Background: The mechanisms underlying slow kinetics of the kainate receptor-mediated EPSCs are not totally understood.
Results: Phosphorylation-dependent binding of 14-3-3 proteins to GluK2a subunits modulates the biophysical property of GluK2a-containing kainate receptors.
Conclusion: 14-3-3 is an important regulator of GluK2a-containing kainate receptors.
Significance: The dynamically regulated 14-3-3·GluK2a protein complex may contribute to the diverse functions of kainate receptors in synaptic signaling.
Keywords: Glutamate Receptors, Phosphorylation, Protein Kinase C (PKC), Protein/Protein Interactions, Receptor Desensitization, 14-3-3 Proteins, Channel Kinetics, Kainate Receptors
Abstract
Kainate receptors (KARs) are one of the ionotropic glutamate receptors that mediate excitatory postsynaptic currents (EPSCs) with characteristically slow kinetics. Although mechanisms for the slow kinetics of KAR-EPSCs are not totally understood, recent evidence has implicated a regulatory role of KAR-associated proteins. Here, we report that decay kinetics of GluK2a-containing receptors is modulated by closely associated 14-3-3 proteins. 14-3-3 binding requires PKC-dependent phosphorylation of serine residues localized in the carboxyl tail of the GluK2a subunit. In transfected cells, 14-3-3 binding to GluK2a slows desensitization kinetics of both homomeric GluK2a and heteromeric GluK2a/GluK5 receptors. Moreover, KAR-EPSCs at mossy fiber-CA3 synapses decay significantly faster in the 14-3-3 functional knock-out mice. Collectively, these results demonstrate that 14-3-3 proteins are an important regulator of GluK2a-containing KARs and may contribute to the slow decay kinetics of native KAR-EPSCs.
Introduction
Kainate receptors (KARs)4 are one of the three principal classes of ionotropic glutamate receptors that consist of five subunits termed GluK1 through GluK5. Among them, GluK1 through GluK3 subunits are able to form functional homomeric receptors, but GluK4 and GluK5 have to be coassembled with one of the GluK1 through GluK3 subunits (1–5). In neurons, KARs are present at both presynaptic terminals and postsynaptic sites, where they regulate neurotransmitter release, synaptic plasticity, and neuronal excitability (6–14). KAR-mediated excitatory postsynaptic currents (KAR-EPSCs) exhibit characteristically slow decay kinetics, which might be important for synaptic integration of temporal information (4, 15–19). Despite recent progress, there appears to be a discrepancy between the kinetics of recombinant and native KARs, as homomeric or heteromeric KARs display fast desensitization and deactivation kinetics in heterologous expression systems (20–22). Interestingly, Barberis et al. (41) reported that the heteromeric GluK2/GluK5 receptor exhibits slow deactivation kinetics when activated by brief agonist application mimicking the glutamate transient in the synaptic cleft. Because the heteromeric GluK2/GluK5 receptors likely constitute postsynaptic KARs in the brain, their intrinsic slow deactivation property should play an important role in determining the slow kinetics of KAR-EPSCs.
Additionally, regulatory/auxiliary proteins of KARs have also been implicated in modulating the kinetics of KARs (23–30). Recently, several groups have identified the neuropilin and tolloid-like proteins, NETO1 and NETO2, as novel auxiliary subunits that modulate properties of KARs. Both NETO1 and NETO2 significantly slow desensitization and deactivation of homomeric and heteromeric KARs in heterologous expression systems (30–33). In addition, the decay kinetics of KAR-EPSCs at hippocampal synapses is notably accelerated in NETO1 knock-out mice, demonstrating the importance of regulatory proteins in determining biophysical properties of KARs (32, 34).
We report here that kinetics of KARs is also modulated by 14-3-3, a family of proteins that consist of seven homologous isoforms denoted β, γ, ϵ, η, ζ, σ, and τ. They bind to target proteins containing specific phosphoserine motifs and participate in regulation of a variety of cellular processes (35). In the brain, 14-3-3 proteins are abundantly expressed, with some isoforms being particularly enriched at synapses (36). Previously, certain 14-3-3 isoforms have been identified as potential binding partners of KARs (26, 37). In this study, we carried out detailed analyses of both the biochemical basis and functional significance of the protein/protein interaction between 14-3-3 and the GluK2a subunit. We found that decay kinetics of GluK2a-containing receptors is modulated by PKC phosphorylation-dependent binding of 14-3-3. Moreover, antagonizing 14-3-3 binding in the 14-3-3 functional knock-out mice accelerates the decay kinetics of KAR-EPSCs at hippocampal synapses. Together, our findings establish a novel role of 14-3-3 proteins in regulating biophysical properties of KARs.
EXPERIMENTAL PROCEDURES
Mice
All animal procedures were carried out in accordance with the guidelines for the Care and Use of Laboratory Animals of both Shanghai Jiao Tong University School of Medicine and the Florida State University and were approved by the respective Institutional Animal Care and Use Committees (IACUC). Transgenic 14-3-3 functional knock-out (FKO) mice were generated by expressing the YFP-fused difopein (dimeric fourteen-three-three peptide inhibitor) using a thy-1 promoter. After PCR-based genotyping, positive founders were backcrossed to C57BL/6J mice for at least eight generations. Expression patterns of transgene expression in different founders were examined by fluorescence microscopy.
cDNAs and Antibodies
Two main splice variants of rat GluK2 (GluK2a and GluK2b) were used in our experiments. The full-length cDNAs of rat GluK2a with the three RNA-editing sites encoding VCQ and rat GluK5 were kindly provided by Dr. Peter Seeburg (Max-Plank Institute, Germany). Rat myc-GluK2b with the three RNA-editing sites encoding IYQ was kindly provided by Dr. Christophe Mulle (Institute for Interdisciplinary Neuroscience, France). GluK2a was subcloned into pcDNA3.1+ and pAdTrack-CMV vectors. cDNAs encoding various isoforms of human 14-3-3 were subcloned into either pcDNA3 with an N-terminal FLAG tag or pEBFP-N1 as reported previously (38). The plasmids for enhanced YFP-fused difopein (pSCM138) and the inactive mutant (pSCM174) were kindly provided by Dr. Haian Fu (Emory University). Mutant GluK2a cDNAs (S846A, S856A, S859A, S868A, and 4SA-GluK2a) were generated using the QuikChange site-directed mutagenesis kits (Stratagene) and confirmed by DNA sequencing. Antibodies used were as follows: rabbit anti-GluK2/3 polyclonal antibody (53518) was from AnaSpec; rabbit anti-GluK5 polyclonal antibody (06-315) was from Millipore; mouse monoclonal anti-FLAG M2 (F1804) was from Sigma; rabbit anti-pan14-3-3 (K19) polyclonal antibody (sc-629) was from Santa Cruz Biotechnology; and mouse anti-c-Myc monoclonal antibody (MA003) was from Shanghai Immune Biotech Co. Ltd.
RT-PCR
Total RNAs from the HEK293 cells or rat hippocampi were isolated using TRNzol-A+ reagent (TIANGEN BIOTECH, China). They were then used to generate cDNAs via reverse transcription reaction using PrimeScript RT reagent kits with gDNA Eraser (Takara, China) according to the manufacturer's protocol. The PCR was performed in 50 μl of reaction mixture containing 5 μl of 10× PCR buffer, 4 μl of dNTP mixture (each 2.5 mm), 0.25 μl of Takara TaqDNA polymerase (5 units/μl), 0.4 μm final concentration of both primers (forward and reverse), and 1.5 μl of cDNA. The sequences of the primers used for GluK2a are as follows: forward, GGCACTCCTATGGGCTCT; reverse, TTCTTTACCTGGCAACCT. The size of the PCR product was 417 bp.
Cell Culture, Transfection, and Protein Preparation
HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C in a humidified 5% CO2 incubator. Cells were plated in 60-mm dishes at 1 × 106 cells and allowed to grow to 70–90% confluence prior to transient transfection using the Lipofectamine 2000 reagent (Invitrogen). Two days after transfection, cells were lysed in lysis buffer (20 mm Tris-HCl, pH 8.0, 137 mm NaCl, 1% Nonidet P-40, 10% glycerol, 2 mm EGTA) supplemented with protease and phosphatase inhibitor mixture tablets (Roche Applied Science). In PKC-related experiments, transfected cells were treated with either PMA (5 μm) or calphostin C (CC, 1 μm) for 1 h at 37 °C before harvesting.
Hippocampal Lysates
Dissected hippocampi were homogenized in ice-cold homogenization buffer (20 mm Tris-HCl, pH 8.0, 137 mm NaCl, 1% Nonidet P-40, 10% glycerol, 2 mm EGTA) supplemented with protease and phosphatase inhibitor mixture tablets. The homogenates were centrifuged at 13,000 × g for 15 min at 4 °C. Protein concentrations in collected supernatants were determined by BCA assay.
Immunoprecipitation and Western Blot
Immunoprecipitation was performed using standard approaches. Briefly, lysates containing 800 μg of proteins were first precleared by incubating with 30 μl of protein G-agarose (Roche Applied Science) for 2 h at 4 °C. They were then incubated with 2 μg of antibodies overnight at 4 °C, followed by the incubation with 30 μl of protein G-agarose at 4 °C for 4 h. The beads were then washed five times with lysis buffer, and bound proteins were eluted from the beads by boiling in sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 0.01% bromphenol blue, 10% glycerol, 1% β-mercaptoethanol) for 10 min and analyzed by electrophoresis and immunoblotting. Immunoreactive bands were visualized using horseradish peroxidase (HRP)-conjugated goat anti-mouse/rabbit secondary antibody (1:2000, Bio-Rad). Protein densities on Western blots were analyzed by ImageJ software. Relative band densities were determined by normalizing the immunoprecipitated band density to that of the lysate band. All experiments were repeated at least three times.
Crude Membrane Preparations
Cell membranes were prepared as described (40) with modifications. Briefly, cells were washed twice with ice-cold PBS 48 h post-transfection and were homogenized by ultrasonic treatment in buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 320 mm sucrose, 1 mm MgCl2) containing both protease and phosphatase inhibitor mixtures. The cell homogenate was centrifuged at 1000 × g for 10 min at 4 °C to remove nuclei and unbroken cells. The volume of 2.5% supernatant fraction was stored as total protein used for input control in Western blot. The resulting supernatant was centrifuged at 200,000 × g for 1 h at 4 °C. The membrane pellet was solubilized in lysis buffer (20 mm Tris-HCl, pH 8.0, 137 mm NaCl, 2 mm EGTA, 10% glycerol, 1% Nonidet P-40) containing both protease and phosphatase inhibitor mixtures for a minimum of 1 h at 4 °C. Insoluble material was removed by centrifugation at 14,000 × g for 10 min at 4 °C, and then the coimmunoprecipitation was performed as described above.
