Highlights
-
•
miR-21 targets KRIT1.
-
•
miR-21 and KRIT1 expression anticorrelate in human breast tumors.
-
•
KRIT1 is involved in miR-21-mediated tumor cell growth.
Keywords: KRIT1, miR-21, Tumor growth
Abstract
miR-21 is overexpressed in tumors and it displays oncogenic activity. Here, we show that expression of miR-21 in primary tumors anticorrelates with KRIT1/CCM1, an interacting partner of the Ras-like GTPase Rap1, involved in Cerebral Cavernous Malformations (CCM). We present evidences that miR-21 silences KRIT1 by targeting its mRNA 3′UTR and that this interaction is involved in tumor growth control. In fact, miR-21 over-expression or KRIT1 knock-down promote anchorage independent tumor cell growth compared to controls, whereas the opposite is observed when anti-miR-21 or KRIT1 overexpression are employed. Our findings suggest that miR-21 promotes tumor cell growth, at least in part, by down-modulating the potential tumor suppressor KRIT1.
1. Introduction
Tumorigenesis is a complex multi-step process, which includes cellular and stroma alterations leading to the formation of a primary tumor mass and eventually metastasis in distant organs [1]. In addition to protein-coding genes, microRNAs (miRNAs) have been found to participate to cancer development [2].
miRNAs are endogenous small-non coding RNAs that occur naturally and down-regulate gene expression post-transcriptionally by binding to the 3′UTRs of their target mRNAs causing translation inhibition or promoting RNA decay [3,4]. Over 1000 human miRNAs exist and they act as fine tuners of protein-coding gene expression through the regulation of up to 50% of protein-coding genes [5]. Expression of miRNAs is tissue specific and it is often altered (up- or down) in tumors compared to normal tissues [2,6]. Examples of deregulated miRNAs, proven to control tumorigenesis, are let-7, miR-9, miR-10b, miR-21, miR-31, miR-34, miR-126, miR-146, miR-155, miR-200, miR-214, miR-221/222, miR-373, and 520c [7].
miR-21 is one of the early discovered small non-coding RNAs in human cells and one of the most overexpressed small RNAs in a variety of solid cancers, examples are breast, colon, melanoma, cervix, ovarian, lung, pancreas, prostate and stomach cancers [8]. Forced miR-21 overexpression in tumor cells coincide with increased tumor growth and invasion, while miR-21 knock down induces cell cycle arrest, increased apoptosis, increased chemosensitivity to anticancer agents and reduced invasion [8]. The importance of miR-21 in tumorigenesis is well demonstrated in genetically engineered mice that overexpress miR-21. These animals develop pre-B cell lymphomas which are addicted to miR-21 and in fact, miR-21 inactivation leads to complete lymphoma regression in a few days [9]. At the same time, overexpression or ablation of miR-21 in a K-Ras mouse model of lung cancer respectively, accelerates or suppresses tumorigenesis [10]. All the experimental evidences strongly suggest that miR-21 acts as an oncogene and it is, therefore, essential to unravel the molecular pathway coordinated by miR-21 in tumors. A number of tumor suppressor genes have been proven to be targeted by miR-21. Examples are various components of p53 [11] or PTEN/AKT networks [12] or RAS pathway antagonists such as programmed cell death protein-4 (PDCD4) [13] and sprouty 1 or 2 (SPRY1 or 2) [14,15]. However, it is reasonable to hypothesize that many more genes involved in neoplasia are under miR-21 control and it is essential to identify them.
Relevantly, miR-21 is reported to have a role not only in cancerous tissues but also in the cardiovascular systems, including cardiomyocytes, cardiac fibroblasts in infarcted hearts, endothelial and smooth muscle cells however its function appears to be even more complex than in tumors [15–17].
In this work, we evaluated the potential link between miR-21 and one of its predicted targets, KRIT1/CCM1, a gene with a well known function in endothelial cells and in the cardiovascular system [18] and a putative role in tumorigenesis [19].
2. Materials and methods
2.1. Cell cultures
MC-1, melanoma cells, were provided by Xu [20] and maintained as described in [21]. MDAMB231, HEK293T (293T) and HeLa were from American Type Culture Collection and kept in standard conditions.
2.2. Reagents and antibodies
miR precursors and inhibitors: Pre-miR™ miRNA Precursor Negative Control #1, Pre-miRTM miRNA Precursor Hsa-miR-21 (PM10206), Anti-miR™ miRNA Inhibitor Negative Control #1, Anti-miR™ miRNA Inhibitor Hsa-miR-21 (AM10206), (all from Applied Biosystems, Foster City, CA). TaqMan® MicroRNA assays for miRNA detection: Hsa-miR-21 ID 000397, U6 snRNA ID001973 (from Applied Biosystems, Foster City, CA). pLKO.1-empty or pLKO.1-shKRIT1 lentiviral expression vectors were purchased from Sigma (St. Louis, MO, cat. No. MISSION shRNA: NM_004912/TRCN0000072879, TRCN0000072880, TRCN0000072881, TRCN0000072882) and indicated in the text pLKO.1-shKRIT1 #1–4. Primary antibodies: anti-KRIT1 pAb [22], anti-Actin pAb, (Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies: HRP-conjugated goat anti-rabbit IgG, donkey anti-goat IgG (all from Santa Cruz Biotechnology, Santa Cruz, CA).
