Background: Peptide modulation of MHC flexibility can affect recognition by immune receptors.
Results: Different peptides alter the flexibility of the MHC protein at sites around the peptide-binding groove.
Conclusion: Peptide modulation of MHC flexibility is not limited to specific peptides or isolated regions.
Significance: Peptide modulation of MHC flexibility indicates an extension of antigenicity from the peptide to the MHC.
Keywords: Fluorescence, Mass Spectrometry (MS), Major Histocompatibility complex (MHC), Peptides, Protein Dynamics
Abstract
T cells use the αβ T cell receptor (TCR) to recognize antigenic peptides presented by class I major histocompatibility complex proteins (pMHCs) on the surfaces of antigen-presenting cells. Flexibility in both TCRs and peptides plays an important role in antigen recognition and discrimination. Less clear is the role of flexibility in the MHC protein; although recent observations have indicated that mobility in the MHC can impact TCR recognition in a peptide-dependent fashion, the extent of this behavior is unknown. Here, using hydrogen/deuterium exchange, fluorescence anisotropy, and structural analyses, we show that the flexibility of the peptide binding groove of the class I MHC protein HLA-A*0201 varies significantly with different peptides. The variations extend throughout the binding groove, impacting regions contacted by TCRs as well as other activating and inhibitory receptors of the immune system. Our results are consistent with statistical mechanical models of protein structure and dynamics, in which the binding of different peptides alters the populations and exchange kinetics of substates in the MHC conformational ensemble. Altered MHC flexibility will influence receptor engagement, impacting conformational adaptations, entropic penalties associated with receptor recognition, and the populations of binding-competent states. Our results highlight a previously unrecognized aspect of the “altered self” mechanism of immune recognition and have implications for specificity, cross-reactivity, and antigenicity in cellular immunity.
Introduction
αβ T cell receptors (TCRs)4 on the surfaces of CD8+ T cells recognize antigenic peptides bound and presented by class I major histocompatibility complex proteins (class I pMHC complexes) to initiate and propagate an antigen-dependent immune response. Both TCR and peptide conformational properties play key roles in antigen recognition and discrimination. Conformational changes in the TCR complementarity-determining region binding loops often accompany binding (1), and measurements of complementarity-determining region loop dynamics have directly linked loop flexibility to TCR cross-reactivity (2, 3). Likewise, conformational changes in peptides often accompany TCR binding (e.g. Refs. 4 and 5), and peptide flexibility can be altered through peptide modifications or MHC polymorphisms (6–8).
Less appreciated, however, is the role that MHC flexibility plays in TCR recognition. Examples of conformational changes in MHC peptide-binding domains occurring upon TCR binding are mostly limited to small shifts in the shapes and positions of the α1 or α2 helices (e.g. Ref. 9). In one telling case, however, the conformation of the α2 helix of the human class I MHC protein HLA-A*0201 (HLA-A2) was shown to undergo a large reorganization upon binding of the A6 TCR (10). Notably, this reorganization is dependent on the peptide as it has not been observed upon recognition of other HLA-A2-presented peptides by the same TCR (11, 12).
In protein binding, crystallographically observed conformational differences have been shown to occur in regions with enhanced motions as the lower energy barriers that facilitate conformational changes translate into faster rates of conformational exchange (13). Indeed, with HLA-A2, the mobility of the region that undergoes a structural change upon binding of the A6 TCR was shown to vary with different peptides (10). This may not be a unique observation as similar effects are believed to contribute to differential T cell recognition of native and modified MART-1 peptides (6), and MHC flexibility can be altered by micropolymorphisms (7, 14, 15). As demonstrated by the A6 TCR, peptide-dependent MHC dynamics can impact TCR recognition by altering the barriers for conformational adjustments, influencing the entropic costs for receptor binding and shifting the populations of binding-competent states. Although altered MHC flexibility has not been apparent structurally, differences in flexibility are generally difficult to ascertain from examination of structure alone.
Here, we explore the extent to which different peptides modify the flexibility of the peptide-binding domain of the class I MHC protein in detail. Using hydrogen/deuterium exchange, fluorescence anisotropy, and an analysis of the class I pMHC structural database, we found a surprising degree of peptide dependence to the motional properties of the HLA-A2 peptide-binding groove. The results were not limited to one region, but extended throughout both α-helices as well as the β-sheet floor of the groove. Regions affected included those that interact with TCRs as well as other activating and inhibitory receptors of the immune system, such as the CD8 coreceptor and natural killer receptors. Our observations emphasize that association of peptides with HLA-A2 proteins physically alters the properties of the protein. Indeed, our results encompass yet broaden the “altered self” model of immune recognition, which emerged from the finding that receptors of the cellular immune system recognize MHC proteins altered through the binding and presentation of antigens (16). Our findings have implications for immunological specificity, cross-reactivity, and ultimately, the determinants of antigenicity for class I MHC-presented peptides.
EXPERIMENTAL PROCEDURES
Proteins and Peptides
pMHC complexes were produced by refolding bacterially expressed inclusion bodies of the HLA-A2 heavy chain and the β2-microglobulin subunit in the presence of excess peptide as described previously (17). Refolded complexes were purified by a combination of ion exchange and size exclusion chromatography. Peptides were synthesized commercially (GenScript). Cysteine mutations in HLA-A2 were performed by PCR mutagenesis and confirmed by sequencing.
Hydrogen/Deuterium Exchange-Mass Spectrometry
Hydrogen/deuterium exchange-mass spectrometry (HDX-MS) was performed as described previously (18). Briefly, exchange was initiated by diluting protein samples at 25 °C in 25 mm HEPES, 50 mm NaCl (pH 7.4) 10-fold with the same buffer made with 99.9% 2H2O. pMHC concentrations were 30 μm after dilution. Excess free peptide was present at a concentration of 30 μm. Exchange was performed within 1 h after purification over a size exclusion column. After dilution into 2H2O, 5-μl aliquots were removed at various time points. For each aliquot, exchange was quenched at 0 °C with 100 μl of 0.1% trifluoroacetic acid (TFA) at pH 2.4. Pepsin (8 μl at 1 mg/ml) was added and digestion performed for 5 min before freezing in liquid nitrogen.
