Background: Controlling host phosphoinositide metabolism is critical in bacterial infection.
Results: A Legionella effector, SidP, has been identified as a phosphoinositide phosphatase, and its crystal structure was determined.
Conclusion: SidP is a PI(3)P phosphatase, which may play a role in controlling bacterial phagosomal lipid composition.
Significance: Identification of a novel PI phosphatase suggests the importance of exploiting host PI lipids in many bacterial infections.
Keywords: Bacterial Pathogenesis, Lipid Metabolism, Membrane Trafficking, Phagocytosis, Phosphoinositides
Abstract
Bacterial pathogen Legionella pneumophila is the causative agent of Legionnaires' disease, which is associated with intracellular replication of the bacteria in macrophages of human innate immune system. Recent studies indicate that pathogenic bacteria can subvert host cell phosphoinositide (PI) metabolism by translocated virulence effectors. However, in which manner Legionella actively exploits PI lipids to benefit its infection is not well characterized. Here we report that L. pneumophila encodes an effector protein, named SidP, that functions as a PI-3-phosphatase specifically hydrolyzing PI(3)P and PI(3,5)P2 in vitro. This activity of SidP rescues the growth phenotype of a yeast strain defective in PI(3)P phosphatase activity. Crystal structure of SidP orthologue from Legionella longbeachae reveals that this unique PI-3-phosphatase is composed of three distinct domains: a large catalytic domain, an appendage domain that is inserted into the N-terminal portion of the catalytic domain, and a C-terminal α-helical domain. SidP has a small catalytic pocket that presumably provides substrate specificity by limiting the accessibility of bulky PIs with multiple phosphate groups. Together, our identification of a unique family of Legionella PI phosphatases highlights a common scheme of exploiting host PI lipids in many intracellular bacterial pathogen infections.
Introduction
The facultative intracellular pathogen Legionella pneumophila is the causative agent of Legionnaires' disease. This disease often involves acute lung injury caused by the intracellular replication of the bacteria in macrophages of human innate immune system (1, 2). Due to the development of artificial water systems, such as air conditioning cooling towers, Legionnaires' disease has emerged in the second half of last century (3). It has been a human health threat with recent devastating outbreaks in Edinburgh, UK in June 2012 and in Quebec, Canada in July 2012. Upon transmission to the human lung via contaminated aerosols, the bacterial pathogen is then internalized through phagocytosis by alveoli macrophages. Inside the alveoli macrophage, L. pneumophila transports a large array of bacterial proteins into the host through its Dot/Icm type IV secretion system (4, 5). The translocated effectors take control of a variety of host cellular processes to establish a replication-permissive compartment called the Legionella-containing vacuole (LCV)2 (6, 7).
Among the cellular processes, lipid signaling and metabolism have recently emerged as critical targets for bacterial virulence factors. Cellular lipids, particularly phosphoinositides (PIs), are pivotal cellular regulators and play major roles in a broad spectrum of cellular processes including defining intracellular organelle identity, cell signaling, proliferation, cytoskeleton architecture, and membrane trafficking (8–10). Several bacterial pathogens have been shown to target host cell PI metabolism and signaling for their infection and survival. For example, Mycobacterium tuberculosis secretes a lipid phosphatase, SapM, that clears PI(3)P on the bacterial phagosome to inhibit phagosomal maturation (11, 12). Salmonella typhimurium, which is responsible for most food-borne gastroenteritis (13), delivers the PI phosphatase SigD/SopB into the host. SigD/SopB plays multiple roles in bacterial entry and intracellular survival (14). S. flexneri, the causative agent of human dysentery, modifies PI metabolism in host cells to favor its internalization through the phosphoinositide-4-phosphatase activity of the virulent factor IpgD (15). In Legionella pneumophila, several effectors have been reported to interact with specific PI species for their proper localization and function. For example, the L. pneumophila effector SidC has been shown to anchor on the LCV through binding to PI(4)P and promotes the fusion of endoplasmic reticulum-derived vesicles with LCV (16). SidM, a Rab1 guanine nucleotide exchange factor, has also been shown to bind to PI(4)P on the LCV through its C-terminal P4M (PI(4)P binding of SidM/DrrA) domain with nanomolar range affinity (17, 18). Other PI binding effectors have also been documented, such as LidA (18), LpnE (19), and SetA (20). Thus, subverting host PI signaling and metabolism appears to be a general scheme used in the intracellular life cycle of bacterial pathogens (21, 22). Despite these observations, our knowledge about the role of host PI signaling and metabolism in the pathogenesis process of these bacterial pathogens is still largely limited (23). We recently conducted bioinformatics analyses and identified Legionella effectors that may function as PI-metabolizing enzymes. We reported that the Legionella effector SidF is a bona fide PI phosphatase that specifically hydrolyzes PI(3,4)P2 and PI(3,4,5)P3 at the D3 position (24). This activity may facilitate the enrichment of PI(4)P on the LCV and consequently the anchoring of other Legionella effector proteins on the LCV via the binding to PI(4)P.
Here we report a new L. pneumophila Dot/Icm substrate Lpg0130, which we named SidP, a PI-3-phosphatase that specifically hydrolyzes PI(3)P and PI(3,5)P2 in vitro. Structural characterization of the SidP orthologue from Legionella longbeachae strain NSW150 revealed a multidomain organization of the protein. The crystal structure of SidP further revealed unique features of its catalytic pocket that likely afford substrate selectivity. Our new findings of SidP as a PI phosphatase together with our previously reported results on SidF (24) strongly support that L. pneumophila actively exploits host PI metabolism via its own PI phosphatases during infection.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
SidP (Lpg0130) gene and its longbeachae orthologue were PCR amplified from the L. pneumophila strain Philadelphia 1 and L. longbeachae strain NSW150 genomic DNA, respectively. The PCR products were digested with BamH1 and Xho1 or Sal1 restriction enzymes and inserted into a pET28a-based vector in-frame with an N-terminal His-SUMO tag. The same digested SidP fragments were inserted into pRS416-pGPD-GFP vector (25) for the expression of GFP fused SidP and its catalytically inactive mutant in yeast. Point mutations were generated by site-directed mutagenesis. All constructs were confirmed by DNA sequencing.
Protein Expression and Purification
All recombinant proteins were expressed in the Escherichia coli Rosetta strain, which were grown in LB medium supplemented with 50 μg/ml kanamycin. Protein expression was induced at A600 = 0.8 overnight at 18 °C with 0.1 mm isopropyl-β-d-thiogalactopyranoside. The selenomethionine-substituted proteins were expressed in M9 minimum media supplied with synthetic drop-out mix minus methionine (USBiological). Selenomethionine powder (Affymetrix) was added with a final concentration of 120 mg/liter 15 min before isopropyl-β-d-thiogalactopyranoside induction. Harvested cells were resuspended in a buffer containing 50 mm Tris-HCl at pH 8.0, 500 mm NaCl, and protease inhibitor mixture (Roche Applied Science) and lysed by sonication. Soluble fractions were collected by centrifugation at 40,000 × g for 40 min at 4 °C and incubated with cobalt resins (Clontech) for 2 h at 4 °C. Protein-bound resins were extensively washed with lysis buffer. The His-SUMO tag was removed by incubating the protein-bound resin with the SUMO-specific protease Ulp1 at 4 °C for overnight. Eluted protein samples were further purified by FPLC size exclusion chromatography. The peak corresponding to SidP and the homologue were pooled and concentrated to 7–8 mg/ml in a buffer containing 50 mm HEPES at pH 7.4 and 150 mm NaCl.