2.3. Primers
Oligonucleotides employed in this study were: cloning miR-21: TGCTTGGGAGGAAAATAAACA and TTTCAAAACCCACAATGCAG; cloning KRIT1 3′UTR: GGTGACACAGCAAGACTCCA and TTTTTCCAAACTAATAAACTGATGAA Hsa_KRIT1_1 QuantiTect Primer Assay QT00090552 GAPDH QuantiTect Primer Assay QT00079247 (all from Qiagen, Stanford, CA).
2.4. qRT-PCRs for miRNA or mRNA detection
Total RNA was isolated from cells using TRIzol® Reagent (Invitrogen Life Technologies, Carlsbad, CA). KRIT1 and miR-21 detection was carried out as described in [21]. Quantitative normalization was performed on RNU6 or GAPDH, for miR or mRNA detection, respectively and the relative expression levels calculated as in [23].
2.5. Northern Blots
20 μg of total RNA isolated as above were resolved on 12.5% (w/v) TBE–Urea–polyacrylamide gel electrophoresis and transferred to a Hybond N+ membrane (GE Healthcare Life Sciences, Piscataway, NJ, USA). The filter was hybridized overnight at 45 °C with a specific miR-21 digoxigenin-labeled LNA Detection probe (Exiqon, Vedbaek, Denmark Cat. No.: 38102-01), washed and visualized with a specific DIG antibody (1:10,000) using the DIG Nucleic Acid Detection kit (all from Roche, GmbH). The filter was then stripped and re-probed overnight at 45 °C using a specific U6 digoxigenin-labeled LNA Detection probe (Exiqon, Vedbaek, Denmark Cat. No.: 99002-01).
2.6. Protein preparation and Immunoblottings
Protein extracts were obtained using RIPA buffer [1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 150 mM NaCl, 50 mM Tris–HCl (pH 7.2), 0.4 mM Na3VO4, 10 g/ml leupeptin, 4 g/ml pepstatin, 0.1 TIU/ml aprotinin]. Immunoblotting performed as in [21].
2.7. Vectors construction
pCLL-KRIT1 overexpression vector was obtained as described in [22]. pLVX-KRIT1 overexpression vector was obtained by subcloning the KRIT1 cDNA cassette [24] into the pLVX lentiviral expression vector (Clontech Laboratories, Mountain View, CA). The human miR-21 gene was amplified from HeLa genomic DNA and cloned into the pWPT lentivirus vector (Addgene, Cambridge, MA) to obtain pWPT-miR-21.The luciferase reporter vector containing a partial portion (973 nts, positions 340–1313) of the human KRIT1 3′UTR was generated by PCR amplification from HeLa genomic DNA, cloned downstream of the luciferase coding sequence into the pMIR-REPORT™ vector (Ambion, Austin, TX). To generate the miR-21 sensor construct, an oligonucleotide containing three miR-21 binding sequences was annealed with its complementary sequence and cloned into the pMIR-REPORT™ (Ambion, Austin, TX, USA) at SacI and SpeI sites.
2.8. Transfections, infections and luciferase assays
Transfections or infections with pre- or anti-miRs or overexpression constructs or pMIR-REPORT™ vectors and luciferase assays were carried out as described in [21].
2.9. Soft agar assay
Transiently transfected or stably transduced cells were subjected to soft agar colony formation assays as in [21,25].
2.10. Prediction of miR-21 targets
Performed by Targetscan 6.2 [26].
2.11. Statistical analyses
Data are presented as mean ± Standard Deviation (SD) or as mean ± Standard Error of Mean (SEM), as indicated, and two tailed Student’s t test was used for comparison, with ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 considered to be statistically significant. ns indicates a non-statistically significant p-value.
2.12. Analysis of human tumor data sets
Expression data for mRNA and microRNA of human primary breast tumors were obtained from [27] and Pearson correlation used to evaluate KRIT1 and miR-21 relative expression. p value <0.05 statistically significant.
3. Results
3.1. miR-21 and KRIT1 expression anticorrelates in primary human tumors
Krev Interaction Trapped 1 (KRIT1) is a putative target of miR-21 as predicted by various algorithms, for instance Targetscan 6.2, www.targetscan.org, [26] and shown in Table 1. To investigate a possible link between miR-21 and KRIT1 we first analyzed KRIT1 and miR-21 expression in a large dataset [27] of primary breast tumors (Table 2) and found a strong anticorrelation when all (n = 521) the tumor samples were considered or when only Luminal A n = 208 or Luminal B n=113 subtypes were investigated. No anticorrelation was revealed in HER2+ (n = 53) or Basal n = 81 tumors or in normal (n = 8) mammary gland tissue samples.