The identities of pepsin-generated fragments of HLA-A2 were previously established in our laboratory by LC/MS/MS (18). The analysis of the exchange behavior of these fragments was performed by MALDI mass spectrometry as described previously (18). Briefly, a matrix solution of 2,5-dihydroxybenzoic acid in 50:50 0.1% TFA (pH 2.4) and acetonitrile was mixed 1:1 with the acidified protein digest on a chilled MALDI plate. The plate was dried under vacuum to minimize back exchange; experiments with fully deuterated fragments indicated that back exchange never exceeded 5%. Spectra were acquired on a Bruker Autoflex III Smartbeam MALDI-TOF mass spectrometer. Data were collected out to hours, although only data to 10 min were selected for comparison as shown in Fig. 1D. This timescale was chosen as it shows clear differences among the samples, and each sample reached an apparent plateau after 10 min, permitting an exponential fit and an estimate of error.
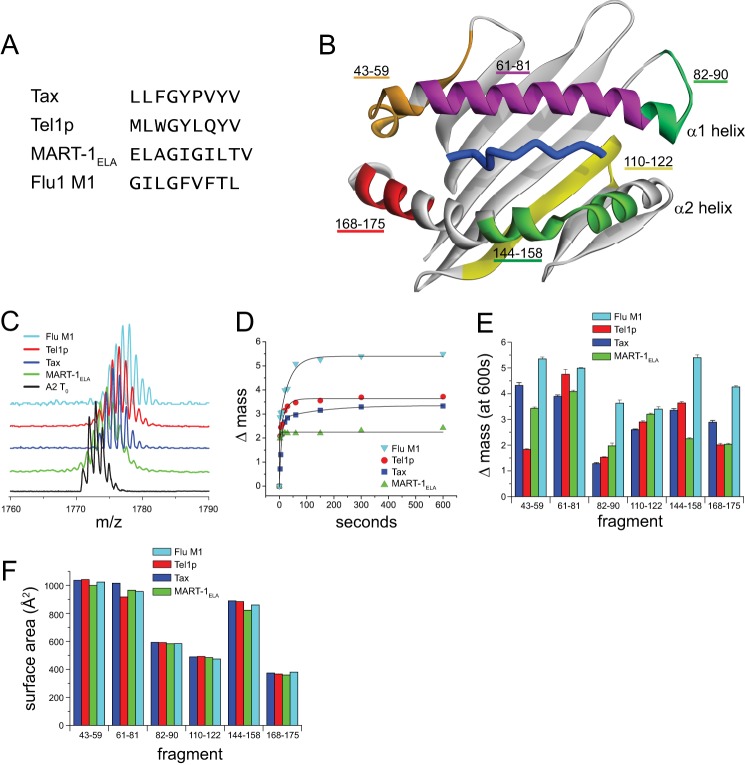
FIGURE 1.
Variation in hydrogen/deuterium exchange properties suggests a peptide dependence to HLA-A2 molecular flexibility. A, sequences of the four peptides whose complexes with HLA-A2 were examined. Each peptide has optimal primary anchors for HLA-A2. B, the different peptidic fragments of the peptide-binding groove identified by HDX-MS mapped onto the HLA-A2 peptide-binding domain. Only residues 1–180 of the HLA-A2 protein are shown. C, mass spectrometric envelopes for the fragment comprising residues 144–158 of the α2 helix at time 0 and after 10 min of exchange for the different peptide·HLA-A2 complexes studied. D, centers of mass for the 144–158 fragment as a function of time out to 10 min of exchange for each of the complexes. Solid lines represent fits to either a single or a double exponential equation. E, mass increase after 10 min of exchange for each of the peptic fragments of the HLA-A2-binding groove in the various complexes. Error bars reflect S.E. at 10 min of exchange from exponential fits as in panel D. F, solvent-accessible surface area of each of the peptide fragments calculated from the crystallographic structures of the different peptide·HLA-A2 complexes. The variation in the structures is small when compared with the variation in hydrogen/deuterium exchange, and there is no correlation between the surface area and isotope exchange datasets.
Fluorescence Anisotropy
Protein labeling and analysis by fluorescence anisotropy were performed as described previously (10). Briefly, purified single-cysteine mutants in 20 mm Na2HPO4, 75 mm NaCl (pH 7.4) were mixed with a 10-fold molar excess of Alexa Fluor 488 C5-maleimide or BODIPY-FL N-(2-aminoethyl) maleimide along with 10-fold excess tris(2-carboxyethyl)phosphine. Parallel reactions with wild-type protein were performed to investigate nonspecific labeling. After mixing protein and label for 1 h at room temperature, excess label was removed by dialysis followed by chromatography. Labeling efficiencies measured spectrophotometrically varied between 92 and 97%. UV images of reduced and nonreduced SDS-PAGE gels verified that fluorescence emanated only from the chain with the cysteine mutant, and fluorescence measurements confirmed that wild-type protein had insignificant amounts of nonspecifically incorporated label.
Steady-state anisotropy measurements were performed in a Beacon 2000 instrument (Invitrogen) in 20 mm Na2HPO4, 75 mm NaCl (pH 7.4) buffer with excess free peptide (between 20- and 100-fold molar excess). Anisotropy measurements on individual samples were repeated 50 times, and samples were measured in duplicate utilizing two independently prepared samples. The values in Fig. 2 and Tables 1 and 2 were collected at 100 nm protein concentration and 25 °C. Additional data for most samples were also collected at concentrations of 10 nm, 500 nm, and 1 μm and at 4 and 35 °C.