Crystallization and Preliminary X-ray Crystallographic Analysis
Initial crystallization trials were set up with a PHOENIX liquid handling system (Art Robbins Instruments). SidP crystals were improved by hanging drop vapor diffusion at 4 °C by mixing 1 μl of protein (7.5 mg/ml) with an equal volume of reservoir solution containing 0.1 m succinic acid, 0.1 m HEPES at pH 7.0, 6.5% (w/v) PEG3350, 5 mm DTT. Plate shape crystals were formed within 7–10 days. Selenomethionine-substituted SidP crystals were grown under similar conditions. For data collection, SidP crystals were step-wise-transferred into the reservoir solution supplemented with 5–20% glycerol before flash-cooling to 100 K in liquid nitrogen. The crystal diffracted up to 2.58 Å at the Cornell synchrotron light source MacCHESS beam line A1. All data sets were indexed, integrated, and scaled with HKL-2000 (26). The crystals belong to space group P21 with unit cell parameters of a = 89.28 Å, b = 119.65 Å, c = 133.53 Å, α = 900, β = 101.330, γ = 900 (Table 1). The calculated Matthews coefficient Vm = 3.9, with 68.4% of solvent in the crystal and two protein molecules in an asymmetric unit (27).
TABLE 1.
X-ray data collection, phasing, and structural refinement statistics
| Data collection and phasing | |
| Space group | P21 |
| Cell dimensions | a = 89.28 Å, b = 119.65 Å, c = 133.53 Å, α = 900, β = 101.330, γ = 900 |
| Se SAD data | |
| Synchrotron beam lines | MCCHESS A1 |
| Wavelength (Å) | 0.978 |
| Maximum resolution (Å) | 2.58 |
| Observed reflections | 536,737 |
| Unique reflections | 164,803 |
| Completeness (%)a | 98.5 (95.5) |
| 〈I〉/〈σ〉a | 10.96 (1.21) |
| Rsyma,b (%) | 9.9 |
| Number of molecules in an ASU | 2 |
| Number of selenium sites | 20 |
| Refinement statistics | |
| Resolution (Å)a | 49.44-2.58 (2.65-2.58) |
| Rcrys/Rfree (%)a,c | 19.8/23.7 (30.8/37.5) |
| Root mean square bond length (Å) | 0.016 |
| Root mean square bond angles (°) | 1.729 |
| Ramachandran plot | |
| Most favored/allowed (%) | 97.04/2.96 |
| Outliers (%) | 0 |
a Values in parenthesis are for the highest resolution shell.
b Rsym = ΣhΣi|II(h) − <I(h)>|/ΣhΣiII(h).
c Rcrys = Σ(|Fobs| − k|Fcal|)/Σ|Fobs|. Rfree was calculated for 5% of reflections randomly excluded from the refinement.
Structure Determination and Refinement
Twenty selenium sites were identified in the asymmetric unit by single anomalous scattering method using the program HKL2MAP (28). The initial phase was calculated by the single anomalous scattering method and was improved by solvent flattening in HKL2MAP. The ab initio protein model was then built with COOT (29). Iterative cycles of model building and refinement were carried out with the refmac5 program (30) in the CCP4 suite (31) to complete the final model. The final SidP structure consists of 1–825 amino acids with excellent stereochemistry and good crystallographic statistics (Table 1).
Small-angle X-ray Scattering (SAXS) Analysis of SidP
SAXS experiments were done at the Cornell High Energy Synchrotron Source (CHESS, beamline F2) at x-ray of 9.881 keV at 25 °C. Scattering data were collected at 4 different protein concentrations (0.6, 1.25, 2.5, and 5 mg/ml) in a buffer containing 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm DTT. BioXTAS RAW program was used for data reduction, averaging, and scaling to provide one-dimensional intensity profiles as function of q (4πsinθ/λ, where 2θ is the scattering angle). CRYSOL (32) was used to calculate the theoretical scattering for the two possible models in the SidP crystal and was fitted to the experimental scattering data. The final averaged scattering profile, covering a q-range from 0.01 to 0.25 Å−1 was further analyzed using the programs GNOM (33) to calculate distance distribution function P(r). Low resolution shapes were calculated and averaged from solution-scattering data using the programs DAMMIF (34) and DAMAVER (35). The crystal structure of SidP was docked in the low resolution solution structure using SUPCOMB (36).
Enzymatic Assays
All diC8-phosphoinositides were purchased from Cell Signals, Inc. All reactions were performed in a polystyrene 96-well plate for 20 min at 37 °C with a total volume of 50 μl that contained reaction buffer (50 mm Tris-HCl, pH 8.0, and 150 mm NaCl, 1 mm DTT), 1 nmol concentrations of lipids, and 0.1 μg of purified enzymes. Phosphate release was measured at A620 absorbance with the addition of malachite green reagent as described by Maehama et al. (37).
Yeast in Vivo Experiments and PI Analysis
All yeast strains used in this study are in the SEY6210.1 background. Strain YTS1 (ymr1Δsjl2Δsjl3Δ[CEN ymr1ts]; a temperature-sensitive allele of the YMR1 gene) (38) and strain YCS215 (sac1Δsjl2Δsjl3Δ[CEN sac1ts]; a temperature-sensitive allele of the SAC1 gene) (39) were transformed with pRS416-pGPD-GFP plasmids expressing corresponding SidP WT and mutant proteins. For growth assay, yeast transformants were grown to midlog (A600 = 0.5) at permissive temperature (26 °C), adjusted to 1 A/ml, serially diluted 1:10 five times, and spotted onto −Leu−Ura selection plates. The viability of these transformed cells was analyzed by incubating the plates in permissive and restricted temperatures for 3–4 days. PI levels were analyzed as previously described (40). Briefly, cells (5 A600) were harvested, washed in media lacking inositol, and labeled with 50 μCi of myo-[2-3H]inositol (PerkinElmer Life Sciences) in synthetic media lacking inositol for 1 h. Lipids were deacylated. [3H]Glycerophosphoinositides were extracted, and 5 × 106 cpm of samples were separated by HPLC.