Table 1.
Top 25 predicted miR-21 target genes as for Targetscan 6.2.
| Target gene | Representative transcript | Gene name | Conserved sites | Poorly conserved sites |
|---|---|---|---|---|
| ZNF367 | NM_153695 | Zinc finger protein 367 | 2 | 1 |
| GPR64 | NM_001079858 | G protein-coupled receptor 64 | 2 | 0 |
| YOD1 | NM_018566 | YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae) | 2 | 2 |
| PHF14 | NM_014660 | PHD finger protein 14 | 1 | 2 |
| PLEKHA1 | NM_001001974 | Pleckstrin homology domain containing, family A member 1 | 1 | 1 |
| PIKFYVE | NM_015040 | Phosphoinositide kinase, FYVE finger containing | 1 | 1 |
| PBRM1 | NM_018165 | Polybromo 1 | 1 | 0 |
| GATAD2B | NM_020699 | GATA zinc finger domain containing 2B | 2 | 0 |
| SCML2 | NM_006089 | Sex comb on midleg-like 2 (Drosophila) | 1 | 1 |
| VCL | NM_003373 | Vinculin | 2 | 0 |
| BMPR2 | NM_001204 | Bone morphogenetic protein receptor, type II (serine/threonine kinase) | 2 | 1 |
| C17orf39 | NM_024052 | Chromosome 17 open reading frame 39 | 1 | 1 |
| TIAM1 | NM_003253 | T-cell lymphoma invasion and metastasis 1 | 1 | 1 |
| FGF18 | NM_003862 | Fibroblast growth factor 18 | 1 | 0 |
| FRS2 | NM_001042555 | Fibroblast growth factor receptor substrate 2 | 1 | 1 |
| KRIT1 | NM_001013406 | KRIT1, ankyrin repeat containing | 1 | 1 |
| SKI | NM_003036 | V-ski sarcoma viral oncogene homolog (avian) | 1 | 0 |
| ST3GAL6 | NM_006100 | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 1 | 0 |
| PELI1 | NM_020651 | Pellino homolog 1 (Drosophila) | 1 | 0 |
| PGRMC2 | NM_006320 | Progesterone receptor membrane component 2 | 1 | 1 |
| RP2 | NM_006915 | Retinitis pigmentosa 2 (X-linked recessive) | 1 | 2 |
| MTMR12 | NM_001040446 | Myotubularin related protein 12 | 1 | 1 |
