Background: Although widespread, the significance of Rap1GAP down-regulation in human tumors is unknown.
Results: Genetic silencing of Rap1GAP in human colon cancer cells confers a mesenchymal mode of cell migration and enhances invasive behavior.
Conclusion: Rap1GAP regulates the mechanism of cell motility.
Significance: Down-regulation of Rap1GAP endows tumor cells with more aggressive properties.
Keywords: Cancer Biology, Cell Invasion, Cell Motility, Colon Cancer, Migration, Rac, Rap, Rap1GAP
Abstract
The functional significance of the widespread down-regulation of Rap1 GTPase-activating protein (Rap1GAP), a negative regulator of Rap activity, in human tumors is unknown. Here we show that human colon cancer cells depleted of Rap1GAP are endowed with more aggressive migratory and invasive properties. Silencing Rap1GAP enhanced the migration of confluent and single cells. In the latter, migration distance, velocity, and directionality were increased. Enhanced migration was a consequence of increased endogenous Rap activity as silencing Rap expression selectively abolished the migration of Rap1GAP-depleted cells. ROCK-mediated cell contractility was suppressed in Rap1GAP-depleted cells, which exhibited a spindle-shaped morphology and abundant membrane protrusions. Tumor cells can switch between Rho/ROCK-mediated contractility-based migration and Rac1-mediated mesenchymal motility. Strikingly, the migration of Rap1GAP-depleted, but not control cells required Rac1 activity, suggesting that loss of Rap1GAP alters migratory mechanisms. Inhibition of Rac1 activity restored membrane blebbing and increased ROCK activity in Rap1GAP-depleted cells, suggesting that Rac1 contributes to the suppression of contractility. Collectively, these findings identify Rap1GAP as a critical regulator of aggressive tumor cell behavior and suggest that the level of Rap1GAP expression influences the migratory mechanisms that are operative in tumor cells.
Introduction
RapGTPases (Rap1A/B, Rap2A/B/C) play pivotal roles in the ability of cells to respond to environmental cues. Rap regulates cell polarity and actin-based morphology in yeast and slime mold (for review, see Refs. 1–3), cell/cell adhesion in Drosophila (4–6), and morphogenesis in Xenopus embryos (7, 8). In mammalian cells, Rap regulates cytoskeletal dynamics stimulated by growth factors, hormones, cytokines, and tension. Rap interacts directly with actin (9–11) and activators and inhibitors of Rho family GTPases (12–17). Rap regulates the balance in cell/matrix and cell/cell adhesion through effects on integrin activation (2, 18, 19) and cadherin-mediated cell/cell (20–24) adhesion.
Alterations in the cytoskeleton, cell/cell adhesion, and integrin activation are critical nodes in the transition from benign to invasive carcinomas. Stable expression of activated Rap enhanced metastasis in prostate cancer cells and the infiltration of breast cancer cells into the vasculature (25, 26). However, the significance of these studies to human tumors is unclear in that activating mutations in Rap have not been reported (27). Down-regulation of Rap1GAP2 is widespread in human tumors (28–33). Overexpression of Rap1GAP in human tumor cells impaired cell migration and invasion in vitro (28, 29, 31, 33–35) and metastasis in vivo (36, 37). Intriguingly, the expression of ectopically expressed Rap1GAP was lost from disseminated tumors but retained in those that formed at the sites of subcutaneous injection (36). This supports the existence of selective pressure to decrease Rap1GAP expression, which appears to be operative in human tumors where the expression of Rap1GAP decreases with tumor progression (30, 31, 33, 35).
The cellular processes that are sensitive to the levels of Rap1GAP are unknown. Importantly, whether the widespread down-regulation of Rap1GAP observed in human tumors alters the behavior of tumor cells has not been determined. We previously reported that silencing the expression of Rap1GAP in human colon cancer cells weakened cell/cell adhesion and enhanced spreading on collagen, changes that are reminiscent of those that take place during the early stages of tumor cell dissemination (32). We demonstrate that silencing Rap1GAP endows cells with a Rap- and Rac1-dependent mechanism of cell motility that was inactive in parental cells. Invasive behavior was profoundly up-regulated in Rap1GAP-depleted cells. Collectively, these findings suggest that down-regulation of Rap1GAP in human tumors harbors the potential to increase migratory and invasive behaviors that promote tumor progression.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HT29 and LoVo cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS). The isolation of the Rap1GAP-depleted HT29 cell lines was described previously (32). In brief, SMARTvector human Rap1GAP and nonspecific control were purchased from Thermo Scientific (Dharmacon). Viruses expressing two different Rap1GAP-directed shRNAs were used, and multiple clones expressing each were isolated and analyzed. Cell lines E11 and C10 were isolated using shRNA#2 and the C4 and F3 lines isolated using shRNA#3. Control cell lines (Con) were isolated from cells expressing a nonspecific shRNA (32). All experiments were conducted in three or more Rap1GAP-deficient cell lines, including one made with each shRNA with similar results and compared with control and parental HT29 cells. For acute silencing, cells were transfected with Rap1/2 or Rap1GAP-directed and nonspecific siRNAs using Amaxa-mediated electroporation as described previously (32).
Wound Closure Assays
Cells were grown to confluence on 6-well plates marked with a line down the center and coated with 2 μg/ml collagen IV. Cells were starved for 24 h, and wounds (5–6/well) were made perpendicularly to the line. After wounding, cells were stimulated with serum-supplemented medium. Images were acquired using a Nikon Eclipse TE2000 microscope and Northern Eclipse software (Empix Imaging). The distances between the intersection of the mark and the border of the wound were measured using Multi Gauge software (Fujifilm). The distance between the wound borders at 20 h was subtracted from that at 0 h and the difference expressed as percentage closure. The percentage closure from 5–6 wounds/dish was averaged in each experiment.
Phagocytic Tracking Assays
Fluorescent beads (Cellomics Cell motility kit; Thermo Scientific) were plated on collagen IV-coated glass slips. Starved cells (104 cells/ml) were plated under conditions where the cells were subconfluent, allowed to attach for 2 h, and then stimulated with serum-supplemented medium for 24 h. Where inhibitors were used, cells were plated in the absence of inhibitor and stimulated with inhibitor-supplemented medium. Cells were fixed and stained for F-actin, and the tracks of individual cells were analyzed on a Zeiss Axiophot fluorescence microscope using Zeiss Axiovision software.
Time Lapse Microscopy
Cells (20,000/well) were plated on collagen IV-coated chamber slides. Phase contrast images of cells in serum-supplemented medium were acquired every 10 min over 16 h using a Deltavision Deconvolution Microscope (Applied Precision, Inc.). Tracks of randomly selected single cells were analyzed by manual tracking using ImageJ software (National Institutes of Health). The distance, velocity, and directionality (calculated as net displacement/total path length) of migration were determined using chemotaxis software.
Immunostaining
Cells plated on collagen IV-coated coverslips were starved overnight and stimulated with 10% FBS for varying times. Cells were fixed in 3.7% formaldehyde in PBS and stained with rhodamine-phalloidin or antibodies to FAK-Y397p followed by an Alexa Fluor 488-conjugated secondary antibody. Within an experiment, images for a given antibody were acquired for the same times.
Antibodies and Western Blotting
Rap1 (sc-65), Rap1GAP (sc-28189), and actin antibodies were obtained from Santa Cruz Biotechnology. Rap2 (610215) and FAK-Y397p (611723) antibodies were from BD Transduction Laboratories. MLC2-T18/S19p antibody was from Cell Signaling. For total cell lysates, cells were disrupted in Triton buffer (20 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1% Triton X-100, 2 mm EDTA, 1 mm NaF, plus protease inhibitors). Protein concentrations were determined by DC protein assay (Bio-Rad) and equal micrograms of protein analyzed.