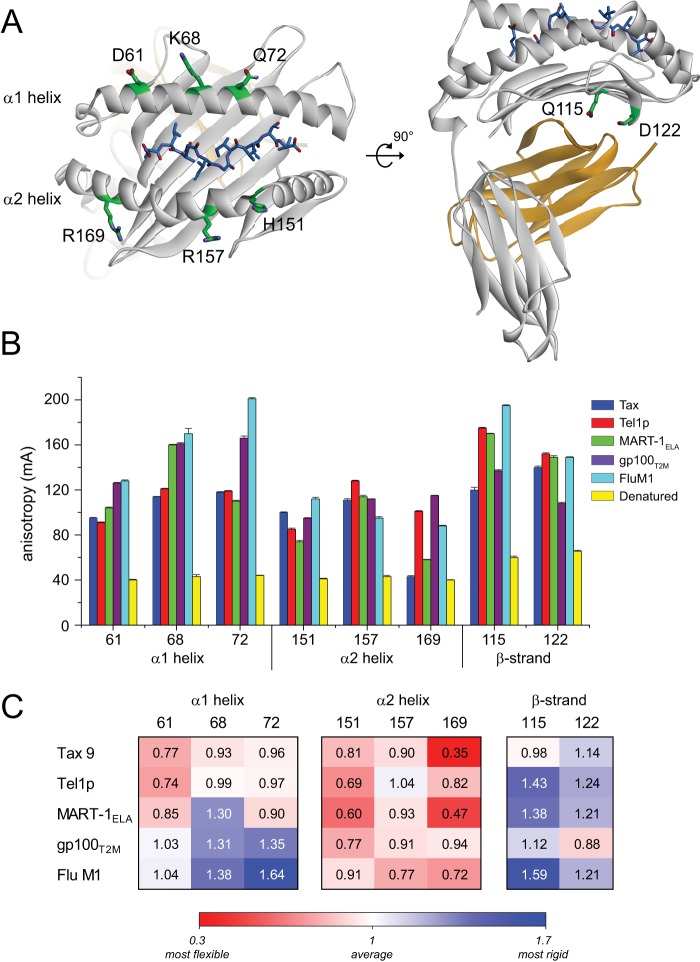
FIGURE 2.
Fluorescence anisotropy indicates a peptide dependence to HLA-A2 molecular flexibility. A, location of the fluorescently labeled sites in the HLA-A2 molecule. In the structures of the wild-type protein, the sites chosen for labeling were all solvent-exposed, polar residues whose side chains made no contacts to the peptide. B, anisotropy values (in mA, or anisotropy reading × 10−3) measured at each site for the various labeled peptide·HLA-A2 complexes. Values plotted are the means and standard deviations of 50 measurements on two independently prepared samples. C, anisotropy values in panel B recast as the percentage of deviation from the average. Higher numbers and blue shading reflect less than average flexibility. Lower numbers and red shading reflect greater than average flexibility.
TABLE 1.
Denatured samples refolded again with a different peptide are indistinguishable from fresh samples
| Tax | Tel1p | MART-1ELA | gp100T2M | Flu M1 | Denatured | |
|---|---|---|---|---|---|---|
| Fresh sample labeled at position 151 | 100.3 ± 0.5 | 85.2 ± 1.1 | 74.4 ± 1.2 | 95.1 ± 0.4 | 111.8 ± 1.5 | 40.9 ± 0.6 |
| Re-refolded sample | 101.1 ± 0.9 | 86.1 ± 1.1 | 75.2 ± 1.5 | 95.9 ± 0.5 | 112.1 ± 0.9 | 41.0 ± 0.7 |
TABLE 2.
Differential peptide dynamics are independent of fluorescent probe
| Position 68 |
Position 72 |
Position 151 |
||||
|---|---|---|---|---|---|---|
| gp100T2M | Tel1p | Flu M1 | Tax | Tel1p | Tax | |
| Alexa Fluor 488 | 170.2 ± 4.7 | 121.2 ± 0.5 | 200.9 ± 1.0 | 117.9 ± 0.5 | 85.2 ± 1.1 | 100.3 ± 0.5 |
| BODIPY-FL | 186.9 ± 0.4 | 154.8 ± 0.8 | 233.1 ± 0.3 | 141.8 ± 0.2 | 110.1 ± 0.6 | 130.2 ± 0.7 |
Measurements on denatured samples were performed by diluting protein in 8 m urea (pH 10) (19). Refolding of fluorescently labeled heavy chain cysteine mutants with new peptides was performed by denaturing a 1-ml solution of fully assembled, labeled Tax·HLA-A2 as above, dialyzing against 500 ml of denaturant over 24 h with three buffer changes to remove peptide, and then using the labeled heavy chain in new small scale refolding reactions with different peptides.
Structural Analyses
Solvent-accessible surface areas were calculated from peptide·HLA-A2 crystallographic structures using Discovery Studio 3.5 (Accelrys), using a grid-based algorithm with 960 points/Å2 and a probe radius of 1.4 Å. Analyses of helix geometry for peptide·HLA-A2 and peptide·H-2Kb structures were performed with HELANAL+ (20). Two-dimensional plots of peptide-protein interactions were generated with LigPlot+ (21). When multiple molecules were present in the asymmetric unit, calculations were performed on only the first molecule.
RESULTS
Peptides Alter the Hydrogen/Deuterium Exchange Behavior of the HLA-A2 Peptide-binding Domain
Although NMR is the most powerful approach for characterizing protein flexibility, the size and complexity of pMHC complexes has made their characterization by NMR challenging. We thus began examining whether and how different peptides alter HLA-A2 flexibility by monitoring hydrogen/deuterium exchange via proteolysis followed by mass spectrometry (HDX-MS). In HDX-MS, protein prepared in aqueous solution is incubated in 2H2O and digested into fragments and deuteron incorporation for each fragment is determined via mass spectrometry (22). Increased levels of deuteration indicate that backbone amides are more accessible to exchange through greater sampling of more open, solvent-exposed states, whereas lower levels of incorporation indicate that backbone amides are protected from exchange through less frequent excursions into solvent-exposed states. HDX-MS has been utilized in a number of cases to explore protein motions, including a recent study of conformational lability in class II MHC proteins (23) as well as our recent study of the effects of binding on the properties pMHC complexes and TCRs (18). Changes in hydrogen/deuterium exchange upon events such as ligand binding result from alterations in exchange kinetics for backbone amides, which manifest as a different number of deuterons incorporated as a function of incubation time.
We performed HDX-MS on four different peptide·HLA-A2 complexes. The peptides chosen were 9- or 10-mers for which crystallographic structures of the pMHC complexes were available (Fig. 1A) (10, 24–26). Care was taken to ensure that the data were not influenced by peptide dissociation. Each peptide chosen was a well characterized peptide with optimal anchors for HLA-A2, and exchange was performed on freshly purified samples at micromolar concentrations well above the nanomolar KD values believed to characterize the interaction of optimal peptides with class I MHC proteins. (Note that accurate KD values are difficult to measure for class I MHC proteins as the peptide-free molecule is unstable, rapidly aggregates, and cannot be directly purified. However, the half-life of the Tax·HLA-A2 complex is 8 h at 25 °C with an estimated KD of 18 nm (19), and the Tm values of the Tax and Flu M1 complexes measured by thermal denaturation are identical within error (24, 27). Further, as indicated below, identical results were obtained with and without the presence of excess peptide in the exchange reactions. Due to its instability, measurements on the peptide-free protein could not be performed.)