| AIM1L | NM_001039775 | Absent in melanoma 1-like | 1 | 0 |
| FAM13A | NM_001015045 | Family with sequence similarity 13, member A | 1 | 1 |
| GRAMD3 | NM_001146319 | GRAM domain containing 3 | 1 | 0 |
Table 2.
Analysis of KRIT1 and miR-21 expression in primary breast tumors.
| Samples | Number of cases | Pearson correlation miR-21/KRIT1 | p value |
|---|---|---|---|
| All breast tumors | 521 | −0.23 | 6.967e−08 |
| Luminal A | 208 | −0.29 | 1.69e−051 |
| Luminal B | 113 | −0.203 | 0.03111551 |
| HER2+ | 53 | −0.258 | 0.06239282 |
| Basal | 81 | −0.1132 | 0.3144468 |
| Normal | 8 | +0.2003 | 0.56343428 |
3.2. KRIT1 is directly modulated by miR-21
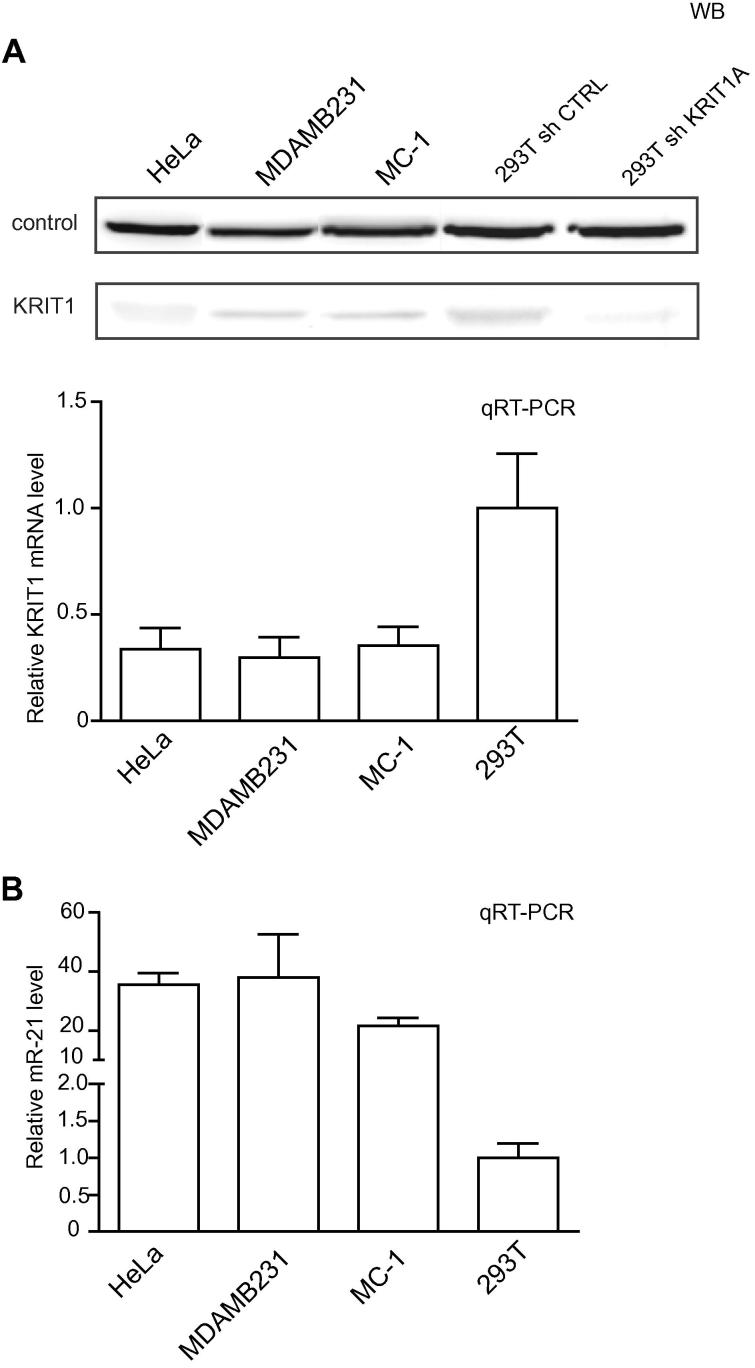
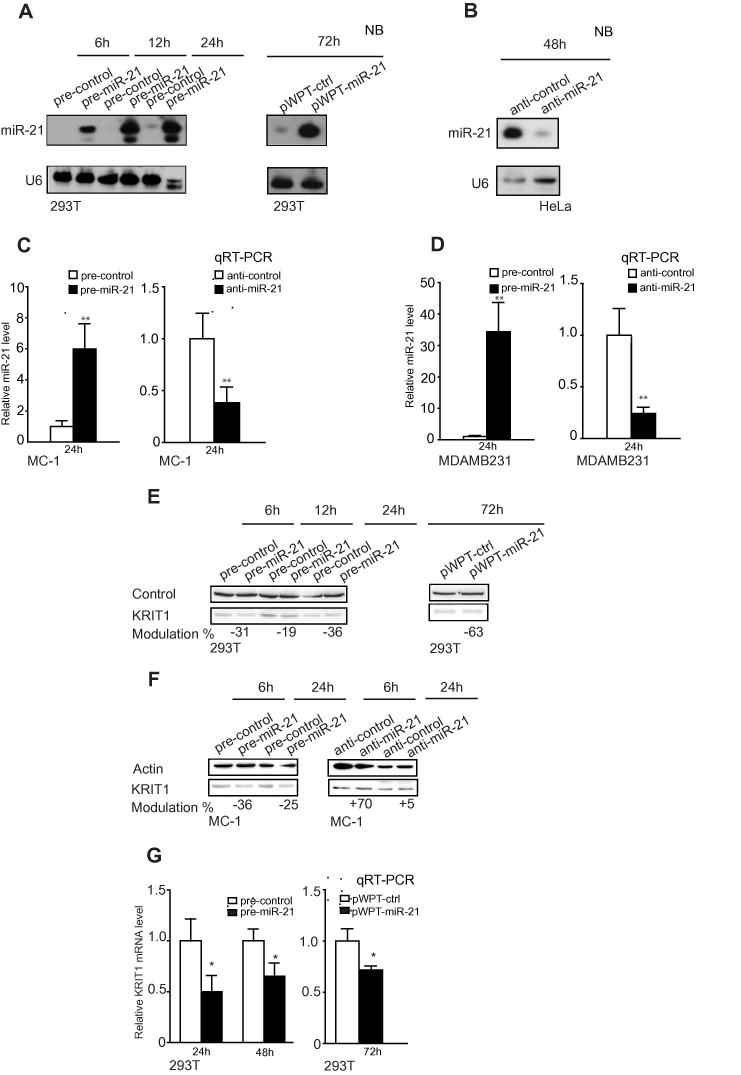
Consequently to the anticorrelation found in human tumors for KRIT1 and miR-21, we investigated a possible functional direct link between these two genes, following miR-21 modulation in various tumor cell lines, HeLa, MDAMB231, MC-1 and 293T. The endogenous levels of miR-21 and KRIT1 were evaluated by Western Blot (WB) and qRT-PCR analyses and shown in Fig. 1A and B. 293T cells stably silenced (sh-KRIT1) or not (sh-CTRL) for KRIT1 were used in WB analysis to show anti-KRIT1 Ab specificity (80KDa band). miR-21 expression was increased or decreased by transfecting the cells with pre- or anti-miRs or their controls (pre-miR-21, anti-miR-21, pre- or anti-control) or with pWPT-empty or miR-21 overexpression lentivirus vectors (pWPT-ctrl, pWPT-miR-21). miR-21 expression was verified at different time points, 6–72 h post-transfection, by Northern Blot–NB (Fig. 2A and B) or qRT-PCR (Fig. 2C and D) analyses. When KRIT1, protein or mRNA expression, was evaluated, important inverse modulations were found, as shown and quantitated by WB or qRT-PCR (Fig. 2E–G).
