Background: Diphtheria toxin inhibits translation by ADP-ribosylation of elongation factor 2.
Results: Diphtheria toxin expression results in accumulation of cells that fail to separate following mitosis.
Conclusion: Diphtheria toxin expression has unique effects on translocation and the production of specific proteins.
Significance: Understanding how ADP-ribosylated elongation factor 2 affects translocation will increase knowledge of translation and potentially lead to new treatments for toxin exposure.
Keywords: ADP-ribosylation, Bacterial Toxins, Cell Cycle, Protein Synthesis, Translation Elongation Factors, Diphthamide
Abstract
Eukaryotic translation elongation factor 2 (eEF2) facilitates the movement of the peptidyl tRNA-mRNA complex from the A site of the ribosome to the P site during protein synthesis. ADP-ribosylation (ADPR) of eEF2 by bacterial toxins on a unique diphthamide residue inhibits its translocation activity, but the mechanism is unclear. We have employed a hormone-inducible diphtheria toxin (DT) expression system in Saccharomyces cerevisiae which allows for the rapid induction of ADPR-eEF2 to examine the effects of DT in vivo. ADPR of eEF2 resulted in a decrease in total protein synthesis consistent with a defect in translation elongation. Association of eEF2 with polyribosomes, however, was unchanged upon expression of DT. Upon prolonged exposure to DT, cells with an abnormal morphology and increased DNA content accumulated. This observation was specific to DT expression and was not observed when translation elongation was inhibited by other methods. Examination of these cells by electron microscopy indicated a defect in cell separation following mitosis. These results suggest that expression of proteins late in the cell cycle is particularly sensitive to inhibition by ADPR-eEF2.
Introduction
Translation elongation is a cycle of repeated aminoacyl-tRNA (aa-tRNA)2 binding, peptide bond formation, and ribosomal movement catalyzed in most systems by two soluble elongation factors (for review, see Ref. 1). Eukaryotic elongation factor 1A (eEF1A) binds and delivers aa-tRNA to the A site of the ribosome in a GTP-dependent manner. When a codon:anticodon match is detected by the ribosome, the GTPase activity of eEF1A is stimulated leading to aa-tRNA release into the A site. Following peptide bond formation the peptidyl-tRNA:mRNA hybrid is moved from the A site to the P site to position the next codon for decoding. This translocation event is catalyzed by another G protein, eukaryotic elongation factor 2 (eEF2). Both EFs are highly conserved and essential for protein synthesis.
Crystal structures of eEF2 revealed that it consists of five domains, where the three N-terminal domains are similar to eEF1A, aside from a unique insertion in the GTP binding domain, whereas domains IV and V form an extension (2). Interestingly, the overall structure of eEF2 shows molecular mimicry to the eEF1A-aa-tRNA complex (2). Cryo-EM structures of eEF2 bound to the ribosome demonstrated that the tip of domain IV, which constitutes the anticodon mimic site of eEF2, is located near the ribosomal A site and plays an important role in translocation and translational fidelity (3, 4). Positioned within this region of domain IV is a unique diphthamide residue that results from the multistep enzymatic conversion of a single histidine (His-715 and His-699 in humans and yeast, respectively) (5–7). The diphthamide modification is found only on eEF2 and is unique to eukaryotes (8). The role of this residue in the normal process of translation elongation and cell growth is unclear because Saccharomyces cerevisiae with mutations in the diphthamide synthesis pathway are viable (6, 9, 10). However, the absence of diphthamide-modified eEF2 has been associated with altered translational fidelity in both yeast and mammals (4, 11).
Translation elongation is the target of several human bacterial pathogens. Corynebacterium diphtheria, Pseudomonas aeruginosa, and Vibrio cholerae each produce a toxin that ADP-ribosylates (ADPR) eEF2 on the diphthamide residue in domain IV, and this modification inhibits eEF2 activity (12, 13). The presence of the diphthamide modification on eEF2 is essential for its ADPR by bacterial toxins (4, 14, 15). In fact, attempts to isolate diphtheria toxin (DT)-resistant mutants have thus far identified only eEF2 mutants that are no longer diphthamide-modified or mutations in the genes responsible for the enzymatic conversion of histidine to diphthamide (for review, see Ref. 12). The mechanism through which ADPR inhibits eEF2 function and results in cell death, however, remains unknown.
Several studies have shown that ADPR-eEF2 is significantly inhibited in its ability to support in vitro translation (16, 17); however, determining which individual step(s) in the eEF2 cycle are affected has proven difficult. For example, the position of the toxin-induced modification at the tip of domain IV suggests that it may influence ribosome binding. Studies have shown that ADPR-eEF2 binds ribosomes as well as the unmodified protein (18–20), and cryo-EM studies of ADPR-eEF2 bound to ribosomes containing a P site tRNA support these results (3). However, others have shown that ADPR-eEF2 displays a specific defect in binding to pretranslocation ribosomes (21). Furthermore, early studies reported that ADPR-eEF2 showed a decrease in GTP binding (22) whereas more recent fluorescent analyses showed no difference in GTP binding by ADPR-eEF2 (18). Taken together, these biochemical experiments suggest that ADPR-eEF2 is able to bind GTP and the ribosome but is unable either to efficiently or accurately promote translocation.
