggested that the Elongin ABC-Cul5/Rbx2 ubiquitin ligase contributes to ubiquitination and degradation of RNAPII stalled at sites of DNA damage (26, 27).
We previously carried out a comprehensive microarray analysis that revealed that expression of only a small fraction of genes (∼3–4%) is affected in Elongin A-deficient mouse ES cells, raising the possibility that Elongin A is not a global regulator of gene expression but rather controls the expression of select genes (28). More recently, we have generated Elongin A-deficient mice (29) and showed that Elongin A plays a crucial role in expression of a variety of genes for neural development (19). We also observed that mouse embryonic fibroblasts (MEFs) derived from Elongin A-null embryos displayed both increased apoptosis and senescence-like growth defects (29). These phenotypes were accompanied by activation of p38 mitogen-activated protein kinase and p53, suggesting a possible role for Elongin A in stress responses.
Typically, stress response genes must be activated rapidly in reaction to stimuli. To guarantee the swiftness of activation, RNAPIIs that are paused near the 5′-ends of genes are in a “poised” state. Increasing evidence indicates that release from this pausing is the rate-limiting step for the regulation of gene activation in vivo (30). For example, P-TEFb (31, 32), NELF and DSIF (33), FACT (34), and Elongin A (35) are reported to be involved during the activation of HSP70 gene. Furthermore, P-TEFb and NELF are implicated in epidermal growth factor-mediated c-fos (36, 37) and junB induction (38), respectively. However, functional details of Elongin A in in vivo stress response are still elusive.
In this study, we explored further the role of Elongin A in stress responses. Our results provide evidence for a role for Elongin A in controlling expression of the leucine zipper transcription factor and stress response regulator activating transcription factor 3 (ATF3). Arguing that Elongin A directly regulates ATF3 transcription, we show that it is recruited to the transcribed region of ATF3 gene in response to various stimuli. Furthermore, domains of Elongin A essential for maximal induction of the stress response were also identified.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
A mammalian expression plasmid vector, pCI-neo-hElongin A, was prepared by subcloning a human Elongin A cDNA containing an N-terminally tagged FLAG sequence between the NheI and XbaI sites of pCI-neo vector (Promega). Mammalian expression plasmids for rat Elongin A deletion mutants were as described (22). Rabbit polyclonal anti-RNAPII (N-20), anti-TFIIE-α (C-17), and anti-FCP1 (C-16) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rabbit polyclonal anti-Leo1 (A300-175A) was purchased from Bethyl Laboratories, Inc. Rabbit anti-RAP74 antibody was prepared as described (13), and anti-human Elongin A antibody was generated against the 15 amino acids from 758 to 772 (MAKTIKAFKNRFSRR) and affinity-purified using Affi-Gel-102 (Bio-Rad) conjugated with Elongin A peptide. This antibody was highly specific in Western blots. Antibodies against Elongins B and C were kindly provided by Dr. Takumi Kamura (Nagoya University). Monoclonal anti-RNAPII (8WG16; Covance), anti-Ser2-phosphorylated form of the RNAPII C-terminal domain (CTD) (H5; Covance), anti-Ser5-phosphorylated form of the RNAPII CTD (H14; Covance), anti-TBP (Promega), anti-TFIIB (Promega), anti-FLAG M2 (Sigma), and anti-α-tubulin (Sigma) antibodies were used. Thapsigargin and tunicamycin were from Sigma, and doxorubicin and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) were from Calbiochem. 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) and hydrogen peroxide (H2O2) were from Nacalai Tesque (Japan). Other chemicals were reagent grade.
Cell Culture, Cell Extract Preparation, and Western Blot
HeLa cells were cultured in DMEM supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in 5% CO2 at 37 °C. Cells treated as indicated were harvested, washed in phosphate-buffered saline (PBS), and resuspended in lysis buffer (50 mm Hepes-KOH, pH 7.5, 150 mm NaCl, 1% TritonX-100, 1.5 mm MgCl2, 1 mm EGTA, 0.1 mm PMSF, 10 μg/ml each leupeptin and aprotinin, 200 μm sodium vanadate, 100 mm NaF, 10% glycerol). After incubation on ice for 10 min, the cells were centrifuged at 10,000 rpm for 10 min, and the supernatants were taken as whole cell extract. Protein concentrations were measured by the Lowry method using bovine serum albumin as standard (39). Cell extracts (20 μg of protein) were separated by SDS-PAGE, transferred onto PVDF membranes, subjected to Western blotting, and detected using ImmunoCruz (Santa Cruz Biotechnologies, Inc.) according to the manufacturer's instructions.
Preparation of MEFs
MEFs were prepared from E10.5 embryos as described (29). Briefly, fetal tissue samples were rinsed in PBS and mechanically dissociated in DMEM with 10% FBS. All animal experiments were done with the approval of the Institutional Animal Care Committees of the Tokyo Medical and Dental University and Kochi Medical School.
HeLa Cell Lines Stably Expressing Elongin A or RAP74 Subunit of TFIIF
HeLa cells were transfected with pCI-neo-hElongin A using Superfect (Qiagen) and cultured in the presence of 800 μg/ml neomycin for 9 days. Drug-resistant clones were isolated and assayed for FLAG-tagged Elongin A expression using reverse transcription (RT)-PCR and Western blot. To establish cell lines expressing FLAG-RAP74, HeLa cells stably expressing mouse ecotropic retrovirus receptor were infected with retrovirus vector pMX-puro encoding FLAG-tagged human RAP74, and clonal cell lines positive for FLAG-RAP74 expression were selected in medium containing 20 μg/ml puromycin.
Cell Treatment, RNA Isolation, and RT-PCR
Control or Elongin A-knocked down HeLa cells were treated as indicated. After treatment, total RNA was isolated by the acid-guanidinium method and assayed for mRNAs of various genes by quantitative RT-PCR using a kit from TaKaRa (Japan) as described (40). Primers used were as follows: ATF3, 5′-ATGATGCTTCAACACCCAGGCCA-3′ (forward) and 5′-ATACCACGACTGCTTAGCTCTGC-3′ (reverse); GAPDH, 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ (forward) and 5′-CATGTGGGCCATGAGGTCCACCAC-3′ (reverse); p21, 5′-AGACTCTCAGGGTCGAAAACG-3′ (forward) and 5′-GACACACAAACTGAGACTAAG-3′ (reverse). Averaged results are presented as mean ± S.E.
Immunoprecipitation and Complex Analysis
Nuclear extract of HeLa cells stably expressing FLAG-Elongin A or FLAG-RAP74 or control cells (1 × 107 cells) were prepared as described by Dignam et al. (41). Extracts were then incubated with 0.5 μg of anti-FLAG antibody or control IgG at 4 °C for 3 h in buffer containing 20 mm Hepes-KOH, pH 7.9, 0.2 mm EDTA, 20% glycerol, 0.2% Nonidet P-40, 1 mm PMSF, 0.5 mm DTT, 80 mm NaCl or 150 mm NaCl followed by incubation with 30 μl of Protein G-agarose (Roche Applied Science) for 2 h. The resulting immunocomplexes were washed with the same buffer and subjected to Western blot analysis.
Immunostaining and Labeling of RNA
Cells were fixed with 2% formaldehyde in PBS for 20 min and 0.5% Triton-X for 20 min and then incubated with 100 mm glycine for 10 min. After blocking in PBS containing 3% BSA, cells were incubated with primary antibody at room temperature for 1.5 h or at 4 °C overnight, washed, and sequentially incubated with anti-mouse or anti-rabbit IgG antibody conjugated with Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes). Cell nuclei were stained by treatment with DAPI (Nacalai Tesque). For in vivo RNA labeling, cells were incubated in the presence of 2 mm bromouridine at 37 °C for 20 min and then subjected to immunostaining using mouse monoclonal anti-BrdU antibody (Roche Applied Science). Images were captured with an LSM510 confocal microscope (Carl Zeiss, Jena, Germany). For the evaluation of the colocalization, Pearson's correlation coefficient and van Steensel's cross-correlation coefficient (correlation coefficient and dx) were computed using ImageJ software with the JaCoP plug-in (42).
Isolation of HeLa Cell Lines Stably Expressing Elongin A siRNAs
HeLa cells were transfected with piGENE-neo plasmids (Clontech) encoding siRNA sequences for human Elongin A under the control of the human tRNAVal gene promoter. After selection of cells with 800 μg/ml neomycin for 14 days, each colony was assayed for Elongin A expression by Western blot. Sequences for siRNAs that target nucleotides +1420 to 1439 (siElA6) and nucleotides 2216 to 2235 (siElA4) were 5′-AGgAAGGTGCtTGgTGTGTTCTTCCTGTCAAACACATCAGGCACCTTTCT-3′ and 5′-GtAGCAtTGTTTCtTATGgTCTTCCTGTCAATCATAGGAAACAGTGCTGC-3′, respectively. For rescue experiments, FLAG-tagged rat Elongin A was introduced into these cell lines by transfecting pcDNA3.1 vector encoding rat FLAG-Elongin A and selected by hygromycin. Elongin A knockdown and introduction of rescue constructs were also accomplished using retroviral vectors. Production of retrovirus vector using Plat E cells and pMX-puroII-U6 vector was described elsewhere (43).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed as described (44) according to the protocol supplied by Upstate. Briefly, HeLa cells (4 × 107 cells) were cross-linked with 1% formaldehyde for 10 min at room temperature. After washing twice with PBS, the cells were collected, lysed with 2 ml of SDS lysis buffer (50 mm Tris-HCl, pH 8.1, 10 mm EDTA, 1% SDS) containing a protease inhibitor mixture (Roche Applied Science), and sonicated to DNA lengths of ∼250–500 bp. After centrifugation at 13,000 rpm for 10 min, supernatants were diluted 10-fold in ChIP dilution buffer (16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA). Immunoprecipitations were then performed with the control IgG or the indicated antibodies, and the chromatin immunoprecipitated DNA was assessed by quantitative PCR (qPCR) using primer sets (Ref. 45 and Table 1). Fluorescence intensities were calculated by (Immunoprecipitate average − IgG average)/(Input average). Averaged results are presented as mean ± S.E.