RESULTS
Legionella Effector SidP Is a Phosphoinositide 3-Phosphatase
Previously, we used a sequence pattern-based method to identify proteins in the genome of L. pneumophila strain Philadelphia 1 that contains the CX5R motif, a signature sequence of the catalytic residues in a number of enzymes, including lipid phosphatases (41), protein-tyrosine phosphatases (42), and arsenate reductases (43). More than 400 hypothetical proteins were found possessing this motif, and 29 of these proteins have been identified as substrates of the Dot/Icm secretion system (24, 44). Recombinant proteins of these 29 candidates were then expressed, purified, and examined for in vitro PI phosphatase activities by a malachite green-based assay (37). In addition to our recently published L. pneumophila PI phosphatase, SidF (24), we found that the Legionella effector Lpg0130, which we named SidP, has PI phosphatase activities. SidP from L. pneumophila strain Philadelphia 1 contains 822 amino acids with a peptide sequence CVSGKDR that matches the consensus CX5R motif (Fig. 1A). Although SidP is conserved in all genome sequenced Legionella species as well as some related pathogenic bacterium species (supplemental Fig. S1), it has no significant sequence homology to any known PI phosphatase, including SidF, besides the catalytic motif. Wild type recombinant protein of SidP was shown to specifically hydrolyze PI(3)P and PI(3,5)P2 in vitro (Fig. 1, B and C), whereas this activity was completely abolished for the catalytic cysteine to serine (C554S) mutant. Mutations of the conserved residues within the catalytic P-loop (D559N, R560K) also abrogated their catalytic activity (Fig. 1, B and C). We also analyzed the enzymatic activity of SidP orthologue from L. longbeachae strain NSW150 (locus tag LLO_3270), which had a sequence identity of 54% and similarity of 68% to the L. pneumophila orthologue. Similar to Lpg0130, LLO_3270 protein showed specific enzymatic activity against PI(3)P but no detectable activity against PI(3,5)P2 (Fig. 1D). This finding demonstrated that Legionella species encodes a novel Dot/Icm effector that functions as a PI-3-phosphatase.
FIGURE 1.
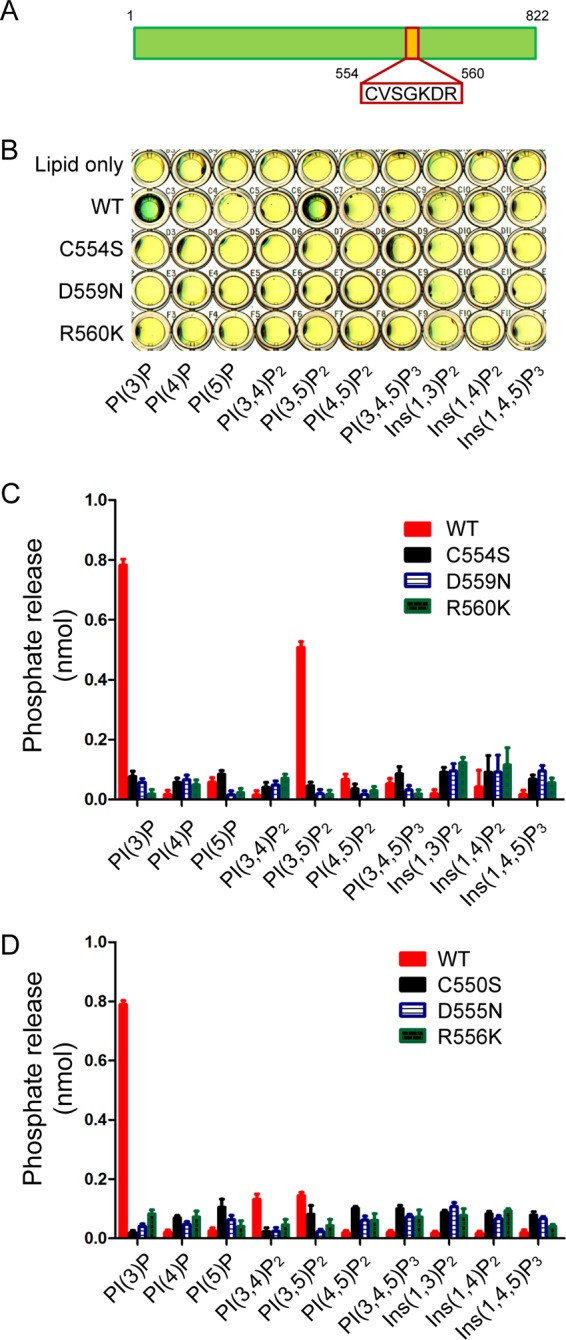
Legionella effector SidP is a phosphoinositide phosphatase. A, shown is a schematic structure of SidP (Lpg0130). The CX5R is highlighted. B, shown is phosphoinositide substrate specificity of purified wild type and C554S, D559N, and R560K mutant SidP as determined by the malachite green assay (green indicates the release of free phosphate). PI(3)P and PI(3,5)P2 are the preferred substrates. C, quantification of the amount of released phosphate is shown. Data are from three replicate experiments (mean ± S.E.). D, shown is quantification of the amount of released phosphates from the phosphatase assay of SidP L. longbeachae orthologue LLO_3270. Data are from three replicate experiments (means ± S.E.). Ins, inositol.
SidP Rescues the Growth Phenotype of Yeast Defective of PI(3)P Metabolism
To further assay the in vivo activity of SidP, we took advantage of yeast mutant strains that have growth defects due to mutations in PI-metabolizing enzymes. The YTS1 strain harboring ymr1tssjl2Δsjl3Δ mutations (protein products of these three genes hydrolyze PI(3)P in yeast) can grow at permissive temperature but is lethal at restrictive temperature due to the increased levels of PI(3)P (25). At restrictive temperature, cells transformed with vector or SidP carrying the catalytically inactive Cys to Ser mutation were not viable. However, the growth phenotype could be rescued by the expression of Ymr1p or wild type SidP (Fig. 2A), suggesting that SidP is able to hydrolyze PI(3)P in vivo. We further tested the substrate specificity of SidP in vivo using the YCS215 strain containing sac1tssjl2Δsjl3Δ mutations (the only three proteins that hydrolyze PI(4)P in yeast). Cells carrying sac1tssjl2Δsjl3Δ mutations are lethal at restrictive temperatures due to elevated levels of PI(4)P (39). Exogenous expression of SidP in sac1tssjl2Δsjl3Δ cells did not rescue the growth phenotype (Fig. 2B), suggesting PI(4)P is not the substrate for SidP. The in vivo activity of SidP orthologue from L. longbeachae (LLO_3270) was also analyzed by yeast complementation assays. Similar to SidP, exogenous expression of GFP-tagged LLO_3270 in ymr1tssjl2Δsjl3Δ strain rescued the growth phenotype but not the sac1tssjl2Δsjl3Δ strain at restrictive temperatures (Fig. 2, A and B). In agreement with the growth phenotype, PI analysis experiments showed that the endogenous PI(3)P levels were reduced by nearly 3-fold in the presence of SidP compared with cells transformed with empty vector or the SidP CS mutant in ymr1tssjl2Δsjl3Δ cells (Fig. 2C). However, the in vivo PI(4)P and PI(4,5)P2 levels were not changed. These results demonstrated that SidP specifically functions as a PI-3-phosphatase in vivo (Fig. 2C).