Our analysis focused on fragments that encompass the HLA-A2 peptide-binding domain, including three fragments that span the α1 helix, two that span the α2 helix, and one that forms a strand of the C-terminal half of the β-sheet floor of the binding groove (Fig. 1B). These regions were chosen as they are the primary sites of interaction with TCRs, and in the case of the β-sheet region, they interact with other activating and inhibitory receptors of the immune system. These regions are not glycosylated in class I MHC proteins and are not likely to be impacted by the potential formation of TCR-pMHC clusters in an immunological synapse.
The exchange behavior of each fragment followed single or double exponential kinetics as typically seen for HDX-MS data (Fig. 1, C and D). Exchange in each fragment varied with peptide, which we quantified as the degree of deuteration after 10 min of exchange (Fig. 1E). The results were reproducible with repeated measurements on separate samples (supplemental Fig. S1a). The values reported in Fig. 1 included 30 μm excess peptide, although experiments without additional peptide yielded identical results (supplemental Fig. S1b).
The variation in exchange behavior with different peptides is consistent with a peptide dependence on the motions of the HLA-A2-binding domain. However, the results could also be explained by structural variations in the different complexes. The latter seems unlikely as the peptide-binding domains of the four peptide·HLA-A2 complexes superimpose with an average root mean square deviation of 1.2 Å. However, an influence from the different structures and compositions of the peptides is possible. To explore both possibilities, we calculated the solvent-accessible surface areas of the seven fragments in the four pMHC structures (Fig. 1F). The surface areas of each fragment were very similar in all four structures, and there was no correlation between the degree of deuteration and solvent accessibility. Thus, the peptide-dependent variation in the HDX-MS data cannot easily be explained by differences in the crystallographic structures or in the compositions of the peptides.
Fluorescence Anisotropy Reveals Substantial Local Variation in HLA-A2 Flexibility with Different Peptides
To more directly assess the peptide dependence of HLA-A2 flexibility, we examined protein motions using fluorescence anisotropy. When a fluorophore is excited with polarized light, emission is depolarized by the molecular motion that occurs over the fluorescence lifetime. In a protein covalently labeled with a fluorescent probe, motional modes leading to depolarization are protein tumbling, rotation of the probe around its tether, and the influence of backbone and neighboring side chain motion on probe dynamics. As different peptides do not alter the size and shape of the pMHC complex, protein tumbling will be invariant with peptide. Thus, provided a probe attached to the MHC does not interact directly with a peptide, differences in fluorescence depolarization can be attributed to the influence that peptides have on MHC solution structure and dynamics.
We studied the peptide-dependent variation of MHC flexibility by introducing cysteine mutations at six positions within the α-helices of the HLA-A2 heavy chain groove and two positions within the β-sheet that forms the floor of the groove. The six positions on the α-helices encompass regions contacted by TCRs and were within the fragments studied by HDX-MS. The positions on the β-sheet were also in a fragment identified by HDX-MS and are in proximity to the binding sites for the β2-microglobulin subunit, the CD8 coreceptor, and, for murine class I MHC proteins, the Ly49 class of inhibitory receptors (28, 29). The positions chosen for labeling have side chains in the wild-type protein that are polar or charged and, in the structures of the various pMHC complexes, point away from the protein and make no contacts to peptide (Fig. 2A and supplemental Fig. S2).
Each mutant pMHC complex was independently labeled with Alexa Fluor 488 C5-maleimide. We examined the four peptides studied by HDX-MS as well as gp100T2M, another tight binding peptide whose structure bound to HLA-A2 is available (30). Single-cysteine mutants were refolded with the five peptides, fluorescently labeled, and repurified prior to collecting data. Excess peptide was included to ensure stability of the complex, and each set of measurements was performed on two independently prepared samples. To help benchmark the measurements, we also collected data for chemically denatured samples as well as unconjugated fluorescein.
We observed a dramatic variation in anisotropy with the position of the probe (Fig. 2B). For example, with the Flu M1 peptide, the anisotropy ranged from 128 to 201 mA for probes on the α1 helix and from 88 to 112 mA for probes on the α2 helix. To help place the numbers in perspective, the theoretical maximum anisotropy for a fully constrained molecule is 400 mA, and we measured a value of 9 mA for unconjugated Alexa Fluor 488. Measurements on the denatured samples ranged from 40 to 64 mA. The measurements in Fig. 2 were reproducible and independent of concentration over a 100-fold range (10 nm to 1 μm). The general variations in anisotropy with peptide and position were maintained at 4 and 35 °C. The independence of the results with both concentration and temperature further rules out any influence of complex instability.
More striking than the positional variation in the anisotropy was how it varied with different peptides. These data are also in Fig. 2B and recast as percentages of deviation from the mean in Fig. 2C. Although in general the α2 helix was more flexible than the α1 helix, the flexibilities at sites within both helices varied considerably. Large (∼2-fold) variations were seen at the center of the α1 helix, as well as the end of the α2 helix. Variations of 20–40% were seen at the other positions. Our previous studies demonstrated that a 25–30% shift was associated with a large MHC conformational rearrangement and higher entropic penalty upon TCR binding (10). Thus, the variation in flexibility within the HLA-A2 peptide-binding groove is significant. The variation in the region of the β-sheet floor was also significant; of particular interest is the wide variation for position 115, an amino acid critical for recognition of HLA-A2 by the CD8 coreceptor (31).
We examined in detail the possibility that nonspecific labeling could influence the results. No nonspecific covalent labeling of wild-type HLA-A2 heavy chains was observed in parallel labeling reactions. To rule out any noncovalent nonspecific labeling, a sample of purified protein labeled at position 151 and refolded with the Tax peptide was dialyzed under denaturing conditions to remove peptide. After extensive dialysis, the sample was recovered and split into five aliquots, and the labeled HLA-A2 heavy chain in each aliquot was refolded a second time with each of the peptides studied. Measurements on these samples were essentially identical to the measurements on samples that had not undergone this process (Table 1).
We also repeated select measurements using BODIPY-FL, a fluorophore with structural and chemical properties very different from Alexa Fluor 488. Although the absolute anisotropy numbers differed as expected due to the different fluorescence lifetimes and linker arms, the variation seen with different peptides was very similar with the two probes (Table 2), ruling out any probe-specific effects.