Measurement of KAR Kinetics
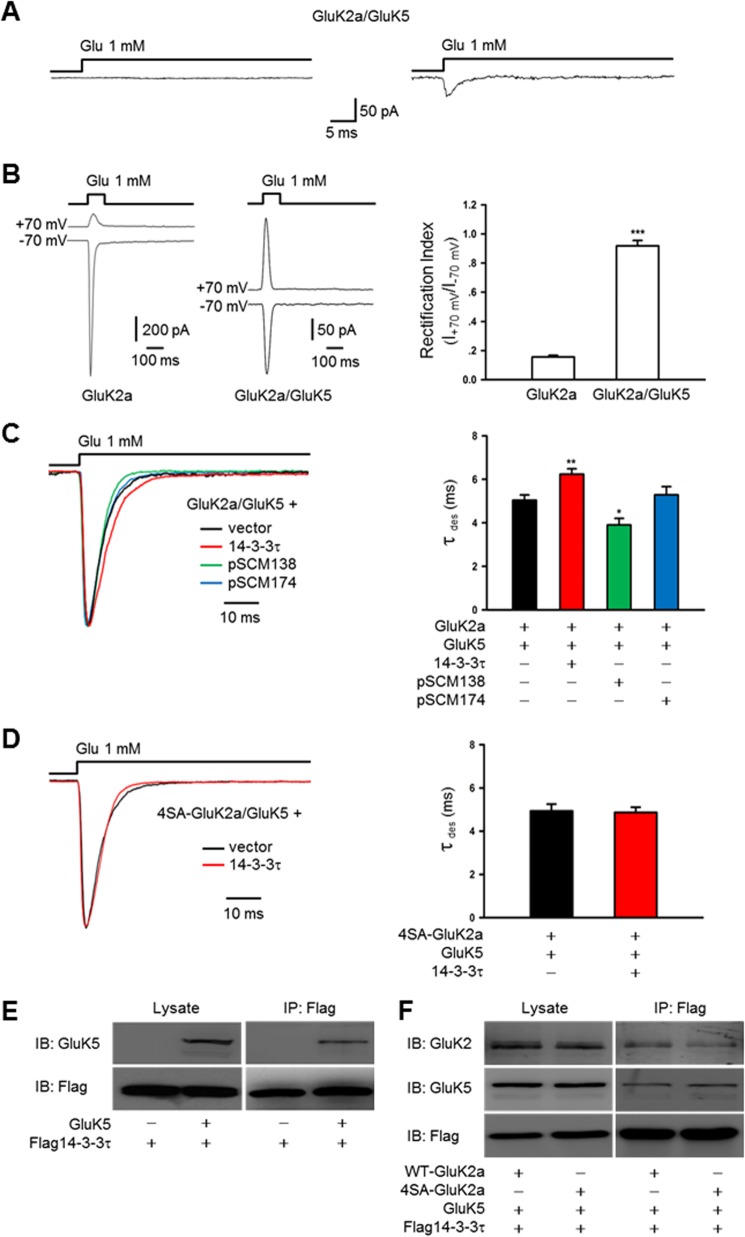
To record recombinant KAR-mediated currents, HEK293 cells were transfected with cDNAs of KAR subunits (GluK2a, GluK2b, GluK5, or 4SA-GluK2a), either alone or together with 14-3-3 isoforms, pSCM138 or pSCM174. At least 48 h after transfection, currents were recorded from transfected cells identified by tagged fluorescent protein (enhanced GFP, enhanced BFP, or enhanced YFP) in lifted whole-cell or outside-out patch configuration with an EPC10 amplifier (HEKA) at room temperature (22–25 °C). Recording microelectrodes with resistances of 3–5 megohms were pulled from thin-walled borosilicate glass with inner filament (Sutter Instrument) and filled with the intracellular solution containing 135 mm CsCl, 10 mm EGTA, 4 mm Mg-ATP, and 10 mm HEPES (pH adjusted to 7.2 with CsOH, 290–295 mosm). The coverslips were placed in a perfusion chamber and continuously perfused with a standard extracellular solution containing 150 mm NaCl, 5 mm KCl, 10 mm HEPES, 1 mm MgCl2, 2 mm CaCl2, and 10 mm glucose (pH adjusted to 7.35 with NaOH, 305–315 mosm). To ensure cell dialysis, measurements were obtained at least 3–5 min after the whole-cell configuration was established. Channel currents were recorded in outside-out patch or lifted whole-cell configuration at a holding potential of −60 mV, and signals were analog low pass-filtered at 2.9 kHz (four-pole Bessel-type, 23 db) and sampled at 20 kHz. In experiments involving recording currents from heteromeric GluK2a/GluK5 receptors, we utilized the following established criteria (41, 42). First, the cells were cotransfected with GluK2a and GluK5 cDNAs in a 1:3 ratio, which yielded a largely homogeneous population of GluK2a/GluK5 heteromeric KARs at the cell membrane surface (50); Second, glutamate pulses (1 mm, 100 ms) delivered to GluK2a/GluK5 cotransfected cells elicited current responses of smaller amplitude than that of GluK2a singly transfected cells. Third, the rectification index of heteromeric receptors is greater than 0.9 (Fig. 6B).
FIGURE 6.
14-3-3 proteins modulate the kinetics of heteromeric GluK2a-containing receptors in a heterologous system. A, GluK2a/GluK5 currents recorded in outside-out configuration from cotransfected HEK293 cells. In our experimental conditions, most of the patches showed either no detectable current (left panel, n = 20) or small currents ranging from 30 to 50 pA (right panel, n = 3). B, representative whole-cell currents recorded from HEK293 cells transfected with GluK2a, alone or together with GluK5 subunits. GluK2a and GluK5 cDNAs were transfected at a ratio of 1:3. Glutamate pulses (1 mm, 100 ms) delivered to cells expressing homomeric GluK2 subunits elicited current responses of large amplitude at a holding potential of −70 mV, whereas the peak current amplitude was considerably smaller in GluK2a/GluK5-cotransfected cells (left panel). The rectification index (ratio between the current amplitudes at membrane potential +70 mV and −70 mV) of currents recorded from cells transfected with GluK2a alone or together with GluK5. Quantification data show means ± S.E. of GluK2a (n = 9) and GluK2a/GluK5 (n = 11) groups. ***, p < 0.001 (right panel). For recordings from GluK2a/GluK5-cotransfected cells, data with rectification index of <0.9 were discarded. C, representative whole-cell current traces from HEK293 cells cotransfected with GluK2a and GluK5 subunits, in combination with other proteins as indicated. Desensitization kinetics of heteromeric GluK2a/GluK5 receptors is also slowed by exogenous 14-3-3τ (red trace) and accelerated by cotransfection of pSCM138 (green trace) but not pSCM174 (blue trace). Right panel summarizes group data for vector- (n = 14), 14-3-3τ- (n = 28), pSCM138- (n = 8), and pSCM174 (n = 8)-cotransfected cells. Data represent mean ± S.E.; statistical significance is denoted as follows: *, p < 0.05; **, p < 0.01. D, representative whole-cell current traces from HEK293 cells cotransfected with 4SA-GluK2a mutant and GluK5 in combination without or with 14-3-3τ. Cotransfection of 14-3-3τ (red trace) has no effect on the desensitization of heteromeric 4SA-GluK2a/GluK5 receptors. Right panel shows the summary data (n = 12 and 13 for without and with 14-3-3τ, respectively). E and F, 14-3-3τ associates with GluK5 subunits. Western blot analyses of immunoprecipitates (IP) and cell lysates from HEK293 cells transfected with 14-3-3τ and other proteins as indicated. E, 14-3-3τ and GluK5 subunits coimmunoprecipitate in cotransfected cells. F, interaction between 14-3-3τ and GluK5 subunits is not dependent on 14-3-3 binding to the GluK2a subunit. The results shown are representative of at least three independent experiments. IB, immunoblot.
Drug Application
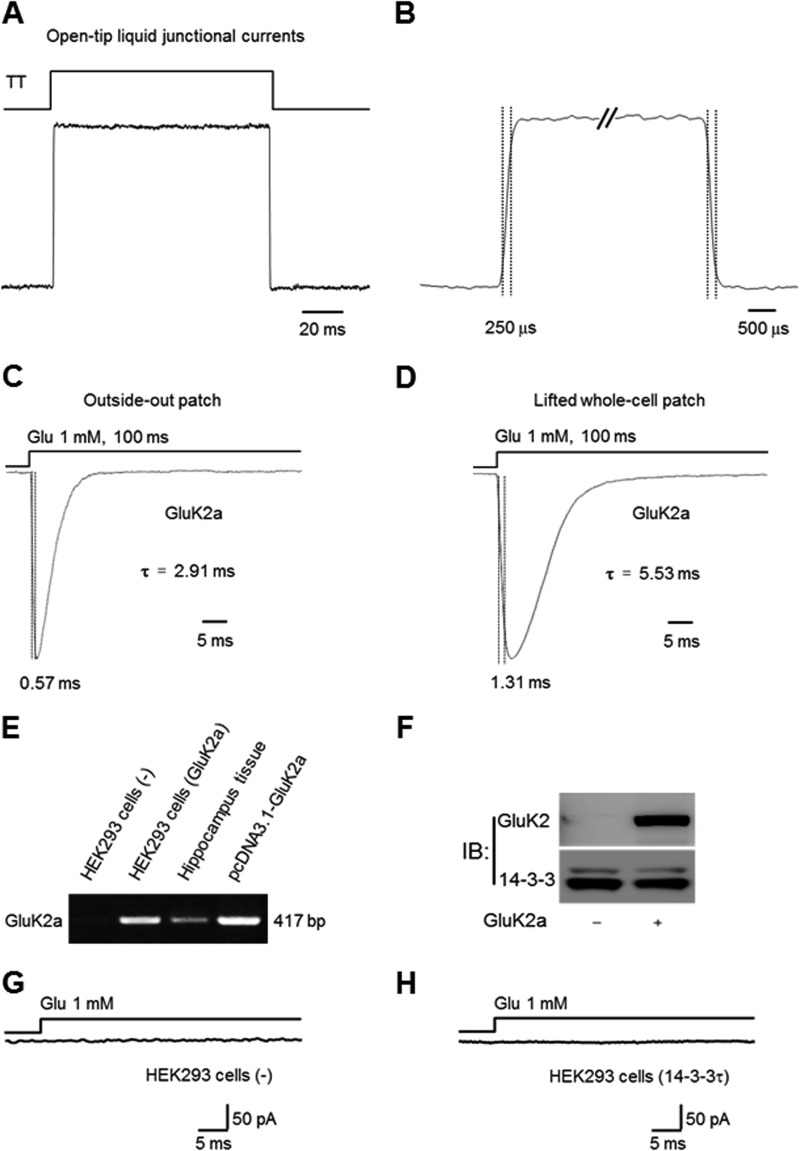
Glutamate was applied to HEK293 cells using a fast step perfusion system (Warner Instruments, SF-77B) through a θ-tube with a tip diameter about 200 μm. Patches of outside-out or lifted whole cells were positioned near the solution interface, and the interface was moved by applying voltage across digital to analog converter output on EPC10 amplifier to SF-77B. The open-tip recordings of the liquid junction potentials revealed that the 10–90% exchange of solution occurred within 300 μs. For lifted whole-cell recording, HEK293 cells were lifted from the coverslip and placed within 100 μm of the tip of a θ tube (41, 43, 44). To minimize the solution exchange time in the whole-cell configuration, currents were recorded from small HEK293 cells, showing a membrane capacitance of ∼8–16 picofarads. Agonist was applied for 100 ms at 60-s intervals to monitor the peak current amplitude and desensitization time constant. Desensitization time constants were determined by adjusting cursors in FITMASTER software (HEKA Electronik) to find the best (visual) fit of the current decay to single exponential function (Fig. 3, A–D) (45).