TABLE 1.
Primer sequences for ChIP assay
Primer sets used for ChIP combined with qPCR are shown.
| Forward | Reverse | |
|---|---|---|
| ATF3 | ||
| −5000 | TGGACACACACACGGAAACT | GAAGCAGGCTGGAAGTCAAG |
| P1 | AGGATGATGGAAGGCTGTC | GTCACATCTTCCCATCTGATC |
| +20000 | AATGCCCTCACAGAAACACC | GTTAGGCAGGAAGGGGAAAG |
| +40000 | GCTCTTCAACCTAGCGGAG | GCCCTCTCTCTCCATATCAG |
| +55000 | ACCTGTTCCCCATGGATGTA | TGTGTCAGGGAGCCCAAATA |
| HSP70 | ||
| P | CCGACCCTTCCTGTCAATTA | TCCTCAGGCTAGCCGTTATC |
| 5′ | AGGTGATCAACGACGGAGAC | GCTGCGAGTCGTTGAAGTAG |
| 3′ | GGAGTCCTACGCCTTCAACA | GCTCAAACTCGTCCTTCTCG |
| p21 | ||
| −1984 | CAGAAGTCCTCCCTTAGAGTGTGTCT | GCAACCATGCACTTGAATGTGTA |
| −20 | TATATCAGGGCCGCGCTG | GGCTCCACAAGGAACTGACTTC |
| +507 | CCAGGAAGGGCGAGGAAA | GGGACCGATCCTAGACGAACTT |
| +4001 | AGTCACTCAGCCCTGGAGTCAA | GGAGAGTGAGTTTGCCCATGA |
| +7878 | CCAGCTGGGCTCTGCAATT | GCTGAGAGGGTACTGAAGGGAAA |
| +8566 | CCTCCCACAATGCTGAATATACAG | AGTCACTAAGAATCATTTATTGAGCACC |
| c-Myc | ||
| −2000 | AAGACGCTTTGCAGCAAAATC | AGGCCTTTGCCGCAAAC |
| +15 | GGGCTTCTCAGAGGCTTG | CTGGAATTACTACAGCGAGTTAGAT |
| +190 | GCCGCATCCACGAAACTTT | TCCTTGCTCGGGTGTTGTAAG |
| +2018 | TGCCCCTCAACGTTAGCTTC | GGCTGCACCGAGTCGTAGTC |
| +6028 | TCCCCATATTAGAAGTAGAGAGGGAA | CTTGGGCATGTGGATGAGTCT |
| +7028 | ATCGGGAAGGTGTTAGTCTGAATC | CACTCTCTCCTATTCTGAGGGCTT |
| c-Jun | ||
| −1773 | GAACCCAAGGCTGAATTCCAAG | GATCTTTCCCAAGCCTCTGTC |
| −55 | GGCCTTGGGGTGACATCATGG | CTATACTGCCGACCTGGCTGG |
| +1913 | GGCACAGCTTAAACAGAAAG | CTCAAGTCTGTCTCTCTGTG |
| +3027 | AGCCCACTGAGAAGTCAAAC | GTAAAATCTGCCACCAATTCC |
| +6822 | GTCTAGAGCCCACCTGTTC | GTGAAGACCAGTTCTGTGC |
| RPL29 | ||
| −426 | GTGGTGGGCGCCTGTAATCC | CGGAGTTTCGCTCTTCCACC |
| −84 | TTCGCGGAGCCTGGGGAAGC | CAACACTCACCATAAGCCGC |
| +954 | ACAGCCTAACTGGCCCACCC | CCTCAGGAGGCAGGTTACAC |
| +1888 | CCTCGTAAAGCCCAAGGAGG | TGAGCCCCTTGGCAATACGG |
| +2288 | TCTATGATCCTGGGGATGGG | TCCTCCTCTTGCACTTCAGG |
| +2794 | GCCCAAAGGGAGAAATCTTA | AAATCTGGGACACCTCAGGC |
| GAPDH | ||
| −300 | CAATTCCCCATCTCAGTCGT | GCAGCAGGACACTAGGGAGT |
| +55 | CTCCTGTTCGACAGTCAGC | TTCAGGCCGTCCCTAGC |
| +1019 | GCCCGATTTCTCCTCCG | GGACCTCCATAAACCCACTT |
| +3882 | CCCTGTGCTCAACCAGT | CTCACCTTGACACAAGCC |
| +5747 | TTCACATTCCTGATACGTGGACAA | CGTGGGTGTCTGCTACT |
Luciferase Assay
pLuc-ATF3 (−384) containing the human ATF3 gene promoter was as described (46, 47). HeLa cells (5 × 104 cells), either wild type or siElA6 cells, were co-transfected with pLuc-ATF3 (−384) and 1 μg each of expression vectors encoding rat Elongin A deletion mutants as well as sea pansy control vector. Cells were treated with 1 μm doxorubicin and incubated for 8 h. Cells were then harvested, and their extracts were assayed for luciferase activity as described (40). Both firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System according to the manufacturer's protocol (Promega). pRL-TK (Toyo Ink, Tokyo, Japan) containing the sea pansy luciferase gene was used as an internal control of transfection and expression. Averaged results are presented as mean ± S.E.
Microarray Analysis
siElA or control (siGFP) HeLa cells were treated with 1 μm doxorubicin for 2 h. Total RNA was extracted using an RNeasy kit (Qiagen) according to the manufacturer's instructions. RNA was transcribed to Cy3-labeled cRNA using a Low Input Quick Amp Labeling kit (Agilent). Hybridization, processing, imaging, and analysis were performed according to the Agilent protocols using an Agilent DNA Microarray Scanner and GeneSpring GX software (Platform GPL4133). Data were deposited in the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) under accession number GSE30432.
RESULTS
Elongin A Associates with RNAPII
Results of a previous study suggested that the SII-like N-terminal region of Elongin A when present at high concentration as the bait protein during protein affinity chromatography can bind a complex or complexes containing RNAPII and general initiation factors (48). Furthermore, TFIIS has been reported to function not only as a transcription elongation factor but also as a component of RNAPII preinitiation complexes (49, 50). A domain similar to the Elongin A SII-like region is also found in the human Mediator subunit MED26 and was recently reported to play a role as a docking site for both TFIID and P-TEFb- and ELL/EAF-containing super elongation complexes (51). We therefore began these studies by investigating the in vivo association between the transcription apparatus and Elongin A expressed at physiologically relevant levels. To do so, we established HeLa cells stably expressing FLAG-tagged human Elongin A. As shown in Fig. 1A, FLAG-tagged Elongin A is expressed in these cells at levels similar to the endogenous protein. Nuclear extracts from this cell line and from control cells not expressing a FLAG-tagged protein were subjected to immunoprecipitation with anti-FLAG antibody, and immunoprecipitated proteins were detected by Western blotting. As expected, FLAG-Elongin A bound Elongins B and C in vivo, suggesting that FLAG-tagged Elongin A is functional in this cell line (Fig. 1B). To determine whether Elongin A bound RNAPII, general initiation factors, or other transcriptional regulatory proteins, we performed additional immunoprecipitations at two salt conditions (80 and 150 mm). As a control, proteins associated with the general initiation and elongation factor TFIIF were studied in parallel using cells stably expressing FLAG-tagged TFIIF subunit RAP74. As shown in Fig. 1C, RAP74 copurified with RNAPII and initiation factors TBP, TFIIB, and TFIIE and with the CTD phosphatase FCP1. These interactions were stable in both low and high salt conditions. Elongin A could not be detected in the TFIIF complex at these conditions. On the other hand, an immunoprecipitation of FLAG-Elongin A revealed that Elongin A bound RNAPII but not the other initiation factors examined (Fig. 1C). We next addressed whether these complexes had different binding affinity to RNAPIIO. Fig. 1D shows that the amount of RNAPIIO phosphorylated at Ser5 or Ser2 in the TFIIF complex was significantly reduced in the high salt condition. In contrast, RNAPIIO (Ser5 or Ser2) in the Elongin A complex was more stable under the same salt condition, indicating that Elongin A has stronger affinity to bind RNAPIIO. Thus, our data suggest that Elongin A forms relatively stable complexes with phosphorylated RNAPIIO; however, we could not detect the interaction of Elongin A with components of the PIC when it was expressed at physiologically relevant levels in vivo.
FIGURE 1.