In this paper, we examine the effects of ADPR-eEF2 on its function in the cellular environment. Because eEF2 is highly conserved and both yeast and mammalian cells are sensitive to intracellular DT, we utilized an inducible expression system for the catalytic fragment of DT in S. cerevisiae to study the effects of ADPR-eEF2. Strains expressing DT were unable to grow on agar but able to grow in liquid culture, albeit at a significantly slower growth rate. This finding allowed us to assess the effects of ADPR on translation elongation. Induction of the toxin led to a decrease in protein synthesis consistent with an elongation defect. Analysis of eEF2 localization in polyribosome gradients showed no significant defects in ribosome binding. Prolonged culture of yeast expressing DT led to an increase in DNA content which was shown to be consistent with a defect in cell separation. This effect was not seen with other eEF2 mutants that reduce total translation. Together, these data suggest that protein synthesis is important for the final stages of cell division and that this synthesis is particularly sensitive to inhibition by ADPR-eEF2.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
S. cerevisiae strains used in this study are listed in Table 1. DPH2 was disrupted in DBY12100 by homologous recombination using a PCR fragment amplified from the Open Biosystems null collection. Recombination was confirmed by PCR using the primers 5′-GCTTCGTCATCACCACCTTT-3′ and 5′-CTGCAGCGAGGAGCCGTAAT-3′. DNA was transformed by the lithium acetate method (23). For galactose induction of DT, liquid cultures of the indicated strains were pregrown at 30 °C to exponential phase in complete medium lacking the appropriate amino acids and containing either 2% raffinose or 2% sucrose. The culture was then split, and the cells were pelleted and resuspended in medium containing either 2% glucose or 2% galactose. The induction was carried out at 30 °C for the indicated time with dilution as required. For DT induction using β-estradiol, liquid cultures of the indicated strains were grown to exponential phase in complete medium lacking uracil at 30 °C. The cultures were then split in half, and either 50 nm β-estradiol (Tocris Bioscience) or an equal amount of 100% ethanol was added. The induction was carried out at 30 °C for the indicated time with dilution as required. For cycloheximide treatment, cells were grown in the presence of 175 ng/ml cycloheximide in complete medium at 30 °C for 24 h with dilution.
TABLE 1.
S. cerevisiae strains and plasmids
| S. cerevisiae | Genotype | Ref. |
|---|---|---|
| Strain | ||
| W303 | Matα, leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15 | 41 |
| DBY12100 | Matα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, ura3Δ::HphMX4, HAP1+ | 30 |
| TKY1713 | Matα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, ura3Δ::HphMX4, dph2Δ::KanMX, HAP1+ | This study |
| TKY675 | MATa ade2 leu2 ura3 his3 leu2 trp1 eft1::HIS3 | 25 |
| eft2::TRP1 pEFT2-His6 LEU2 CEN | ||
| TKY825 | MATa ade2 leu2 ura3 his3 leu2 trp1 eft1::HIS3 | 4 |
| eft2::TRP1 pEFT2-His6 D696A LEU2 CEN | ||
| TKY1367 | MATa ade2 leu2 ura3 his3 leu2 trp1 eft1::HIS3 | This study |
| eft2::ADE2, pEFT2-His6 TRP1 CEN, pEFT2-HA I698A LEU2 CEN | ||
| pGAL1-DT URA3 2μ | ||
| Plasmid | ||
| pLMY101 | pGAL1-DT URA3 2μ | 24 |
| pTKB1122 | pGAL1-DT URA3 CEN | This study |
| pTKB1050 | pEFT2-HA I698A LEU2 CEN | This study |
| pTKB1051 | pEFT2-His6 TRP1 CEN | This study |
Plasmids and DNA Manipulations
Plasmids used in this study are listed in Table 1. The catalytic fragment of DT was cloned from pLMY101 (24) as a BamHI fragment into pTKB372 under control of the GAL1 promoter to produce pTKB1121. The SacI/XhoI fragment containing GAL1-DT was cloned into pRS316 to create DTIND (pTKB1122).
eEF2 Purification, Mass Spectrometry, and Native Gel Electrophoresis/Immunoblotting
eEF2-His6 was purified from the indicated strains and analyzed by mass spectrometry using established methods (4, 25). Total protein extracts were collected by glass bead lysis in buffer containing 100 mm Tris, pH 8.0, 20% glycerol (v/v), 1 mm DTT, and 1 mm PMSF. Extracts were resolved on a 4–20% Tris-glycine native gel (Novex) and transferred to nitrocellulose membranes using standard techniques. Immunoblotting was performed using a rabbit anti-eEF2 antibody raised against S. cerevisiae eEF2.
Total Protein Synthesis and Polyribosome Profile Analysis
Incorporation of [35S]methionine into total cellular protein was measured as described (26). At 1-h intervals, the optical density (A600) of the cultures was determined, and 1-ml aliquots were removed to monitor [35S]methionine incorporation by cold trichloroacetic acid precipitation. Samples from all time points were analyzed in duplicate. Polyribosomes were harvested and analyzed as described (27). Fractions (0.9 ml) were collected using a Frac-920 (GE Healthcare), run on 8% SDS-polyacrylamide gels, and analyzed by immunoblotting with the indicated antibodies. ImageJ software (National Institutes of Health) was used to quantitate the area under the 80S and polyribosome peaks.
Flow Cytometry and Light Microscopy
Cultures were grown and treated with β-estradiol as described above. A600 of the cultures at the time of harvest ranged from 0.3 to 1.0. At the indicated time points, samples were sonicated (8 pulses, 35% duty cycle, Misonix ultrasonic processor XL) and then fixed by incubation overnight at 4 °C in 70% ethanol (v/v). Approximately 1 × 107 fixed cells were washed with 50 mm sodium citrate buffer, pH 7.4, resuspended in 50 mm sodium citrate buffer plus 0.25 mg/ml boiled RNase A, and incubated at 50 °C for 1 h. Proteinase K was added to a final concentration of 1 mg/ml, and incubation was continued at 50 °C for 1 h. Propidium iodide was added to a final concentration of 8 μg/ml, and samples were analyzed on a Beckman Coulter Cytomics FC500 Flow Cytometer in the EOHSI Analytical Cytometry Facility. For light microscopy, cultures were grown and sonicated as above, and the number of cells (∼250 cells counted) with multiple buds was determined using phase contrast microscopy. A representative experiment is presented.