Invasion Assays
Cells (100,000) were mixed with 100 μl of ice-cold growth factor-reduced Matrigel (BD 356230), plated onto Transwell filters (Corning, 8 μm, 3422) and incubated for 30 min at 37 °C until hardened. Filters were placed into a well containing 600 μl of growth medium (± inhibitors) and 100 μl of serum free medium (± inhibitors) added to the cells in the upper chamber. After 48 h, Matrigel was removed from the upper well with a cotton swab, and cells on the bottom of the filter were fixed in 3.7% formaldehyde, stained with 0.1% crystal violet, washed with water, and counted. For outgrowth in Matrigel, cells (10,000) were mixed with 160 μl of Matrigel and plated in 35-mm Mattek dishes containing a glass coverslip. After hardening, cells were overlaid with 2 ml of growth medium and incubated at 37 °C for up to 11 days.
Statistical Analysis
All experiments were repeated at least three times. The bar graphs shown represent the combined results from at least three experiments unless otherwise noted. Statistical significance was determined by Student's t test.
RESULTS
Down-regulation of Rap1GAP Enhances Cell Motility
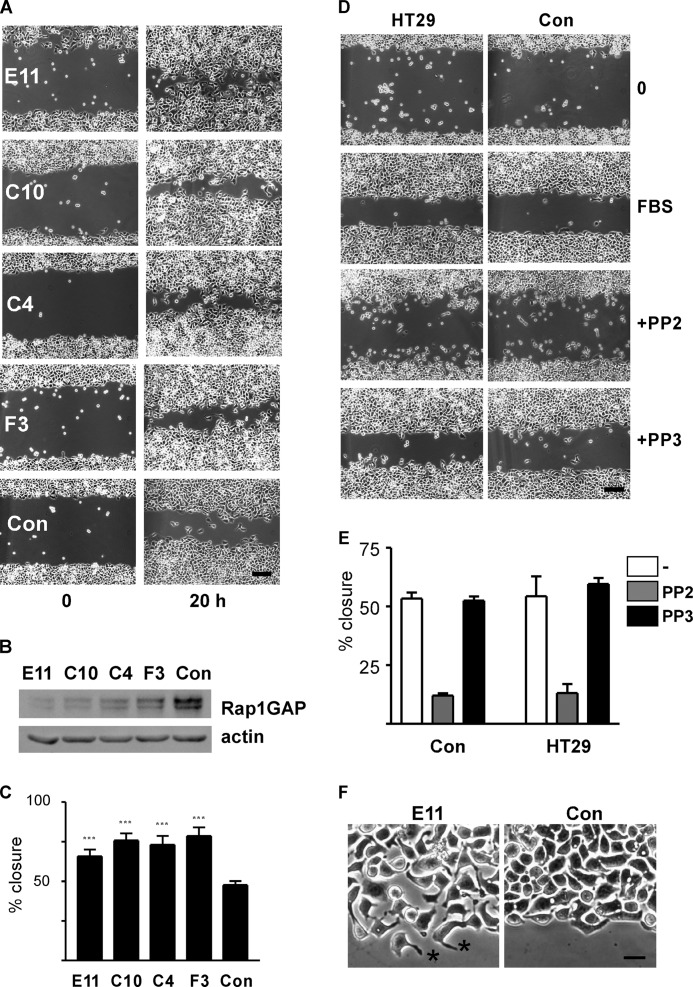
To explore the consequences of Rap1GAP down-regulation on tumor cell motility, wound closure assays were conducted in four independent lines of Rap1GAP-depleted HT29 cells (32). Confluent monolayers of cells, plated onto collagen IV to mimic adhesion to the basement membrane, were grown to confluence, starved for 18 h, wounded, and stimulated with serum-containing medium. Images were acquired immediately after wounding and after 20 h in serum-supplemented medium. Down-regulation of Rap1GAP significantly accelerated wound closure compared with cells selected to express a nonspecific shRNA (control) (Fig. 1, A–C). Wound closure in control cells was unchanged in the absence or presence of inhibitors (Src inhibitor, PP2 shown here) compared with parental HT29 cells (Fig. 1, D and E).
FIGURE 1.
Silencing Rap1GAP accelerates wound closure. A, photomicrographs of Rap1GAP-depleted cell lines (E11, C10, C4, F3) and a control (Con) cell line selected to express a nonspecific shRNA immediately after wounding (0) and after stimulation with FBS for 20 h are shown. Scale bar, 20 μm. B, total cell lysates prepared from Rap1GAP-depleted and control cells were subjected to Western blotting for Rap1GAP and actin to confirm equal protein loading. C, increased wound closure in Rap1GAP-depleted cells was statistically significant (***, p < 0.0001). Error bars, S.E. D, photomicrographs show parental HT29 and control cells in wound closure experiments conducted in the absence and presence of the Src inhibitor PP2 (10 μm, gray bars) or inactive analog PP3 (10 μm, black bars). Inhibitors were added for 60 min prior to the addition of serum-supplemented medium. Scale bar, 20 μm. E, results from five wound closure experiments comparing control and HT29 cells are shown. F, photomicrographs show Rap1GAP-depleted (E11) and control (Con) cells at the wound border. Asterisks show examples of elongated protrusions on Rap1GAP-depleted cells. Scale bar, 20 μm.
We previously observed that silencing Rap1GAP impaired cell/cell adhesion (32). Indeed, Rap1GAP-depleted cells along the border of the wound were more dispersed than control cells, which maintained cell/cell contacts (Fig. 1F). Rap1GAP-depleted cells elaborated elongated membrane protrusions that were not observed in control cells (examples marked by asterisks in Fig. 1F). These findings prompted us to investigate whether depleting Rap1GAP altered migratory mechanisms.
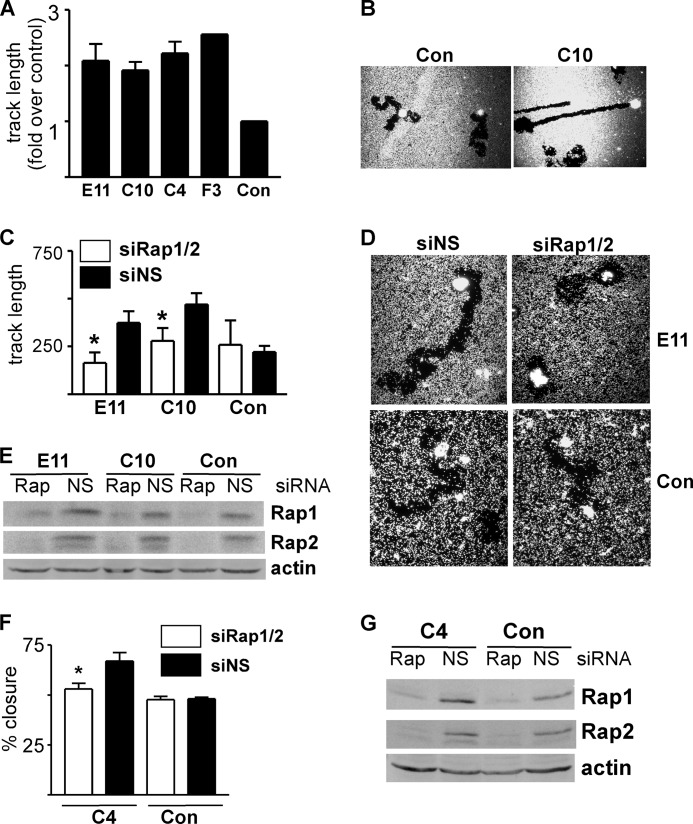
Because weakened cell/cell adhesion could enhance wound closure, we determined whether the migratory activity of single cells was enhanced by Rap1GAP depletion using phagocytic tracking assays. Cells were plated on a lawn of beads on collagen IV, and migration in the presence of serum was analyzed. The tracks made by all four lines of Rap1GAP-depleted cells were longer (Fig. 2A) and less circuitous (C10 shown in Fig. 2B; E11 and C4 in Fig. 6F) than those made by control cells. Rap1 and Rap2 activity are durably increased in Rap1GAP-depleted cells (32). Therefore, we determined whether enhanced migration was a consequence of increased Rap activity. Silencing Rap1 and Rap2 expression significantly impaired the migration of Rap1GAP-depleted cells in phagocytic tracking (Fig. 2, C–E) and in wound closure assays (Fig. 2, F and G). The migration of control cells was not affected by silencing Rap (Fig. 2, C and F). The selective requirement for Rap activity in Rap1GAP-depleted cells implies that cells acquire additional or altered means of cell motility when endogenous Rap activity is increased. This is the first demonstration that down-regulation of Rap1GAP, an event widely observed in human tumors, enhances migratory behavior.