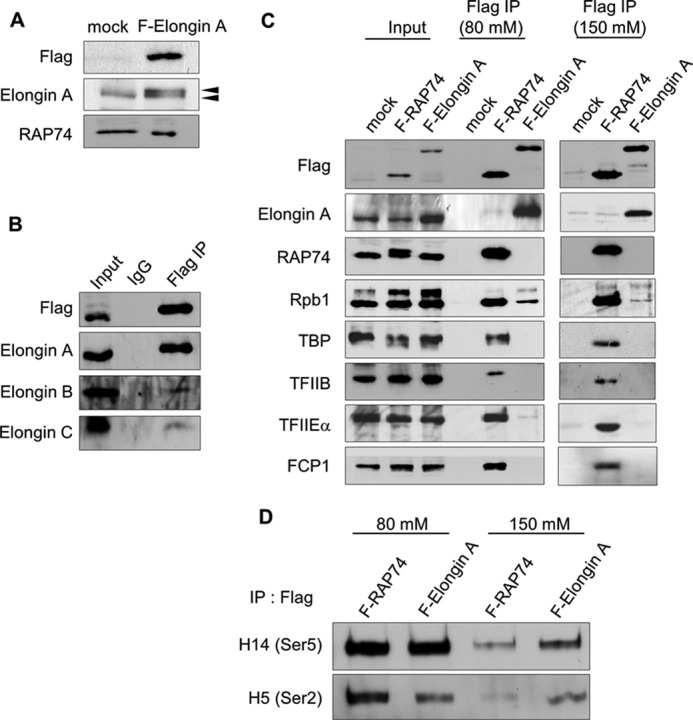
Elongin A complexes immunopurified under different salt conditions. A, nuclear extracts were prepared from HeLa cells stably transfected with pCI-neo-FLAG-hElongin A (F-Elongin A) or with empty pCI-neo (mock) and subjected to Western blotting using anti-FLAG, anti-Elongin A, or anti-RAP74 antibody. Note that dual bands (arrowheads) in FLAG-hElongin A cell extract in the Elongin A blot indicate both endogenous and FLAG-tagged hElongin A. B, nuclear extracts from hElongin A-expressing cells were subjected to immunoprecipitation (IP) using the control IgG or anti-FLAG antibody, and the resultant immunocomplexes were assayed for the Elongin trimer using anti-FLAG, anti-Elongin A, anti-Elongin B, or anti-Elongin C antibody. C, nuclear extracts from HeLa cells stably expressing FLAG-hElongin A, FLAG-RAP74, or the control mock vector were immunoprecipitated by anti-FLAG antibody or control IgG under low (80 mm) or high (150 mm) salt conditions. Resulting immunocomplexes containing Elongin A or RAP74 were assayed by Western blotting using anti-Rpb1, -TBP, -TFIIB, -TFIIEα, and -FCP1 antibodies. D, immunocomplexes in C were also analyzed by Western blot using monoclonal H14 or H5 antibody recognizing the RNAPII CTD phosphorylated on Ser5 or Ser2, respectively.
Colocalization of Elongin A and RNAPIIO during Active Transcription
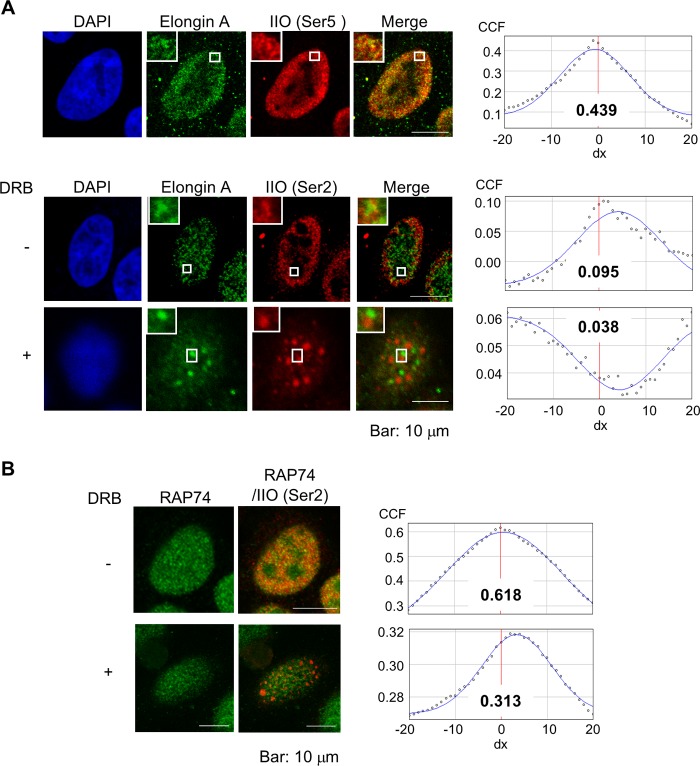
The above data are most consistent with the idea that Elongin A associates not with components of the PIC but rather with actively transcribing RNAPIIO. In cells undergoing active transcription, RNAPII is stained broadly throughout the nucleoplasm in a meshwork pattern (52, 53). To compare the localization of Elongin A and RNAPII in vivo, we performed an immunocytochemical study. We found the majority of Elongin A staining to be localized in the nucleoplasm in a meshwork pattern that exhibits partial overlap with those of Ser5- and Ser2-phosphorylated RNAPIIO (Fig. 2A, upper and lower panels). The degree of overlap between Elongin A and Ser5-phosphorylated RNAPII appeared to be greater than the overlap between Elongin A and Ser2-phosphorylated RNAPII (Pearson's correlation coefficient, 0.439 versus 0.095). Notably, Ser5 phosphorylation occurs concomitantly with transcription initiation and continues during the early elongation stage. Thus, Elongin A may associate more stably with Ser5-phosphorylated RNAPII and play a role in the release from transient pausing rather than in the early elongation stage. Next, we cultured cells in the presence of DRB, an inhibitor of P-TEFb kinase and transcriptional elongation. As in Fig. 2A (lower panel), RNAPIIO (Ser2) relocated to dotlike domains, consistent with the previous report that it localizes to speckles in the nucleus not actively involved in the transcription (54). Remarkably, Elongin A also relocated to speckle-like domains distinct from those associated with RNAPIIO (Ser2). Note that cross-correlation analysis indicated a mutually exclusive pattern of RANPIIO (Ser2) and Elongin A signals in the presence of DRB (42). RAP74 also exhibited partial colocalization with RNAPIIO, but its distribution did not appear to be significantly altered in DRB-treated cells perhaps because TFIIF appears to function predominantly during and shortly after initiation at steps that are not affected by DRB (55–57) (Fig. 2B).
FIGURE 2.
Colocalization of Elongin A and RNAPIIO and their relocation in the presence of DRB. A, HeLa cells stably expressing FLAG-tagged Elongin A grown in the absence or presence of 100 μm DRB for 3 h were immunostained with anti-FLAG (Elongin A), monoclonal H14 (RNAPIIO (IIO) at Ser5), or H5 (RNAPIIO at Ser2) antibody as described under “Experimental Procedures.” Magnified views are shown at the upper left-hand corner. Right panels indicate the results of van Steensel's cross-correlation analysis. Each value of Pearson's correlation coefficient (CCF) (dx, 0) is shown. B, HeLa cells stably expressing FLAG-tagged RAP74 were treated with DRB and immunostained using anti-FLAG (RAP74) and monoclonal H5 (RNAPIIO at Ser2) antibodies as in A. Cross-correlation analysis was performed as well (right panels).
Mammalian Elongin A Partly Colocalizes with Nascent RNA, and Global RNA Synthesis Is Not Impaired in Elongin A-null MEFs
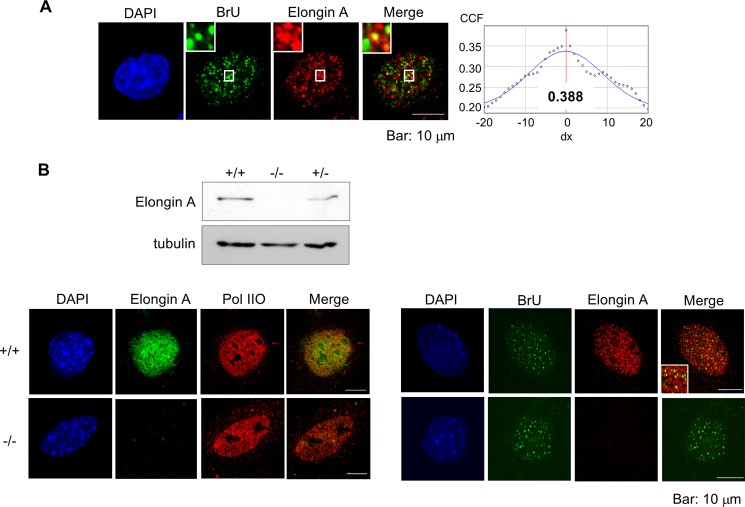
To further explore the role of Elongin A in transcription in cells, we compared the localization of FLAG-tagged Elongin A in HeLa cells or endogenous Elongin A in MEFs with that of newly synthesized RNA using an in vivo RNA labeling technique in which nascent transcripts are labeled with bromouridine (53). As shown in Fig. 3, A and B (lower panel), Elongin A colocalized in both cell types with bromouridine incorporated into newly transcribed RNA at some but not all loci, consistent with the idea that Elongin A contributes to transcription of only a subset of genes. Bromouridine staining was abolished after treatment by RNase or 0.1 μm α-amanitin (data not shown), indicating that most of the detectable signals were indeed derived from nascent RNA. To directly test the contribution of Elongin A to global transcription, Elongin A-null MEFs were immunostained with antibodies against Elongin A and RNAPIIO (phospho-Ser2) (Fig. 3B, lower left panel). As expected, there was no apparent difference in the level or distribution of RNAPIIO in wild type or Elongin A-null MEFs. Moreover, the level of bromouridine incorporated into nascent RNA in Elongin A-null cells appeared to be similar to that in wild type cells (Fig. 3B, lower right panel). Taken together, these data support the idea that mammalian Elongin A is involved in transcription in vivo but is not essential for transcription of all genes.
FIGURE 3.
Colocalization of Elongin A with newly synthesized RNA. A, HeLa cells stably expressing FLAG-tagged Elongin A were cultured in the presence of 2 mm bromouridine (BrU) at 37 °C for 20 min and then immunostained with anti-BrdU and anti-FLAG antibodies. Magnified views are also shown. Colocalizations were analyzed as in Fig. 2. The result of the van Steensel's cross-correlation analysis is shown. CCF, correlation coefficient. B, Western blot of Elongin A in the wild type (+/+) and Elongin A-deficient MEFs (+/− and −/−) are shown in the top panel. MEFs from the wild type and Elongin A-null mouse (−/−) were immunostained with monoclonal H5 (RNAPIIO (Pol IIO) at Ser2) and anti-Elongin A antibodies (lower left panel). In the right panel, MEFs were treated as in A and immunostained with anti-BrdU or anti-Elongin A antibody.