FIGURE 3.
Characterization of our rapid solution exchange and heterologous expression systems. A, open tip recordings were used to assess the solution exchange time of our θ-glass application pipette system. The solution is applied using SF-77B perfusion step controlled by PatchMaster software through digital to analog converter output in EPC10 amplifier. The 10–90% rise time of the liquid junction currents (resulting from the application of a 1:10 diluted extracellular solution) is ∼250 μs (B). A and B are the same current trace shown in different time scales. C and D, representative traces of outside-out (C) and whole-cell (D) currents recorded from HEK293 cells expressing homomeric GluK2a receptors, evoked by rapid application of glutamate using the rapid solution exchange system. The time courses of current onset and desensitization are comparable with data reported by other laboratories. E–H, no endogenous GluK2a expression in HEK293 cells. E, GluK2a mRNA was not detected by RT-PCR assay in nontransfected HEK293 cells (left lane). F, Western blot analyses confirm the lack of endogenous GluK2a protein in nontransfected HEK293 cells (left lane). G and H, in the absence of heterologously expressed GluK2a, no whole-cell current was evoked by 1 mm glutamate in either vector (G) or 14-3-3τ (H)-transfected HEK293 cells. IB, immunoblot.
Synaptic Electrophysiology
Acute hippocampal slices (250 μm) were prepared from male 14-3-3 FKO mice and their wild-type (WT) littermates (4–6 weeks old). The procedures for preparing brain slices were as described previously (32). Briefly, the mouse was sacrificed after being deeply anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg). The whole brain was removed and placed in iced and 95% O2, 5% CO2 oxygenated cutting medium including the following (in mm): 230 sucrose, 2.5 KCl, 10 MgSO4, 1.25 Na2HPO4, 26 NaHCO3, 0.5 CaCl2, 10 d-glucose. The sagittal or coronal slices were cut with a VT1200S vibratome (Leica) and moved to the artificial cerebrospinal fluid (ACSF) saturated with 95% O2, 5% CO2. The ACSF contained the following (in mm): 124 NaCl, 5 KCl, 2.5 CaCl2, 1.3 MgSO4, 1.2 KH2PO4, 26 NaHCO3, and 10 glucose. The slices were incubated at 37 °C for at least 45 min in oxygenated ACSF before use.
Brain slice was transferred to the recording chamber and kept in place with a slice anchor (Warner Instruments) under a microscope (ECLIPSE FN1, Nikon) equipped with infrared differential interference contrast and water immersion objective for visualization of neurons in live tissue. The slice in the recording chamber was continuously perfused with oxygenated ACSF (2 ml/min) at room temperature. The recording solution contained the following (in mm): 145 potassium gluconate, 7.5 KCl, 9 NaCl, 1 MgSO4, 10 HEPES, 0.2 EGTA, 2 sodium ATP, 0.25 sodium GTP, adjusted to pH 7.4 with KOH, osmolality 290–300 mosm. The concentric bipolar stimulating electrode was placed in the dentate gyrus cell body layer to activate mossy fibers or in the CA3 striatum radiatum to activate associational commissural fibers. The single stimulating pulse (100 μs duration) was produced by Master-8 stimulator (AMPI, Israel) with the frequency of 0.1 Hz. As reported previously (32), to assess the contribution of mossy fiber-CA3 transmission, 1 μm (1R,2R)-3-[(1S)-1-amino-2-hydroxy-2-oxoethyl]cyclopropane-1,2-dicarboxylic acid (DCG-IV), a group II mGluR agonist that selectively blocks mossy fiber synaptic transmission, was applied at the end of every experiment, and the data were accepted only if synaptic responses were reduced by more than 90%. The synaptic response remaining in DCG-IV was then subtracted from all previous responses before further analysis to isolate mossy fiber-specific synaptic activity. KAR-EPSCs were monitored in the presence of the selective AMPAR antagonist GYKI 53655 (30 μm), and cells were voltage-clamped to −60 mV or +30 mV. The mossy fiber-CA3 EPSCs were evoked by single stimulation in the dentate gyrus. NMDAR-EPSCs were monitored in 10 μm 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxaline-2,3-dione, 100 μm picrotoxin, and 3 μm CGP55845 while voltage-clamping to +30 mV. Given the variability and potential polysynaptic contamination of AMPAR-mediated EPSCs at mossy fiber-CA3 synapses (46), AMPAR-mediated transmission was evaluated by activating associational-commissural CA3 synapses (Vh = −60 mV) in the presence of 50 μm D-AP5, 100 μm picrotoxin, and 3 μm CGP55845. Analysis of decay kinetics was carried out by fitting trace averages of 20 sweeps with a single exponential. The EPSCs were recorded with a MultiClamp 700B amplifier (Molecular Devices) and filtered at 2 kHz with a low pass filter, and data were digitized at 10 kHz and stored on line using the pClamp 10 software.
Statistical Analyses
Data are expressed as mean ± S.E. with statistical significance assessed by Student's t test for two group comparisons or one-way analysis of variance tests for multiple comparisons. The value of *, p < 0.05, was considered statistically significant difference.
RESULTS
14-3-3 Proteins Associate with GluK2a-containing KARs
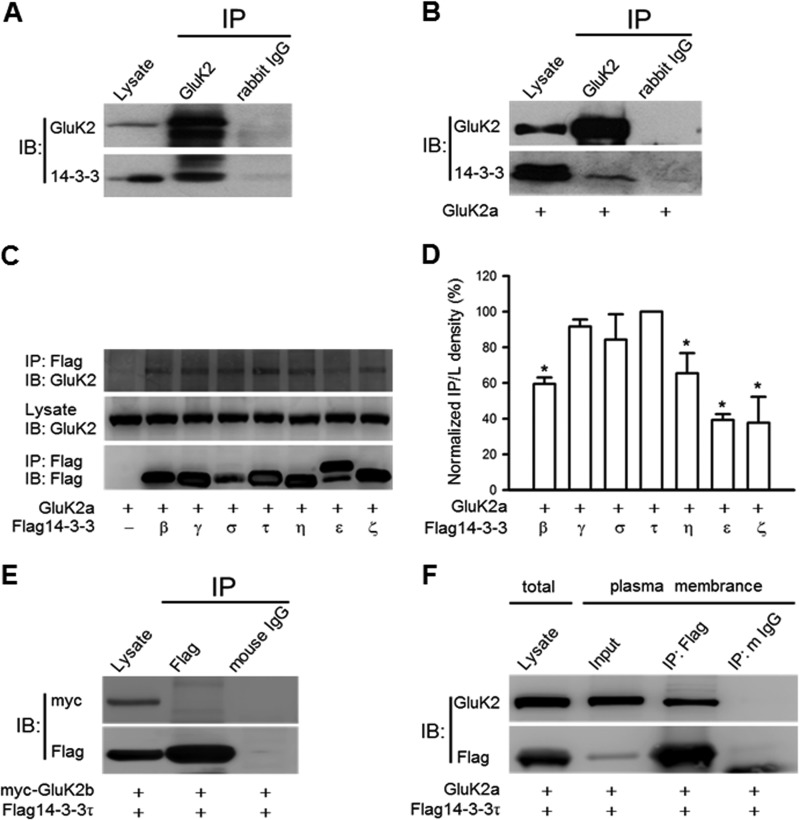
A previous proteomic analysis has identified 14-3-3γ as a putative binding partner of the GluK2a subunit (26). Here, we directly assessed the interaction of GluK2-containing receptors with 14-3-3 proteins using coimmunoprecipitation approaches. As shown in Fig. 1A, 14-3-3 proteins and GluK2 subunits were effectively coimmunoprecipitated from rat hippocampal homogenates, indicating the existence of a GluK2-containing KAR and 14-3-3 protein complex in the brain (Fig. 1A). This was further supported by our finding that exogenously expressed GluK2a coimmunoprecipitated with endogenous 14-3-3 proteins in HEK293 cells (Fig. 1B). In contrast, 14-3-3 proteins were unable to coimmunoprecipitate with GluK2b in cotransfected HEK293 cells (Fig. 1E), which is in agreement with a published study (26).
FIGURE 1.
14-3-3 proteins associate with GluK2a-containing KARs. A, GluK2 subunits coimmunoprecipitate with 14-3-3 proteins from rat hippocampal lysates. B, GluK2 coimmunoprecipitates with endogenous 14-3-3 proteins in HEK293 cells transfected with GluK2a cDNA. The anti-pan 14-3-3 antibody was used for immunoblotting (IB) in A and B; images shown are representative Western blots from three independent experiments. C, coimmunoprecipitation of GluK2a and different FLAG14-3-3 isoforms. The identities of the transfected cDNAs are indicated below each lane. The exogenously expressed 14-3-3 proteins were immunoprecipitated (IP) with the FLAG antibody. D, normalized levels of coimmunoprecipitation between GluK2a and various 14-3-3 isoforms. These values were determined by measuring the relative intensity of immunoprecipitated bands and their corresponding lysate (L) bands on Western blots and then normalized to and compared with that of 14-3-3τ (n = 3, *, p < 0.05). E, no interaction between GluK2b and 14-3-3τ is detected by coimmunoprecipitation in cotransfected HEK293 cells. F, GluK2a and FLAG14-3-3τ coimmunoprecipitate in the crude plasma membrane fraction prepared from cotransfected HEK293 cells. Image shown is representative Western blots from three independent experiments. Data represent mean ± S.E.; IB, antibody used for immunoblot analysis; IP, antibody used for immunoprecipitation.