FIGURE 2.
Depletion of Rap1GAP induces Rap-dependent cell motility. A, results from three experiments comparing track length in Rap1GAP-depleted cells with control cells. Track length in control cells was set to 1.0. B, photomicrographs of control (Con) and Rap1GAP-depleted (C10) cells in phagocytic tracking assays. Additional Rap1GAP-depleted cells (E11, C4) are shown in Fig. 6F. C, Rap1GAP-depleted and control cells transiently transfected with siRNAs directed to Rap1 and Rap2 (open bars) or nonspecific (NS) siRNAs (filled bars). Phagocytic tracking assays were performed at 48 h after transfection. Silencing Rap significantly (*, p < 0.05) decreased migration in Rap1GAP-depleted cells. Error bars, S.E. D, photomicrographs of siRNA-transfected cells in tracking assays. E, total cell lysates prepared from the cells used in the tracking assays analyzed for Rap1/2 expression by Western blotting. Western blotting for actin confirmed equal protein loading. F, control and Rap1GAP-depleted (C4) cells transiently transfected with siRNAs directed to Rap1/2 (open bars) or nonspecific (NS) siRNAs (filled bars) and analyzed in wound closure experiments. Silencing Rap significantly (*, p < 0.05) impaired wound closure in Rap1GAP-depleted cells. G, total cell lysates prepared from the cells used in the wound closure assays analyzed for Rap1/2 expression by Western blotting. Western blotting for actin confirmed equal protein loading.
FIGURE 6.
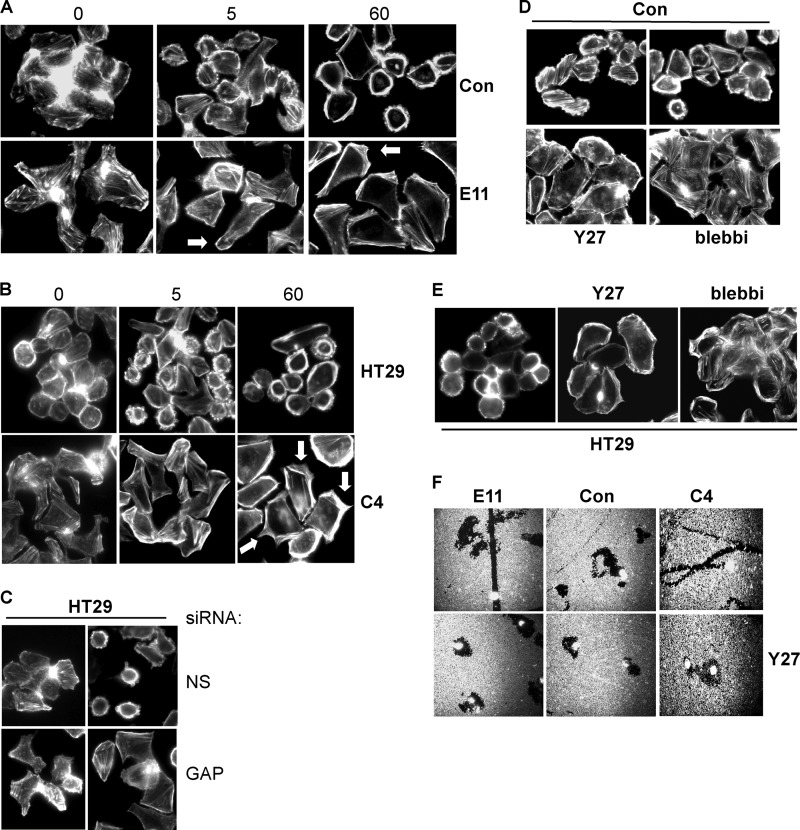
Loss of Rap1GAP suppresses ROCK-mediated contractility. A and B, starved Rap1GAP-depleted (E11, C4), control (Con) and HT29 cells (0) were stimulated with 10% FBS for 5 and 60 min, fixed, and stained for F-actin. Arrows show examples of actin-rich protrusions in Rap1GAP-depleted cells. C, HT29 cells acutely transfected with Rap1GAP-directed (GAP) or nonspecific (NS) siRNAs were starved overnight and stimulated with 10% FBS for 5 min. Cells were fixed and stained for F-actin. D and E, starved control and HT29 cells were stimulated with 10% FBS for 5 min in the absence or presence of Y27632 (10 μm) or blebbistatin (10 μm). F, phagocytic tracking assays were conducted in Rap1GAP-depleted (E11, C4) and control cells in the absence or presence of Y27632 (10 μm).
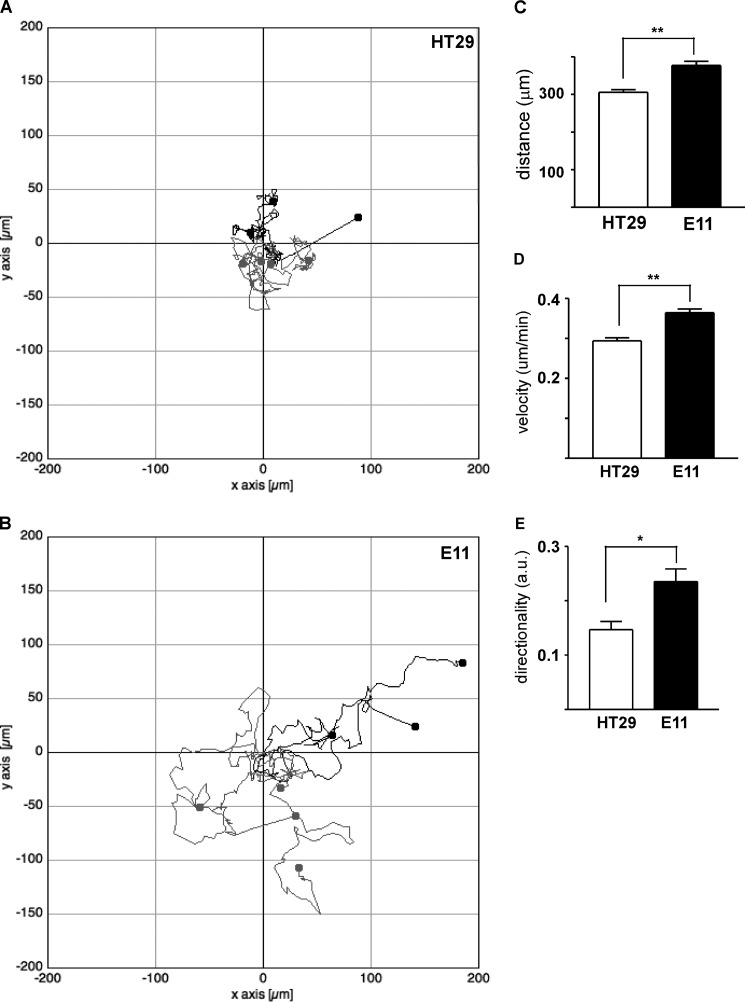
Individual cell migration was further analyzed in time lapse microscopy experiments. HT29 cells migrate in a circuitous fashion, characterized by frequent alterations in the direction of cell migration (Fig. 3A). Rap1GAP-depleted cells made fewer turns and migrated over greater distances (Fig. 3B). Migration distance, velocity, and directionality were increased in Rap1GAP-depleted cells (Fig. 3, C–E).
FIGURE 3.