Mammalian Elongin A Promotes Efficient Induction of Stress Genes
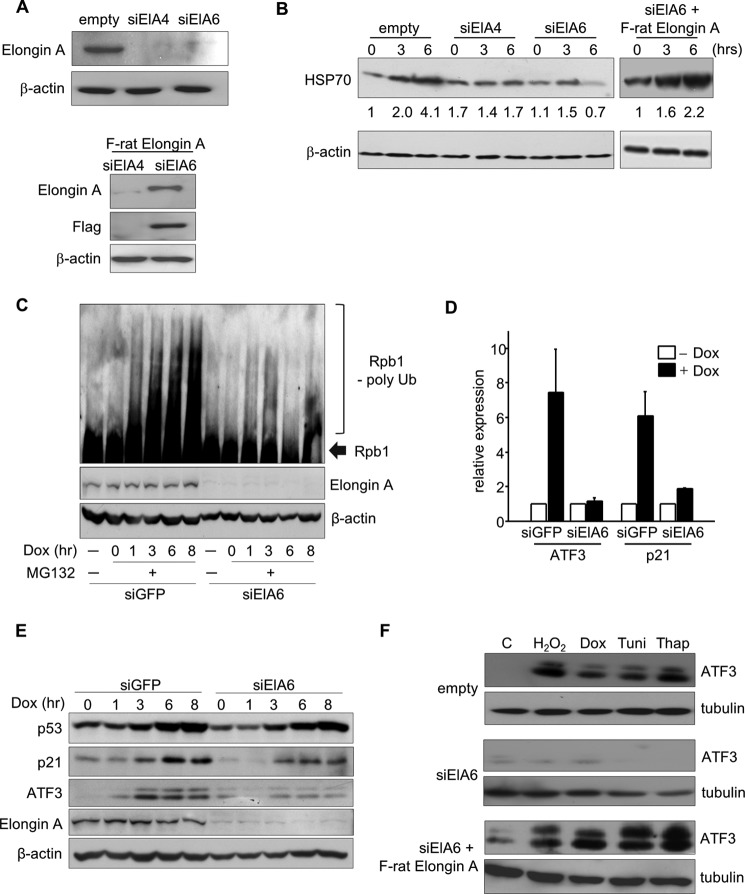
Our previous study showed that mammalian Elongin A deficiency causes sensitization of cells to stress stimuli (29), and Gerber et al. (35) demonstrated that Elongin A is essential for HSP70 gene induction in Drosophila. Thus, we examined whether mammalian Elongin A is also involved in the heat shock response using HeLa cells in which expression of Elongin A is stably down-regulated by either of two distinct shRNAs, siElA4 or siElA6 (Fig. 4A, upper panel). Induction of HSP70 protein after heat shock was significantly reduced in these two cell lines (Fig. 4B). To confirm that this reduction was not due to off-target effects of the shRNAs, rat Elongin A, the nucleotide sequence of which differs in six of 20 target nucleotides from human Elongin A, was introduced into siElA6-expressing HeLa cells. In this cell line, rat Elongin A was well expressed (Fig. 4A, lower panel) and restored the heat shock response (Fig. 4B), confirming that HSP70 gene activation depends on Elongin A. In addition, consistent with previous evidence that Elongin A is required for DNA damage-induced polyubiquitination of Rpb1 (26, 27), we observed that depletion of Elongin A led to a substantial decrease in Rpb1 polyubiquitination in cells treated with the DNA-damaging agent doxorubicin (Fig. 4C and Ref. 26).
FIGURE 4.
Mammalian Elongin A is required for the induction of several stress response genes. A, whole cell extract of Elongin A, siElA4, or siElA6 was subjected to Western blotting using anti-Elongin A antibody (upper panel). In the lower panel, an expression vector encoding FLAG-tagged rat Elongin A (F-rat Elongin A) was reintroduced into the siElA4 or siElA6 cells, and cells stably expressing rat Elongin A were selected by hygromycin. Cell extracts were analyzed by Western blotting using anti-Elongin A or anti-FLAG antibody. B, wild type, siElA6, siElA4, or siElA6 cells with FLAG-rat Elongin A were subjected to heat shock at 43 °C for 30 min and returned to 37 °C. After the indicated times of incubation, whole cell extracts were prepared and subjected to Western blot analysis. Relative intensities of each band are indicated. C, HeLa cells stably expressing siElA6 were treated with MG132 for 1 h followed by 1 μm doxorubicin stimulation. Cells were harvested at the indicated time points, and polyubiquitination (poly Ub) of Rpb1 was assessed by Western blot with H14 antibody. The amount of β-actin is shown as a loading control. D, Elongin A-knocked down cells were treated with 1 μm doxorubicin, and total RNA was extracted after 2-h stimulation. ATF3 and p21 mRNA levels were measured by quantitative RT-PCR. The means of three independent experiments with error bars (S.E.) are shown. E, cells treated as in D were harvested at the indicated time points, and levels of p53, p21, and ATF3 proteins were assessed by Western blotting. The level of β-actin is also shown. F, HeLa cells stably expressing siElA6 or the control cells (C) were treated for 3 h with 50 μm H2O2, 1 μm doxorubicin (Dox), 1 μm tunicamycin (Tuni), or 2 μm thapsigargin (Thap), and the induction of the ATF3 gene was measured by Western blot.
To identify other stress-induced genes whose expression depends on Elongin A, we performed genome-wide microarray analysis of doxorubicin-treated siElA cells (GEO accession number GSE30432). A total of 431 genes were up-regulated, and 1061 genes were down-regulated. Among the down-regulated genes (Table 2), ATF3, a member of the ATF/cAMP response element family transcription factors, is a stress response gene whose expression is maintained at a low or undetectable level but is rapidly induced in response to several stimuli at the transcriptional level (46, 58). Because ATF3 is a target of the tumor suppressor p53 (47), induction by doxorubicin of another p53-dependent, stress-inducible gene, p21, was analyzed as well. As expected, induction of both p21 and ATF3 was significantly reduced at both the mRNA (Fig. 4D) and protein levels (Fig. 4E) in HeLa cells transduced with retrovirus encoding siElA6. To determine whether Elongin A is generally required for ATF3 induction or whether it functions specifically in the response to DNA damage, various reagents were used to stimulate HeLa cells, and the induction of ATF3 was examined. As shown in Fig. 4F, the induction of ATF3 protein in response to oxidative (H2O2), DNA damage (doxorubicin), or endoplasmic reticulum stress stimuli (tunicamycin and thapsigargin) was significantly reduced in Elongin A- knocked down cells stably expressing siElA6, indicating that ATF3 induction is Elongin A-dependent. This repression of ATF3 expression was restored by reintroduction of rat Elongin A (Fig. 4F) as observed for HSP70 gene expression (Fig. 4B). Collectively, these data demonstrate that Elongin A plays a significant role in promoting fully efficient induction of ATF3 in response to multiple stresses.
TABLE 2.
Genome-wide RNA expression analysis comparing control and Elongin A knockdown HeLa cells under doxorubicin treatment
siElA or control (siGFP) HeLa cells were treated with 1 μm doxorubicin for 2 h. Microarray analysis was carried out as described under “Experimental Procedures.” Representative genes that are up-regulated and down-regulated by Elongin A depletion are shown. Indicated values are the relative expression levels in siEIA cells compared with the control. ATF3 expression was decreased to 0.555 in siElA cells. Data were deposited in the Gene Expression Omnibus (GEO; (www.ncbi.nlm.nih.gov/geo/) under accession number GSE30432.