FIGURE 2.
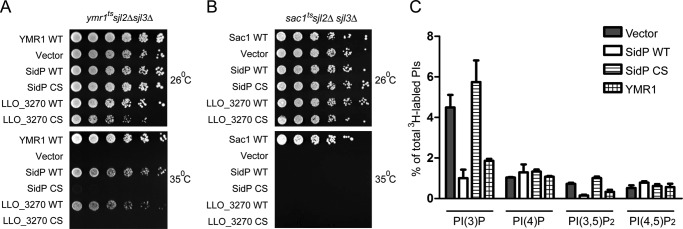
Functional complementation assays of SidP and its L. longbeachae orthologue (LLO_3270) in yeast. A, shown is an in vivo growth rescue assay of SidPs in ymr1tssjl2Δsjl3Δ stain. B, shown is an in vivo growth rescue assay of SidPs in sac1tssjl2Δsjl3Δ strain. In both A and B, corresponding yeast cells were transformed with the indicated constructs or vector control. The cells were spotted onto selection plates with four serial of 10× dilutions from left to right and grown at the indicated temperatures for 3 days. C, shown is quantitative analysis of in vivo PI levels. ymr1tssjl2Δsjl3Δ cells were transformed with vectors expressing GFP (negative control), wild type SidP, SidP C554S mutant, or YMR1 (positive control) and labeled with myo-[2-3H]inositol for 1 h at 37 °C. Total lipids were extracted and analyzed by HPLC.
Crystal Structure Determination and Overall Structure of SidP
To further characterize the molecular mechanism of this novel PI-3-phosphatase, we set out to determine the atomic structure of SidP by x-ray crystallography. Although the full-length protein of SidP from L. pneumophila strain Philadelphia 1 could be highly expressed and purified to homogeneity, extensive crystallization trials failed to yield any crystal hits. SidP orthologues from other Legionella species were then cloned for crystallization screens. Protein crystals of SidP orthologue from L. longbeachae (LLO_3270) were successfully obtained using the hanging drop vapor diffusion method with the reservoir solution containing 0.1 m HEPES, pH 7.0, 0.1 m succinic acid, and 6.5% (w/v) PEG3350. The plate-shaped crystals belong to the space group P21 with unit cell dimensions of a = 89.28 Å, b = 119.65 Å, c = 133.53 Å, α = 900, β = 101.330, γ = 900. There are two molecules in the asymmetric unit. The structure was solved by selenium single wavelength anomalous diffraction method using the program HKL2MAP (28). The final structure was refined against a 2.58 Å resolution data set with Rwork = 19.8 and Rfree = 23.7, respectively (Table 1).
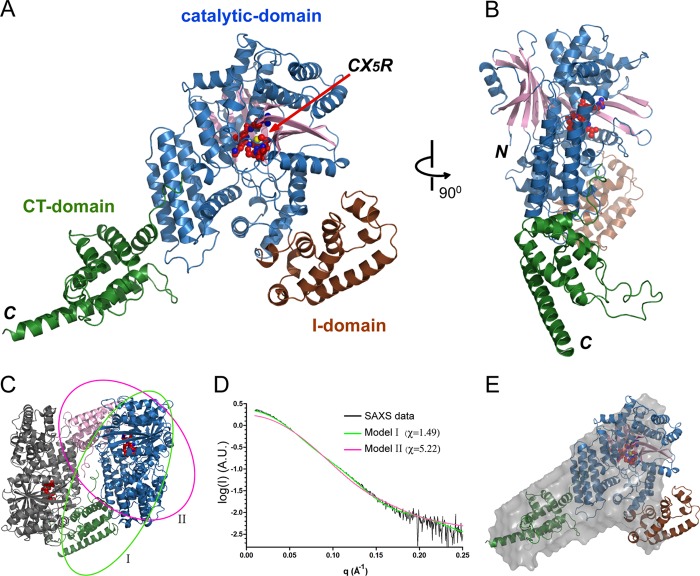
Full-length SidP from L. longbeachae contains 825 amino acids. The crystal structure of SidP revealed that SidP is composed of three distinct domains (Fig. 3, A and B). The N-terminal portion (residue 1–666) contains the catalytic domain (colored in blue for α-helices and pink for β-strands) and an appendage domain (I-domain shown in brown), which is inserted within the catalytic domain between residue 189 and 306. The C-terminal part (residue 667–825) folds in an all α-helix domain (the C-terminal (CT) domain is colored in green). Because the electron density for the linker peptide between the catalytic domain and the C-terminal domain is missing, there are two possible ways to assign the C-terminal domain to either one of the two SidP molecules in the asymmetric unit (Fig. 3C). To circumvent this ambiguity, we used the SAXS method to obtain the solution properties of SidP. Shape reconstruction algorithms using the SidP SAXS data allowed us to distinguish between the two possible atomic models obtained from our crystal structure. The model presented in Fig. 3A (labeled as model I in Fig. 3C) had a significantly better fit with the SAXS data than the other model (Fig. 3, D and E); thus, the overall domain configuration of SidP was ascertained.
FIGURE 3.
Crystal structure of SidP. A and B, shown are two orthogonal views of the crystal structure of SidP, represented in ribbons. The catalytic CX5R motif is shown in spheres and indicated by an arrow. SidP consists of three domains. The N-terminal catalytic domain is colored in blue, and the I-domain, which is inserted within the catalytic domain, is shown in brown. The CT domain is colored in green. C--E, shown is BioSAXS analysis of SidP. C, shown are the two protein molecules in the asymmetric unit of the SidP crystal. The green (model I)- and pink (model II)-colored ovals indicate two distinct monomer models of SidP. D, shown is CRYSOL fitting of SidP crystal structure models to the experimental data (black curve) on a plot of intensity versus q. The theoretic scattering from the SidP crystallographic model I is shown in green and in pink for model II. The chi (√χ2) values are 1.49 and 5.22 for the fitting of model I and II, respectively. The chi values indicate that model I is acceptable and has a significant better fit with the experimental data than model II. E, shown is SAXS-based shape reconstruction for SidP. The crystal structure of Model I is docked in the low resolution envelope using the SUPCOMB program.