DAPI Staining, Confocal Microscopy, and EM
At the indicated time points, liquid cultures grown overnight at 30 °C in complete medium lacking uracil to an A600 of 0.5–0.6 in the presence or absence of β-estradiol were sonicated and fixed in 3.7% formaldehyde for 1 h at room temperature with shaking. Ethanol was added dropwise to washed cells to a final concentration of 70%. Cells were stained by addition of 0.1 mg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Sigma) and viewed using a Leica TCS SP5 Confocal Microscope with LAS AF Lite version 2.3.0 software in the EOHSI Image Analysis Core Facility. For EM, ∼2 × 108 cells grown overnight at 30 °C to an A600 of 0.3–0.5 in the presence or absence of β-estradiol were washed with water and sonicated. The cells were fixed overnight at 4 °C by resuspension in 1.5 ml of freshly prepared 2.5% glutaraldehyde in phosphate-buffered saline (PBS). Samples were washed three times with PBS and incubated for 30 min at room temperature in 4% KMnO4/0.5×PBS. Cells were washed in PBS and prepared for EM by the RWJMS Core Imaging Laboratory as described (28).
RESULTS
The diphthamide residue resulting from post-translational modification of a conserved histidine in eukaryotic EF2 is the known site of action of DT. Mutations in the S. cerevisiae eEF2 anticodon mimicry domain at or near this His-699 confer dominant resistance to DT expression in S. cerevisiae (4). The DT resistance (DTR) of these strains is due to the absence of the diphthamide modification on His-699 in the eEF2 mutants. This dominant resistance to toxin suggests that DTR-eEF2 is functional despite the presence of nonfunctional ADPR-eEF2; however, the expression and ADP-ribosylation of wild-type eEF2 was not confirmed under these conditions. To address this limitation, wild-type (eEF2-His6) and DTR-eEF2 (eEF2-I698A-HA) were expressed with different epitope tags from plasmids in an eEF2-null background (lacking the EFT1 and EFT2 genes), and the catalytic fragment of DT was expressed from the galactose-inducible GAL1 promoter (pLMY101). Both tagged versions of eEF2 were previously shown to be functional as the only form of eEF2 (25, 29), and dominant DTR was observed in this strain (data not shown). Immunoblotting of extracts prepared from glucose- and galactose-grown cells showed similar expression of both wild-type and DTR forms of eEF2 (Fig. 1A). To demonstrate ADPR of the wild-type protein, eEF2-His6 was purified from extracts of this strain grown in galactose and subjected to LC-tandem MS. Peptides with fragmentation patterns corresponding to ADPR-eEF2 were observed in eEF2-His6 purified from galactose-grown cells (Fig. 1B) and not in the absence of DT. Together, these data suggest that ADPR-eEF2 is produced in the presence of DTR-eEF2 mutants and that ADPR-eEF2 does not block the function of DTR-eEF2 mutant proteins in translocation in vivo.
FIGURE 1.
ADPR of wild-type eEF2 in the presence of the dominant DTR eEF2 mutant, I698A. A, strain TKY1367 containing the Gal-inducible DT plasmid (pLMY101), eEF2-His6 (pTKB1051), and eEF2-I698A-HA (pTKB1050) was grown for 24 h in C-Ura medium with either 2% glucose or 2% galactose. Whole cell extracts were prepared, separated by SDS-PAGE, and immunoblotted with the indicated antibodies. B, tandem MS spectra of the ADPR tryptic fragment 686VNILDVTLHADAIHR700 from eEF2-His6 purified from the strain described in A. y ions are marked. Peaks that are marked with an asterisk are peaks of neutral loss of 601 Da.
Next, strains expressing both wild-type and DTR-eEF2 were used to study translation elongation in more detail. It became apparent during the course of these experiments that a control strain expressing two differentially tagged wild-type copies of eEF2 (eEF2-His6 and eEF2-HA) was capable of growth at a reduced rate in liquid media upon DT induction (data not shown). This growth obviated the need to employ DTR-eEF2 mutants to maintain growth. Therefore, to analyze the effect of ADPR-eEF2 on translation in vivo, galactose-inducible DT (pTKB1122) was expressed in the wild-type W303 yeast strain. This strain was unable to grow on galactose-containing agar plates but still proliferated in galactose-containing liquid media albeit at a much reduced rate relative to a wild-type strain (data not shown), thus permitting analysis of translation in the presence of ADPR-eEF2. Total protein synthesis was measured in glucose media as a control and in galactose media to induce DT. In the W303 strain background, the carbon source itself affected overall protein synthesis in control cells with growth in galactose causing an approximate 3-fold decrease in translation (Fig. 2A). Expression of DT resulted in further reduction in total protein synthesis (Fig. 2A). To determine whether elongation was specifically affected by ADPR-eEF2, polyribosome analysis was performed in the presence and absence of DT. As shown in Fig. 2B (upper panels), growth of control cells in galactose-containing media had a significant effect on the polyribosome profile. Growth in galactose resulted in an increase in the amount of 80S ribosomes detected in cells containing the empty vector. An increase in 80S ribosomes was also detected in DT-expressing cells in galactose although to a lesser extent. Immunoblotting of eEF2 in fractions from the polyribosome gradients suggested that the majority of eEF2 was associated with the 80S ribosome peak in galactose grown cells (Fig. 2C). Consequently, the observed changes in total protein synthesis, polyribosome profile, and eEF2 localization in galactose media in the absence of DT suggest that the induction conditions used in these experiments may mask some of the effects of ADPR-eEF2 on translation elongation.
FIGURE 2.