| Up-regulated genes | |
| Glucosidase, β; acid (includes glucosylceramidase) (GBA) | 20.39 |
| Hypoxia up-regulated 1 (HYOU1) | 12.81 |
| Sulfatase-modifying factor 2 (SUMF2) | 10.38 |
| Cathepsin D (CTSD) | 9.979 |
| Eukaryotic translation elongation factor 1α2 (EEF1A2) | 8.146 |
| G protein-coupled receptor 108 (GPR108) | 6.002 |
| Nitric-oxide synthase-interacting protein (NOSIP) | 5.891 |
| Protein kinase N1 (PKN1) | 5.079 |
| Ubiquitin-specific peptidase 5 (isopeptidase T) (USP5) | 5.125 |
| Down-regulated genes | |
| Heat shock 10-kDa protein 1 (chaperonin 10) (HSPE1) | 0.084 |
| Heat shock 70-kDa protein 1A (HSPA1A) | 0.106 |
| Hypothetical protein HSPC111 (HSPC111) | 0.116 |
| Jun proto-oncogene (c-Jun) | 0.145 |
| Heat shock factor-binding protein 1 (HSBP1) | 0.154 |
| Nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor, α (NFKBIA) | 0.186 |
| TBC1 domain family, member 13 (TBC1D13) | 0.215 |
| Serine/threonine kinase receptor-associated protein (STRAP) | 0.230 |
| N-Myristoyltransferase 1 (NMT1) | 0.253 |
| v-myc myelocytomatosis viral oncogene homolog (avian) (c-Myc) | 0.336 |
| Mediator of RNA polymerase II transcription, subunit 19 homolog (Saccharomyces cerevisiae) (MED19) | 0.379 |
| Structure-specific recognition protein 1 (SSRP1) | 0.384 |
| ATF3 | 0.555 |
RNAPII-transcribed ATF3 Gene Is Associated with Elongin A
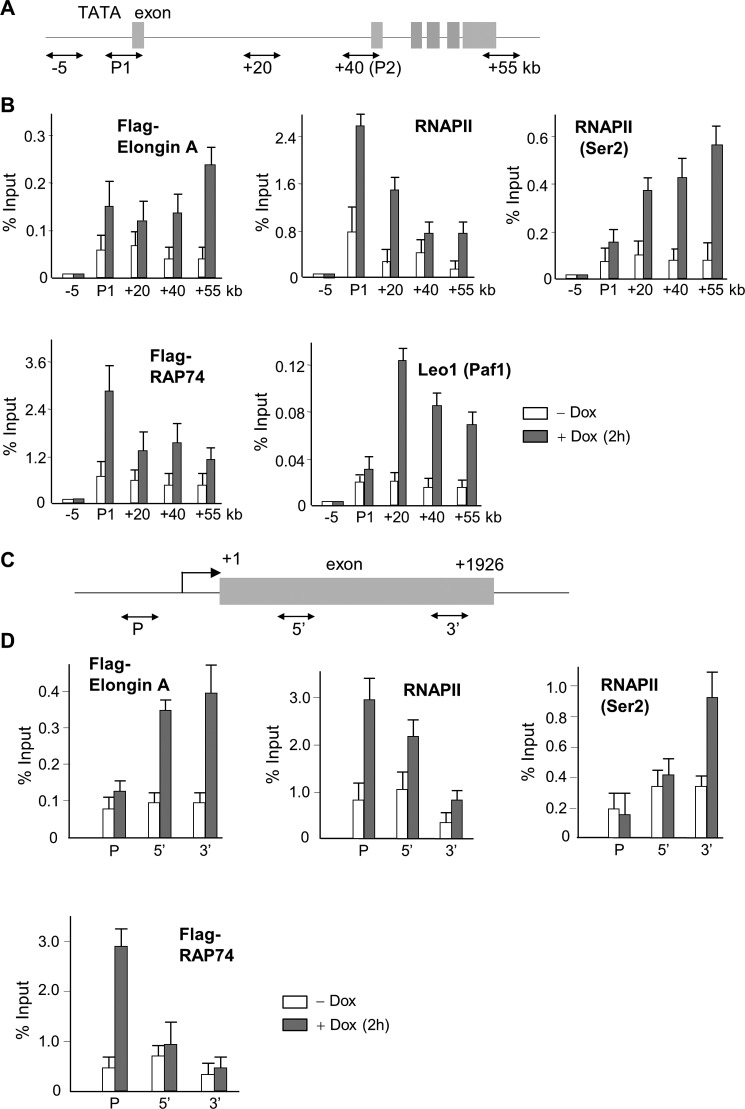
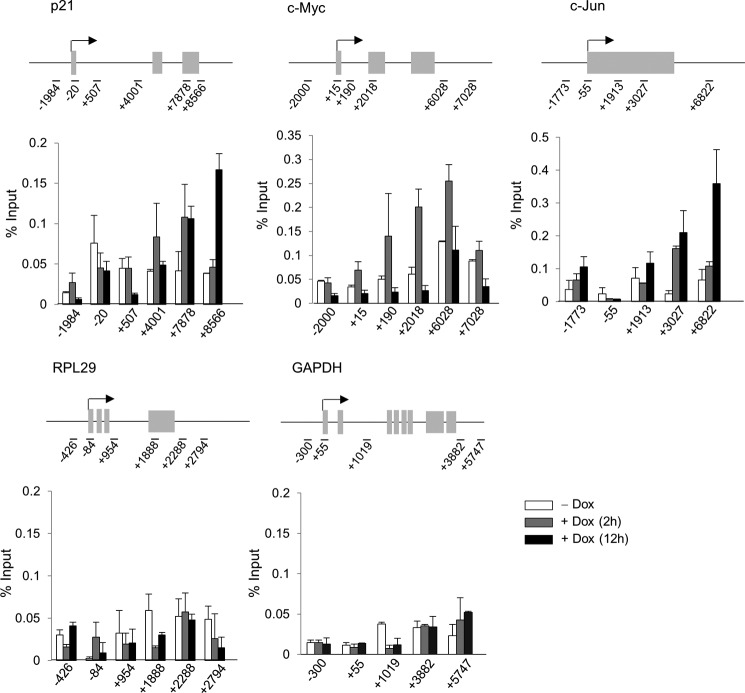
To determine whether Elongin A affects ATF3 transcription directly or via an indirect mechanism, we used ChIP combined with qPCR (Fig. 5A) to assess the occupancy of Elongin A, RNAPII, and other transcription regulators at the ATF3 gene before and after doxorubicin treatment. As shown in Fig. 5B (upper left panel), Elongin A ChIP signals were increased across the entire transcribed region of the ATF3 gene after stimulation but remained at near background levels upstream of the ATF3 gene promoter (−5 kb), indicating stress-induced recruitment of Elongin A onto the ATF3 gene. We also observed an increase in the association of total RNAPII as well as CTD Ser2-phosphorylated RNAPII across the ATF3 gene after stimulation. It should be noted here that some poised RNAPIIs appeared to be present at the P1 promoter before treatment, consistent with the recent report by Lin et al. (59). Moreover, Leo1 and RAP74 were also recruited to the ATF3 gene along with Elongin A; however, their ChIP signals were different from one another, implying that they may play different roles in ATF3 gene expression. Gerber et al. (35) reported previously that Elongin A and RNAPII colocalize at heat shock puffs on Drosophila polytene chromosomes; however, they did not test recruitment of Elongin A to heat shock genes by ChIP. As shown in Fig. 5D, following heat shock, we observed a substantial increase in ChIP of Elongin A and CTD Ser2-phosphorylated RNAPII at the transcribed region of the human HSP70 gene and increased RAP74 near the promoter. These observations support a direct role of Elongin A in regulating heat shock-induced transcription of HSP70. We further assessed the recruitment of Elongin A to a collection of stress-induced and constitutively active genes, all of which have been reported to be associated with promoter-proximally paused RNAPII (60, 61). The p21, c-myc, and c-jun genes are all activated by doxorubicin in an Elongin-dependent manner (Table 2 and Fig. 4, D and E). As shown in Fig. 6, Elongin A occupancy was increased on the p21, c-myc, and c-jun genes after doxorubicin treatment. Although it is well established that promoter-proximal pausing is a key step in regulation of these three genes (60), the increase in ChIP signal was much more pronounced in more 3′-regions of these genes than at their promoters. Substantially lower levels of Elongin A ChIP signal were detected at the highly expressed, Elongin A-independent RPL29 and GAPDH genes again with a 3′ bias; however, Elongin A ChIP on these genes was not increased by doxorubicin. Taken together, these data imply (i) that Elongin A contributes preferentially to the expression of stress response genes and (ii) that it associates broadly with the transcribed region of genes with higher recruitment to 3′-regions.
FIGURE 5.
Mammalian Elongin A is associated with the entire ATF3 gene under doxorubicin treatment. A, diagram of the ATF3 gene. Positions of the major two promoters are indicated (P1 and P2; Ref. 40). Boxes denote exons. Double headed arrows denote the locations of regions amplified by qPCR. B, HeLa cell lines stably expressing FLAG-tagged Elongin A were treated with or without 1 μm doxorubicin (Dox) for 2 h. Then a ChIP assay combined with qPCR was carried out as described under “Experimental Procedures” using anti-FLAG (Elongin A), anti-RNAPII, H5 (anti-RNAPIIO Ser2-phosphorylated), and anti-Leo1 antibodies. RAP74 association was also investigated using FLAG-RAP74 stable cell lines and anti-FLAG antibody. Primer sets used for qPCR are depicted in A. Data represent the means of three independent experiments with error bars representing S.E. C, diagram of the HSP70 gene. Double headed arrows denote the locations of regions amplified by qPCR. D, ChIP combined with qPCR was performed as in B with the indicated primer sets in C.
FIGURE 6.
Elongin A association is increased on the p21, c-myc, and c-jun genes after doxorubicin (Dox) treatment. A FLAG (Elongin A) ChIP assay was performed for the five indicated genes. The positions of primer sets are depicted above each bar graph. Data represent the means of three independent experiments with error bars representing S.E.
Both the C-terminal and SII Similarity Regions of Mammalian Elongin A Are Required for the Proper Response of the ATF3 Gene Promoter
Because Elongin A appeared to play a critical role in the rapid induction of ATF3, we sought to identify the Elongin A functional domain(s) required for induction of the ATF3 gene in vivo. Colgan and Manley (62) previously used luciferase reporter assays to evaluate the function of TFIID, one of the general transcription factors. Thus, we used this assay to assess the transcriptional activity of several mutants of Elongin A using pLuc-ATF3 (−384) in which ATF3 promoter sequences from −384 to +34 drive expression of a luciferase reporter gene (47). We first confirmed that (i) doxorubicin-dependent induction of this reporter gene was impaired in siElA6 cells and that (ii) it could be rescued by reintroduction of wild type rat Elongin A (Fig. 7C, empty and 1–773). Thus, ATF3-luciferase reporter gene behaves similarly to the endogenous ATF3 gene and provides a good model with which to study Elongin A-dependent transcription.
FIGURE 7.