Fig. 1.
KRIT1 and miR-21 expression in tumor cell lines. KRIT1 (A) and miR-21 (B) expression was evaluated in HeLa, MDAMB231, MC-1 and 293T (wild type or silenced-sh- for KRIT1 and its sh-control-ctrl) human tumor cells by Western Blot (WB) and qRT-PCR analyses. An aspecific band was used as loading control in WBs as in [22]. For the qRT-PCRs, results are shown as fold changes (mean ± SEM) relative to expression in 293T cells, normalized on GAPDH for KRIT1 or U6 for miR-21 levels. Two independent analyses were performed in triplicate and a representative one is shown.
Fig. 2.
miR-21 directly modulates KRIT1 expression. miR-21 expression was analyzed by Northern Blot-NB- (A, B) or qRT-PCR (C, D) analysis in 293T or HeLa or MC-1 or MDAMB231 cells transfected with miR-21 precursors or inhibitors or their negative controls (pre- and anti-miR-21 or -control) or with pWPT-ctrl or miR-21 overexpression (pWPT-miR-21) vectors at the indicated time points (h). U6 was used for normalization in NB and qRT-PCR analyses. KRIT1 protein (WB: E, F) or mRNA (qRT-PCR: G) expression in cells manipulated as in (A and C). Protein modulations in (E, F) were calculated relative to controls, normalized on controls (aspecific band [22] or actin), and expressed as percentages (%) of reduction. mRNA modulations in (G) were calculated as fold changes (mean ± SEM) relative to controls, normalized on GAPDH. (A–G) three independent analyses were performed in triplicate and representative results are shown.
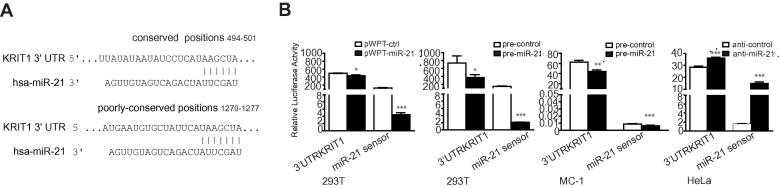
Bioinformatics analysis revealed that KRIT1 3′UTR contained two putative miR-21 binding sites for miR-21 (Fig. 3A). To examine whether miR-21 binds at its putative binding site, 293T or MC-1 or HeLa cells were transiently transfected with a Luciferase reporter vector containing a 973 bps portion (including both sites) of the 3′UTR for KRIT1 in miR-21 overexpression (pre-miR-21 or pWPT-miR-21) or silencing (anti-miR-21) conditions and luciferase activity measured and compared to the negative controls (pre- or anti-controls; pWPT-ctrl). As shown in Fig. 3B, luciferase activity was significantly reduced in presence of miR-21 overexpression in 293T and MC-1 cells. Conversely, it was increased following miR-21 silencing in HeLa cells. As a positive control, a miR-21-sensor construct, containing 3 perfect bindings for miR-21, was used for each experiment. Results were normalized on Renilla activity.
Fig. 3.
miR-21 directly modulates KRIT1 3′UTR. (A) Scheme showing the two putative miR-21 binding sites in human KRIT1 3′UTR at positions 494 (highly conserved) and 1270 (poorly conserved) paired with miR-21 seed. (B) Luciferase assays in 293T or MC-1 or HeLa cells cotransfected with pMIR-luciferase constructs containing either a 973 nt-long portion of KRIT1 3′UTR (position 340–1313) or a synthetic sequence, which includes three perfect miR-21 binding sites (miR-21 sensor), together with miR-21 precursors or inhibitors or their negative controls (pre- and anti-miR-21 or control) or with pWPT-ctrl or miR-21 overexpression (pWPT-miR-21) vectors. Results are shown as mean ± SEM of Firefly Luciferase activity relative to controls, normalized on Renilla Luciferase activity. Three independent analyses were performed in triplicate and representative results are shown.