Migration distance, velocity, and directionality are enhanced by Rap1GAP depletion. A and B, graphical representations show tracks made by seven individual (A) HT29 and (B) Rap1GAP-depleted (E11) cells as assessed by time lapse microscopy. C–E, the migration of 21 HT29 and E11 cells was analyzed for distance (C), velocity (D), and directionality (E) as described under “Experimental Procedures.” Distance (**, p < 0.005), velocity (**, p < 0.005), and directionality (*, p < 0.05) were significantly increased in Rap1GAP-depleted cells. Error bars, S.E.
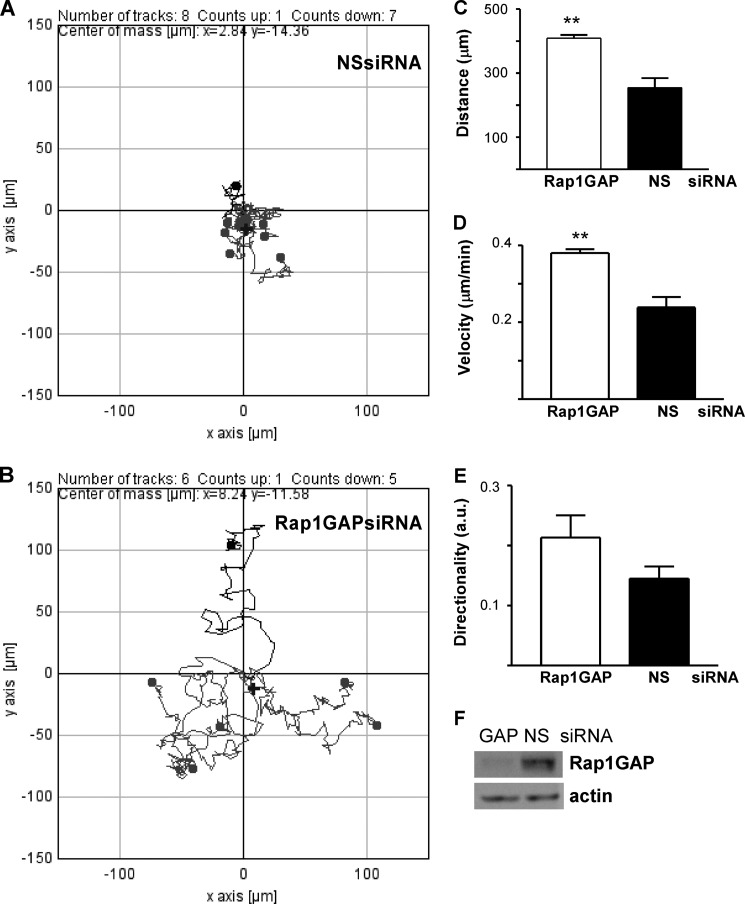
To confirm that increased migration was due to depletion of Rap1GAP rather than secondary events that occurred during the isolation of stable cell lines, Rap1GAP expression was acutely silenced in HT29 cells. Cells were transfected with Rap1GAP-directed or nonspecific siRNAs and cell migration analyzed at 48 h after transfection. The Rap1GAP siRNA targeted a region different from either of the two shRNAs used to isolate the Rap1GAP-depleted cell lines. Cells in which Rap1GAP expression was acutely silenced exhibited an increase in the distance, velocity, and directionality of migration (Fig. 4, A–F). Therefore, enhanced migration is likely a primary consequence of Rap1GAP depletion.
FIGURE 4.
Acute silencing of Rap1GAP enhanced cell migration. A and B, graphical representation of the tracks made by eight HT29 cells transfected with a nonspecific siRNA (NSsiRNA; A) versus those made by six cells transfected with a Rap1GAP-directed siRNA (B). Cells were transfected and after 48 h plated and migration analyzed by time lapse microscopy. C–E, migration distance (**, p < 0.005) (C), velocity (**, p < 0.005) (D), and directionality (E) were increased in Rap1GAP-depleted cells. Results shown are from 29 cells transfected with Rap1GAP-directed siRNAs and 20 cells transfected with nonspecific siRNAs. F, Western blot showing decreased Rap1GAP expression in cells transfected with Rap1GAP-directed siRNAs. Actin was analyzed to confirm equal protein loading.
Enhanced Invasive Activity in Rap1GAP-depleted Cells
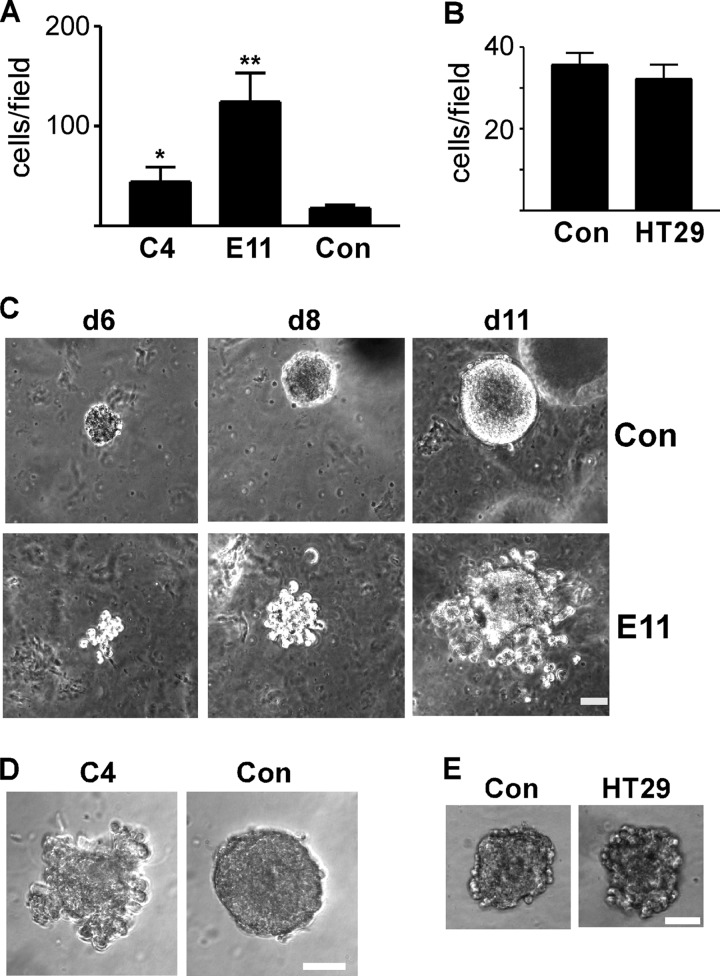
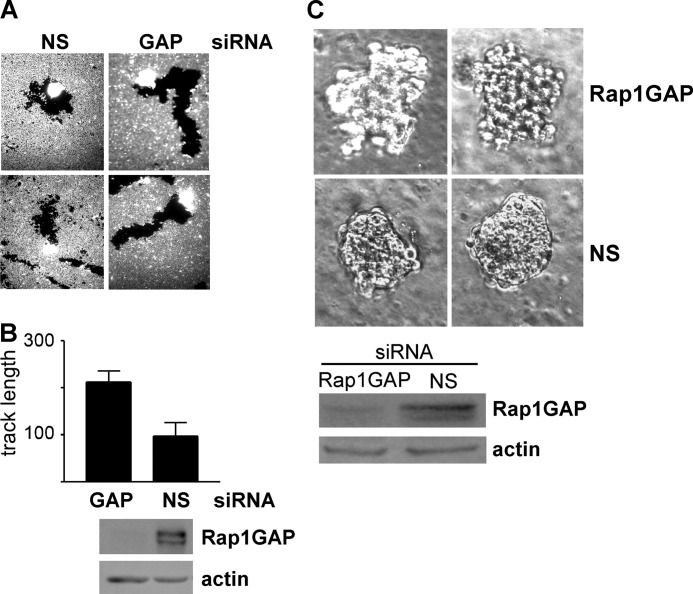
Rap1GAP-depleted cells exhibited weakened cell/cell adhesion, enhanced spreading on collagen (32) and increased migratory activity, changes that resemble those seen in invasive cancer cells. To explore the consequences of Rap1GAP depletion on invasive behavior, cells were plated in Matrigel in the upper well of Transwell chambers and exposed to serum in the lower chamber. Control and HT29 cells were poorly invasive (Fig. 5, A and B). Invasion was significantly increased in Rap1GAP-depleted cells.