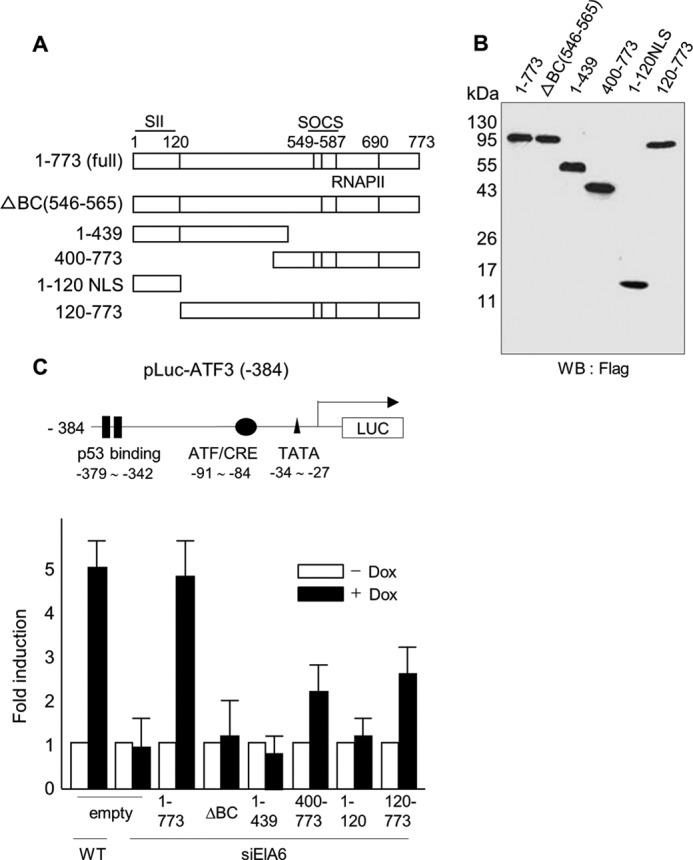
The BC box of mammalian Elongin A is essential for ATF3 induction. A and B, structures and expressions of FLAG-tagged rat Elongin A mutants are shown (upper and lower right panels). SII, SOCS, and RNAPII denote the SII homology domain, SOCS box, and RNAPII binding domain, respectively (22). C, wild type or siElA6 HeLa cells were co-transfected with reporter plasmid pLuc-ATF3 (−384) along with expression vectors encoding various mutants of rat Elongin A. Cells were then treated with doxorubicin (Dox), and the reporter activity was assayed as described under “Experimental Procedures.” The bar graph represents the -fold induction of activity after treatment (solid columns) compared with that without treatment (open columns). The means of five independent experiments with error bars (S.E.) are shown. LUC, luciferase; WB, Western blot; CRE, cAMP response element. NLS, nuclear localization signal.
As described earlier, Elongin A has an N-terminal SII similarity region that is dispensable for Elongin A transcriptional activity in vitro and a C-terminal elongation stimulatory domain that is required for Elongin A transcriptional activity in vitro. The latter C-terminal domain includes a SOCS box composed of a BC box (Elongin BC binding site) and a Cul5 box (required for Cul5 binding) (Figs. 7A and 8A). We explored whether these domains have a functional role in the stress-induced activation of the ATF3 gene promoter. As shown in Fig. 7C, mutants 1–439 and 1–120 NLS (nuclear localization signal), which lack the C-terminal elongation stimulatory domain, did not support doxorubicin-induced activity of the ATF3 promoter, indicating that the C-terminal elongation stimulatory domain is essential for in vivo transcription activity. The Elongin A mutant lacking the BC domain (ΔBC) was also inactive for the induction of the ATF3 reporter. By contrast, both mutants 400–773 and 120–773, which contain the C-terminal elongation stimulatory domain but lack the SII similarity region, only weakly stimulated reporter activity. These mutants behaved very similarly when assayed for their ability to stimulate an HSP70 promoter after heat shock (data not shown). These data indicate that the C-terminal elongation stimulatory domain including the Elongin BC interaction region is essential for efficient transcription in vivo, whereas the N-terminal SII similarity region is not required but has a positive regulatory role.
FIGURE 8.
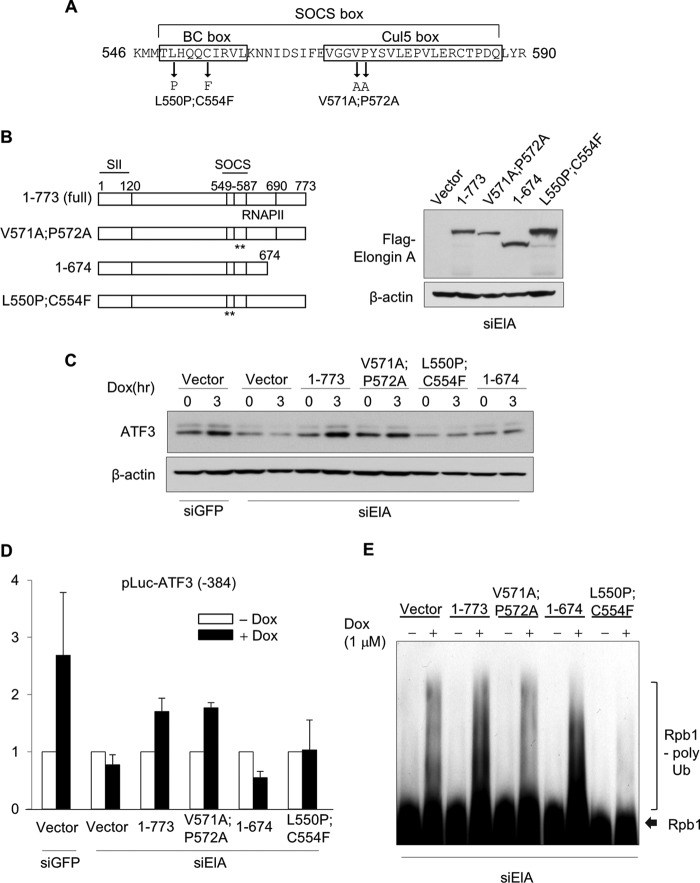
The C terminus of mammalian Elongin A is essential, but the Cul5 box is dispensable for ATF3 induction. A, amino acid sequence of SOCS box in human Elongin A from 546 to 590. Locations of BC box and Cul5 box are depicted. Arrows indicate the positions of amino acid substitution to abolish each functional domain. B, structures and expressions of several variants of Elongin A are shown. Elongin A full length and L550P/C554F were derived from rat Elongin A. Elongin A V571A/P572A and 1–674 were derived from mouse Elongin A. Each construct was reintroduced into the Elongin A-knocked down cells with retrovirus vectors. The levels of each variant expression were assessed by Western blot using anti-FLAG antibody. The amount of β-actin is shown as a loading control. The asterisks indicate the positions of amino acid substitution detailed in A. C, inductions of ATF3 by doxorubicin (Dox) were analyzed by Western blot. Elongin A mutants shown in B were reintroduced into Elongin A-knocked down HeLa cells. Cells were harvested at 3 h following treatment with 1 μm doxorubicin. As a control, HeLa cells stably expressing siGFP with empty vector reintroduction were also investigated. D, Elongin A mutants shown in C were transfected with ATF3 reporter into HeLa cells expressing siElA6, and then cells were treated with doxorubicin. Reporter activity was measured as in Fig. 6C. Data represent the means of three independent experiments with error bars representing S.E. E, cells as in B were treated with MG132 followed by 1 μm doxorubicin stimulation. After 8 h, polyubiquitination (poly Ub) of Rpb1 was assessed by Western blot as in Fig. 4C.
The E3 Ligase Activity of Elongin A Is Dispensable for Sufficient Inductions of ATF3
From the results of the reporter assay, the BC box is likely to be essential for the sufficient induction of ATF3. The BC box, which provides a platform for binding of Elongins B and C, has been shown to be essential for the ability of Elongin A to stimulate transcriptional elongation in vitro (25) and to support its E3 ubiquitin ligase activity (26). On the other hand, an intact Cul5 box, which is adjacent to the BC box (Fig. 8A), is reported to be required for E3 ligase activity but to be dispensable for stimulation of transcript elongation. To further define regions of Elongin A needed for rapid activation of the ATF3 gene, we reintroduced several mutants of mouse or rat Elongin A into the siElA6 HeLa cells. In addition to those mutants, we introduced mouse Elongin A deletion mutant 1–674, which contains a complete SOCS box but lacks more C-terminal sequences. This mutant is active in RNAPII ubiquitination but does not stimulate transcript elongation in vitro (19). Each mutant was expressed in siElA6 cells (Fig. 8B), and the induction of the endogenous ATF3 gene was analyzed after doxorubicin treatment by Western blotting using anti-ATF3 antibody. As shown in Fig. 8C, full-length Elongin A(1–773) and the Cul5 box mutant (V571A/P572A) partly but significantly rescued the induction of ATF3. By contrast, BC box mutant (L550P/C554F) and Elongin A(1–674) did not promote ATF3 induction. Similar results were obtained when the abilities of wild type and mutant Elongin A to support ATF3 promoter activity were assessed using the luciferase reporter assay (Fig. 8D). As expected, Elongin A(1–674)-reintroduced cells exhibit the same degree of ubiquitination of Rpb1 after doxorubicin treatment as full-length Elongin A-reintroduced cells. On the other hand, Cul5 box mutant (V571A/P572A) and BC box mutant (L550P/C554F) exhibit only background level of ubiquitination activity, which likely is due to the presence of residual Elongin A or another E3 ubiquitin ligase that targets Rpb1 (Fig. 8E). Taken together, these experiments argue that proper activation of ATF3 in vivo depends on the presence of an intact Elongin A elongation stimulatory domain that can bind Elongins B and C but that the E3 ubiquitin ligase activity of Elongin A is dispensable.
DISCUSSION
The results presented in this study indicate that mammalian Elongin A is essential for rapid and full induction of the immediate early response gene ATF3 and of the heat shock-induced HSP70 gene. Previous studies suggested that Elongin A is not generally required for transcription in vivo (19, 28). Supporting this idea, shRNA-mediated depletion of Elongin A or deletion of the Elongin A gene neither altered the distribution of RNAPII in cell nuclei nor impaired production of the majority of nascent RNA (Fig. 3B), indicating that Elongin A is not required for transcription of all genes but rather regulates expression of select genes. With regard to the in vivo function of Elongin A, Gerber et al. (35) reported that Drosophila Elongin A plays a key role for HSP70 gene induction, whereas Yasukawa et al. (19) very recently demonstrated that Elongin A is needed for retinoic acid-induced expression of several genes during neuronal differentiation. In neither case, however, did the authors demonstrate by ChIP that these genes are direct, rather than indirect, targets for regulation by Elongin A. In this study, for the first time, we have provided evidence that Elongin A directly binds to the coding regions of target genes ATF3 and HSP70 along with elongating RNAPII, consistent with the model that it directly regulates their transcription during elongation. Moreover, we identified domains of Elongin A essential for fully efficient activation of stress-induced gene expression in vivo.