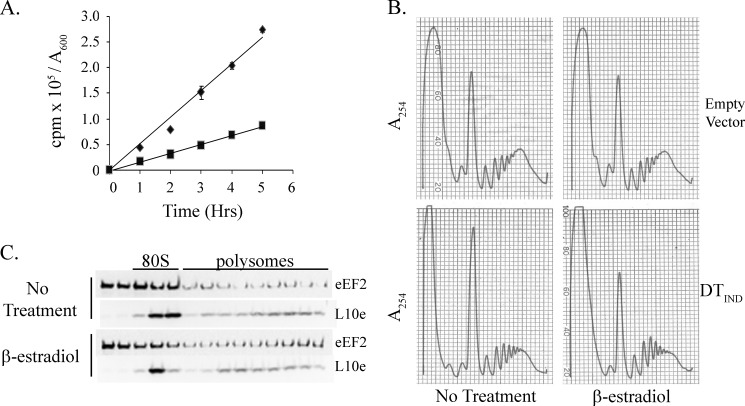
Growth in galactose has significant effects on translation in W303 cells. A, W303 cells containing either an empty vector or galactose-inducible DT plasmid (DTIND) were grown to log phase in C-Met-Ura with 2% sucrose at 30 °C. DT expression was induced for 2 h by growth in medium containing 2% galactose. [35S]Methionine was then added and total protein synthesis measured by trichloroacetic acid precipitation. Incorporation (counts per minute) is expressed per A600 unit. Each time point was performed in duplicate, and results include ±S.D. (error bars). A representative experiment is shown. Empty vector in glucose (♦), empty vector in galactose (■), DTIND in glucose (▴) and DTIND in galactose (×) are indicated. B, strains were grown in C-Ura medium as in A and fixed following induction by incubation in 1% formaldehyde for 1 h on ice. Ribosome extracts were collected and centrifuged on a sucrose gradient. Representative A254 traces are shown. C, fractions were collected from the polyribosome gradients in B, and eEF2 was localized by immunoblotting.
To avoid metabolic induced effects on translation, hormone-inducible expression of DT was employed (30). In this system, a chimeric transactivator drives rapid expression from the GAL1 promoter upon addition of β-estradiol, which has been shown to have minimal effects on the cellular transcriptional program (30). β-Estradiol treatment of this strain had no observable effects on growth rate or polyribosome profile thus demonstrating that, on its own, β-estradiol has little effect on translation (Figs. 3B and 4B, upper panels). Transformation of the GAL1-DT plasmid pTKB1122 into DBY12100 resulted in the absence of growth on agar plates containing β-estradiol and reduced growth in liquid media containing the hormone (Fig. 3, A and B). Addition of the ADP-ribose moiety to eEF2 results in the addition of two negative charges derived from the phosphate groups. It has been shown that this change in charge can be used to separate ADPR-eEF2 from unmodified eEF2 by native gel electrophoresis and immunoblotting (31). Using this method, ADPR-eEF2 could be detected as early as 15 min following the addition of β-estradiol (Fig. 3C). Whereas the majority of eEF2 was modified to the ADPR form following hormone addition, a small portion of eEF2 that appeared to remain unmodified could be detected. This observation was not unexpected given that previous mass spectrometric analysis of wild-type eEF2 has shown that ∼20% of the purified protein lacks the diphthamide modification and thus would not be ADPR by DT (4, 32). In summary, β-estradiol induction of DT provides a clean and rapid system for the analysis of the effects of ADPR-eEF2 on translation without the metabolic effects on translation seen with galactose induction.
FIGURE 3.
β-Estradiol-mediated induction of DT inhibits cell growth. A, DBY12100 containing either an empty vector (pRS316) or DTIND (pTKB1122) was struck on C-Ura plates with and without 50 nm β-estradiol. B, strains described in A were grown to log phase in C-Ura medium at 30 °C. 50 nm β-estradiol was added, and A600 was measured at the indicated time points. Each time point was measured in triplicate and is represented as a -fold increase in growth compared with the 0 h time point. S.D. (error bars) are indicated. A representative experiment is shown. Empty vector (♦), empty vector with 50 nm β-estradiol (■), DTIND (▴), and DTIND with 50 nm β-estradiol (●) are indicated. C, DBY12100 containing DTIND (pTKB1122) was grown as in B. 50 nm β-estradiol was added to the cultures for the indicated time. Whole cell extracts were prepared, run on native PAGE gels, and immunoblotted using a polyclonal anti-eEF2 antibody. The asterisk marks ADPR-eEF2.
FIGURE 4.
β-Estradiol-mediated induction of DT inhibits translation elongation. A, strain DBY12100 containing DTIND (pTKB1122) was grown to log phase in C-Met-Ura medium at 30 °C. [35S]Methionine and either 50 nm β-estradiol (■) or ethanol (♦) were added at the 0 h time point. Total protein synthesis was measured at the indicated time points as in Fig. 2A. Each time point was performed in duplicate, and the S.D. (error bars) is indicated. A representative experiment is shown. B, DBY12100 strains containing either empty vector (pRS316) or DTIND (pTKB1122) were grown to log phase in C-Ura medium at 30 °C. 50 nm β-estradiol was added for 1 h at which point ribosome extracts were collected without formaldehyde fixation and analyzed as in Fig. 2B. C, fractions were collected from the polyribosome gradients in B, and eEF2 was localized by immunoblotting. L10e is a component of the large ribosomal subunit which serves as a control for ribosome localization and distribution.
To analyze translation in this system, total protein synthesis was measured in the presence and absence of β-estradiol. ADPR of eEF2 by DT led to a 2–3-fold decrease in total protein synthesis which persisted over at least 5 h of growth (Fig. 4A). This decrease in overall protein synthesis corresponded with an approximate 2-fold increase in the polyribosome to monoribosome ratio (Fig. 4B, lower panels). These data are consistent with ADPR of eEF2 leading to an elongation defect. Addition of cycloheximide to the cultures prior to polyribosome harvest was able to further increase the accumulation of polyribosomes (data not shown), which suggests that the elongation block induced by ADPR-eEF2 is not complete. Finally, eEF2 localization within the polyribosome gradient was analyzed by Western blotting with an eEF2 polyclonal antibody (Fig. 4C). No significant change in eEF2 localization within the polyribosome gradient was observed upon expression of DT. These data suggest that ADPR of eEF2 does not significantly affect the ability of eEF2 to bind or dissociate from the ribosome in vivo.