FIGURE 5.
Loss of Rap1GAP enhances invasive behavior. A, RapGAP-depleted (C4, E11) and control (Con) cells were plated in Matrigel in the upper well of Transwell chambers, and cells that crossed the membrane in response to 10% FBS after 48 h were scored. Depletion of Rap1GAP significantly increased invasion (*, p < 0.05; **, p < 0.005). B, results are from a representative experiment of two performed with similar results that compared invasion in control and HT29 cells. Error bars, S.E. C–E, equal numbers of control (Con), Rap1GAP-depleted (E11, C4) and HT29 cells were embedded in Matrigel and imaged at days 6, 8, and 11 (C) or at day 8 (D and E). Scale bar, 20 μm.
To explore effects on invasive activity under conditions that more closely mimic those in vivo, cells were embedded in Matrigel, and their ability to invade the matrix in three dimensions was analyzed. Control and HT29 cells formed compact, dense spheres under these conditions (Fig. 5, C–E). The boundaries of the colonies were smooth, indicative of robust cohesive activity. Single cells or dispersed colonies were rarely seen. In contrast, Rap1GAP-depleted cells actively invaded the surrounding matrix (Fig. 5, C and D). Single cells and small clusters of cells separate from the large colonies were routinely observed. In conclusion, depletion of Rap1GAP endows cells with enhanced migratory and invasive activity. These results suggest that tumors in which Rap1GAP expression is down-regulated harbor the potential for more aggressive behavior.
Loss of Rap1GAP Suppresses ROCK-mediated Actomyosin Contractility
Cell migration and invasion entail dramatic reorganization of the actin cytoskeleton. To explore the consequences of Rap1GAP depletion on cytoskeletal dynamics under the same conditions used to monitor cell motility, the acute effects of serum on the actin cytoskeleton were investigated in cells plated on collagen IV. Serum induced profound membrane blebbing in control (Fig. 6, A and D) and HT29 (Fig. 6, B and C) cells. Membrane blebbing was most pronounced at 5 min and maintained for up to 120 min after serum stimulation. Membrane blebbing was suppressed in Rap1GAP-depleted cells (Fig. 6, A and B). Similar results were observed when Rap1GAP was acutely silenced in HT29 cells (Fig. 6C).
Cancer cells are endowed with multiple mechanisms of cell motility. The migration of single tumor cells has been characterized as amoeboid (also called rounded) or mesenchymal (38–41). Amoeboid/rounded cell migration is driven by high levels of ROCK-mediated contractility that underlie profound membrane blebbing. To assess whether membrane blebbing reflected actomyosin-mediated contractility, cells were exposed to inhibitors of ROCK (Y27632) and myosin ATPase (blebbistatin) activity. Membrane blebbing in control and parental HT29 cells required ROCK and myosin ATPase activity (Fig. 6, D and E). To assess whether migration was ROCK-dependent, the effects of Y27632 on individual cell migration were analyzed. The migration of control and Rap1GAP-depleted cells required ROCK activity (Fig. 6F).
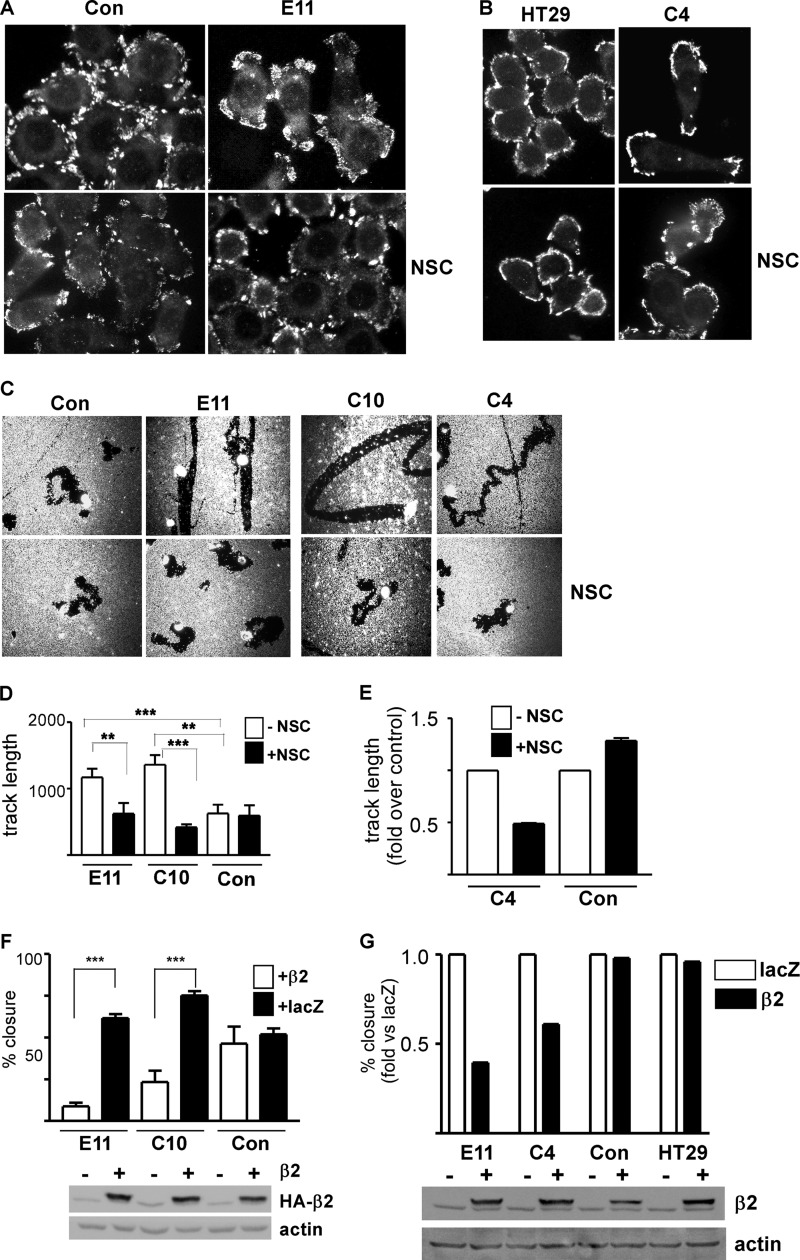
Depletion of Rap1GAP Confers Rac1-dependent, Mesenchymal Cell Migration
Cells that migrate via a mesenchymal mechanism exhibit shared features including a spindle-shaped morphology, the elaboration of actin-rich membrane protrusions, and dependence on Rac1 activity (for review, see Ref. 40). Unlike control and HT29 cells, which were round, Rap1GAP-depleted cells were elongated and exhibited actin-rich protrusions (examples indicated by arrows in Fig. 6, A and B). Focal adhesions were uniformly distributed around the circumference of control and HT29 cells (Fig. 7, A and B). In Rap1GAP-depleted cells, focal adhesions were less uniform in distribution. To assess whether migration was Rac1-dependent, cells were treated with the Rac1 inhibitor, NSC23766 (42). Strikingly, the migration of Rap1GAP-depleted, but not of control cells, required Rac1 activity (Fig. 7, C–E). Moreover, in the presence of the Rac1 inhibitor, the tracks made by Rap1GAP-depleted cells reverted to a circuitous pattern more similar to that in control cells (Fig. 7C). The selective requirement for Rac1 activity in the migration of Rap1GAP-depleted cells was confirmed using β2-chimaerin, a highly specific Rac1GAP (43). Expression of β2-chimaerin impaired wound closure in Rap1GAP-depleted, but not control or parental HT29 cells (Fig. 7, F and G). Inhibition of Rac1 also restored a rounded morphology and a more uniform distribution of focal adhesions to Rap1GAP-depleted cells (Fig. 7, A and B). Together, these findings demonstrate that depletion of Rap1GAP endows cells with an altered mechanism of cell motility that requires Rap and Rac1 activity.