Evidence for Differential MHC Dynamics from Crystallographic Structures
Although our HDX-MS and fluorescence anisotropy data show that different peptides alter the motional properties of HLA-A2 protein, as indicated above, the overall architecture of class I MHC proteins is relatively invariant with peptides. Nonetheless, some variation in the position and geometry of the α1 and α2 helices with peptide has been noted (e.g. Refs. 10, 12, and 32). These variations are similar to those that have been observed in comparative molecular dynamics simulations of different peptides bound to the same class I MHC protein (6, 7, 10).
To more precisely quantify the structural differences in the same class I MHC protein crystallized with different peptides, we assembled a database of 51 nonredundant peptide·HLA-A2 structures (supplemental Table S1A) and examined how the geometry of the HLA-A2 α1 and α2 helices varies with peptide. We computed the average bending angle of the helices (defined as the angle formed by least squared-fit lines between the (i-3)-i and i-(i+3) α-carbons of a helix) (20) as well as the width of the peptide-binding groove. The results indicated small but significant peptide-dependent variations (Fig. 3, A and B). We also compared binding groove geometry in all cases in which two peptide·HLA-A2 complexes were present in the crystallographic asymmetric unit. Although the database was smaller (26 complexes), the differences between the two molecules were similar to those observed in the larger dataset (supplemental Fig. S3a). Lastly, we examined cases in which structures of the same peptide·HLA-A2 complex were available both free and bound to a TCR. Although the size of the bound dataset was even smaller (only 12 different TCR-peptide·HLA-A2 structures were available, with three pairs of TCRs bound to the same pMHC), differences of the same magnitude were again observed between free and bound (supplemental Fig. S3b).
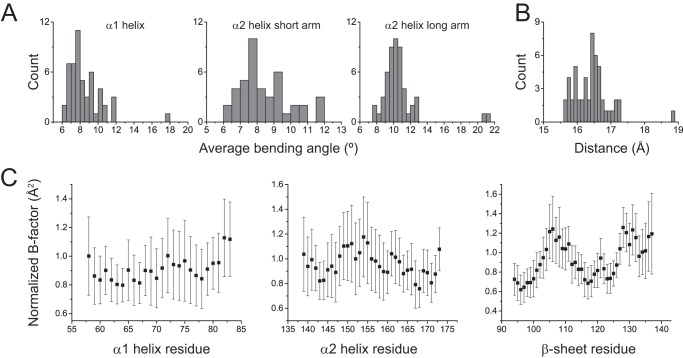
FIGURE 3.
Variation in HLA-A2-binding groove geometry with different peptides. A, distributions of the average bending angles for the α1 helix and the short and long arms of the α2 helix in a database of 51 nonredundant peptide·HLA-A2 structures. B, distribution of the width of the HLA-A2 peptide-binding groove as measured from the α-carbons of Ala-69 and Gln-155 in the 51 peptide·HLA-A2 complexes. C, average normalized B factors in the 51 peptide·HLA-A2 complexes for α-carbons of the α1 helix, α2 helix, and C-terminal half of the β-sheet of the peptide-binding groove. Error bars show the standard deviations, highlighting the considerable variation in B factor with peptide.
To further examine the structures, we examined the variation in crystallographic B-factors (or temperature factors). Although B-factors are impacted by model error and lattice imperfections and report more on static disorder within the crystalline environment as opposed to motions in solution (33, 34), we reasoned that an analysis of a large number of pMHC complexes would allow some of these limitations to be circumvented.
The average and standard deviations for normalized B-factors of α-carbons of the α1/α2 helices and the C-terminal half of the β-sheet floor of the groove are shown in Fig. 3C. In general, the average normalized B-factors fit with expected constraints on protein secondary structure, increasing toward the termini of the helices and in the turns of the β-sheet. Importantly however, for all three elements of structure, the variation in the average values was considerable. The standard deviations for the values in the α1 helix for example ranged from 19 to 30% of the mean values, with an average of 24%. Similar results were seen for the α2 helix and the β-sheet. These results are consistent with the peptide dependence of flexibility detected by HDX-MS and fluorescence anisotropy.
To assess whether our results are unique to HLA-A2, we repeated the B-factor analysis on structures of different peptides bound to the murine H-2Kb class I MHC protein (supplemental Table S1B). Although the number of complexes was smaller (28 nonredundant H-2Kb structures when compared with 51 for HLA-A2), the same general results were seen; the average standard deviations of the B-factors of the α-carbons of the H-2Kb α1 helix, α2 helix, and β-sheet were 30, 32, and 24%, respectively. Thus, the results observed with HLA-A2 may be generalizable to other class I MHC proteins.
DISCUSSION
Studying recognition of the Tel1p and Tax peptides by the A6 TCR, we previously observed that peptide-dependent motions of the HLA-A2 peptide-binding groove could influence TCR recognition, specificity, and cross-reactivity (10). Here, we provide concrete evidence that the “tuning” of the flexibility of class I MHC proteins by different peptides is a general phenomenon and not localized to certain peptides or a single region of the protein.