To determine isoform-specific interactions of 14-3-3 proteins with the GluK2a subunit, we carried out coimmunoprecipitation and Western blot analyses in HEK293 cells transiently coexpressing GluK2a and different 14-3-3 isoforms. GluK2a subunits were present in 14-3-3 immunoprecipitates from all 14-3-3-cotransfected cells but not from vector-cotransfected cells (Fig. 1C), demonstrating that these 14-3-3 isoforms are all capable of binding to GluK2a. However, quantification of Western blots revealed differences in the extent of interactions between GluK2a and various isoforms of 14-3-3 proteins (Fig. 1D). Among them, 14-3-3τ exhibited quantitatively the strongest binding to GluK2a, and this particular isoform was thus utilized in most of the subsequent experiments.
Furthermore, we asked where in the cell the interaction between KARs and 14-3-3 takes place, as this is a question with important functional implications. To address this, we performed the coimmunoprecipitation experiment with crude membranes prepared from cells coexpressing 14-3-3 and GluK2a subunits. As shown in Fig. 1F, 14-3-3 coimmunoprecipitated GluK2a in the membrane fraction, demonstrating that this protein/protein interaction can happen after trafficking of the KARs to the plasma membrane.
14-3-3 Binds to GluK2a C-terminal Tail in a Phosphorylation-dependent Manner
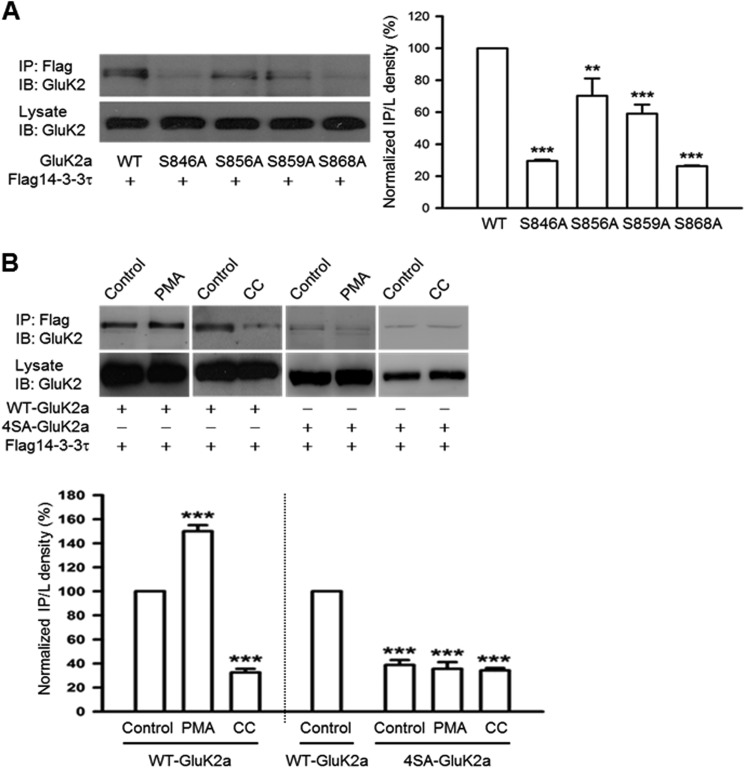
14-3-3 generally interacts with its target proteins through binding to consensus phosphoserine-containing motifs (35, 47, 48). To identify potential 14-3-3-binding sites in the GluK2a subunit, we introduced site-directed mutations to each of the four serine residues (S846A, S856A, S859A, and S868A) located at the GluK2a C-terminal tail, a region that was previously reported to be important for 14-3-3 binding (26). As assessed by coimmunoprecipitation and Western blot analysis in a heterologous expression system, the interaction between 14-3-3 and GluK2a was significantly attenuated in all four mutants compared with the wild-type (WT) GluK2a (Fig. 2A). Based on quantification of relative band intensity for 14-3-3-coimmunoprecipitated GluK2a, mutations of Ser-846 and Ser-868 residues at the GluK2a subunit resulted in more pronounced reductions in 14-3-3 binding than the S856A and S859A mutations (Fig. 2A, bar graph). Thus, the GluK2a C-terminal serine residues, especially Ser-846 and Ser-868, play a significant role in mediating 14-3-3 binding.
FIGURE 2.
14-3-3 proteins bind to the C terminus of GluK2a in a PKC phosphorylation-dependent manner. Western blot analyses of immunoprecipitates and cell lysates from HEK293 cells were transfected with 14-3-3τ and other proteins as indicated. A, mutation of single serine residues at the GluK2a C terminus impairs interaction between 14-3-3 and GluK2a. Quantification of Western blots indicates that the S846A and S868A mutants have more significant reduction in 14-3-3 binding (n = 3, **, p < 0.01; ***, p < 0.001, comparison with WT) (right panel). B, coimmunoprecipitation of 14-3-3 and WT GluK2a is enhanced by PMA (5 μm) (a PKC activator) and reduced by calphostin C (1 μm) (a PKC inhibitor, CC). Right panel shows that the mutant 4SA-GluK2a subunit has impaired 14-3-3 binding, which is not affected by either PMA or calphostin C treatment. Lower panel summarizes quantified Western blot results for groups data of PMA (n = 4), CC (n = 3) in WT GluK2a, and PMA (n = 3), CC (n = 3) in 4SA-GluK2a. Data represent mean ± S.E., ***, p < 0.001, comparison with WT control. IB, immunoblot; IP, immunoprecipitation.
Interestingly, Ser-846 and Ser-868 have previously been identified as the major phosphorylation sites of protein kinase C (PKC) within the GluK2a C-terminal tail (49). To determine whether the GluK2a and 14-3-3 interaction is regulated by PKC phosphorylation, we treated the 14-3-3τ and GluK2a cotransfected HEK293 cells with either a PKC activator (PMA) or a PKC inhibitor (calphostin C), and we assessed their effects on the GluK2a and 14-3-3 interaction. As shown in Fig. 2B, 14-3-3 binding to the WT GluK2a was markedly enhanced by PMA while notably inhibited by calphostin C. Quantification of Western blots indicated that these changes were statistically significant (Fig. 2B).
Next, we asked whether the effect of PKC on the 14-3-3/GluK2a interaction is mediated by phosphorylation of the serine residues at the GluK2a C-terminal tail. Here, we constructed the mutant 4SA-GluK2a (S846A, S856A, S859A, and S868A). As expected, 14-3-3 binding was nearly abolished by the 4SA-GluK2a mutation (Fig. 2B). Moreover, activation of PKC by PMA treatment or inhibition of PKC by calphostin C did not result in any enhancement or reduction of 14-3-3 binding to the mutant 4SA-GluK2a subunit (Fig. 2B). Taken together, these data clearly demonstrated that 14-3-3 binding is regulated by PKC-mediated phosphorylation of serine residues at the GluK2a C-terminal tail.
14-3-3 Modulates Desensitization Kinetics of GluK2a Homomeric Receptors
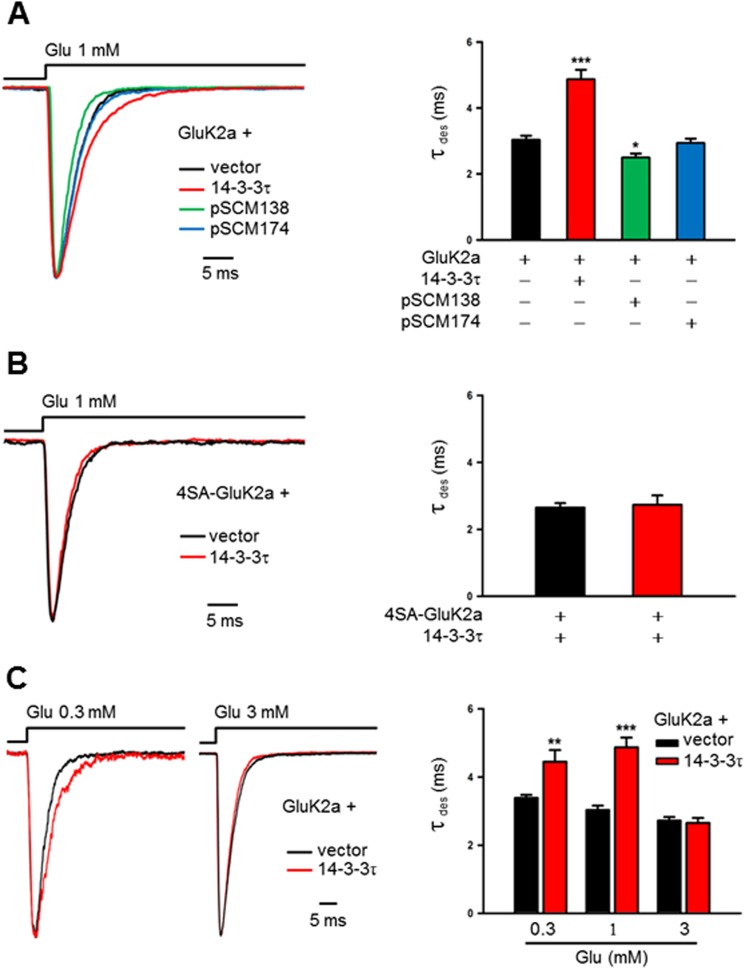
To determine whether 14-3-3 binding modulates the biophysical properties of GluK2a receptors, we recorded homomeric GluK2a receptor currents from transfected HEK293 cells and assessed the changes in channel properties induced by exogenously expressed 14-3-3 proteins. In these experiments, GluK2a currents were elicited by 1 mm glutamate in outside-out patches at a holding potential of −60 mV. Note that no current was evoked by this concentration of glutamate in the absence of heterologously expressed GluK2a, corresponding to a lack of endogenous GluK2a transcript or protein in these cells (Fig. 3, E–H). Nevertheless, coexpression 14-3-3τ resulted in a significant slowing of desensitization kinetics of GluK2a (Fig. 4A). As analyzed by a single exponential fitting of the GluK2a currents, the desensitization time constant (τdes) was significantly larger in cells cotransfected with 14-3-3τ than the corresponding values for GluK2a alone (Fig. 4A, bar graph). Consistent with our biochemical results, coexpression of other 14-3-3 isoforms also slowed desensitization of GluK2a homomeric receptors, but 14-3-3τ had the biggest quantitative effect (data not shown).