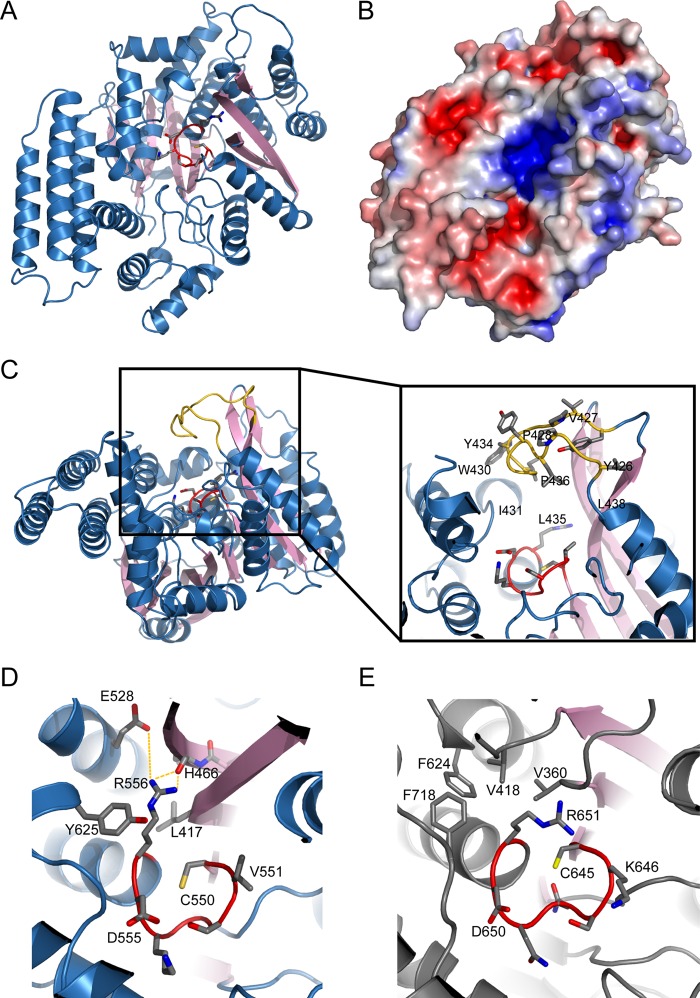
The Catalytic Domain of SidP
The catalytic domain of SidP consists of an 11-pleated β-sheet as the structural core that is surrounded by 18 α-helices. These secondary structure elements pack in a bird nest-like structure with a deep invaginated pocket at the center of the catalytic domain (Fig. 4, A and B). Similar to other PI phosphatases, this pocket, at the bottom of which the catalytic CX5R motif (residue 554–560) is aligned, is highly positively charged (Fig. 4B). The positive electrostatic potentials at the catalytic site are contributed partially by several conserved arginine and lysine residues (labeled with a blue oval in supplemental Fig. S1) and the electric dipole of the α-helix, where the catalytic P-loop ends. Like other CX5R-containing phosphatases, this charge distribution is ideal for the recognition of the negatively charged head group of PI lipids. Interestingly, the catalytic pocket of SidP is tightly covered by a long loop enriched with hydrophobic residues between β8 and α20 (colored in gold in Fig. 4C and supplemental Fig. S1). This structural architecture suggests that this hydrophobic loop may play a role in controlling the accessibility of substrates.
FIGURE 4.
The catalytic domain of SidP. A, shown is a ribbon diagram of the catalytic domain of SidP. The catalytic motif is shown in red in sticks. For clarity, a hydrophobic loop that covers on top of the catalytic pocket is removed. B, the catalytic domain at the same orientation as in A is represented on the surface. The surface is colored based on electrostatic potential with positively charged regions in blue (+4 kcal per electron) and the negatively charged regions in red (-4 kcal per electron). Surface potential calculations were determined using APBS plug-in on PyMOL (DeLano Scientific, LLC). C, shown is a hydrophobic loop that covers the catalytic pocket. The loop is colored in gold and is shown in sticks in the zoomed-in view. D, shown is a close view of the catalytic P-loop of SidP. Note that CX5R arginine is away from the catalytic cysteine (Cys-550). The hydrophobic part of Arg-556 side chain is sandwiched between two hydrophobic residues, Tyr-625 and Leu-417, whereas the guanidinium group interacts with the main chain carbonyl group of His-466 through hydrogen bonding and residue Glu-528 through electrostatic attraction. E, the catalytic site of SidF is shown. The presence of a cluster of hydrophobic residues (Val-360, Val-418, Phe-624, Phe-718) retains the CX5R arginine Arg-651 in a conformation in which its guanidinium group is close to the catalytic cysteine.
The crystal structure of SidP also revealed a unique conformation of the CX5R arginine (Arg-556). In SidP, Arg-556 is away from the catalytic cysteine (Cys-550). The hydrophobic part of Arg-556 side chain is sandwiched between two hydrophobic residues, Tyr-625 and Leu-417, whereas the guanidinium group interacts with the main chain carbonyl group of His-466 through hydrogen bonding and with residue Glu-528 through electrostatic attraction (Fig. 4D). In contrast to SidP, the presence of a hydrophobic “wall” formed by four residues (Val-360, Val-418, Phe-624, and Phe-718) in SidF may restrict the CX5R arginine (Arg-651) in a conformation that holds the releasing phosphate group of the substrate in the position ready for nucleophilic attack by the catalytic Cys-645 (Fig. 4E). The functional role of this unique conformation of Arg-556 in SidP is not clear. However, in SidP, a conformational change of Arg-556 to assume a similar conformation as seen in SidF seems necessary in substrate binding and hydrolysis. Conformational change of the catalytic P-loop has been proposed to regulate the activity of other CX5R-based phosphatases, such as Sac1 (45). It is possible that this unique conformation of Arg-556 may play a role in regulating SidP activity. It is also worth noting that although most protein-tyrosine phosphatases and some lipid phosphatases have a WPD loop featuring an aspartic residue, which acts as a general acid/base during catalysis (42), this loop is not present in SidP. Instead, the conserved Asp-555 within the P-loop functions as the general acid/base, similar to SidF, MTMR2, and Sac1 (Fig. 4E). D559N in SidP or D555N in LLO_3270 mutation resulted in a complete loss of the catalytic activity in both enzymes (Fig. 1).