Characteristic Feature of Elongin A in Transcriptional Event in Mammalian Cells
Upon doxorubicin treatment, Elongin A becomes physically associated with the entire ATF3 gene (Fig. 5). Moreover, Elongin A clearly binds to RNAPIIO phosphorylated at Ser5 or Ser2 in vivo (Fig. 1). Importantly, this binding does not appear to be due to direct interactions with the CTD of RNAPII because we were unable to detect appreciable in vitro binding of Elongin A to a GST-CTD fusion protein that was either unphosphorylated or phosphorylated by MAPK (data not shown). Accordingly, it seems most likely that Elongin A binds to RNAPIIO through sequences in its C-terminal elongation activation domain between residues 590 and 690 (Fig. 1C and Ref. 23).
It is intriguing to note that Elongin A relocated to large speckle-like domains in the presence of DRB (Fig. 2A). Although RNAPII is also known to alter its nuclear localization in transcriptionally inactive cells (52, 53), Elongin A and RNAPII relocalized to distinct domains. Because DRB is a well known inhibitor of protein kinases such as casein kinase II (61) and P-TEFb (62) and strongly inhibits transcriptional elongation, relocation following DRB treatment may suggest that phosphorylation of Elongin A controls its activity during transcription elongation or that decreased CTD phosphorylation interferes with the ability of Elongin A to bind RNAPII. Alternatively, Elongin A could have a unique unknown activity when transcription is repressed.
Our reporter assays using various Elongin A mutants demonstrated that the C-terminal elongation activation domain including the BC box is crucial for the response of ATF3 gene, consistent with the activity in vitro (22, 25, 63). In addition, the N-terminal SII similarity region has a mild but positive regulatory role for the Elongin A-driven transcription activity in vivo (Fig. 7C). This region is well conserved among human, rodents, and Drosophila (64) and shares sequence similarity with the N-terminal domains of SII and Mediator subunit MED26 (65). The N-terminal region of yeast SII has been reported to interact with Med13 and Spt8, which belong to the Cdk8 module of the Mediator and to a subform of the SAGA (Spt-Ada-Gcn5-acetyltransferase) co-activator, respectively (66). Thus, this region of Elongin A may also interact with Mediator(s) or co-activator(s) in the PIC or poised RNAPII complex, thereby helping to determine the Elongin A target genes. Although we did not detect PIC components in our immunoprecipitations (Fig. 1C), our results do not exclude a role for Elongin A in the PIC; indeed, although low, the Elongin A ChIP signal around the transcription start site is detectable (Fig. 5).
These analyses altogether support a view that when cells are subjected to stress Elongin A, through interaction with Ser5- and/or Ser2-phosphorylated RNAPII, is recruited to elongation complexes where it can enhance transcription throughout the entire coding region (Figs. 1 and 5). On the other hand, our immunocytochemical assay indicated that Elongin A colocalizes with RNAPII (Ser5) more than RNAPII (Ser2) (Fig. 2A). Considering that Elongin A is more strongly associated with more 3′-regions than with the promoters of all the genes we tested (Figs. 5 and 6), the elongation activity of Elongin A seems to be exerted mainly after the release of promoter-proximal pausing and thus differs from the super elongation complex (51). Further biochemical analysis of Elongin A after the transition from the paused to elongating RNAPIIO will be required to characterize the active elongation complex in vivo.
Elongin A Has Two Properties in Vivo: Elongation and Ubiquitination
Elongin A has at least two biochemical activities. (i) It stimulates transcriptional elongation by RNAPII, and (ii) it is a substrate recognition subunit of a ubiquitin ligase that supports RNAPIIO ubiquitination. Although several roles of ubiquitination have been reported, including proteolysis, regulation of protein activity, and regulation of protein localization (67), Elongin A-dependent Rpb1 ubiquitination promotes degradation of Rpb1. Moreover, assembly of an active Elongin A ubiquitin ligase that supports this degradation requires Elongin A to bind both Elongin BC and Cul5 (26). Our rescue experiments provide strong evidence that Elongin BC binding is required, but Cul5 binding is dispensable for the activation of ATF3 transcription (Fig. 8). Thus, during rapid induction of stress genes, the elongation activity of Elongin A is totally independent of its ubiquitination activity in vivo as is the case for retinoic acid induction of several genes for neuronal development in ES cells (19). Therefore, the elongation and ubiquitination activity might be more generally independent functions.
Interestingly, these two activities are apparently opposing forces in terms of the regulation of the transcript amount. Polyubiquitination is the early step to degrade the RNAPII itself. By contrast, elongation is the activity that assists the primary role of RNAPII. One of the possible explanations for this issue is that ubiquitination and degradation of paused or stalled RNAPII free the DNA template of RNAPII that would otherwise block passage of the next polymerase, resulting in an overall increase of the amount of fully elongated transcript. In this regard, however, our new data do not completely fit this model because the ubiquitination activity of Elongin A is unnecessary for the rapid induction of ATF3. In parallel with this, E3 ligase activity of Elongin A is not essential for the neurogenesis-related gene activation upon retinoic acid stimulation in mouse embryonic stem cells (19). We predict that this might be due to the RNAPII pausing feature. In the case of physiological pausing during mid to late elongation phase, Elongin A might facilitate the RNAPII elongation without ubiquitination activity. However, when RNAPII is arrested for a prolonged time because of profound DNA damage, the ubiquitination activity of Elongin A might be indispensable by cooperating with Cul5 to modify or degrade arrested RNAPIIO to ensure efficient gene expression (68).
Finally, rapid and sufficient induction of stress response genes is unambiguously an essential process for the survival of living cells. The role of Elongin A in activation of stress response genes is therefore likely to be of extraordinary importance.
Acknowledgments
We greatly appreciate Dr. Kamura T. for the generous gift of anti-Elongin B and C antibodies, Drs. Wada T. and Handa H. for advice on stable cell establishment, and Dr. Nakai A. for valuable advice on heat shock treatment. We also thank A. Nakamura, Y. Hosaka, and M. Watanabe for technical assistance.
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grants 18012015, 18055008, and 21590302 (to S. K.), 21590311 (to T. A.), and 24118002 (to J. K.) and by the Joint Usage/Research Program of Medical Research Institute, Tokyo Medical and Dental University.
- RNAPII
- RNA polymerase II
- Rpb1
- polymerase (RNA) II (DNA-directed) polypeptide A
- TF
- transcription factor
- SII
- stimulatory factor for RNA polymerase II
- CTD
- C-terminal domain
- MEF
- mouse embryonic fibroblast
- ATF
- activating transcription factor
- HSP
- heat shock protein
- DRB
- 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- SOCS
- suppressor of cytokine signals
- TBP
- TATA-binding protein
- PIC
- preinitiation complex
- DSIF
- DRB sensitivity-inducing factor
- NELF
- negative elongation factor
- P-TEFb
- positive transcription elongation factor b
- ELL/EAF
- eleven-nineteen lysine-rich in leukemia/ELL-associated factor
- FACT
- facilitates chromatin transcription
- Cul5
- Cullin 5
- qPCR
- quantitative PCR
- hElongin A
- human Elongin A.