FIGURE 4.
14-3-3 proteins modulate the kinetics of homomeric GluK2a receptors in a heterologous system. A and B, GluK2a current traces are evoked by glutamate (1 mm, 100 ms) in outside-out configuration from HEK293 cells cotransfected with KAR subunits and other proteins as indicated. Desensitization time constants (τdes) are determined by single exponential fits to the decaying phase of the currents. A, representative current traces recorded from outside-out patches in HEK293 cells transfected with GluK2a subunits, together with either corresponding vector (dark trace), 14-3-3τ (red trace), pSCM138 (green trace), or pSCM174 (blue trace). Cotransfection of pSCM138, but not pSCM174, significantly accelerates the desensitization of homomeric GluK2a receptors, whereas cotransfection 14-3-3τ significantly slows the desensitization of homomeric GluK2a receptors. Right panel summarizing group data for vector (n = 12), 14-3-3τ (n = 10), pSCM138 (n = 9), and pSCM174 (n = 11) cotransfected cells. Data represent mean ± S.E., *, p < 0.05; ***, p < 0.001. B, representative current traces from outside-out patches in HEK293 cells transfected with the 14-3-3-binding deficient mutant 4SA-GluK2a, together with either the vector (dark trace) or 14-3-3τ (red trace). Desensitization of the mutant GluK2a receptor is not affected by coexpressed 14-3-3τ. Right panel shows the summary data (n = 10 and 9 for without and with 14-3-3τ, respectively). C, representative currents evoked by either 0.3 or 3 mm glutamate on outside-out patches from HEK293 cells transfected with GluK2a subunits, together with either corresponding vector (dark trace) or 14-3-3τ (red trace). Cotransfection of 14-3-3τ slows the desensitization of homomeric GluK2a currents evoked by low (0.3 mm) but not the high (3 mm) concentration of glutamate. Right panel shows the summary data (0.3 mm group, n = 11 and 9; 1 mm group, n = 12 and 10; 3 mm group, n = 16 and 15 for without and with 14-3-3τ, respectively). Data represent mean ± S.E., **, p < 0.01; ***, p < 0.001.
Next, we tested whether 14-3-3 binding is necessary for its modulation of GluK2a homomeric receptors. Our strategy was to cotransfect cells with pSCM138, encoding the YFP-fused difopein that binds to 14-3-3 proteins with a very high affinity and thus disrupting 14-3-3s' interaction with their endogenous partners (38, 39). As shown in Fig. 4A, cotransfection of pSCM138, but not its inactive form pSCM174, significantly accelerated desensitization kinetics of GluK2a homomeric receptors. Quantitatively, τdes of GluK2a currents in pSCM138 cotransfected cells was significantly smaller than that of vector or pSCM174-cotransfected cells, revealing the modulation conferred by endogenous 14-3-3 proteins in HEK293 cells.
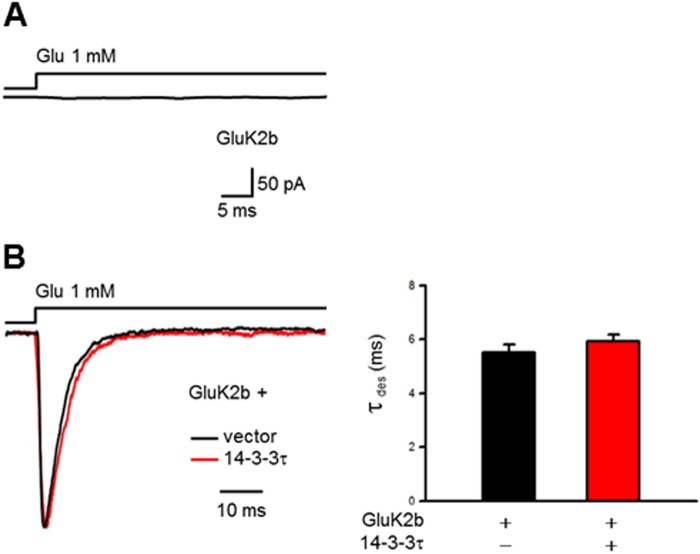
To further define the role of 14-3-3/GluK2a interactions in regulating the kinetics of GluK2a homomeric receptors, we recorded channel currents in outside-out patches from cells expressing the mutant 4SA-GluK2a receptor that is deficient in 14-3-3 binding. In agreement with our biochemical data (Fig. 2B), the desensitization kinetics of 4SA-GluK2a homomeric receptors was not significantly affected by exogenously expressed 14-3-3τ in HEK293 cells (Fig. 4B). Moreover, we assessed the effect of 14-3-3 on the GluK2b subunit that did not interact with 14-3-3 proteins (Fig. 1E). Because of the poor surface expression of GluK2b homomeric receptors (26), no current was elicited by 1 mm glutamate in outside-out patches from cells expressing GluK2b subunits (Fig. 5A). Thus, we recorded the homomeric GluK2b currents in lifted whole-cell configuration and found that coexpression of 14-3-3τ did not affect the desensitization kinetics of GluK2b homomeric receptors (Fig. 5B). Together, these results demonstrated that 14-3-3 modulates desensitization kinetics of GluK2a homomeric receptors through its interaction with GluK2a.
FIGURE 5.
14-3-3 proteins do not modulate the kinetics of homomeric GluK2b receptors in a heterologous system. A, in outside-out configuration, no detectable current was evoked by glutamate (1 mm, 100 ms) from HEK293 cells expressing GluK2b subunits. Shown is a representative trace of 10 independent experiments. B, representative whole-cell current traces from HEK293 cells transfected with GluK2b, together with either corresponding vector (dark trace) or 14-3-3τ (red trace). Cotransfection of 14-3-3τ (red trace) has no effect on the desensitization of homomeric GluK2b receptors. Right panel shows the summary data for control (n = 7) and 14-3-3τ (n = 10) group, respectively.
To explore the potential underlying mechanisms, we examined the impact of 14-3-3 on the desensitization kinetics of GluK2a receptors using different concentrations of glutamate. When evoked by 0.3 mm glutamate, a concentration that is lower than the EC50 (∼0.5 mm) of GluK2a homomeric receptors (41, 63), desensitization of GluK2a currents was significantly slowed by coexpressed 14-3-3 (Fig. 4C, left panel). Interestingly, no such effect of exogenous 14-3-3 was observed at a glutamate concentration of 3 mm (Fig. 4C, right panel) that reaches the saturation level of the dose-response curve (41, 63). These data fit well with previous publications showing that GluK2a receptors display slow desensitization kinetics only at partially agonist-bound states (41, 63, 64). Under such conditions, gating properties of the GluK2a receptor are likely altered as a result of 14-3-3 binding.
Effect of 14-3-3 on Desensitization Kinetics of GluK2a-containing Heteromeric Receptors
KARs composed of GluK2a and GluK5 are the most abundant KAR subtype in the brain. We therefore assessed the effect of 14-3-3 on the kinetics of receptors formed by GluK2a/GluK5 heteromers. In heterologous cells, cotransfection of GluK2a and GluK5 cDNAs at an appropriate ratio is expected to yield surface expression of functional GluK2a/GluK5 heteromeric receptors (41, 50). However, the amplitude of heteromeric GluK2a/GluK5 receptor currents is considerably smaller than that of homomeric GluK2a receptors (41). Under our experimental conditions, most of the outside-out patches showed no detectable current (Fig. 6A). Thus, we examined the properties of GluK2a/GluK5 currents in lifted whole-cell configuration. To ensure that recorded whole-cell currents were primarily derived from heteromeric GluK2a/GluK5 receptors, we utilized the established criteria to distinguish between homomeric and heteromeric receptor currents (Fig. 6B) (41, 42). Coexpression of 14-3-3τ significantly slowed desensitization of heteromeric GluK2a/GluK5 receptors (Fig. 6C). Conversely, desensitization kinetics of GluK2a/GluK5 heteromers was accelerated by antagonizing 14-3-3 binding in cells with cotransfection of pSCM138 (Fig. 6C, bar graph). Thus, 14-3-3 also modulated the desensitization kinetics of GluK2a-containing heteromeric receptors.
14-3-3 was previously shown to interact with GluK5 subunits as well (37). This was confirmed by our biochemical data showing a direct binding of 14-3-3 to GluK5 in the HEK293 expression system (Fig. 6, E and F). To dissect the biochemical basis underlying 14-3-3-dependent modulation of the GluK2a/GluK5 heteromers, we recorded whole-cell currents in cells expressing heteromeric receptors composed of GluK5 and the 14-3-3-binding deficient mutant 4SA-GluK2a. As shown in Fig. 6D, 4SA-GluK2a/GluK5 heteromers desensitized similarly in cells with or without exogenously expressed 14-3-3τ proteins, suggesting that 14-3-3 binding to GluK2a plays a critical role in regulating desensitization kinetics of GluK2a-containing receptors.