FIGURE 7.
Depletion of Rap1GAP endows cells with Rac1-dependent morphological changes and motility. A and B, starved control, HT29, and Rap1GAP-depleted (E11, C4) cells were stimulated with 10% FBS for 120 min in the absence or presence of NSC23766 (10 μm), fixed, and stained for autophosphorylated FAK (FAK-Y397P). C, photomicrographs show control and Rap1GAP-depleted (E11, C10, C4) cells after 24 h in the absence or presence of NSC23766 (10 μm). D and E, inclusion of NSC23766 (10 μm) (filled bars) selectively impaired the migration of Rap1GAP-depleted cells (**, p < 0.01; ***, p < 0.005). Error bars, S.E. E, track length in cells treated with vehicle (open bars, −NSC) was set to 1.0. F, Rap1GAP-depleted (E11, C10) and control cells were infected with adenoviruses expressing LacZ or β2-chimaerin at equal multiplicities of infection. At 48 h after infection, confluent cells were wounded and wound closure assessed after 20 h. The expression of HA-tagged β2-chimaerin in total cell lysates is shown. Actin was used to document equal protein loading. Expression of β2-chimaerin (β2) selectively impaired the migration of Rap1GAP-depleted cells (***, p < 0.001). G, representative experiment from two wound closure assays in which Rap1GAP-depleted cells (E11, C4) were compared with control and HT29 cells in the same experiment. Migration in LacZ-infected cells was set to 1.0. The expression of HA-tagged β2-chimaerin in total cell lysates is shown.
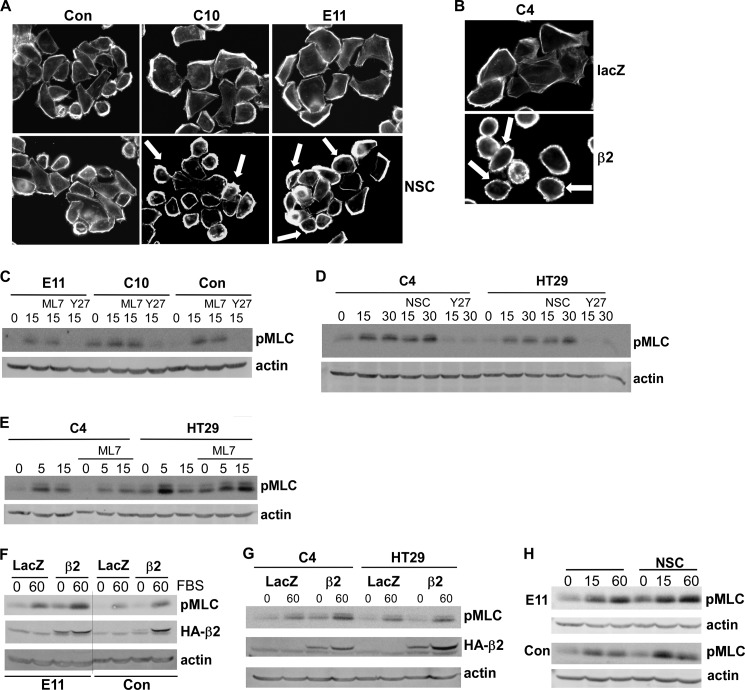
Rac1 Suppresses ROCK-dependent Contractility in Rap1GAP-depleted Cells
Overexpression of activated Rac1 in human tumor cells induced a switch from an amoeboid/rounded mechanism of cell migration to a mesenchymal mechanism (44). To explore the contribution of Rac1 to the suppression of cell contractility in Rap1GAP-depleted cells, the effects of Rac1 inhibition on membrane blebbing were examined. Treatment with NSC (Fig. 8A) or expression of β2-chimaerin (Fig. 8B) induced membrane blebbing in Rap1GAP-depleted cells. Because membrane blebbing was ROCK-dependent, we assessed whether Rac1 impaired ROCK activity. The phosphorylation of two ROCK substrates, the myosin-binding subunit of myosin phosphatase (MYPT1) and myosin 2 light chain (MLC), were examined. Western blotting with two different antibodies revealed that MYPT1 was basally phosphorylated in Rap1GAP-depleted and control cells and that phosphorylation was unaffected by serum stimulation or ROCK inhibition (data not shown). Serum stimulated MLC phosphorylation on serine 18/threonine 19 (Fig. 8, C–H). Although this site is phosphorylated by ROCK and myosin light chain kinase, only inhibition of ROCK blocked MLC phosphorylation (Fig. 8, C–E). Expression of β2-chimaerin increased basal and serum-stimulated MLC phosphorylation in Rap1GAP-depleted cells (Fig. 8, F and G). Similar effects were observed using NSC (Fig. 8H). We conclude that the suppression of contractility in Rap1GAP-depleted cells is not due to global inhibition of ROCK activity, but that the increase in Rac1 signaling suppresses contractility at least in part through inhibition of ROCK activity.
FIGURE 8.
Rac1 suppresses ROCK-mediated contractility in Rap1GAP-depleted cells. A, starved control and Rap1GAP-depleted (C10, E11) cells were stimulated with 10% FBS for 120 min in the absence or presence of NSC23766 (10 μm), fixed, and stained for F-actin. Examples of blebbing cells are illustrated by arrows. B, Rap1GAP-depleted cells (C4) infected with adenoviruses expressing LacZ or β2-chimaerin were stimulated with 10% FBS for 120 min, fixed, and stained for F-actin. C–E, starved cells were stimulated with 10% FBS for the times indicated (minutes) in the absence or presence of ML7, Y27632, or NSC23766 (each at 10 μm). Total cell lysates were analyzed for MLC phosphorylation on serine 18/threonine 19 (pMLC) and for actin to document equal protein loading. F and G, Rap1GAP-depleted (E11, C4), control, and HT29 cells infected with LacZ or β2-chimaerin adenoviruses were starved and stimulated with FBS for 60 min. Total cell lysates were analyzed for MLC phosphorylation and expression of HA-β2-chimaerin. Western blotting for actin confirmed equal protein loading. H, Rap1GAP-depleted (E11) and control cells were stimulated with 10% FBS in the absence or presence of NSC23766 (10 μm) and total cell lyastes analyzed for pMLC and actin.
Down-regulation of Rap1GAP Enhances Migration and Invasion in Other Colon Cancer Cell Lines
To confirm that the effects of Rap1GAP depletion on migratory and invasive behavior were not restricted to a single cell line, the expression of Rap1GAP was acutely silenced in the human colon cancer cell line, LoVo. LoVo cells were selected for analysis as these cells express Rap1GAP (Fig. 9, B and C). Silencing Rap1GAP increased the migration of individual LoVo cells in phagocytic tracking assays (Fig. 9, A and B). Furthermore, acute silencing of Rap1GAP promoted outgrowth in Matrigel (Fig. 9C). Thus, the consequences of Rap1GAP depletion on tumor cell behavior are likely to be general.
FIGURE 9.
Acute silencing of Rap1GAP enhances cell migration and invasion in LoVo cells. A, LoVo cells transiently transfected with Rap1GAP-directed (GAP) or nonspecific (NS) siRNAs were analyzed in phagocytic tracking assays. B, quantitation of track length in a representative experiment of three experiments conducted with similar results is shown. Western blot shows decreased Rap1GAP expression in cells transfected with Rap1GAP-directed siRNAs. Actin was analyzed to confirm equal protein loading. C, LoVo cells transiently transfected with Rap1GAP-directed or nonspecific (NS) siRNAs were embedded in Matrigel and analyzed after 8 days. Two representative images from the same experiment are shown. Western blot shows Rap1GAP expression in transfected cells at the time of plating in Matrigel. Actin was analyzed to confirm equal protein loading.