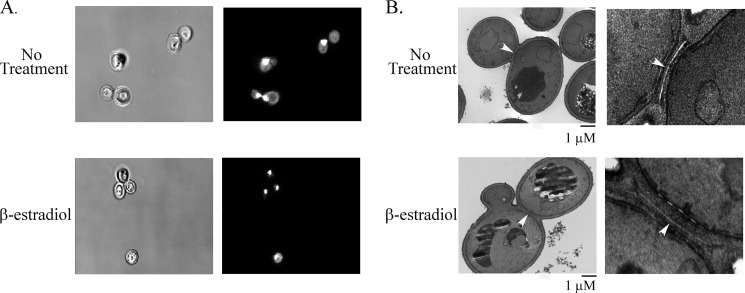
The residual slow growth observed in DBY12100 cells expressing DT could be due to either a reduction in growth rate or an increase in cell death. To address this question, cells exposed to β-estradiol for 24 h were analyzed by phase contrast microscopy. Interestingly, an increase in abnormal cell morphology was observed in strains expressing DT compared with uninduced cells. Approximately 19% of the cells expressing DT appeared to have two buds with occasional cells having more than two buds (see Fig. 6A, lower panels). These abnormal cells were detected at low levels in uninduced strains containing the DTIND plasmid (∼4%) and at very low levels in β-estradiol-treated control strains containing an empty vector (∼1%). Thus, the cell morphology defect is not caused by hormone exposure. The presence of these multibudded cells suggested a defect in cell cycle progression. β-Estradiol-treated cells were also analyzed for DNA content by flow cytometry. Cells with >2 n DNA content were readily detectable by flow cytometry after induction of DT and were absent in untreated cells and β-estradiol-treated control strains (Fig. 5A and data not shown). These data suggest that ADPR of eEF2 leads to specific defects in cell cycle progression.
FIGURE 6.
DT expression results in defects in cell separation. A, strain DBY12100 containing DTIND (pTKB1122) was grown in the presence or absence of β-estradiol for 24 h in C-Ura medium at 30 °C. Cells were then fixed, stained with DAPI, and imaged by confocal microscopy. Left panels, brightfield; right panels, DAPI. B, strain DBY12100 was grown as in A and fixed for EM. The right panels show a magnified view of the budneck. Arrowheads indicate the primary septa.
FIGURE 5.
Prolonged DT exposure leads to cell cycle defects. A, strain DBY12100 containing DTIND (pTKB1122) was grown to log phase in C-Ura medium at 30 °C, and 50 nm β-estradiol or no compound was added. At the indicated time points, cells were sonicated, fixed, stained with propidium iodide, and analyzed as described under “Experimental Procedures.” B, wild-type (TKY675) and eEF2-D696A (TKY825) mutant strains were analyzed as in A. C, DBY12100 was grown to log phase in complete medium at 30 °C. Cells were grown in the presence or absence of 175 ng/ml cycloheximide for 24 h. D, strain TKY1713 (dph2Δ) containing DTIND (pTKB1122) was grown to log phase in C-Ura medium at 30 °C. Cells were grown in the presence or absence of 50 nm β-estradiol for 24 h before harvest as in A.
It is possible that perturbation in translation elongation itself may lead to similar changes in cell morphology. To address this question, DNA content was analyzed in a strain containing the eEF2 mutant, eEF2-D696A, which is known to cause significant defects in growth rate and total protein synthesis (4) and in cells treated with a known translation elongation inhibitor, cycloheximide. No change in total DNA content was observed under either condition (Fig. 5, B and C). To demonstrate that the effect of DT on cell morphology is dependent on the ADPR of eEF2, DPH2 was deleted in the DBY12100 strain background. In the absence of DPH2, eEF2 lacks diphthamide and is therefore not able to be ADPR by DT. As expected, this dph2Δ strain was resistant to DT expression (data not shown). It was also resistant to the formation of the abnormal cell morphology (data not shown) and the altered flow cytometry distribution (Fig. 5D) induced by DT. In summary, these data show the abnormal cell morphology in response to DT is specific and requires ADPR-eEF2.
The increase in DNA content observed in the presence of ADPR-eEF2 may result from defects in DNA separation during mitosis or alternatively may be the result of cytokinesis defects. To determine whether mitosis was compromised, cells expressing DT were fixed and stained with DAPI to visualize DNA. Each bud of the large multibudded cells contained a single nucleus as shown in Fig. 6A, suggesting that nuclear separation was not affected by DT expression. Smaller secondary buds did not contain nuclear DNA as would be expected because nuclear division is linked by bud size (33). To analyze cytokinesis, cells expressing ADPR-eEF2 were examined by transmission EM to visualize the bud neck. During cytokinesis in budding yeast, the actin myosin ring that is formed in the late stages of mitosis contracts and the membrane ingresses (for review, see Ref. 34). As the ring contracts, a layer of chitin, known as the primary septum, is laid down from either end of the bud neck, effectively separating the cytoplasm of the mother and daughter cell. The secondary septum composed of cell wall components then forms on either side of the primary septum. Finally, the cells are separated by the action of chitinase and glucanases. In cells expressing DT, the primary and secondary septa appeared to form normally, suggesting that it is the very last step in the cell division process, which is compromised in strains with ADPR-eEF2 (Fig. 6B). Together, these data suggest that the mother cell initiates a second bud in the presence of ADPR-eEF2 despite failure of the primary daughter cell to be released and that this failure to separate may be due to an inability to degrade the primary septa.