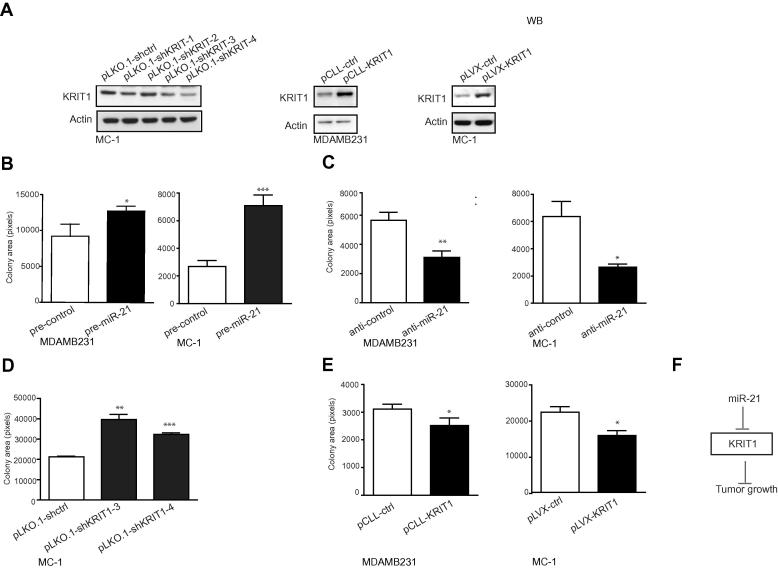
3.3. miR-21 promotes anchorage independent growth opposite to KRIT1
To evaluate the functions of miR-21 and KRIT1 in tumorigenesis, we performed anchorage independent growth experiments with MDAMB231 and MC-1 cells following miR-21 (Fig. 2C and D) or KRIT1 (Fig. 4A) modulations. miR-21 expression up- or down-regulations were obtained as presented above (Fig. 2C and D). Instead, to obtain KRIT1 modulations of expression, MC-1 or MDAMB231 cells were stably transduced with lentiviral vectors containing either specific shKRIT1 (pLKO.1-shKRIT1 #1–4) or a KRIT1 cDNA lacking its 3′UTR (pCLL-KRIT1 or pLVX-KRIT1) or empty controls (pLKO.1-shctrl or pCLL-ctrl or pLVX-ctrl) and expression was evaluated by Western Blot (WB) analysis; actin was used as loading control (Fig. 4A). miR-21 overexpression (pre-miR-21) resulted in increased (up to 3-folds) colony-formation compared to controls (pre-control), as shown in Fig. 4B, whereas inhibition of miR-21 led to decreased (2–3-folds) colony formation as in Fig. 4C. In parallel, when MC-1 cells were transduced with either specific shKRIT1 or control lentiviral vectors (pLKO-shKRIT1 #3 or #4 or pLKO-shctrl), the results obtained phenocopied miR-21 overexpression. Indeed, 1.5–2-fold increase in the area occupied by colonies (Fig. 4D) was observed. Instead, the opposite results were obtained when KRIT1 was overexpressed in MDAMB231 and MC-1 cells following transduction of KRIT1 cDNA or control (pCLL-ctrl and pCLL-KRIT1 or pLVX-ctrl and pLVX-KRIT1) expressing lentivirus vectors (Fig. 4E). In conclusion, we propose that KRIT1 is a functional player able to control tumor growth, downstream of miR-21 (Fig. 4F).
Fig. 4.
miR-21 regulates anchorage-independent growth opposite to KRIT1. (A) KRIT1 expression modulations were analyzed in MC-1 or MDAMB231 cells stably transduced with lentiviral vectors containing either specific shKRIT1 (pLKO.1-sh-KRIT1 #1 to #4) or a KRIT1 cDNA lacking its 3′UTR (pCLL-KRIT1 or pLVX-KRIT1) or empty controls (pLKO.1-sh-ctrl or pCLL-ctrl or pLVX-ctrl) by Western Blot (WB) analysis and actin was used as loading control. 3 independent analyses were performed in triplicate and representative results are shown. (B–E) Anchorage-independent growth of MDAMB231 and MC-1 cells transfected with miR-21 precursors or inhibitors or their negative controls (pre- and anti-miR-21 or control) or stably transduced with lentiviral vectors containing either specific shKRIT1 (pLKO.1-sh-KRIT1-3, -4) or a KRIT1 cDNA lacking its 3′UTR (pCLL-KRIT1 or pLVX-KRIT1) or empty controls (pLKO.1-sh-control or pCLL-ctrl or pLVX-ctrl). All results are shown as mean ± SEM of the area covered by colonies (pixels). Three independent experiments were performed in triplicate and representative results are shown. (F) Cartoon showing the proposed model for the role of KRIT1 in miR-21-mediated tumorigenesis.
4. Discussion
Our work establishes a link between miR-21 [28–31] and one of its targets, KRIT1, in tumorigenesis. In fact, we evidenced an anticorrelation for miR-21 and KRIT1 in primary human tumors and unravelled that KRIT1 is able to oppose miR-21 pro-oncogenic functions in tumor cells. In addition, we showed that miR-21 modulations of expression lead to opposite cellular KRIT1 fluctuations.