DISCUSSION
The significance of the widespread down-regulation of Rap1GAP expression in human tumors is unknown. Our previous study revealed that depleting Rap1GAP from human colon cancer cells impaired cell/cell adhesion and enhanced adhesion and spreading on collagen (32). As these changes are reminiscent of those observed at the early stages of tumor cell dissemination, we set out to explore whether silencing Rap1GAP altered migratory properties. Wound closure was accelerated in four independent Rap1GAP-depleted cell lines compared with cells selected to express a nonspecific shRNA or parental HT29 cells. To exclude the possibility that increased wound closure was a consequence of impaired cell/cell adhesion and enhanced spreading, the motility of individual cells was analyzed. Phagocytic tracking experiments, conducted in four Rap1GAP-depleted cell lines, revealed that the migration of individual Rap1GAP-depleted cells was enhanced. Time lapse microscopy experiments demonstrated increased velocity, distance, and directionality of migration in Rap1GAP-depleted cells. Importantly, acute silencing of Rap1GAP using an siRNA directed to a region on Rap1GAP distinct from those targeted by either of the Rap1GAP-directed shRNAs used to isolate stable cell lines was sufficient to enhance migration in HT29 cells. Moreover, acute silencing of Rap1GAP increased migration in a second human colon cancer cell line, LoVo. Therefore, alterations in cell motility are most likely a direct consequence of Rap1GAP down-regulation. As evidence for this, silencing Rap1/Rap2 expression selectively impaired the migration of Rap1GAP-depleted cells. Thus, enhanced migration is a consequence of increased Rap activity. These are the first data showing that up-regulation of endogenous Rap activity promotes cell migration in human tumor cells.
Manipulating the balance in Rac1 and Rho activity in tumor cells induced a switch from a contractility-based, rounded mechanism of cell motility to Rac-1-dependent mesenchymal cell migration (44). Intriguingly, control and parental HT29 cells exhibited high levels of serum-stimulated contractility, which was suppressed by acute or chronic silencing of Rap1GAP. Moreover, the migration of Rap1GAP-depleted, but not control or parental cells, was Rac1-dependent. Activation of Rap has been shown to stimulate Rac1 activity (12, 45) and to inhibit Rho activity (16, 46, 47). Rap1 and Rap2 activity are stably increased in Rap1GAP-depleted cells (32). Nonetheless, total Rac1 activity was not increased, and global levels of ROCK activity were not decreased by Rap1GAP depletion. Inhibition of Rac1 restored contractility, as evidenced by membrane blebbing, and enhanced MLC phosphorylation in Rap1GAP-depleted cells. Additionally, the tracks made by Rap1GAP-depleted cells reverted to a more circuitous pattern similar to that observed in control cells in the face of Rac1 inhibition. These results suggest that increases in endogenous Rap activity harbor the potential to alter migratory mechanisms, likely through the regulation of discrete pools of Rac1.
The alterations conferred by depleting Rap1GAP, impaired cell/cell adhesion, enhanced adhesion and spreading on collagen (32), and enhanced mesenchymal motility are similar to those observed in invasive cancer cells. Depletion of Rap1GAP enhanced invasion through Matrigel-coated filters in response to serum. The differences in invasion were more striking in cells embedded in Matrigel. HT29 and LoVo cells formed highly compact colonies that retained cell/cell adhesion. By contrast, cells acutely or stably silenced for Rap1GAP invaded the matrix in all directions. Collectively, these data suggest that, at least in vitro, loss of Rap1GAP endows human tumor cells with more aggressive migratory and invasive properties. Consistent with this notion, overexpression of Rap1GAP in tumor cell lines impaired migration in vitro (28, 29, 31, 33–35) and metastasis in vivo (36, 37). Whether depletion of Rap1GAP is sufficient to enhance metastasis remains to be determined.
The precise mechanism through which silencing Rap1GAP confers Rac1-dependent cell motility remains to be elucidated. Activated Rap recruits RacGEFs to the membrane (12). A role for Rap1 in the membrane translocation of Rac1 has been reported (48). It is conceivable that Rap activates specific pools of Rac1 that suppress ROCK-mediated contractility.
In conclusion, alterations in the level of Rap1GAP expression and of endogenous Rap activity appear to be important determinants of migratory and invasive behavior in human tumor cells. Although it is well documented that the migration of tumor cells is highly plastic, the endogenous signals that dictate the selection of migratory mechanisms are poorly understood. Our data suggest a previously unknown role for Rap1GAP in the regulation of migratory mechanisms.
Footnotes
- Rap1GAP
- Rap1 GTPase-activating protein
- MLC
- myosin 2 light chain.
REFERENCES
- 1. Bos J. L., de Rooij J., Reedquist K. A. (2001) Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2, 369–377 [DOI] [PubMed] [Google Scholar]
- 2. Caron E. (2003) Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J. Cell Sci. 116, 435–440 [DOI] [PubMed] [Google Scholar]
- 3. Kortholt A., van Haastert P. J. (2008) Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell. Signal. 20, 1415–1422 [DOI] [PubMed] [Google Scholar]
- 4. Asha H., de Ruiter N. D., Wang M. G., Hariharan I. K. (1999) The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J. 18, 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hariharan I. K., Carthew R. W., Rubin G. M. (1991) The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell 67, 717–722 [DOI] [PubMed] [Google Scholar]
- 6. Knox A. L., Brown N. H. (2002) Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295, 1285–1288 [DOI] [PubMed] [Google Scholar]
- 7. Choi S. C., Han J. K. (2005) Rap2 is required for Wnt/β-catenin signaling pathway in Xenopus early development. EMBO J. 24, 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsai I. C., Amack J. D., Gao Z. H., Band V., Yost H. J., Virshup D. M. (2007) A Wnt-CKIvarϵ-Rap1 pathway regulates gastrulation by modulating SIPA1L1, a Rap GTPase activating protein. Dev. Cell 12, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torti M., Bertoni A., Canobbio I., Sinigaglia F., Lapetina E. G., Balduini C. (1999) Interaction of the low-molecular-weight GTP-binding protein Rap2 with the platelet cytoskeleton is mediated by direct binding to the actin filaments. J. Cell. Biochem. 75, 675–685 [DOI] [PubMed] [Google Scholar]
- 10. Torti M., Ramaschi G., Sinigaglia F., Lapetina E. G., Balduini C. (1994) Glycoprotein IIb-IIIa and the translocation of Rap2B to the platelet cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 91, 4239–4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. White G. C., 2nd, Fischer T. H., Duffy C. M. (1998) Rap1b association with the platelet cytoskeleton occurs in the absence of glycoproteins IIb/IIIa. Thromb. Haemost. 79, 832–836 [PubMed] [Google Scholar]
- 12. Arthur W. T., Quilliam L. A., Cooper J. A. (2004) Rap1 promotes cell spreading by localizing Rac guanine nucleotide exchange factors. J. Cell Biol. 167, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boettner B., Van Aelst L. (2009) Control of cell adhesion dynamics by Rap1 signaling. Curr. Opin. Cell Biol. 21, 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoshino T., Sakisaka T., Baba T., Yamada T., Kimura T., Takai Y. (2005) Regulation of E-cadherin endocytosis by nectin through afadin, Rap1, and p120ctn. J. Biol. Chem. 280, 24095–24103 [DOI] [PubMed] [Google Scholar]