REFERENCES
- 1. Shilatifard A., Conaway R. C., Conaway J. W. (2003) The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72, 693–715 [DOI] [PubMed] [Google Scholar]
- 2. Sims R. J., 3rd, Belotserkovskaya R., Reinberg D. (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18, 2437–2468 [DOI] [PubMed] [Google Scholar]
- 3. Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G. A., Winston F., Buratowski S., Handa H. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 [DOI] [PubMed] [Google Scholar]
- 5. Saunders A., Core L. J., Lis J. T. (2006) Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7, 557–567 [DOI] [PubMed] [Google Scholar]
- 6. Zhou Q., Li T., Price D. H. (2012) RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116 [DOI] [PubMed] [Google Scholar]
- 8. Otero G., Fellows J., Li Y., de Bizemont T., Dirac A. M., Gustafsson C. M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109–118 [DOI] [PubMed] [Google Scholar]
- 9. Shilatifard A., Lane W. S., Jackson K. W., Conaway R. C., Conaway J. W. (1996) An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271, 1873–1876 [DOI] [PubMed] [Google Scholar]
- 10. Kong S. E., Banks C. A., Shilatifard A., Conaway J. W., Conaway R. C. (2005) ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc. Natl. Acad. Sci. U.S.A. 102, 10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Price D. H., Sluder A. E., Greenleaf A. L. (1989) Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 9, 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tan S., Aso T., Conaway R. C., Conaway J. W. (1994) Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J. Biol. Chem. 269, 25684–25691 [PubMed] [Google Scholar]
- 13. Kitajima S., Chibazakura T., Yonaha M., Yasukochi Y. (1994) Regulation of the human general transcription initiation factor TFIIF by phosphorylation. J. Biol. Chem. 269, 29970–29977 [PubMed] [Google Scholar]
- 14. Selby C. P., Sancar A. (1997) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 94, 11205–11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bradsher J. N., Jackson K. W., Conaway R. C., Conaway J. W. (1993) RNA polymerase II transcription factor SIII. I. Identification, purification, and properties. J. Biol. Chem. 268, 25587–25593 [PubMed] [Google Scholar]
- 16. Bradsher J. N., Tan S., McLaury H. J., Conaway J. W., Conaway R. C. (1993) RNA polymerase II transcription factor SIII. II. Functional properties and role in RNA chain elongation. J. Biol. Chem. 268, 25594–25603 [PubMed] [Google Scholar]
- 17. Wind M., Reines D. (2000) Transcription elongation factor SII. BioEssays 22, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fish R. N., Kane C. M. (2002) Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577, 287–307 [DOI] [PubMed] [Google Scholar]
- 19. Yasukawa T., Bhatt S., Takeuchi T., Kawauchi J., Takahashi H., Tsutsui A., Muraoka T., Inoue M., Tsuda M., Kitajima S., Conaway R. C., Conaway J. W., Trainor P. A., Aso T. (2012) Transcriptional elongation factor Elongin A regulates retinoic acid-induced gene expression during neuronal differentiation. Cell Rep. 2, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 20. Garrett K. P., Tan S., Bradsher J. N., Lane W. S., Conaway J. W., Conaway R. C. (1994) Molecular cloning of an essential subunit of RNA polymerase II elongation factor SIII. Proc. Natl. Acad. Sci. U.S.A. 91, 5237–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aso T., Lane W. S., Conaway J. W., Conaway R. C. (1995) Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science 269, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 22. Yasukawa T., Sugimura K., Fukuda M., Yamazaki K., Kitajima S., Okumura K., Aso T. (2007) Functional characterization of a mammalian transcription factor, Elongin A. Biochem. Biophys. Res. Commun. 352, 237–243 [DOI] [PubMed] [Google Scholar]
- 23. Aso T., Yamazaki K., Amimoto K., Kuroiwa A., Higashi H., Matsuda Y., Kitajima S., Hatakeyama M. (2000) Identification and characterization of Elongin A2, a new member of the Elongin family of transcription elongation factors, specifically expressed in the testis. J. Biol. Chem. 275, 6546–6552 [DOI] [PubMed] [Google Scholar]
- 24. Yamazaki K., Guo L., Sugahara K., Zhang C., Enzan H., Nakabeppu Y., Kitajima S., Aso T. (2002) Identification and biochemical characterization of a novel transcription elongation factor, Elongin A3. J. Biol. Chem. 277, 26444–26451 [DOI] [PubMed] [Google Scholar]
- 25. Aso T., Haque D., Barstead R. J., Conaway R. C., Conaway J. W. (1996) The inducible elongin A elongation activation domain: structure, function and interaction with the elongin BC complex. EMBO J. 15, 5557–5566 [PMC free article] [PubMed] [Google Scholar]
- 26. Yasukawa T., Kamura T., Kitajima S., Conaway R. C., Conaway J. W., Aso T. (2008) Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 27, 3256–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harreman M., Taschner M., Siqurdsson S., Anindya R., Reid J., Somesh B., Kong S. E., Banks C. A., Conaway R. C., Conaway J. W., Svejstrup J. Q. (2009) Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl. Acad. Sci. U.S.A. 106, 20705–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamazaki K., Aso T., Ohnishi Y., Ohno M., Tamura K., Shuin T., Kitajima S., Nakabeppu Y. (2003) Mammalian elongin A is not essential for cell viability but is required for proper cell cycle progression with limited alteration of gene expression. J. Biol. Chem. 278, 13585–13589 [DOI] [PubMed] [Google Scholar]
- 29. Miyata K., Yasukawa T., Fukuda M., Takeuchi T., Yamazaki K., Sakumi K., Tamamori-Adachi M., Ohnishi Y., Ohtsuki Y., Nakabeppu Y., Kitajima S., Onishi S., Aso T. (2007) Induction of apoptosis and cellular senescence in mice lacking transcription elongation factor, Elongin A. Cell Death Differ. 14, 716–726 [DOI] [PubMed] [Google Scholar]
- 30. Levine M. (2011) Paused RNA polymerase II as a developmental checkpoint. Cell 145, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lis J. T., Mason P., Peng J., Price D. H., Werner J. (2000) P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14, 792–803 [PMC free article] [PubMed] [Google Scholar]
- 32. Andrulis E. D., Guzmán E., Döring P., Werner J., Lis J. T. (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14, 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu C. H., Yamaguchi Y., Benjamin L. R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D. (2003) NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17, 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saunders A., Werner J., Andrulis E. D., Nakayama T., Hirose S., Reinberg D., Lis J. T. (2003) Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301, 1094–1096 [DOI] [PubMed] [Google Scholar]
- 35. Gerber M., Tenney K., Conaway J. W., Conaway R. C., Eissenberg J. C., Shilatifard A. (2005) Regulation of heat shock gene expression by RNA polymerase II elongation factor, Elongin A. J. Biol. Chem. 280, 4017–4020 [DOI] [PubMed] [Google Scholar]
- 36. Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H. (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21, 227–237 [DOI] [PubMed] [Google Scholar]
- 37. Ryser S., Fujita T., Tortola S., Piuz I., Schlegel W. (2007) The rate of c-fos transcription in vivo is continuously regulated at the level of elongation by dynamic stimulus-coupled recruitment of positive transcription elongation factor b. J. Biol. Chem. 282, 5075–5084 [DOI] [PubMed] [Google Scholar]
- 38. Aida M., Chen Y., Nakajima K., Yamaguchi Y., Wada T., Handa H. (2006) Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 26, 6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 40. Hua B., Tamamori-Adachi M., Luo Y., Tamura K., Morioka M., Fukuda M., Tanaka Y., Kitajima S. (2006) A splice variant of stress response gene ATF3 counteracts NF-κB-dependent anti-apoptosis through inhibiting recruitment of CREB-binding protein/p300 coactivator. J. Biol. Chem. 281, 1620–1629 [DOI] [PubMed] [Google Scholar]
- 41. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Steensel B., van Binnendijk E. P., Hornsby C. D., van der Voort H. T., Krozowski Z. S., de Kloet E. R., van Driel R. (1996) Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 109, 787–792 [DOI] [PubMed] [Google Scholar]
- 43. Morita S., Kojima T., Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 44. Tamura K., Hua B., Adachi S., Guney I., Kawauchi J., Morioka M., Tamamori-Adachi M., Tanaka Y., Nakabeppu Y., Sunamori M., Sedivy J. M., Kitajima S. (2005) Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. EMBO J. 24, 2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miyazaki K., Inoue S., Yamada K., Watanabe M., Liu Q., Watanabe T., Adachi M. T., Tanaka Y., Kitajima S. (2009) Differential usage of alternate promoters of the human stress response gene ATF3 in stress response and cancer cells. Nucleic Acids Res. 37, 1438–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cai Y., Zhang C., Nawa T., Aso T., Tanaka M., Oshiro S., Ichijo H., Kitajima S. (2000) Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH2-terminal kinase and promoter response element. Blood 96, 2140–2148 [PubMed] [Google Scholar]
- 47. Zhang C., Gao C., Kawauchi J., Hashimoto Y., Tsuchida N., Kitajima S. (2002) Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem. Biophys. Res. Commun. 297, 1302–1310 [DOI] [PubMed] [Google Scholar]
- 48. Pan G., Aso T., Greenblatt J. (1997) Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J. Biol. Chem. 272, 24563–24571 [DOI] [PubMed] [Google Scholar]
- 49. Kim B., Nesvizhskii A. I., Rani P. G., Hahn S., Aebersold R., Ranish J. A. (2007) The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl. Acad. Sci. U.S.A. 104, 16068–16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guglielmi B., Soutourina J., Esnault C., Werner M. (2007) TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 16062–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takahashi H., Parmely T. J., Sato S., Tomomori-Sato C., Banks C. A., Kong S. E., Szutorisz H., Swanson S. K., Martin-Brown S., Washburn M. P., Florens L., Seidel C. W., Lin C., Smith E. R., Shilatifard A., Conaway R. C., Conaway J. W. (2011) Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Grande M. A., van der Kraan I., de Jong L., van Driel R. (1997) Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J. Cell Sci. 110, 1781–1791 [DOI] [PubMed] [Google Scholar]
- 53. Zeng C., Kim E., Warren S. L., Berget S. M. (1997) Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16, 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bregman D. B., Du L., van der Zee S., Warren S. L. (1995) Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129, 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Roeder R. G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 [PubMed] [Google Scholar]
- 56. Yan Q., Moreland R. J., Conaway J. W., Conaway R. C. (1999) Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 274, 35668–35675 [DOI] [PubMed] [Google Scholar]
- 57. Pokholok D. K., Hannett N. M., Young R. A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809 [DOI] [PubMed] [Google Scholar]
- 58. Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., Sivaprasad U. (1999) ATF3 and stress responses. Gene Expr. 7, 321–335 [PMC free article] [PubMed] [Google Scholar]
- 59. Lin C., Garrett A. S., De Kumar B., Smith E. R., Gogol M., Seidel C., Krumlauf R., Shilatifard A. (2011) Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 25, 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Larochelle S., Amat R., Glover-Cutter K., Sansó M., Zhang C., Allen J. J., Shokat K. M., Bentley D. L., Fisher R. P. (2012) Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 19, 1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheng B., Li T., Rahl P. B., Adamson T. E., Loudas N. B., Guo J., Varzavand K., Cooper J. J., Hu X., Gnatt A., Young R. A., Price D. H. (2012) Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell 45, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Colgan J., Manley J. L. (1992) TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 6, 304–315 [DOI] [PubMed] [Google Scholar]
- 63. Takagi Y., Conaway R. C., Conaway J. W. (1996) Characterization of elongin C functional domains required for interaction with elongin B and activation of elongin A. J. Biol. Chem. 271, 25562–25568 [DOI] [PubMed] [Google Scholar]
- 64. Gerber M., Eissenberg J. C., Kong S., Tenney K., Conaway J. W., Conaway R. C., Shilatifard A. (2004) In vivo requirement of the RNA polymerase II elongation factor elongin A for proper gene expression and development. Mol. Cell. Biol. 24, 9911–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Booth V., Koth C. M., Edwards A. M., Arrowsmith C. H. (2000) Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70. J. Biol. Chem. 275, 31266–31268 [DOI] [PubMed] [Google Scholar]
- 66. Wery M., Shematorova E., Van Driessche B., Vandenhaute J., Thuriaux P., Van Mullem V. (2004) Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 23, 4232–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bhat K. P., Greer S. F. (2011) Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. Biophys. Acta 1809, 150–155 [DOI] [PubMed] [Google Scholar]
- 68. Selth L. A., Sigurdsson S., Svejstrup J. Q. (2010) Transcript elongation by RNA polymerase II. Annu. Rev. Biochem. 79, 271–293 [DOI] [PubMed] [Google Scholar]