KRIT1 is an intracellular protein with ankyrin repeats and a FERM domain that was originally found through its interaction with the Ras-family GTPase Krev1/Rap1a inferring a role in GTPase signaling cascades [32]. While KRIT1 is a positive and indispensable regulator of Krev1 [33], the latter has been considered as a negative regulator of oncogenesis by competing with Ras for downstream targets [34]. KRIT1 is also able to interact with the integrin cytoplasmic domain associated protein-1-alpha (ICAP1a) [35,36], and it is mostly known because of its involvement in Cerebral Cavernous Malformations (CCMs), vascular lesions of the central nervous system (CNS) that can arise sporadically or may be inherited as autosomal dominant conditions with incomplete penetrance [37,38]. For this reason the KRIT1 gene is also called CCM1.
Even if most of the findings linked KRIT1 function to endothelial cells and mouse genetics studies proved its involvement in CCMs [18] and in cardiovascular development, KRIT1 is ubiquitously expressed in many cells and tissues [39], suggesting a possible function in various cells. KRIT1 has been revealed associated with microtubules, membranes, adherens junctions and present in nuclei as well [40]. Modulations of KRIT1 expression (overexpression or silencing) in endothelial cells showed that KRIT1 is an antiangiogenic protein able to keep the human endothelium quiescent and to inhibit proliferation, migration, lumen formation and sprouting angiogenesis [41]. This by inducing DLL4-NOTCH signaling, which respectively promotes and reduces phosphorylation of AKT and ERK [42]. KRIT1 negatively coordinates cell cycle also in mouse embryo fibroblasts (MEFs) by controlling FoxO-mediated downregulation of cyclin D1 and upregulation of p27Kip1 levels through the modulation of intracellular ROS levels [22]. At the same time KRIT1 is able to stabilize beta-catenin-containing endothelial or epithelial cell–cell junctions downstream of the Rap1 GTPase, therefore inhibiting beta-catenin-driven transcription of genes involved in proliferation, transformation and tumor progression [33]. Relevantly, when KRIT1 ± mice were crossed onto the ApcMin/+ model of familial APC, more polyps were observed compared to control animals, suggesting a control of KRIT1 in epithelial cell proliferation [33]. KRIT1 was also one of the three genes re-expressed in CHO cells during chemical reverse transformation, suggesting a potential tumor suppressor function, as hypothesized for its direct interactor Rap1 [19]. The fact, that we observed increased anchorage independent growth in tumor cells silenced for KRIT1 or overexpressing miR-21 and that miR-21 and KRIT1 expressions anticorrelate in a large data set of human tumor samples, shed light on the connection between miR-21 and KRIT1 and reinforce the potential tumor suppressor role of KRIT1.
In conclusion, we presented a relevant link between miR-21 and KRIT1 in the control of tumor growth.
Acknowledgments
We thank L. Xu, R. Hynes and L. Primo for providing cell lines, D. Corà for helping with initial bioinformatics, V. Monica and L. Goitre for participating to the preparation of some tools. Grant support: This work was supported by Grants from the University of Torino (Local Research Funding 2007/DT, 2008/D. T., 2007/SFR, 2008/S. F. R.), Compagnia di San Paolo (D.T.), Torino, PRIN 2008 (D.T.), AIRC 2010 (IG 10104 D. T.), FIRB giovani (RBFR08F2FS-002 F. O.), PRIN 2008 (2008BP25KN to S.F.R.) and Fondazione Telethon (GGP06222 to S.F.R.).
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inui M., Martello G., Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 6.Jansson M.D., Lund A.H. MicroRNA and cancer. Mol. Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valastyan S., Weinberg R.A. MicroRNAs: crucial multi-tasking components in the complex circuitry of tumor metastasis. Cell Cycle. 2009;8:3506–3512. doi: 10.4161/cc.8.21.9802. [DOI] [PubMed] [Google Scholar]
- 8.Pan X., Wang Z.X., Wang R. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol. Ther. 2010;10:1224–1232. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 9.Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 10.Hatley M.E., Patrick D.M., Garcia M.R., Richardson J.A., Bassel-Duby R., van Rooij E., Olson E.N. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papagiannakopoulos T., Shapiro A., Kosik K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 12.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel L.B., Christoffersen N.R., Jacobsen A., Lindow M., Krogh A., Lund A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 14.Sayed D. MicroRNA-21 targets sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jazbutyte V., Thum T. MicroRNA-21: from cancer to cardiovascular disease. Curr. Drug Targets. 2010;11:926–935. doi: 10.2174/138945010791591403. [DOI] [PubMed] [Google Scholar]
- 16.Bonauer A., Boon R.A., Dimmeler S. Vascular microRNAs. Curr. Drug Targets. 2010;11:943–949. doi: 10.2174/138945010791591313. [DOI] [PubMed] [Google Scholar]
- 17.Weber C., Schober A., Zernecke A. MicroRNAs in arterial remodelling, inflammation and atherosclerosis. Curr. Drug Targets. 2010;11:950–956. doi: 10.2174/138945010791591377. [DOI] [PubMed] [Google Scholar]