DISCUSSION
Eukaryotic elongation factor 2 is the target of a class of bacterial mono-ADP-ribosyltransferase toxins which include the prototype, DT, exotoxin A from P. aeruginosa, and cholix toxin from V. cholera. Exposure of eukaryotic cells to these toxins leads to inhibition of protein synthesis and cell growth. Despite intensive investigation, the detailed mechanism of how ADPR of eEF2 affects its ability to function as a translocase and kill cells remains unknown. We have established a conditional DT expression system in S. cerevisiae which allows this question to be addressed in a genetically tractable model organism. This system in which DT expression is rapidly induced upon addition of hormone has several advantages over other methods previously used for regulated toxin expression. For example, the galactose-inducible expression system commonly used in yeast has significant effects on protein synthesis upon changing carbon source making it difficult to isolate the specific downstream effects of ADPR-eEF2 (Fig. 2 and Ref. 35). Induction of DT by β-estradiol, however, had no observable effects on growth or translation (Fig. 3). In addition, leaky expression of DT in the β-estradiol system is minimal. Other vectors containing either a copper-inducible or a tetracycline-repressible DT expression construct were prepared and resulted in severe inhibition of growth even in the absence of induction which is consistent with leaky expression.3
Using the hormone-inducible DT expression system, we demonstrated that yeast could grow at a reduced rate in liquid media in the presence of ADPR-eEF2, allowing us to examine its effects on translation in vivo. This result was not anticipated as previous work had suggested that a single molecule of DT was sufficient to arrest growth of mammalian cells (36), and other experiments in yeast had shown an inability to form colonies upon DT induction (24, 37). In addition, we found that protein synthesis was not completely inhibited in this system, and the reduction in total protein synthesis correlated well with the reduction in cell growth. Growth of DT-expressing cells under these conditions can be accounted for in several ways. Because all assays were performed on populations rather than at the single cell level, it is possible that the growth we observe is due to specific subpopulations that maintain lower or no ADPR-eEF2. This result is unlikely, however, given that the cultures have been maintained for over 24 h with no apparent change in overall growth rate. Alternatively, within each cell a fraction of eEF2 may remain unmodified, and this low level of eEF2 activity is able to support reduced growth. In support of this possibility, non-ADPR-eEF2 is present in Western blots following DT induction (Fig. 3C), and previous work has shown that that ∼20% of purified eEF2 lacks the diphthamide modification required for DT to ADPR eEF2 (Ref. 4 and data not shown). Therefore, a minor pool of unmodified eEF2 may allow for the limited growth observed in this system.
It has been proposed that ADPR of eEF2 interferes with its ability to bind the ribosome. This effect could manifest as an inability to bind pretranslocation ribosomes or an inability to dissociate from post-translocation ribosomes. Ribosome binding by ADPR-eEF2 has been studied extensively in vitro with mixed results (3, 18–22, 42). Using an in vivo approach, we examined ADPR-eEF2 binding to polyribosomes in cellular extracts and found no significant difference in the steady-state level of eEF2 bound to ribosomes in DT-expressing cells compared with uninduced cells. Because ADPR-eEF2 was not distinguished from unmodified eEF2 in these experiments, it is possible that binding by ADPR-eEF2 is not being detected. However, the shift of the majority of eEF2 in native gel electrophoresis upon expression of DT and its ability to be detected by our eEF2 polyclonal antibody suggest that we are primarily observing polyribosome binding by ADPR-eEF2. In addition, the dominant DT resistance and therefore cell viability conferred by eEF2 mutations in the anticodon mimicry domain observed in this and our previous work argue against an inability of ADPR-eEF2 to dissociate from polyribosomes following translocation (4). If ADPR-eEF2 remained bound to the polyribosomes a dominant interfering effect of ADPR eEF2 would be expected rather than the dominant resistance observed. Therefore, our data suggest that ribosome binding by eEF2 is unaffected by ADPR, but its ability to act as a translocase is compromised. Although the exact mechanism of translocation is not completely understood, biochemical, structural, and dynamic single molecule experiments in bacteria have suggested that translocation involves a series of conformational changes of the ribosome (for review, see Ref. 43). ADPR of eEF2 may either inhibit some of these conformational changes or promote the formation of inactive conformations through its interaction with the A site of the ribosome.
Prolonged exposure to DT led to an accumulation of yeast cells with abnormal morphology and distribution by flow cytometry. Nuclear staining and EM of the bud neck indicate that this abnormal morphology is the result of a failure of the mother and daughter cell to separate prior to re-entry into the cell cycle. Previous experiments have shown using cycloheximide and synchronized cell populations that protein synthesis is required for cytokinesis/cell separation (44). However, in our experiments the cell separation defect was seen only upon DT expression and not when translation elongation was inhibited by eEF2 mutations or sublethal doses of cycloheximide. The difference between these results in the presence of cycloheximide is likely due to the dose used and/or the time at which the cells were examined. We used ∼5 times less cycloheximide such that the culture continued to grow but at a reduced growth rate so as to be similar to what we observed with DT induction and not a complete inhibition of protein synthesis. Unsynchronized cells were also observed 24 h after treatment and therefore, may arrest at other points in the cell cycle. We also demonstrated that the diphthamide modification of eEF2 was required for the DT effect on morphology confirming that it is downstream of ADPR-eEF2. These results suggest a model in which ADPR of eEF2 reduces its ability to function as a translocase, slowing the rate of elongation and inhibiting the synthesis of specific proteins required for cell cycle progression. This inhibition of translocation by ADPR of eEF2 is unique, however, in that similar effects are not seen when translocation is inhibited by other means. Control of gene expression at the level of translation elongation is supported by an increasing body of experimental evidence (45–47) whereas the effects of elongation defects on the expression of particular mRNAs is just beginning to be explored (48).
The persistence of the primary septum separating the mother and daughter cells in DT-expressing cells suggests that candidates for such proteins might include chitinase, which degrades the primary septum, as well as glucanases required for cell wall degradation and remodeling. Interestingly, expression of many of the enzymes required for cell separation, including CTS1 (chitinase) and ENG1 (an endo-1,3-β-glucanase) occurs only in the daughter cell (49, 50). Thus, protein synthesis in the daughter cell may be more sensitive to ADPR-eEF2. Whether the daughter cells can eventually separate from the mother and whether they are capable of further cell division or permanently arrested is unknown.