14-3-3-dependent Modulation of Homomeric GluK2a Receptors Is Regulated by PKC Phosphorylation
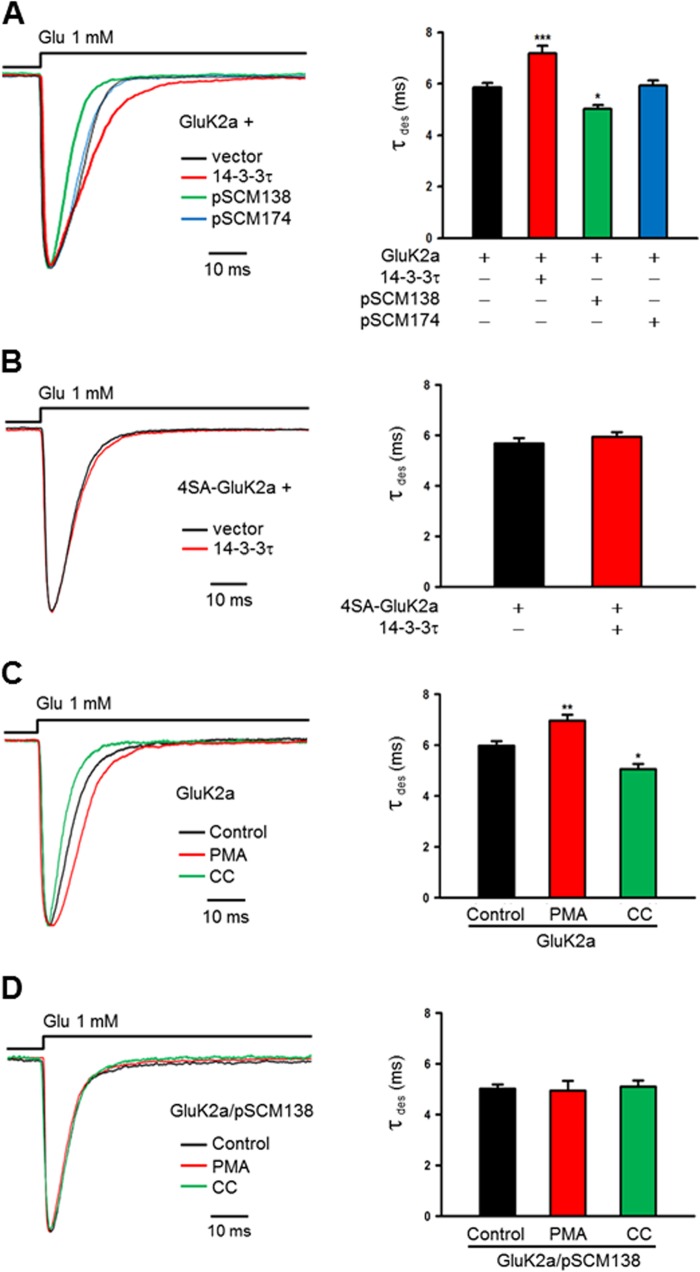
Given that 14-3-3 binding was dependent on PKC phosphorylation of GluK2a subunits, we sought to test whether PKC phosphorylation could dynamically regulate 14-3-3-dependent modulation of GluK2a receptors. To maintain the integrity of cellular contents, we conducted this set of electrophysiological experiments in a lifted whole-cell configuration. At first, we confirmed that the desensitization kinetics of whole-cell WT GluK2a homomeric receptor currents was significantly modulated by altering cellular 14-3-3 levels (Fig. 7A), whereas the whole-cell currents of the mutant 4SA-GluK2a receptors were not affected by exogenously expressed 14-3-3 (Fig. 7B).
FIGURE 7.
PKC regulates the 14-3-3-dependent modulation of GluK2a receptors. A and B, whole-cell current traces are evoked by glutamate (1 mm, 100 ms) from HEK293 cells cotransfected with KAR subunits and other proteins as indicated. Time constant of desensitization (τdes) is determined by single exponential fits to the decaying phase of the currents. A, representative whole-cell currents recorded from HEK293 cells transfected with GluK2a subunits, together with either corresponding vector (dark trace), 14-3-3τ (red trace), pSCM138 (green trace), or pSCM174 (blue trace). Cotransfection of pSCM138, but not pSCM174, significantly accelerates the desensitization of homomeric GluK2a receptors, whereas cotransfection 14-3-3τ significantly slows the desensitization of homomeric GluK2a receptors. Right panel summarizing group data for vector- (n = 9), 14-3-3τ- (n = 15), pSCM138- (n = 8), and pSCM174 (n = 14)-cotransfected cells. Data represent mean ± S.E., *, p < 0.05; ***, p < 0.001. B, representative whole-cell current traces from HEK293 cells transfected with the 14-3-3-binding deficient mutant 4SA-GluK2a, together with either the vector (dark trace) or 14-3-3τ (red trace). Desensitization of the mutant GluK2 receptor is not affected by coexpressed 14-3-3τ. Right panel shows the summary data (n = 10 and 8 for without and with 14-3-3τ, respectively). C, representative whole-cell current traces of GluK2a homomeric receptors from transfected HEK293 cells. PKC activator PMA (5 μm) or inhibitor CC (1 μm) was loaded into cells through the patch electrode. The decay time constant of GluK2a channels is substantially slowed down by PMA (red trace) and notably accelerated by CC (green trace). Summarized data are shown in the right panel for control (n = 11), PMA (n = 19), and CC (n = 15) groups. Data represent mean ± S.E.; statistical significance is denoted as follows: *, p < 0.05; **, p < 0.01. D, representative whole-cell current traces from HEK293 cells cotransfected with GluK2a subunits and pSCM138. The decay kinetics of GluK2 is unchanged by either PMA or CC treatment when 14-3-3 bindings are inhibited by transfection of pSCM138. The right panel show the summarized results for control (n = 8), PMA (n = 8), CC (n = 9) groups. Data represent mean ± S.E.
Next, we loaded either PMA or calphostin C through the patch electrode into HEK293 cells expressing homomeric GluK2a receptors, and we assessed their effect on channel desensitization kinetics. Compared with the control, desensitization of GluK2a was slowed by PMA and accelerated by calphostin C (Fig. 7C). Statistically, there were significant differences in τdes between control and PMA- or calphostin C-treated groups (Fig. 7C, bar graph).
We also examined the effect of PMA and calphostin C on the kinetics of GluK2a homomers in HEK293 cells cotransfected with pSCM138, in which 14-3-3/ligand interactions were inhibited. As analyzed by the decay kinetics of IGlu, desensitization of GluK2a receptors remained unchanged after either PMA or calphostin C treatment (Fig. 7D). Collectively, these results demonstrated that PKC modulated desensitization kinetics of GluK2a receptors by regulating phosphorylation-dependent binding of 14-3-3 to GluK2a.
14-3-3 Proteins Modulate KAR-mediated EPSCs in the Hippocampus
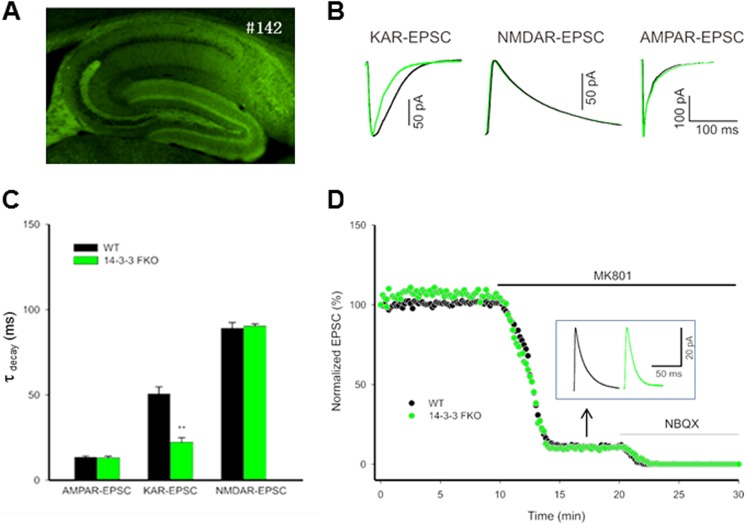
14-3-3 proteins are enriched at synapses (38, 51). We therefore asked whether 14-3-3 has a role in regulating decay kinetics of KAR-mediated EPSCs. These experiments utilized our newly generated 14-3-3 FKO mice, in which the 14-3-3 binding antagonist (YFP-fused difopein) was transgenically expressed in the brain. Although the thy-1-directed transgene expression patterns varied among different founders,5 this study focused on one of the founders (number 142) that has extensive transgene expression in the dentate gyrus and CA3 regions of the hippocampus (Fig. 8A). We recorded KAR-EPSCs in CA3 pyramidal cells on acute mouse hippocampal slices, and we found that the KAR-EPSCs decay kinetics in the number 142 founder of 14-3-3 FKO mice were noticeably faster than that of their WT littermates (Fig. 8B). As assessed by decay time constants (τdecay) of KAR-EPSCs, there was a significant difference between the 14-3-3 FKO and WT mice (Fig. 8C). In contrast, the decay kinetics of either AMPAR- or NMDAR-mediated EPSCs was not altered in this line of 14-3-3 FKO mice (Fig. 8, B and C). In addition, we recorded EPSCs from CA3 pyramidal cells at a positive holding potential (+30 mV) in the presence of the AMPAR antagonist GYKI 53655 (30 μm). After blocking the NMDAR-mediated component of mf-CA3 EPSCs by MK-801, the synaptic current remaining was considered as KAR-mediated EPSCs component, because it was sensitive to the KAR antagonist 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxaline-2,3-dione (NBQX) (Fig. 8D). Consistently, the KAR-EPSCs recorded from the 14-3-3 FKO mice decayed faster than that of WT mice. However, the amplitude and relative contribution (versus NMDAR) of KAR-EPSCs were not different between the 14-3-3 FKO mice and their WT littermates (Fig. 8D). Thus, these results provided evidence that 14-3-3 specifically modulated the kinetics of KAR-EPSCs in the brain.
FIGURE 8.
14-3-3 proteins modulate the kinetics of KAR-mediated synaptic currents. A, fluorescence image of hippocampal slice from the 14-3-3 FKO mice. #142 refers to the founder line that has extensive expression of the transgene (YFP-fused R18 dimer) in specific regions of the hippocampus. B, representative normalized KAR-, NMDAR-, and AMPAR-mediated EPSCs recorded from the 14-3-3 FKO mice (green trace) and their wild-type littermates (dark trace). KAR-EPSCs decay faster in 14-3-3 FKO mice compared with the wild type (left), but there is no difference between 14-3-3 FKO mice and their wild-type littermates in NMDAR-mediated EPSCs (elicited by mossy fiber stimulation; middle), and AMPAR-mediated EPSCs (elicited by stimulation of associational/commissural fibers; right). C, comparison of the decay time constants for KAR-, NMDAR-, and AMPAR-mediated EPSCs, measured by single exponential fit to the decaying phase of the currents. (n = 4, **, p < 0.01). D, representative example of KAR- and NMDAR-EPSCs. Mossy fiber-evoked mixed KAR responses were recorded at +30 mV in the presence of AMPAR blocker. After establishing a base line, 50 μm MK-801 was washed in to isolate pure KAR-EPSCs. Inset traces show the residual KAR-EPSCs after MK-801 block. Green and dark traces are from the 14-3-3 FKO mice and their wild-type controls, respectively.
DISCUSSION
Phosphorylation-dependent Interaction of 14-3-3 and GluK2a-containing Receptors
14-3-3γ has been reported to be a potential binding partner of GluK2a subunits by a previous proteomic analysis (26). Our present results extend this finding and provide both in vitro and in vivo evidence for the interaction of 14-3-3 proteins and GluK2a-containing KARs. Additionally, we determined that phosphorylation of several serine residues at the GluK2a C-terminal tail is required for this protein/protein interaction. Although sequences around those serine residues do not match perfectly the consensus 14-3-3-binding motifs, our findings are consistent with the previous report showing that 14-3-3γ interacts specifically with the full-length subunit GluK2a but not its C-terminal splice variant GluK2b, in which only one of the four crucial serine residues (Ser-846) is present (26, 52).