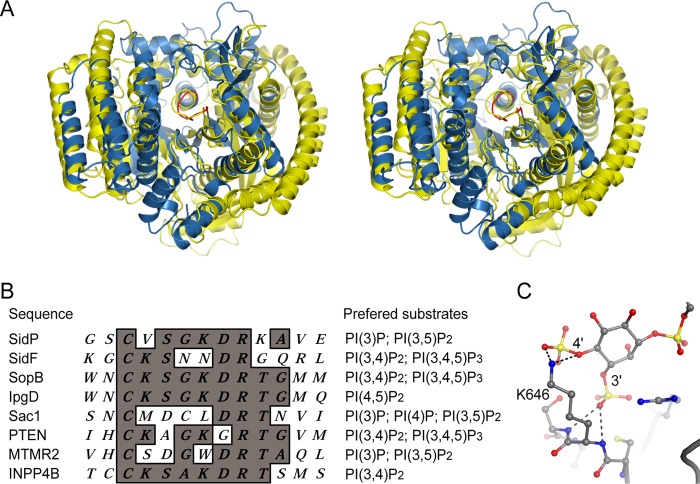
Comparison of the Catalytic Domain of SidP with SidF and Other PI Phosphatases
Despite the lack of primary sequence similarity of SidP with other known PI phosphatases, the three-dimensional structure of the catalytic domain of SidP has a conserved structural core as seen in other phosphatases, which consists of one α-helix and a central β-sheet with four parallel β-strands. Within the structural core, the loop that contains the catalytic CX5R motif connects the carboxyl end of one of the β-strands with the N terminus of the α-helix (Fig. 4D and Fig. 5A) (46–49). Furthermore, structural homology search by the Dali Server (50) revealed that the catalytic domain of SidP has a surprisingly high degree of similarity with SidF, the other L. pneumophila PI phosphatase that we have reported recently to specifically hydrolyze PI(3,4)P2 and PI(3,4,5)P3 (24). Three-dimensional structural superposition calculation of these two structures yielded a Z-score of 20.3 and an root mean square deviation of 2.9 Å for 421 aligned residues (Fig. 5A). An intriguing question arose as to why SidP and SidF have such distinct substrate specificity, whereas their overall structures are highly similar. A close comparison of the catalytic site of SidP with SidF and other PI phosphatases, such as the myotubularin phosphatases (41, 51) and the tumor suppressor PTEN (49), revealed differences in both catalytic P-loop primary sequence and the overall shape of the active site pocket. It is interesting that all the phosphatases that hydrolyze substrates with two adjacent phosphate groups have a lysine residue right after the catalytic cysteine (Fig. 5B). This lysine residue, as revealed in the crystal structure of SidF in complex with its substrate PI(3,4)P2, forms a hydrogen bond interaction with the D3 phosphate group with its main chain amide group. Meanwhile, its ϵ-amide group forms strong electrostatic and hydrogen bond interactions with the D4 phosphate group of the substrate (Fig. 5C). The length of a lysine side chain perfectly matches the distance between two phosphate groups when attached to two neighboring hydroxyl groups of the inositol ring. Thus, this lysine residue seems to function as a “ruler” and plays an important role in recognition and stabilization of substrates with two adjacent phosphate groups. The lack of this lysine residue in SidP as well as in Sac1 and MTMR2 might partially explain why these enzymes do not hydrolyze PI species with two adjacent phosphate groups. The other key substrate specificity determinant is the overall shape of the catalytic pocket. The size of the catalytic pocket in SidP is significant smaller (Fig. 4, A and B) compared with the pocket of SidF (24); thus, it may limit the accessibility of bulky PIs with multiple phosphate groups.
FIGURE 5.
Comparison and substrate specificity of CX5R containing PI phosphatases. A, shown is a stereo view of superposition of the catalytic domain of SidP with SidF. SidP is colored in blue, and SidF (PDB ID 4FYG) is in yellow. The CX5R motif is shown in red. B, shown is sequence alignment of the CX5R motif sequence from representative PI phosphatases. The in vitro preferred substrates are listed for each phosphatase. Enzymes with a non-lysine residue after the catalytic cysteine seem to hydrolyze mono- or non-consecutively di-phosphorylated PI species. However, enzymes with a lysine residue after the cysteine can hydrolyze lipids with two consecutive phosphate groups. C, shown is a close view of the catalytic site lysine (K646) in SidF. The main chain amide group forms hydrogen bond with the D3 phosphate, whereas the ϵ amide group interacts with the D4 phosphate. A lysine residue at the second position of the CX5R motif provides the specificity for PI substrates with two consecutive phosphate groups.
DISCUSSION
Our structural studies not only revealed interesting features of the catalytic domain of SidP but also revealed two additional novel domains in SidP, the I-domain and the CT-domain. Both domains have an all-α structure. Structural homology search by the Dali server does not show any significant homologues. These two domains may function as protein-protein interaction domains for proper targeting of SidP during infection through interactions with host or other Legionella effector proteins. Although we have generated polyclonal antibodies against SidP, which works well in detecting SidP expression by Western blot, nonspecific signals from immuno-staining prevented us from probing the in vivo localization of SidP during Legionella infection. The complete CT-domain structure may also provide a structural model for the understanding of signals that determine the translocation of Legionella effector proteins by the Icm/Dot apparatus. It has been proposed that the extreme C terminus contains the determinants for translocation (52). A region of 6–8 residues called “E-blocks,” which is rich in glutamates, has been proposed to be the signal for translocated substrates (53). Interestingly, the C terminus of SidP forms a long α helix that has an overall anionic charge (Fig. 6, A and B). The glutamate residues in the C-terminal α helix are not completely conserved in terms of their exact positions among different species; however, SidP orthologues/homologues from other Legionella species are all enriched with glutamate residues at their C termini, suggesting the presence of a conversed E-blocks signature with this C-terminal α helix (supplemental Fig. S1).
FIGURE 6.
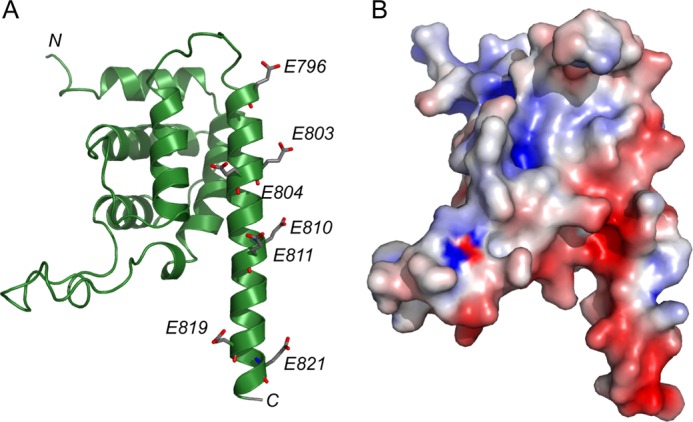
The CT-domain of SidP. A, shown is a ribbon diagram of the CT-domain of SidP. The very C terminus of SidP assumes a long α-helix structure. This α-helix is enriched with glutamate residues (shown in sticks). B, shown is a surface representative of the CT domain. The same color scheme applied in Fig. 4 is used in this figure. Note that the C-terminal α-helix is negatively charged due the cluster of glutamate residues.
Although modulation of host PIs by bacterial virulent factors has been reported in other bacterial pathogens, such as M. tuberculosis (11, 12), S. flexneri (15), and S typhimurium (14, 54), active controlling of host PI system by Legionella species has not been well studied. Our discovery of SidP, a new L. pneumophila PI phosphatase, re-emphasizes the notion that L. pneumophila infection involves active exploiting of host PI metabolism by the pathogen. The presence of SidP homologues in other pathogenic bacteria, such as Fluoribacter dumoffii and Rickettsiella grylli (supplemental Fig. S1), further highlights that controlling the host PI system is a common strategy used by many intracellular bacterial pathogens. An important and interesting future direction is to address the in vivo role of SidP in L. pneumophila infection. It has been shown that PI(3)P, generated from the phosphorylation of phosphatidylinositol by Vps34 (55), appears on the early phagosome membrane and plays a key role during phagosomal maturation (11, 56). Our demonstration that SidP is a PI(3)P phosphatase suggests that through the action of SidP, Legionella may be capable of eliminating the endosomal-like identity and reprogramming the maturation process of bacterial-containing phagosomes.