- 15. Jeon T. J., Lee D. J., Lee S., Weeks G., Firtel R. A. (2007) Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotaxing cells. J. Cell Biol. 179, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krugmann S., Williams R., Stephens L., Hawkins P. T. (2004) ARAP3 is a PI3K- and Rap-regulated GAP for RhoA. Curr. Biol. 14, 1380–1384 [DOI] [PubMed] [Google Scholar]
- 17. Takahashi M., Rikitake Y., Nagamatsu Y., Hara T., Ikeda W., Hirata K., Takai Y. (2008) Sequential activation of Rap1 and Rac1 small G proteins by PDGF locally at leading edges of NIH3T3 cells. Genes Cells 13, 549–569 [DOI] [PubMed] [Google Scholar]
- 18. Bos J. L. (2005) Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17, 123–128 [DOI] [PubMed] [Google Scholar]
- 19. Bos J. L., de Bruyn K., Enserink J., Kuiperij B., Rangarajan S., Rehmann H., Riedl J., de Rooij J., van Mansfeld F., Zwartkruis F. (2003) The role of Rap1 in integrin-mediated cell adhesion. Biochem. Soc. Trans. 31, 83–86 [DOI] [PubMed] [Google Scholar]
- 20. Asuri S., Yan J., Paranavitana N. C., Quilliam L. A. (2008) E-cadherin dis-engagement activates the Rap1 GTPase. J. Cell. Biochem. 105, 1027–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Balzac F., Avolio M., Degani S., Kaverina I., Torti M., Silengo L., Small J. V., Retta S. F. (2005) E-cadherin endocytosis regulates the activity of Rap1: a traffic light GTPase at the crossroads between cadherin and integrin function. J. Cell Sci. 118, 4765–4783 [DOI] [PubMed] [Google Scholar]
- 22. Hogan C., Serpente N., Cogram P., Hosking C. R., Bialucha C. U., Feller S. M., Braga V. M., Birchmeier W., Fujita Y. (2004) Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 24, 6690–6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Price L. S., Hajdo-Milasinovic A., Zhao J., Zwartkruis F. J., Collard J. G., Bos J. L. (2004) Rap1 regulates E-cadherin-mediated cell-cell adhesion. J. Biol. Chem. 279, 35127–35132 [DOI] [PubMed] [Google Scholar]
- 24. Wittchen E. S., van Buul J. D., Burridge K., Worthylake R. A. (2005) Trading spaces: Rap, Rac, and Rho as architects of transendothelial migration. Curr. Opin. Hematol. 12, 14–21 [DOI] [PubMed] [Google Scholar]
- 25. Bailey C. L., Kelly P., Casey P. J. (2009) Activation of Rap1 promotes prostate cancer metastasis. Cancer Res. 69, 4962–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Itoh M., Nelson C. M., Myers C. A., Bissell M. J. (2007) Rap1 integrates tissue polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res. 67, 4759–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuse M., Mitsutake N., Rogounovitch T., Saenko V., Nakazawa Y., Rumyantsev P., Lushnikov E., Suzuki K., Yamashita S. (2009) Mutation analysis of RAP1 gene in papillary thyroid carcinomas. Endocr. J. 56, 161–164 [DOI] [PubMed] [Google Scholar]
- 28. Zhang L., Chenwei L., Mahmood R., van Golen K., Greenson J., Li G., D'Silva N. J., Li X., Burant C. F., Logsdon C. D., Simeone D. M. (2006) Identification of a putative tumor suppressor gene Rap1GAP in pancreatic cancer. Cancer Res. 66, 898–906 [DOI] [PubMed] [Google Scholar]
- 29. Tsygankova O. M., Prendergast G. V., Puttaswamy K., Wang Y., Feldman M. D., Wang H., Brose M. S., Meinkoth J. L. (2007) Down-regulation of Rap1GAP contributes to Ras transformation. Mol. Cell. Biol. 27, 6647–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nellore A., Paziana K., Ma C., Tsygankova O. M., Wang Y., Puttaswamy K., Iqbal A. U., Franks S. R., Lv Y., Troxel A. B., Feldman M. D., Meinkoth J. L., Brose M. S. (2009) Loss of Rap1GAP in papillary thyroid cancer. J. Clin. Endocrinol. Metab. 94, 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng H., Gao L., Feng Y., Yuan L., Zhao H., Cornelius L. A. (2009) Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Res. 69, 449–457 [DOI] [PubMed] [Google Scholar]
- 32. Tsygankova O. M., Ma C., Tang W., Korch C., Feldman M. D., Lv Y., Brose M. S., Meinkoth J. L. (2010) Down-regulation of Rap1GAP in human tumor cells alters cell/matrix and cell/cell adhesion. Mol. Cell. Biol. 30, 3262–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zuo H., Gandhi M., Edreira M. M., Hochbaum D., Nimgaonkar V. L., Zhang P., Dipaola J., Evdokimova V., Altschuler D. L., Nikiforov Y. E. (2010) Down-regulation of Rap1GAP through epigenetic silencing and loss of heterozygosity promotes invasion and progression of thyroid tumors. Cancer Res. 70, 1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vuchak L. A., Tsygankova O. M., Meinkoth J. L. (2011) Rap1GAP impairs cell-matrix adhesion in the absence of effects on cell-cell adhesion. Cell Adhesion Migration 5, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z., Mitra R. S., Henson B. S., Datta N. S., McCauley L. K., Kumar P., Lee J. S., Carey T. E., D'Silva N. J. (2006) Rap1GAP inhibits tumor growth in oropharyngeal squamous cell carcinoma. Am. J. Pathol. 168, 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin K. B., Tan P., Freeman S. A., Lam M., McNagny K. M., Gold M. R. (2010) The Rap GTPases regulate the migration, invasiveness and in vivo dissemination of B-cell lymphomas. Oncogene 29, 608–615 [DOI] [PubMed] [Google Scholar]
- 37. Freeman S. A., McLeod S. J., Dukowski J., Austin P., Lee C. C., Millen-Martin B., Kubes P., McCafferty D. M., Gold M. R., Roskelley C. D. (2010) Preventing the activation or cycling of the Rap1 GTPase alters adhesion and cytoskeletal dynamics and blocks metastatic melanoma cell extravasation into the lungs. Cancer Res. 70, 4590–4601 [DOI] [PubMed] [Google Scholar]
- 38. Friedl P., Wolf K. (2003) Proteolytic and non-proteolytic migration of tumour cells and leucocytes. Biochem. Soc. Symp. 70, 277–285 [DOI] [PubMed] [Google Scholar]
- 39. Friedl P., Wolf K. (2003) Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3, 362–374 [DOI] [PubMed] [Google Scholar]
- 40. Yilmaz M., Christofori G. (2010) Mechanisms of motility in metastasizing cells. Mol. Cancer Res. 8, 629–642 [DOI] [PubMed] [Google Scholar]
- 41. Sanz-Moreno V., Marshall C. J. (2010) The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr. Opin. Cell Biol. 22, 690–696 [DOI] [PubMed] [Google Scholar]
- 42. Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y. (2004) Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang C., Kazanietz M. G. (2007) Chimaerins: GAPs that bridge diacylglycerol signalling and the small G-protein Rac. Biochem. J. 403, 1–12 [DOI] [PubMed] [Google Scholar]
- 44. Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C. J. (2008) Rac activation and inactivation control plasticity of tumor cell movement. Cell 135, 510–523 [DOI] [PubMed] [Google Scholar]
- 45. Maillet M., Robert S. J., Cacquevel M., Gastineau M., Vivien D., Bertoglio J., Zugaza J. L., Fischmeister R., Lezoualc'h F. (2003) Cross-talk between Rap1 and Rac regulates secretion of sAPPα. Nat. Cell Biol. 5, 633–639 [DOI] [PubMed] [Google Scholar]
- 46. Miura K., Jacques K. M., Stauffer S., Kubosaki A., Zhu K., Hirsch D. S., Resau J., Zheng Y., Randazzo P. A. (2002) ARAP1: a point of convergence for Arf and Rho signaling. Mol. Cell 9, 109–119 [DOI] [PubMed] [Google Scholar]
- 47. Myagmar B. E., Umikawa M., Asato T., Taira K., Oshiro M., Hino A., Takei K., Uezato H., Kariya K. (2005) PARG1, a protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2 effector. Biochem. Biophys. Res. Commun. 329, 1046–1052 [DOI] [PubMed] [Google Scholar]
- 48. Suh H. N., Han H. J. (2010) Laminin regulates mouse embryonic stem cell migration: involvement of Epac1/Rap1 and Rac1/cdc42. Am. J. Physiol. Cell Physiol. 298, C1159–C1169 [DOI] [PubMed] [Google Scholar]