In addition, we demonstrated that the formation of the 14-3-3·GluK2a protein complex is dynamically regulated by altering PKC activities in cells (Figs. 2B and 7C). The fact that 14-3-3 binding can be reduced by acute treatment of a PKC inhibitor (calphostin C) suggests that the 14-3-3/GluK2a interaction is reversible and that the bound 14-3-3 proteins can dissociate from the receptors upon their dephosphorylation. Although it has yet to identify the protein phosphatase(s) involved in this process, our mutagenesis study did show that the PKC effect on 14-3-3/GluK2a interaction was mediated by phosphorylation of the serine residues (Ser-846, Ser-856, Ser-859, and Ser-868) at the GluK2a C-terminal tail. This fits well with previous reports that have identified Ser-846 and Ser-868 as the major phosphorylation sites of PKC within the GluK2a C-terminal tail (49). In fact, our site-directed mutagenesis study showed that mutations of Ser-846 and Ser-868 residues resulted in more pronounced reduction of 14-3-3 binding than mutation of the other two serine residues (Fig. 2A).
14-3-3-dependent Modulation of GluK2a-containing KARs
In this study, we also determined that 14-3-3 binding resulted in the modulation of the biophysical property of GluK2a-containing KARs. In the heterologous system, we showed that 14-3-3 binding slows the desensitization kinetics of GluK2a-containing homomeric and heteromeric receptors, and disrupting 14-3-3/GluK2a interactions accelerates desensitization of GluK2a-containing KARs (Figs. 4 and 6). Moreover, we found that the decay kinetics of KAR-EPSCs is accelerated at hippocampal synapses of the 14-3-3 functional knock-out mice (Fig. 8). Taken together, these results support the hypothesis that 14-3-3 proteins are an important regulator of GluK2a-containing KARs and may contribute to the slow decay kinetics of native KAR-EPSCs.
How does 14-3-3 modulate the property of KARs? Our results indicate that 14-3-3 binding to GluK2a is required for the observed modulation. One key piece of evidence was our mutagenesis study showing that mutations of all four serine residues (S846A, S856A, S859A, and S868A) of the GluK2a C-terminal tail not only abolishes the binding of 14-3-3 proteins to GluK2a but also curtails the modulatory effect of 14-3-3 on the receptor biophysical property (Figs. 2B, 4B, and 6D). Interestingly, another study has previously identified an association of 14-3-3 proteins with the GluK5 subunit (37). Although this observation is confirmed by our coimmunoprecipitation and Western blot analysis (Fig. 6, E and F), we found that coexpression of 14-3-3τ did not alter the kinetics of a mutant form of GluK2a/GluK5 heteromeric receptors composed of wild-type GluK5 and the 14-3-3-binding deficient mutant 4SA-GluK2a (Fig. 6D), suggesting that 14-3-3 binding to GluK5 does not seem to play a major role in modulating the kinetics of GluK2a-containing KARs. However, caution has to be exercised in interpreting these results, because our current experimental approaches only permit the analysis of desensitization, but not deactivation kinetics of GluK2a-containing homomeric and heteromeric receptors expressed in heterologous cells. In fact, previous studies have shown that the decay kinetics KARs, particularly those composed of recombinant GluK2 and GluK5 subunits, is much slower when activated by brief (1–2 ms) agonist exposures than those evoked by longer glutamate applications, indicating that the deactivation property is one of the important gating features of KARs and may contribute to the slow decay kinetics of KAR-EPSCs (41). Therefore, it is important for future studies to further determine the action of 14-3-3 binding to GluK2a and/or GluK5 on deactivation properties of KARs, to gain insight into the molecular mechanism underlying 14-3-3-dependent regulation of KAR-EPSC kinetics.
The modulatory effect of 14-3-3 proteins is similar to that of NETO1 and NETO2, which are newly identified auxiliary subunits of native KARs that regulate the key properties of KARs. In heterologous systems, NETO1 and NETO2 slow deactivation and desensitization of GluK1 through GluK3 homomers and GluK2/GluK5 heteromers (30–33). In genetically engineered mice, loss of NETO1 accelerates the decay kinetics of KAR-EPSCs at mf-CA3 synapses (32, 34). However, the underlying molecular mechanisms for 14-3-3 and NETO proteins appear to be different, because NETO1 and NETO2 bind to GluK2-containing KARs through its extracellular CUB (complement C1r/C1s, Uegf, Bmp1) domains (34), although 14-3-3 proteins interact with GluK2a through its intracellular C-terminal domain. In addition, there are no endogenous NETO proteins in the heterologous systems. This excludes the possibility that the NETO proteins are involved in the 14-3-3-dependent modulation.
Functional Implications of the 14-3-3 and GluK2 Regulatory Protein Complex
Phosphorylation of KARs by PKC and its role in regulation of KAR function have been extensively studied (49, 53–57). Here, we propose a novel mechanism by which PKC regulates properties of GluK2a receptors by controlling phosphorylation-dependent binding of 14-3-3 to GluK2a. This hypothesis is supported by several lines of evidence from our biochemical and electrophysiological studies. Most notably, we found that changes of PKC activities no longer alter the kinetics of GluK2a receptors when 14-3-3/ligand interactions were inhibited by cotransfection with pSCM138 (Fig. 7D).
PKC plays a key role in the initial events of signal transduction and is a major modulator of synaptic transmission in the CNS (58). Phosphorylation of KARs by PKC can regulate KAR-mediated synaptic transmission and is involved in long term depression in the hippocampus (25, 54, 55, 59, 60). However, 14-3-3 proteins are known to play a crucial role in the regulation of synaptic transmission and plasticity. Mutations of 14-3-3ζ exhibit impairments in high frequency transmission fidelity and post-tetanic potentiation (51) and result in a deficit in learning and memory in the fruit fly Drosophila (61). In addition, 14-3-3 is required for a presynaptic form of long term potentiation in cerebellar neurons (62). Previously, we have found that 14-3-3 proteins modulate Cav2.2 channel inactivation through its bindings to the carboxyl tail of the pore-forming a1B subunit (38). Given that both 14-3-3 and KARs are localized at pre- and postsynaptic sites, dynamically regulated 14-3-3-GluK2a protein complexes may contribute to the diverse functions of KARs in synaptic signaling.
This work was supported by National Natural Science Foundation of China Grants 81171230 and 30970937 (to Y. L.), National Basic Research Program of China Grant 2010CB912001 (to Y. L.), Major Research Plan of the National Natural Science Foundation of China Grant 91132303 (to T. L. X. and Y. L.), the Shanghai Pujiang Program Grant 11PJ1406100 (to Y. L.), and Shanghai Committee of Science and Technology Grant 11DZ2260200.
M. Foote, Y. Wu, and Y. Zhou, manuscript in preparation.
- KARs
- kainate receptors
- EPSC
- excitatory postsynaptic current
- 4SA
- all four serine residues mutation to alanine (S846A, S856A, S859A and S868A)
- FKO
- functional-knockout
- PMA
- phorbol 12-myristate 13-acetate
- AMPAR
- AMPA receptor
- NMDAR
- NMDA receptor
- CC
- calphostin C
- ACSF
- artificial cerebrospinal fluid.