The yeast system we have employed to study the effects of DT expression on protein synthesis and cell growth shares similar characteristics with studies of mammalian cells intoxicated with DT. For example, analysis of protein synthesis inhibition in individual cells in response to DT showed that prolonged exposure to cytosolic toxin was required to achieve maximal inhibition of protein synthesis (51). This observation suggests that mammalian cells, like yeast, may also be capable of low levels of protein synthesis at early time points following DT exposure. In addition, treatment of purified rat liver eEF2 with DT indicated the presence of a small fraction of eEF2 which could not be ADPR (38). This fraction of eEF2 may represent eEF2 that is already endogenously ADPR (39, 40) or eEF2 that lacks the diphthamide modification as we have observed in S. cerevisiae. Consequently, other observations made in the yeast system may also apply to mammalian cells, including effects of ADPR-eEF2 on the expression of specific proteins.
Acknowledgments
We thank Elizabeth Reuss, Raj Patel, Joan Dubois, Haiyan Zhang, and Theresa Hyejeong for technical assistance and the members of the Kinzy laboratory for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health R21 AI076245.
M. K. Mateyak and T. G. Kinzy, unpublished observations.
- aa-tRNA
- aminoacyl tRNA
- ADPR
- ADP-ribosylation
- DT
- diphtheria toxin
- DTIND
- inducible DT plasmid
- DTR
- diphtheria toxin-resistant
- eEF2
- eukaryotic elongation factor 2.
REFERENCES
- 1. Dever T. E., Green R. (2012) The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 4, a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jørgensen R., Ortiz P. A., Carr-Schmid A., Nissen P., Kinzy T. G., Andersen G. R. (2003) Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 10, 379–385 [DOI] [PubMed] [Google Scholar]
- 3. Taylor D. J., Nilsson J., Merrill A. R., Andersen G. R., Nissen P., Frank J. (2007) Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 26, 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ortiz P. A., Ulloque R., Kihara G. K., Zheng H., Kinzy T. G. (2006) Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 281, 32639–32648 [DOI] [PubMed] [Google Scholar]
- 5. Liu S., Milne G. T., Kuremsky J. G., Fink G. R., Leppla S. H. (2004) Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol. 24, 9487–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen J. Y., Bodley J. W., Livingston D. M. (1985) Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 5, 3357–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Ness B. G., Howard J. B., Bodley J. W. (1980) ADP-ribosylation of elongation factor 2 by diphtheria toxin: isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J. Biol. Chem. 255, 10717–10720 [PubMed] [Google Scholar]
- 8. Dunlop P. C., Bodley J. W. (1983) Biosynthetic labeling of diphthamide in Saccharomyces cerevisiae. J. Biol. Chem. 258, 4754–4758 [PubMed] [Google Scholar]
- 9. Moehring T. J., Danley D. E., Moehring J. M. (1984) In vitro biosynthesis of diphthamide, studied with mutant Chinese hamster ovary cells resistant to diphtheria toxin. Mol. Cell. Biol. 4, 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kimata Y., Kohno K. (1994) Elongation factor 2 mutants deficient in diphthamide formation show temperature-sensitive cell growth. J. Biol. Chem. 269, 13497–13501 [PubMed] [Google Scholar]
- 11. Liu S., Bachran C., Gupta P., Miller-Randolph S., Wang H., Crown D., Zhang Y., Wein A. N., Singh R., Fattah R., Leppla S. H. (2012) Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc. Natl. Acad. Sci. U.S.A. 109, 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng Q., Barbieri J. T. (2008) Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu. Rev. Microbiol. 62, 271–288 [DOI] [PubMed] [Google Scholar]
- 13. Jørgensen R., Purdy A. E., Fieldhouse R. J., Kimber M. S., Bartlett D. H., Merrill A. R. (2008) Cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. J. Biol. Chem. 283, 10671–10678 [DOI] [PubMed] [Google Scholar]
- 14. Foley B. T., Moehring J. M., Moehring T. J. (1995) Mutations in the elongation factor 2 gene which confer resistance to diphtheria toxin and Pseudomonas exotoxin A: genetic and biochemical analyses. J. Biol. Chem. 270, 23218–23225 [DOI] [PubMed] [Google Scholar]
- 15. Phan L. D., Perentesis J. P., Bodley J. W. (1993) Saccharomyces cerevisiae elongation factor 2: mutagenesis of the histidine precursor of diphthamide yields a functional protein that is resistant to diphtheria toxin. J. Biol. Chem. 268, 8665–8668 [PubMed] [Google Scholar]
- 16. Bermek E. (1972) Formation of a complex involving ADP-ribosylated human translocation factor, guanosine nucleotide and ribosomes. FEBS Lett. 23, 95–99 [DOI] [PubMed] [Google Scholar]
- 17. Montanaro L., Sperti S., Testoni G., Mattioli A. (1976) Effect of elongation factor 2 and of adenosine diphosphate-ribosylated elongation factor 2 on translocation. Biochem. J. 156, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jørgensen R., Yates S. P., Teal D. J., Nilsson J., Prentice G. A., Merrill A. R., Andersen G. R. (2004) Crystal structure of ADP-ribosylated ribosomal translocase from Saccharomyces cerevisiae. J. Biol. Chem. 279, 45919–45925 [DOI] [PubMed] [Google Scholar]
- 19. Davydova E. K., Ovchinnikov L. P. (1990) ADP-ribosylated elongation factor 2 (ADP-ribosyl-EF-2) is unable to promote translocation within the ribosome. FEBS Lett. 261, 350–352 [DOI] [PubMed] [Google Scholar]
- 20. Bermek E. (1976) Interactions of adenosine diphosphate-ribosylated elongation factor 2 with ribosomes. J. Biol. Chem. 251, 6544–6549 [PubMed] [Google Scholar]
- 21. Nygård O., Nilsson L. (1990) Kinetic determination of the effects of ADP-ribosylation on the interaction of eukaryotic elongation factor 2 with ribosomes. J. Biol. Chem. 265, 6030–6034 [PubMed] [Google Scholar]