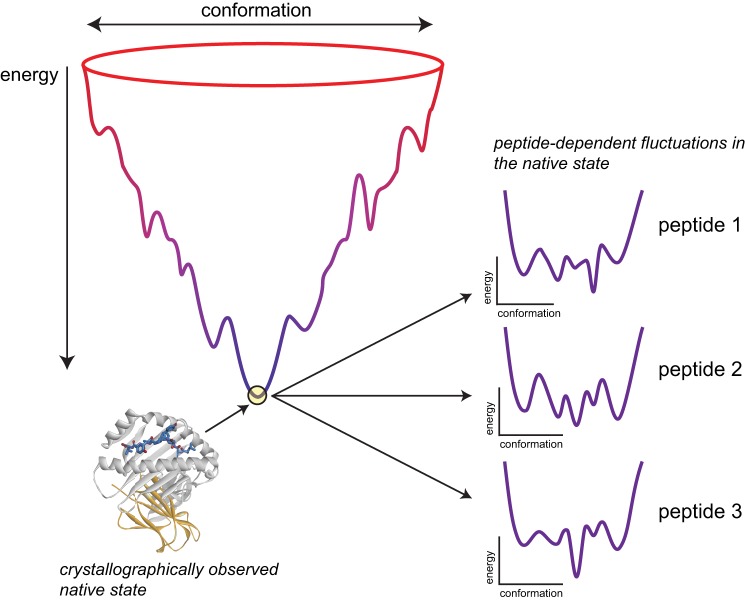
The alteration of protein motions by ligand binding can be interpreted in the context of energy landscapes, which relate protein conformation and energy and define the populations and transition kinetics for the conformational substates that make up the structural ensemble of a protein (35, 36). In altering flexibility, the binding of a ligand to a particular set of substates stabilizes those and destabilizes others, altering the energy landscape and thus entropy and protein dynamics (37). Importantly, the binding of different ligands can stabilize or destabilize different sets of substates, resulting in ligand-dependent changes in protein motions (38–40). In addition to our results here, this behavior has been demonstrated for a variety of proteins, with differential motions impacting processes as diverse as signaling, enzyme activity, and allostery (reviewed in Ref. 41). In structural terms, the stabilization/destabilization of different states by various ligands can arise from formation of different types, strengths, and numbers of noncovalent interactions, as well as different steric limitations on what conformations can be sampled as the protein moves. Given that peptide binding is coupled to class I MHC folding, with the peptide deeply integrated into the structure of the protein, opportunities exist for peptides to impact the stability and thus populations of numerous states (as a simple example, when the structures of the Tax and Flu M1 peptides bound to HLA-A2 are compared, there are 11 amino acids that are uniquely contacted in the two structures (supplemental Fig. S4)). Our results are interpreted in this context in Fig. 4, which shows a stylized class I MHC folding funnel and stylized variations in the folded state energy landscape with different peptides.
FIGURE 4.
Variations in the HLA-A2 energy landscape can explain the peptide dependence of flexibility. Left, a hypothetical, simplified folding funnel for a class I MHC protein, with conformation reflected by the width and free energy reflected by the height. Association of β2-microglobulin and peptide is represented implicitly. The tip of the funnel reflects the crystallographically observed, low energy native state. Right, the sampling of different conformational substates in the native state, or the “breathing” of protein, is dictated by the energy landscape, in which the depths of the wells define the populations of the various substates and thus the entropy, and the heights of the separating barriers give the kinetics for transitioning between substates. Different peptides affect the stability of different substates, as indicated in supplemental Fig. S4, altering the shape of the energy landscape.
It may be notable that beyond showing peptide-specific differences, the correlation between the differences measured by HDX-MS and fluorescence anisotropy is not strong. However, it is important to note that the two experiments probe very different time and length scales. The HDX-MS experiments probe motions indirectly on the millisecond timescale and more slowly, and cannot localize the motions to specific amino acids within the peptidic fragments, which range from 7 to 20 amino acids. Conversely, the fluorescence anisotropy experiments probe motions on the fast, nanosecond timescale directly at the labeled positions. Thus, correlations between the HDX-MS and fluorescence anisotropy results need not be expected. The variation across timescales, however, is important as it speaks to the potential for altered MHC flexibility to impact TCR recognition through multiple ways, including binding entropies (fast timescales), conformational changes (fast and slow timescales), and altered populations of binding-competent states (slow timescales).
A limitation of our experiments is that they do not provide information about what conformational states are sampled. At the nanosecond timescale, some insight can be gained from prior molecular dynamics simulations of the same class I MHC protein with different peptides bound, each of which showed peptide-dependent variations in the overall shape, positions, and stabilities of the α-helices of the peptide-binding groove (6, 7, 10). Although these studies did not address variation in the β-sheet floor of the binding groove, given the tight connectivity between peptides, the helices, and the β-sheet, a peptide dependence to motion within individual β-strands should not be surprising (indeed, the differences in peptide-HLA-A2 contacts in different complexes include multiple residues of the β-sheet (supplemental Fig. S4)). A more direct comparison between our results and these simulations is limited by the finite simulation times of the simulations (50–400 ns); advanced sampling methods that extend the range of molecular simulations should be expected to provide further insight.
Our findings have implications for the determinants of antigenicity and the functional distinctions that are often made between a peptide and a presenting class I MHC protein. When considered alongside our previous findings with the Tax and Tel1p peptides (10), our results imply that there can be an “extension of antigenicity” from the peptide to the MHC and that distinctions commonly made between the two weaken at the atomic level (42). For example, patterns of similar TCR-MHC interatomic interactions in structures of TCRs sharing variable gene segments bound to the same MHC have been used to help argue that TCRs maintain an intrinsic affinity toward MHC proteins (43). However, if the strength of TCR-MHC contacts can vary due to altered MHC dynamics, this limits the extent to which evolution can have imparted an MHC bias onto TCR gene segments. This could help explain why clear patterns of conserved TCR-MHC contacts have been difficult to identify, even in structures where the same TCR is engaged to the same MHC protein presenting different peptides (4).
Our results are also relevant to the surprising degree of specificity seen in many TCR-pMHC interactions. Although TCR cross-reactivity is well appreciated (44), in many cases, TCRs have been shown to be highly sensitive to changes in peptide structure or composition, sometimes with no clear structural explanation. Such fine specificity has been implicated in T cell antagonism as well as the poor performance of modified peptides as vaccine candidates (45, 46). Our findings suggest that in some cases, TCR specificity could arise from dynamical changes resulting from peptide modification.
We focused largely on a single human class I MHC allele, HLA-A*0201. Although the structural analysis suggests similar behavior with the murine class I MHC H-2Kb, an important question to resolve is whether and how MHC micropolymorphisms influence the peptide dependence of dynamics. This may be particularly likely for polymorphic positions whose side chains are embedded within the peptide-binding groove (15). Less clear is the extent to which peptide-dependent dynamics occur in class II MHC proteins. Although peptide association with class II molecules is less of a folding reaction and peptide termini in class II are not embedded in the protein as with class I, a recent examination of a large database of class II pMHC structures revealed structural variations resembling those seen with class I MHC proteins (47), possibly reflecting intrinsic class II MHC dynamics that could be modulated by peptide.