REFERENCES
- 1. Herb A., Burnashev N., Werner P., Sakmann B., Wisden W., Seeburg P. H. (1992) The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron 8, 775–785 [DOI] [PubMed] [Google Scholar]
- 2. Chittajallu R., Braithwaite S. P., Clarke V. R., Henley J. M. (1999) Kainate receptors: subunits, synaptic localization, and function. Trends Pharmacol. Sci. 20, 26–35 [DOI] [PubMed] [Google Scholar]
- 3. Cui C., Mayer M. L. (1999) Heteromeric kainate receptors formed by the coassembly of GluR5, GluR6, and GluR7. J. Neurosci. 19, 8281–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lerma J., Paternain A. V., Rodríguez-Moreno A., López-García J. C. (2001) Molecular physiology of kainate receptors. Physiol. Rev. 81, 971–998 [DOI] [PubMed] [Google Scholar]
- 5. Jaskolski F., Coussen F., Mulle C. (2005) Subcellular localization and trafficking of kainate receptors. Trends Pharmacol. Sci. 26, 20–26 [DOI] [PubMed] [Google Scholar]
- 6. Rodríguez-Moreno A., Herreras O., Lerma J. (1997) Kainate receptors presynaptically down-regulate GABAergic inhibition in the rat hippocampus. Neuron 19, 893–901 [DOI] [PubMed] [Google Scholar]
- 7. Contractor A., Swanson G. T., Sailer A., O'Gorman S., Heinemann S. F. (2000) Identification of the kainate receptor subunits underlying modulation of excitatory synaptic transmission in the CA3 region of the hippocampus. J. Neurosci. 20, 8269–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamiya H., Ozawa S. (2000) Kainate receptor-mediated presynaptic inhibition at the mouse hippocampal mossy fibre synapse. J. Physiol. 523, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Contractor A., Swanson G., Heinemann S. F. (2001) Kainate receptors are involved in short- and long-term plasticity at mossy fiber synapses in the hippocampus. Neuron 29, 209–216 [DOI] [PubMed] [Google Scholar]
- 10. Schmitz D., Mellor J., Nicoll R. A. (2001) Presynaptic kainate receptor mediation of frequency facilitation at hippocampal mossy fiber synapses. Science 291, 1972–1976 [DOI] [PubMed] [Google Scholar]
- 11. Melyan Z., Wheal H. V., Lancaster B. (2002) Metabotropic-mediated kainate receptor regulation of IsAHP and excitability in pyramidal cells. Neuron 34, 107–114 [DOI] [PubMed] [Google Scholar]
- 12. Isaac J. T., Mellor J., Hurtado D., Roche K. W. (2004) Kainate receptor trafficking: physiological roles and molecular mechanisms. Pharmacol. Ther. 104, 163–172 [DOI] [PubMed] [Google Scholar]
- 13. Fisahn A., Heinemann S. F., McBain C. J. (2005) The kainate receptor subunit GluR6 mediates metabotropic regulation of the slow and medium AHP currents in mouse hippocampal neurones. J. Physiol. 562, 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruiz A., Sachidhanandam S., Utvik J. K., Coussen F., Mulle C. (2005) Distinct subunits in heteromeric kainate receptors mediate ionotropic and metabotropic function at hippocampal mossy fiber synapses. J. Neurosci. 25, 11710–11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castillo P. E., Malenka R. C., Nicoll R. A. (1997) Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature 388, 182–186 [DOI] [PubMed] [Google Scholar]
- 16. Vignes M., Collingridge G. L. (1997) The synaptic activation of kainate receptors. Nature 388, 179–182 [DOI] [PubMed] [Google Scholar]
- 17. Frerking M., Ohliger-Frerking P. (2002) AMPA receptors and kainate receptors encode different features of afferent activity. J. Neurosci. 22, 7434–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldin M., Epsztein J., Jorquera I., Represa A., Ben-Ari Y., Crépel V., Cossart R. (2007) Synaptic kainate receptors tune oriens-lacunosum moleculare interneurons to operate at θ frequency. J. Neurosci. 27, 9560–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang E. J., Harris A. Z., Pettit D. L. (2007) Synaptic kainate currents reset interneuron firing phase. J. Physiol. 578, 259–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dingledine R., Borges K., Bowie D., Traynelis S. F. (1999) The glutamate receptor ion channels. Pharmacol. Rev. 51, 7–61 [PubMed] [Google Scholar]
- 21. Erreger K., Chen P. E., Wyllie D. J., Traynelis S. F. (2004) Glutamate receptor gating. Crit. Rev. Neurobiol. 16, 187–224 [DOI] [PubMed] [Google Scholar]
- 22. Contractor A., Mulle C., Swanson G. T. (2011) Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci. 34, 154–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mehta S., Wu H., Garner C. C., Marshall J. (2001) Molecular mechanisms regulating the differential association of kainate receptor subunits with SAP90/PSD-95 and SAP97. J. Biol. Chem. 276, 16092–16099 [DOI] [PubMed] [Google Scholar]
- 24. Coussen F., Normand E., Marchal C., Costet P., Choquet D., Lambert M., Mège R. M., Mulle C. (2002) Recruitment of the kainate receptor subunit glutamate receptor 6 by cadherin/catenin complexes. J. Neurosci. 22, 6426–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirbec H., Francis J. C., Lauri S. E., Braithwaite S. P., Coussen F., Mulle C., Dev K. K., Coutinho V., Meyer G., Isaac J. T., Collingridge G. L., Henley J. M., Couthino V. (2003) Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron 37, 625–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coussen F., Perrais D., Jaskolski F., Sachidhanandam S., Normand E., Bockaert J., Marin P., Mulle C. (2005) Co-assembly of two GluR6 kainate receptor splice variants within a functional protein complex. Neuron 47, 555–566 [DOI] [PubMed] [Google Scholar]
- 27. Laezza F., Wilding T. J., Sequeira S., Coussen F., Zhang X. Z., Hill-Robinson R., Mulle C., Huettner J. E., Craig A. M. (2007) KRIP6: a novel BTB/kelch protein regulating function of kainate receptors. Mol. Cell. Neurosci. 34, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia E. P., Mehta S., Blair L. A., Wells D. G., Shang J., Fukushima T., Fallon J. R., Garner C. C., Marshall J. (1998) SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron 21, 727–739 [DOI] [PubMed] [Google Scholar]
- 29. Bowie D., Garcia E. P., Marshall J., Traynelis S. F., Lange G. D. (2003) Allosteric regulation and spatial distribution of kainate receptors bound to ancillary proteins. J. Physiol. 547, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang W., St-Gelais F., Grabner C. P., Trinidad J. C., Sumioka A., Morimoto-Tomita M., Kim K. S., Straub C., Burlingame A. L., Howe J. R., Tomita S. (2009) A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron 61, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Copits B. A., Robbins J. S., Frausto S., Swanson G. T. (2011) Synaptic targeting and functional modulation of GluK1 kainate receptors by the auxiliary neuropilin and Tolloid-like (NETO) proteins. J. Neurosci. 31, 7334–7340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Straub C., Hunt D. L., Yamasaki M., Kim K. S., Watanabe M., Castillo P. E., Tomita S. (2011) Distinct functions of kainate receptors in the brain are determined by the auxiliary subunit Neto1. Nat. Neurosci. 14, 866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Straub C., Zhang W., Howe J. R. (2011) Neto2 modulation of kainate receptors with different subunit compositions. J. Neurosci. 31, 8078–8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang M., Pelkey K. A., Ng D., Ivakine E., McBain C. J., Salter M. W., McInnes R. R. (2011) Neto1 is an auxiliary subunit of native synaptic kainate receptors. J. Neurosci. 31, 10009–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tzivion G., Avruch J. (2002) 14-3-3 proteins: Active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277, 3061–3064 [DOI] [PubMed] [Google Scholar]
- 36. Foote M., Zhou Y. (2012) 14-3-3 proteins in neurological disorders. Int. J. Biochem. Mol. Biol. 3, 152–164 [PMC free article] [PubMed] [Google Scholar]
- 37. Vivithanaporn P., Yan S., Swanson G. T. (2006) Intracellular trafficking of KA2 kainate receptors mediated by interactions with coatomer protein complex I (COPI) and 14-3-3 chaperone systems. J. Biol. Chem. 281, 15475–15484 [DOI] [PubMed] [Google Scholar]
- 38. Li Y., Wu Y., Zhou Y. (2006) Modulation of inactivation properties of CaV2.2 channels by 14-3-3 proteins. Neuron 51, 755–771 [DOI] [PubMed] [Google Scholar]
- 39. Wang B., Yang H., Liu Y. C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999) Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 [DOI] [PubMed] [Google Scholar]
- 40. Nagamatsu S., Kornhauser J. M., Burant C. F., Seino S., Mayo K. E., Bell G. I. (1992) Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J. Biol. Chem. 267, 467–472 [PubMed] [Google Scholar]
- 41. Barberis A., Sachidhanandam S., Mulle C. (2008) GluR6/KA2 kainate receptors mediate slow-deactivating currents. J. Neurosci. 28, 6402–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fisher J. L., Mott D. D. (2011) Distinct functional roles of subunits within the heteromeric kainate receptor. J. Neurosci. 31, 17113–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robert A., Irizarry S. N., Hughes T. E., Howe J. R. (2001) Subunit interactions and AMPA receptor desensitization. J. Neurosci. 21, 5574–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barberis A. (2012) Fast Perfusion Methods for the Study of Ligand-gated Ion Channels. Neuronal Network Analysis, pp. 173–187, Humana Press Inc., Totowa, NJ [Google Scholar]
- 45. Basiry S. S., Mendoza P., Lee P. D., Raymond L. A. (1999) Agonist-induced changes in substituted cysteine accessibility reveal dynamic extracellular structure of M3-M4 loop of glutamate receptor GluR6. J. Neurosci. 19, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwon H. B., Castillo P. E. (2008) Role of glutamate autoreceptors at hippocampal mossy fiber synapses. Neuron 60, 1082–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 48. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 49. Nasu-Nishimura Y., Jaffe H., Isaac J. T., Roche K. W. (2010) Differential regulation of kainate receptor trafficking by phosphorylation of distinct sites on GluR6. J. Biol. Chem. 285, 2847–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reiner A., Arant R. J., Isacoff E. Y. (2012) Assembly stoichiometry of the GluK2/GluK5 kainate receptor complex. Cell Rep. 1, 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Broadie K., Rushton E., Skoulakis E. M., Davis R. L. (1997) Leonardo, a Drosophila 14-3-3 protein involved in learning, regulates presynaptic function. Neuron 19, 391–402 [DOI] [PubMed] [Google Scholar]
- 52. Jaskolski F., Coussen F., Nagarajan N., Normand E., Rosenmund C., Mulle C. (2004) Subunit composition and alternative splicing regulate membrane delivery of kainate receptors. J. Neurosci. 24, 2506–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martin S., Henley J. M. (2004) Activity-dependent endocytic sorting of kainate receptors to recycling or degradation pathways. EMBO J. 23, 4749–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park Y., Jo J., Isaac J. T., Cho K. (2006) Long-term depression of kainate receptor-mediated synaptic transmission. Neuron 49, 95–106 [DOI] [PubMed] [Google Scholar]
- 55. Rivera R., Rozas J. L., Lerma J. (2007) PKC-dependent autoregulation of membrane kainate receptors. EMBO J. 26, 4359–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Konopacki F. A., Jaafari N., Rocca D. L., Wilkinson K. A., Chamberlain S., Rubin P., Kantamneni S., Mellor J. R., Henley J. M. (2011) Agonist-induced PKC phosphorylation regulates GluK2 SUMOylation and kainate receptor endocytosis. Proc. Natl. Acad. Sci. U.S.A. 108, 19772–19777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chamberlain S. E., González-González I. M., Wilkinson K. A., Konopacki F. A., Kantamneni S., Henley J. M., Mellor J. R. (2012) SUMOylation and phosphorylation of GluK2 regulate kainate receptor trafficking and synaptic plasticity. Nat. Neurosci. 15, 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen H. X., Roper S. N. (2003) PKA and PKC enhance excitatory synaptic transmission in human dentate gyrus. J. Neurophysiol. 89, 2482–2488 [DOI] [PubMed] [Google Scholar]
- 59. Cho K., Francis J. C., Hirbec H., Dev K., Brown M. W., Henley J. M., Bashir Z. I. (2003) Regulation of kainate receptors by protein kinase C and metabotropic glutamate receptors. J. Physiol. 548, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Selak S., Paternain A. V., Aller M. I., Aller I. M., Picó E., Rivera R., Lerma J. (2009) A role for SNAP25 in internalization of kainate receptors and synaptic plasticity. Neuron 63, 357–371 [DOI] [PubMed] [Google Scholar]
- 61. Skoulakis E. M., Davis R. L. (1996) Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron 17, 931–944 [DOI] [PubMed] [Google Scholar]
- 62. Simsek-Duran F., Linden D. J., Lonart G. (2004) Adapter protein 14-3-3 is required for a presynaptic form of LTP in the cerebellum. Nat. Neurosci. 7, 1296–1298 [DOI] [PubMed] [Google Scholar]
- 63. Heckmann M., Bufler J., Franke C., Dudel J. (1996) Kinetics of homomeric GluR6 glutamate receptor channels. Biophys. J. 71, 1743–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fleck M. W., Cornell E., Mah S. J. (2003) Amino acid residues involved in glutamate receptor 6 kainate receptor gating and desensitization. J. Neurosci. 23, 1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]