- 18.Riant F., Bergametti F., Ayrignac X., Boulday G., Tournier-Lasserve E. Recent insights into cerebral cavernous malformations: the molecular genetics of CCM. FEBS J. 2010;277:1070–1075. doi: 10.1111/j.1742-4658.2009.07535.x. [DOI] [PubMed] [Google Scholar]
- 19.Bachrati C.Z., Downes C.S., Rasko I. Chemical reverse transformation of CHO-K1 cells induces changes in expression of a candidate tumour suppressor and of a gene not previously characterised as transformation related. Eur. J. Cell Biol. 1999;78:561–566. doi: 10.1016/s0171-9335(99)80021-1. [DOI] [PubMed] [Google Scholar]
- 20.Xu L. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol. Cancer Res. 2008;6:760–769. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penna E. MicroRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011;30:1990–2007. doi: 10.1038/emboj.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goitre L., Balzac F., Degani S., Degan P., Marchi S., Pinton P., Retta S.F. KRIT1 regulates the homeostasis of intracellular reactive oxygen species. PLoS One. 2010;5:e11786. doi: 10.1371/journal.pone.0011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bookout A.L., Mangelsdorf D.J. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Retta S.F., Avolio M., Francalanci F., Procida S., Balzac F., Degani S., Tarone G., Silengo L. Identification of KRIT1B: a novel alternative splicing isoform of cerebral cavernous malformation gene-1. Gene. 2004;325:63–78. doi: 10.1016/j.gene.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 25.Hynes N.E., Taverna D., Harwerth I.M., Ciardiello F., Salomon D.S., Yamamoto T., Groner B. Epidermal growth factor receptor, but not c-erbB-2, activation prevents lactogenic hormone induction of the beta-casein gene in mouse mammary epithelial cells. Mol. Cell. Biol. 1990;10:4027–4034. doi: 10.1128/mcb.10.8.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Network C.G.A. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang L., Lv X., Li J., Li X., Li W., Li Y. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012;114:582–588. doi: 10.1016/j.acthis.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Yang C.H., Yue J., Pfeffer S.R., Handorf C.R., Pfeffer L.M. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J. Biol. Chem. 2011;286:39172–39178. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grignol V. MiR-21 and miR-155 are associated with mitotic activity and lesion depth of borderline melanocytic lesions. Br. J. Cancer. 2011;105:1023–1029. doi: 10.1038/bjc.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan L.X., Huang X.F., Shao Q., Huang M.Y., Deng L., Wu Q.L., Zeng Y.X., Shao J.Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serebriiskii I., Estojak J., Sonoda G., Testa J.R., Golemis E.A. Association of Krev-1/Rap1a with KRIT1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21-22. Oncogene. 1997;15:1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- 33.Glading A.J., Ginsberg M.H. Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signaling. Dis. Model. Mech. 2010;3:73–83. doi: 10.1242/dmm.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caamano J., DiRado M., Iizasa T., Momiki S., Fernandes E., Ashendel C., Noda M., Klein-Szanto A.J. Partial suppression of tumorigenicity in a human lung cancer cell line transfected with Krev-1. Mol. Carcinog. 1992;6:252–259. doi: 10.1002/mc.2940060406. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J., Clatterbuck R.E., Rigamonti D., Chang D.D., Dietz H.C. Interaction between KRIT1 and icap1alpha infers perturbation of integrin beta1-mediated angiogenesis in the pathogenesis of cerebral cavernous malformation. Hum. Mol. Genet. 2001;10:2953–2960. doi: 10.1093/hmg/10.25.2953. [DOI] [PubMed] [Google Scholar]
- 36.Zawistowski J.S., Serebriiskii I.G., Lee M.F., Golemis E.A., Marchuk D.A. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum. Mol. Genet. 2002;11:389–396. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- 37.Laberge-le Couteulx S. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat. Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 38.Sahoo T. Mutations in the gene encoding KRIT1, a Krev-1/Rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum. Mol. Genet. 1999;8:2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 39.Denier C., Gasc J.M., Chapon F., Domenga V., Lescoat C., Joutel A., Tournier-Lasserve E. KRIT1/cerebral cavernous malformation 1 mRNA is preferentially expressed in neurons and epithelial cells in embryo and adult. Mech. Dev. 2002;117:363–367. doi: 10.1016/s0925-4773(02)00209-5. [DOI] [PubMed] [Google Scholar]
- 40.Francalanci F. Structural and functional differences between KRIT1A and KRIT1B isoforms: a framework for understanding CCM pathogenesis. Exp. Cell Res. 2009;315:285–303. doi: 10.1016/j.yexcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Lampugnani M.G., Orsenigo F., Rudini N., Maddaluno L., Boulday G., Chapon F., Dejana E. CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J. Cell Sci. 2010;123:1073–1080. doi: 10.1242/jcs.059329. [DOI] [PubMed] [Google Scholar]
- 42.Wustehube J. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc. Natl. Acad. Sci. USA. 2010;107:12640–12645. doi: 10.1073/pnas.1000132107. [DOI] [PMC free article] [PubMed] [Google Scholar]