In addition to T cell receptors, a variety of other molecules interact with MHC proteins. These include the CD4/CD8 coreceptors expressed by T cells, and for class I MHC molecules, a range of activating and inhibitory receptors expressed by natural killer and other cell types (48). Structural and binding data indicate that these molecules interact with class I MHC molecules in a way distinct from T cell receptors, making few, if any contacts to peptides (28, 29, 49–51). Key contact regions include the short arm of the α2 helix and the N-terminal end of the α1 helix, as well as the sides or bottoms of the β-sheet floor of the peptide-binding groove. Our data indicate that peptides can modulate the flexibility of each of these regions, suggesting a mechanism for the paradoxical peptide dependence that has been reported for some of these interactions (e.g. Ref. 51). Thus it may be possible that the tuning of MHC dynamics by different peptides reflects a fundamental mechanism for modulating antigenicity.
This work was supported, in whole or in part, by National Institutes of Health Grant GM067079 from the NIGMS (to B. M. B.). This work was also supported by Grant MCB-0448298 from the National Science Foundation.

This article contains supplemental Figs. S1–S4 and Table S1.
- TCR
- T cell receptor
- pMHC
- peptide·MHC complex
- HDX-MS
- hydrogen/deuterium exchange-mass spectrometry.
REFERENCES
- 1. Armstrong K. M., Piepenbrink K. H., Baker B. M. (2008) Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem. J. 415, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scott D. R., Borbulevych O. Y., Piepenbrink K. H., Corcelli S. A., Baker B. M. (2011) Disparate degrees of hypervariable loop flexibility control T-cell receptor cross-reactivity, specificity, and binding mechanism. J. Mol. Biol. 414, 385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hare B. J., Wyss D. F., Osburne M. S., Kern P. S., Reinherz E. L., Wagner G. (1999) Structure, specificity, and CDR mobility of a class II restricted single-chain T-cell receptor. Nat Struct Biol 6, 574–581 [DOI] [PubMed] [Google Scholar]
- 4. Borbulevych O. Y., Santhanagopolan S. M., Hossain M., Baker B. M. (2011) TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J. Immunol. 187, 2453–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tynan F. E., Reid H. H., Kjer-Nielsen L., Miles J. J., Wilce M. C. J., Kostenko L., Borg N. A., Williamson N. A., Beddoe T., Purcell A. W., Burrows S. R., McCluskey J., Rossjohn J. (2007) A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol 8, 268–276 [DOI] [PubMed] [Google Scholar]
- 6. Insaidoo F. K., Borbulevych O. Y., Hossain M., Santhanagopolan S. M., Baxter T. K., Baker B. M. (2011) Loss of T cell antigen recognition arising from changes in peptide and major histocompatibility complex protein flexibility: implications for vaccine design. J. Biol. Chem. 286, 40163–40173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Narzi D., Becker C. M., Fiorillo M. T., Uchanska-Ziegler B., Ziegler A., Böckmann R. A. (2012) Dynamical characterization of two differentially disease associated MHC class I proteins in complex with viral and self-peptides. J. Mol. Biol. 415, 429–442 [DOI] [PubMed] [Google Scholar]
- 8. Pöhlmann T., Böckmann R. A., Grubmüller H., Uchanska-Ziegler B., Ziegler A., Alexiev U. (2004) Differential peptide dynamics is linked to major histocompatibility complex polymorphism. J. Biol. Chem. 279, 28197–28201 [DOI] [PubMed] [Google Scholar]
- 9. Mazza C., Auphan-Anezin N., Gregoire C., Guimezanes A., Kellenberger C., Roussel A., Kearney A., van der Merwe P. A., Schmitt-Verhulst A. M., Malissen B. (2007) How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 26, 1972–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borbulevych O. Y., Piepenbrink K. H., Gloor B. E., Scott D. R., Sommese R. F., Cole D. K., Sewell A. K., Baker B. M. (2009) T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity 31, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garboczi D. N., Ghosh P., Utz U., Fan Q. R., Biddison W. E., Wiley D. C. (1996) Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 [DOI] [PubMed] [Google Scholar]
- 12. Borbulevych O. Y., Piepenbrink K. H., Baker B. M. (2011) Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self molecular mimics. J. Immunol. 186, 2950–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Csermely P., Palotai R., Nussinov R. (2010) Induced fit, conformational selection, and independent dynamic segments: an extended view of binding events. Trends Biochem. Sci. 35, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabian H., Loll B., Huser H., Naumann D., Uchanska-Ziegler B., Ziegler A. (2011) Influence of inflammation-related changes on conformational characteristics of HLA-B27 subtypes as detected by IR spectroscopy. FEBS J. 278, 1713–1727 [DOI] [PubMed] [Google Scholar]
- 15. Fabian H., Huser H., Narzi D., Misselwitz R., Loll B., Ziegler A., Böckmann R. A., Uchanska-Ziegler B., Naumann D. (2008) HLA-B27 subtypes differentially associated with disease exhibit conformational differences in solution. J. Mol. Biol. 376, 798–810 [DOI] [PubMed] [Google Scholar]
- 16. Zinkernagel R. M., Doherty P. C. (1974) Immunological surveillance against altered self components by sensitised T lymphocytes in lymphocytic choriomeningitis. Nature 251, 547–548 [DOI] [PubMed] [Google Scholar]
- 17. Davis-Harrison R. L., Armstrong K. M., Baker B. M. (2005) Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J. Mol. Biol. 346, 533–550 [DOI] [PubMed] [Google Scholar]
- 18. Hawse W. F., Champion M. M., Joyce M. V., Hellman L. M., Hossain M., Ryan V., Pierce B. G., Weng Z., Baker B. M. (2012) Cutting edge: Evidence for a dynamically driven T cell signaling mechanism. J. Immunol. 188, 5819–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Binz A. K., Rodriguez R. C., Biddison W. E., Baker B. M. (2003) Thermodynamic and kinetic analysis of a peptide-class I MHC interaction highlights the noncovalent nature and conformational dynamics of the class I heterotrimer. Biochemistry 42, 4954–4961 [DOI] [PubMed] [Google Scholar]
- 20. Kumar P., Bansal M. (2012) HELANAL-Plus: a web server for analysis of helix geometry in protein structures. J. Biomol. Struct. Dyn. 30, 773–783 [DOI] [PubMed] [Google Scholar]
- 21. Laskowski R. A., Swindells M. B. (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 [DOI] [PubMed] [Google Scholar]