Recently, we have reported the Dot/Icm substrate SidF as a bona fide PI phosphatase that specifically hydrolyzes PI(3,4)P2 and PI(3,4,5)P3 at the D3 position (24). We proposed that the hydrolysis of PI(3,4)P2 and PI(3,4,5)P3 by SidF at the D3 position may play a role in preventing the conversion of these two lipids into PI(3)P by host 5p- or 4p-phosphatases, such as SHIP-1 and Inpp4A, respectively, during endocytic processes (57). The new player, SidP, may further eliminate PI(3)P molecules that have escaped from the action of SidF or that are newly generated by host PI-3 kinase Vps34. The presence of two PI phosphatases in L. pneumophila that have non-overlapping 3-phosphatase activities strongly suggests that removal of the D3 phosphate group of PIs on the phagosomal membrane is critical for bacterial infection. Our findings with SidP allow us to propose a model for the active regulation of PI metabolism by Legionella during infection (Fig. 7). In this model the two Legionella PI phosphatases function in a concerted way on the surface of bacterial phagosomes. SidF anchors on the LCV membrane and hydrolyzes PI(3,4,5)P3 and PI(3,4)P2 to PI(4,5)P2 and PI(4)P. SidP dephosphorylates the PI(3)P on the LCV. By preventing the accumulation of PI(3)P on the LCV, SidF and SidP may play a role in restricting the fusion of host endosomes or lysosomes with the bacterial phagosomes, which may facilitate the escape of the bacterium from the default degradative phagolysosomal pathway. The synergistic action of SidF and SidP may also lead to the enrichment of PI(4)P on the LCV. PI(4)P can then recruit several other bacterial effectors, such as SidC and SidM. Thus, active control of the lipid composition may play a role in programming the LCV for Legionella replication. However, single deletion of either SidP or SidF did not show obvious bacterial infection or growth defects (data not shown); future studies by deleting both SidP and SidF are required to address the specific role of both enzymes during Legionella pathogenesis.
FIGURE 7.
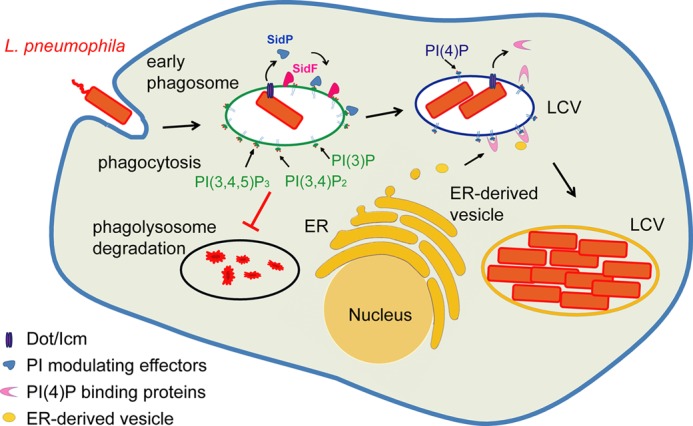
Functional model of modulating host phosphoinositides in Legionella infection. L. pneumophila encodes two PI phosphatases, SidF and SidP. Although SidF specifically hydrolyzes PI(3,4)P2 and PI(3,4,5)P3, SidP removes PI(3)P on the surface of early bacterial phagosomes. By the synergistic action of these two effectors, the LCV will be diverted from the PI(3)P-mediated phagolysosomal pathway into a PI(4)P-enriched compartment that is hospitable for the intracellular replication of the bacterium. ER, endoplasmic reticulum.
Another cellular process that has been proposed to be manipulated by L. pneumophila is autophagy (58). Recent data also showed that host autophagy is inhibited by the Legionella effector RavZ by inactivating Atg8 during infection (59). However, ΔravZ mutant strain can still evade a functional autophagy system, suggesting that additional effectors may be capable of disrupting the recognition of the LCV by the host autophagy system (59). PI(3)P has been demonstrated as one of the key signals to initiate the autophagy process (60). The finding of SidP as a PI(3)P phosphatase indicates that SidP may also play a role in Legionella avoidance of the host autophagy pathway. Legionella strains with double deletion of SidP and ravZ will be created in our future studies to address this hypothesis.
Acknowledgments
We thank Dr. Zhao-Qing Luo for critical discussion and reagents and Dr. Richard Gillilan (MacCHESS, Cornell University) for technical help in BioSAXS data collection and analysis. The x-ray data were collected at MacCHESS beamline A1. CHESS (Cornell High Energy Synchrotron Source) is supported by the National Science Foundation (NSF) and NIGMS, National Institutes of Health (NIH), via NSF Award DMR-0225180, and the MacCHESS resource is supported by Center for Research Resources, NIH Award RR-01646.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-GM094347 (to Y. M.).

This article contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 4JZA) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- LCV
- Legionella-containing vacuole
- PI
- phosphoinositide
- P2
- diphosphate
- P3
- trisphosphate
- SAXS
- small-angle x-ray scattering
- SUMO
- small ubiquitin-like modifier
- CT
- C-terminal.