- 22. Burns G., Abraham A. K., Vedeler A. (1986) Nucleotide binding to elongation factor 2 inactivated by diphtheria toxin. FEBS Lett. 208, 217–220 [DOI] [PubMed] [Google Scholar]
- 23. Gietz R. D., Woods R. A. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 24. Mattheakis L. C., Shen W. H., Collier R. J. (1992) DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 4026–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jørgensen R., Carr-Schmid A., Ortiz P. A., Kinzy T. G., Andersen G. R. (2002) Purification and crystallization of the yeast elongation factor eEF2. Acta Crystallogr. D 58, 712–715 [DOI] [PubMed] [Google Scholar]
- 26. Valente L., Kinzy T. G. (2003) Yeast as a sensor of factors affecting the accuracy of protein synthesis. Cell Mol. Life Sci. 60, 2115–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Esposito A. M., Mateyak M., He D., Lewis M., Sasikumar A. N., Hutton J., Copeland P. R., Kinzy T. G. (2010) Eukaryotic polyribosome profile analysis. J. Vis. Exp. 15, 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Esposito A. M., Kinzy T. G. (2010) The eukaryotic translation elongation factor 1Bγ has a non-guanine nucleotide exchange factor role in protein metabolism. J. Biol. Chem. 285, 37995–38004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Justice M. C., Hsu M. J., Tse B., Ku T., Balkovec J., Schmatz D., Nielsen J. (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 273, 3148–3151 [DOI] [PubMed] [Google Scholar]
- 30. McIsaac R. S., Silverman S. J., McClean M. N., Gibney P. A., Macinskas J., Hickman M. J., Petti A. A., Botstein D. (2011) Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 22, 4447–4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu S., Leppla S. H. (2003) Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol. Cell 12, 603–613 [DOI] [PubMed] [Google Scholar]
- 32. Uthman S., Bär C., Scheidt V., Liu S., ten Have S., Giorgini F., Stark M. J., Schaffrath R. (2013) The amidation step of diphthamide biosynthesis in yeast requires DPH6, a gene identified through mining the DPH1-DPH5 interaction network. PLoS Genet. 9, e1003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Howell A. S., Lew D. J. (2012) Morphogenesis and the cell cycle. Genetics 190, 51–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meitinger F., Palani S., Pereira G. (2012) The power of MEN in cytokinesis. Cell Cycle 11, 219–228 [DOI] [PubMed] [Google Scholar]
- 35. Ashe M. P., De Long S. K., Sachs A. B. (2000) Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11, 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamaizumi M., Mekada E., Uchida T., Okada Y. (1978) One molecule of diphtheria toxin fragment A introduced into a cell can kill the cell. Cell 15, 245–250 [DOI] [PubMed] [Google Scholar]
- 37. Murakami S., Bodley J. W., Livingston D. M. (1982) Saccharomyces cerevisiae spheroplasts are sensitive to the action of diphtheria toxin. Mol. Cell. Biol. 2, 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marzouki A., Lavergne J. P., Reboud J. P., Reboud A. M. (1989) Heterogeneity of native rat liver elongation factor 2. FEBS Lett. 255, 72–76 [DOI] [PubMed] [Google Scholar]
- 39. Lee H., Iglewski W. J. (1984) Cellular ADP-ribosyltransferase with the same mechanism of action as diphtheria toxin and Pseudomonas toxin A. Proc. Natl. Acad. Sci. U.S.A. 81, 2703–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fendrick J. L., Iglewski W. J. (1989) Endogenous ADP-ribosylation of elongation factor 2 in polyoma virus-transformed baby hamster kidney cells. Proc. Natl. Acad. Sci. U.S.A. 86, 554–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas B. J., Rothstein R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 42. Montanaro L., Sperti S., Mattioli A. (1971) Interaction of ADP-ribosylated aminoacyl-transferase II with GTP and with ribosomes. Biochim. Biophys. Acta 238, 493–497 [DOI] [PubMed] [Google Scholar]
- 43. Chen J., Tsai A., O'Leary S. E., Petrov A., Puglisi J. D. (2012) Unraveling the dynamics of ribosome translocation. Curr. Opin. Struct. Biol. 22, 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burke D. J., Church D. (1991) Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3691–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shalgi R., Hurt J. A., Krykbaeva I., Taipale M., Lindquist S., Burge C. B. (2013) Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 49, 439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shenton D., Smirnova J. B., Selley J. N., Carroll K., Hubbard S. J., Pavitt G. D., Ashe M. P., Grant C. M. (2006) Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281, 29011–29021 [DOI] [PubMed] [Google Scholar]
- 47. Liu B., Han Y., Qian S. B. (2013) Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell 49, 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hussey G. S., Chaudhury A., Dawson A. E., Lindner D. J., Knudsen C. R., Wilce M. C., Merrick W. C., Howe P. H. (2011) Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell 41, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baladrón V., Ufano S., Dueñas E., Martín-Cuadrado A. B., del Rey F., Vázquez de Aldana C. R. (2002) Eng1p, an endo-1,3-β-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 1, 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colman-Lerner A., Chin T. E., Brent R. (2001) Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107, 739–750 [DOI] [PubMed] [Google Scholar]
- 51. Falnes P. O., Ariansen S., Sandvig K., Olsnes S. (2000) Requirement for prolonged action in the cytosol for optimal protein synthesis inhibition by diphtheria toxin. J. Biol. Chem. 275, 4363–4368 [DOI] [PubMed] [Google Scholar]