- 22. Marcsisin S. R., Engen J. R. (2010) Hydrogen exchange mass spectrometry: what is it and what can it tell us? Analytical and bioanalytical chemistry 397, 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Painter C. A., Negroni M. P., Kellersberger K. A., Zavala-Ruiz Z., Evans J. E., Stern L. J. (2011) Conformational lability in the class II MHC 310 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange. Proc. Natl. Acad. Sci. U.S.A. 108, 19329–19334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khan A. R., Baker B. M., Ghosh P., Biddison W. E., Wiley D. C. (2000) The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 164, 6398–6405 [DOI] [PubMed] [Google Scholar]
- 25. Sliz P., Michielin O., Cerottini J.-C., Luescher I., Romero P., Karplus M., Wiley D. C. (2001) Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes. J. Immunol. 167, 3276–3284 [DOI] [PubMed] [Google Scholar]
- 26. Madden D. R., Garboczi D. N., Wiley D. C. (1993) The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell 75, 693–708 [DOI] [PubMed] [Google Scholar]
- 27. Bouvier M., Guo H. C., Smith K. J., Wiley D. C. (1998) Crystal structures of HLA-A*0201 complexed with antigenic peptides with either the amino- or carboxyl-terminal group substituted by a methyl group. Proteins 33, 97–106 [DOI] [PubMed] [Google Scholar]
- 28. Gao G. F., Tormo J., Gerth U. C., Wyer J. R., McMichael A. J., Stuart D. I., Bell J. I., Jones E. Y., Jakobsen B. K. (1997) Crystal structure of the complex between human CD8αα and HLA-A2. Nature 387, 630–634 [DOI] [PubMed] [Google Scholar]
- 29. Deng L., Cho S., Malchiodi E. L., Kerzic M. C., Dam J., Mariuzza R. A. (2008) Molecular architecture of the major histocompatibility complex class I-binding site of Ly49 natural killer cell receptors. J. Biol. Chem. 283, 16840–16849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Borbulevych O. Y., Baxter T. K., Yu Z., Restifo N. P., Baker B. M. (2005) Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: implications for vaccine design. J. Immunol. 174, 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wooldridge L., Lissina A., Vernazza J., Gostick E., Laugel B., Hutchinson S. L., Mirza F., Dunbar P. R., Boulter J. M., Glick M., Cerundolo V., van den Berg H. A., Price D. A., Sewell A. K. (2007) Enhanced immunogenicity of CTL antigens through mutation of the CD8 binding MHC class I invariant region. Eur. J. Immunol. 37, 1323–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith K. J., Reid S. W., Stuart D. I., McMichael A. J., Jones E. Y., Bell J. I. (1996) An altered position of the α2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501. Immunity 4, 203–213 [DOI] [PubMed] [Google Scholar]
- 33. Cerutti D. S., Le Trong I., Stenkamp R. E., Lybrand T. P. (2008) Simulations of a protein crystal: explicit treatment of crystallization conditions links theory and experiment in the streptavidin-biotin complex. Biochemistry 47, 12065–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reichert D., Zinkevich T., Saalwächter K., Krushelnitsky A. (2012) The relation of the X-ray B-factor to protein dynamics: insights from recent dynamic solid-state NMR data. J. Biomol. Struct. Dyn. 30, 617–627 [DOI] [PubMed] [Google Scholar]
- 35. Tsai C.-J., Ma B., Nussinov R. (1999) Folding and binding cascades: shifts in energy landscapes. Proc. Natl. Acad. Sci. U.S.A. 96, 9970–9972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller D. W., Dill K. A. (1997) Ligand binding to proteins: the binding landscape model. Protein Sci 6, 2166–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wrabl J. O., Gu J., Liu T., Schrank T. P., Whitten S. T., Hilser V. J. (2011) The role of protein conformational fluctuations in allostery, function, and evolution. Biophys Chem 159, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frederick K. K., Marlow M. S., Valentine K. G., Wand A. J. (2007) Conformational entropy in molecular recognition by proteins. Nature 448, 325–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masterson L. R., Shi L., Metcalfe E., Gao J., Taylor S. S., Veglia G. (2011) Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 108, 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boehr D. D., McElheny D., Dyson H. J., Wright P. E. (2010) Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc. Natl. Acad. Sci. U.S.A. 107, 1373–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smock R. G., Gierasch L. M. (2009) Sending signals dynamically. Science 324, 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baker B. M., Scott D. R., Blevins S. J., Hawse W. F. (2012) Structural and dynamic control of T-cell receptor specificity, cross-reactivity, and binding mechanism. Immunological Reviews 250, 10–31 [DOI] [PubMed] [Google Scholar]
- 43. Garcia K. C., Adams J. J., Feng D., Ely L. K. (2009) The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol 10, 143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mason D. (1998) A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunology Today 19, 395–404 [DOI] [PubMed] [Google Scholar]
- 45. Ding Y. H., Baker B. M., Garboczi D. N., Biddison W. E., Wiley D. C. (1999) Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 11, 45–56 [DOI] [PubMed] [Google Scholar]
- 46. Cole D. K., Edwards E. S., Wynn K. K., Clement M., Miles J. J., Ladell K., Ekeruche J., Gostick E., Adams K. J., Skowera A., Peakman M., Wooldridge L., Price D. A., Sewell A. K. (2010) Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. J. Immunol. 185, 2600–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Painter C. A., Stern L. J. (2012) Conformational variation in structures of classical and non-classical MHCII proteins and functional implications. Immunological Reviews 250, 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Natarajan K., Dimasi N., Wang J., Mariuzza R. A., Margulies D. H. (2002) Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annual Review of Immunology 20, 853–885 [DOI] [PubMed] [Google Scholar]
- 49. Boyington J. C., Motyka S. A., Schuck P., Brooks A. G., Sun P. D. (2000) Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 405, 537–543 [DOI] [PubMed] [Google Scholar]
- 50. Vivian J. P., Duncan R. C., Berry R., O'Connor G. M., Reid H. H., Beddoe T., Gras S., Saunders P. M., Olshina M. A., Widjaja J. M. L., Harpur C. M., Lin J., Maloveste S. M., Price D. A., Lafont B. A. P., McVicar D. W., Clements C. S., Brooks A. G., Rossjohn J. (2011) Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dam J., Guan R., Natarajan K., Dimasi N., Chlewicki L. K., Kranz D. M., Schuck P., Margulies D. H., Mariuzza R. A. (2003) Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2Kb. Nat Immunol 4, 1213–1222 [DOI] [PubMed] [Google Scholar]