REFERENCES
- 1. McDade J. E., Shepard C. C., Fraser D. W., Tsai T. R., Redus M. A., Dowdle W. R. (1977) Legionnaires' disease. Isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203 [DOI] [PubMed] [Google Scholar]
- 2. McKinney R. M., Porschen R. K., Edelstein P. H., Bissett M. L., Harris P. P., Bondell S. P., Steigerwalt A. G., Weaver R. E., Ein M. E., Lindquist D. S., Kops R. S., Brenner D. J. (1981) Legionella longbeachae species nova, another etiologic agent of human pneumonia. Ann. Intern. Med. 94, 739–743 [DOI] [PubMed] [Google Scholar]
- 3. Fields B. S., Benson R. F., Besser R. E. (2002) Legionella and Legionnaires' disease. 25 years of investigation. Clin. Microbiol Rev. 15, 506–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Segal G., Purcell M., Shuman H. A. (1998) Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95, 1669–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vogel J. P., Andrews H. L., Wong S. K., Isberg R. R. (1998) Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 [DOI] [PubMed] [Google Scholar]
- 6. Isberg R. R., O'Connor T. J., Heidtman M. (2009) The Legionella pneumophila replication vacuole. Making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hubber A., Roy C. R. (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 [DOI] [PubMed] [Google Scholar]
- 8. Odorizzi G., Babst M., Emr S. D. (2000) Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25, 229–235 [DOI] [PubMed] [Google Scholar]
- 9. De Matteis M. A., Godi A. (2004) PI-loting membrane traffic. Nat. Cell Biol. 6, 487–492 [DOI] [PubMed] [Google Scholar]
- 10. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 11. Vergne I., Chua J., Deretic V. (2003) Mycobacterium tuberculosis phagosome maturation arrest: selective targeting of PI3P-dependent membrane trafficking. Traffic 4, 600–606 [DOI] [PubMed] [Google Scholar]
- 12. Vergne I., Chua J., Lee H. H., Lucas M., Belisle J., Deretic V. (2005) Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 4033–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. House D., Bishop A., Parry C., Dougan G., Wain J. (2001) Typhoid fever. Pathogenesis and disease. Curr. Opin. Infect. Dis. 14, 573–578 [DOI] [PubMed] [Google Scholar]
- 14. Bakowski M. A., Braun V., Brumell J. H. (2008) Salmonella-containing vacuoles. Directing traffic and nesting to grow. Traffic 9, 2022–2031 [DOI] [PubMed] [Google Scholar]
- 15. Niebuhr K., Giuriato S., Pedron T., Philpott D. J., Gaits F., Sable J., Sheetz M. P., Parsot C., Sansonetti P. J., Payrastre B. (2002) Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S. flexneri effector IpgD reorganizes host cell morphology. EMBO J. 21, 5069–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weber S. S., Ragaz C., Reus K., Nyfeler Y., Hilbi H. (2006) Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schoebel S., Blankenfeldt W., Goody R. S., Itzen A. (2010) High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA. EMBO Rep. 11, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brombacher E., Urwyler S., Ragaz C., Weber S. S., Kami K., Overduin M., Hilbi H. (2009) Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J. Biol. Chem. 284, 4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weber S. S., Ragaz C., Hilbi H. (2009) The inositol polyphosphate 5-phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole, and binds to the bacterial effector LpnE. Cell. Microbiol. 11, 442–460 [DOI] [PubMed] [Google Scholar]
- 20. Jank T., Böhmer K. E., Tzivelekidis T., Schwan C., Belyi Y., Aktories K. (2012) Domain organization of Legionella effector SetA. Cell. Microbiol. 14, 852–868 [DOI] [PubMed] [Google Scholar]
- 21. Pizarro-Cerdá J., Cossart P. (2004) Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat. Cell Biol. 6, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 22. Ham H., Sreelatha A., Orth K. (2011) Manipulation of host membranes by bacterial effectors. Nat. Rev. Microbiol. 9, 635–646 [DOI] [PubMed] [Google Scholar]
- 23. Hilbi H., Weber S., Finsel I. (2011) Anchors for effectors. Subversion of phosphoinositide lipids by Legionella. Front. Microbiol. 2, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu F., Zhu W., Brennan L., Tao L., Luo Z. Q., Mao Y. (2012) Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc. Natl. Acad. Sci. U.S.A. 109, 13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parrish W. R., Stefan C. J., Emr S. D. (2005) PtdIns(3)P accumulation in triple lipid-phosphatase-deletion mutants triggers lethal hyperactivation of the Rho1p/Pkc1p cell-integrity MAP kinase pathway. J. Cell Sci. 118, 5589–5601 [DOI] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 28. Pape T., Schneider T. R. (2004) HKL2MAP. A graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 32. Svergun D. I., Barberato C., Koch M. H. J. (1995) CRYSOL. A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 33. Semenyuk A. V., Svergun D. I. (1991) GNOM. A program package for small-angle scattering and data processing. J. Appl. Crystallogr. 24, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Franke D., Svergun D. I. (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kozin M. B., Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 37. Maehama T., Taylor G. S., Slama J. T., Dixon J. E. (2000) A sensitive assay for phosphoinositide phosphatases. Anal. Biochem. 279, 248–250 [DOI] [PubMed] [Google Scholar]
- 38. Parrish W. R., Stefan C. J., Emr S. D. (2004) Essential role for the myotubularin-related phosphatase Ymr1p and the synaptojanin-like phosphatases Sjl2p and Sjl3p in regulation of phosphatidylinositol 3-phosphate in yeast. Mol. Biol. Cell 15, 3567–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Foti M., Audhya A., Emr S. D. (2001) Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 12, 2396–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stefan C. J., Audhya A., Emr S. D. (2002) The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell 13, 542–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Begley M. J., Taylor G. S., Brock M. A., Ghosh P., Woods V. L., Dixon J. E. (2006) Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc. Natl. Acad. Sci. U.S.A. 103, 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barford D., Das A. K., Egloff M. P. (1998) The structure and mechanism of protein phosphatases. Insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 [DOI] [PubMed] [Google Scholar]
- 43. Mukhopadhyay R., Rosen B. P. (2002) Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Perspect. 110, 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu W., Banga S., Tan Y., Zheng C., Stephenson R., Gately J., Luo Z. Q. (2011) Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6, e17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong S., Hsu F., Stefan C. J., Wu X., Patel A., Cosgrove M. S., Mao Y. (2012) Allosteric activation of the phosphoinositide phosphatase Sac1 by anionic phospholipids. Biochemistry 51, 3170–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barford D., Flint A. J., Tonks N. K. (1994) Crystal structure of human protein-tyrosine phosphatase 1B. Science 263, 1397–1404 [PubMed] [Google Scholar]
- 47. Stuckey J. A., Schubert H. L., Fauman E. B., Zhang Z. Y., Dixon J. E., Saper M. A. (1994) Crystal structure of Yersinia protein-tyrosine phosphatase at 2.5 Å and the complex with tungstate. Nature 370, 571–575 [DOI] [PubMed] [Google Scholar]
- 48. Manford A., Xia T., Saxena A. K., Stefan C., Hu F., Emr S. D., Mao Y. (2010) Crystal structure of the yeast Sac1. Implications for its phosphoinositide phosphatase function. EMBO J. 29, 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee J. O., Yang H., Georgescu M. M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P., Pavletich N. P. (1999) Crystal structure of the PTEN tumor suppressor. Implications for its phosphoinositide phosphatase activity and membrane association. Cell 99, 323–334 [DOI] [PubMed] [Google Scholar]
- 50. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Begley M. J., Taylor G. S., Kim S. A., Veine D. M., Dixon J. E., Stuckey J. A. (2003) Crystal structure of a phosphoinositide phosphatase, MTMR2. Insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol. Cell 12, 1391–1402 [DOI] [PubMed] [Google Scholar]
- 52. Luo Z. Q., Isberg R. R. (2004) Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A. 101, 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang L., Boyd D., Amyot W. M., Hempstead A. D., Luo Z. Q., O'Connor T. J., Chen C., Machner M., Montminy T., Isberg R. R. (2011) The E Block motif is associated with Legionella pneumophila translocated substrates. Cell. Microbiol. 13, 227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Patel J. C., Hueffer K., Lam T. T., Galán J. E. (2009) Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vieira O. V., Botelho R. J., Grinstein S. (2002) Phagosome maturation. Aging gracefully. Biochem J 366, 689–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flannagan R. S., Cosío G., Grinstein S. (2009) Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366 [DOI] [PubMed] [Google Scholar]
- 57. Shin H. W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M. R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., Frankel W. N., Solimena M., De Camilli P., Zerial M. (2005) An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dubuisson J. F., Swanson M. S. (2006) Mouse infection by Legionella, a model to analyze autophagy. Autophagy 2, 179–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Choy A., Dancourt J., Mugo B., O'Connor T. J., Isberg R. R., Melia T. J., Roy C. R. (2012) The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vergne I., Deretic V. (2010) The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett. 584, 1313–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]