Abstract
The Cockayne syndrome complementation group B protein, CSB, plays pivotal roles in transcription regulation and DNA repair. CSB belongs to the SNF2/SWI2 ATP-dependent chromatin remodeling protein family, and studies from many laboratories have revealed that CSB has multiple activities and modes of regulation. To understand the underlying mechanisms of Cockayne syndrome, it is necessary to understand how the biochemical activities of CSB are used to carry out its biological functions. In this review, we summarize our current knowledge of the structure, function and regulation of CSB, and discuss how these properties can impact the biological functions of this chromatin remodeler.
1. CSB: A SNF2/SWI2 ATPASE FAMILY MEMBER
CSB belongs to the SNF2/SWI2 family of ATPases (Figure 1) (Flaus et al., 2006; Hopfner et al., 2012). These enzymes contain a central ATPase domain, which consists of seven conserved helicase motifs, and it is the homology between the ATPase domains that defines this protein family. The regions flanking the central ATPase domain are divergent between family members, and these regions often contain domains that are important for protein localization or auto-regulation (Figure 1). The SNF2/SWI2 family is, in fact, a subfamily of the broader helicase superfamily 2 (SF2), and the hallmark feature of a helicase is the ability to separate strands of duplex nucleic acids; however, a significant helicase activity has not yet been ascribed to any SNF2/SWI2 family member (Pyle, 2008; Selby and Sancar, 1997; Yusufzai and Kadonaga, 2008). Crystal structure studies of Rad54, a member of the SWI2/SNF2 family, revealed the absence of accessory domains believed to be important for strand separating activity, and this may explain the lack of helicase activity (Chen et al., 2005; Thoma et al., 2005). Instead, the SNF2/SWI2 proteins are best known for their ability to regulate chromatin structure, by using the energy from ATP hydrolysis to alter DNA-protein contacts. To date, the majority of eukaryotic SNF2/SWI2 proteins that have been studied alter contacts between DNA and histones, and thus these proteins are often referred to as ATP-dependent chromatin remodelers (Clapier and Cairns, 2009).
Figure 1.
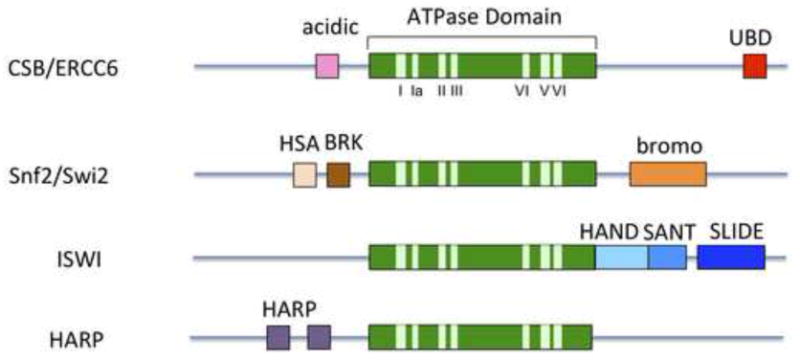
Representative SWI2/SNF2 ATP-dependent chromatin remodeling family members. The central ATPase domain (green) consists of seven conserved helicase motifs (light green) and is the most conserved region among family members. The CSB remodeler contains an acidic-rich region of unknown function and a ubiquitin-binding domain (UBD). Some remodelers have additional domains in their N- or C-terminal regions that are involved in mediating specific chromatin interactions or remodeler function. For example, the bromo domain has an increased affinity for acetylated proteins, including acetylated histones. The HAS domain has been found to mediate interactions with nuclear actin-related proteins (ARPs) as well as actin. The function of the HAND domain is still unclear, but it may be important for histone H3 tail binding (Grüne et al., 2003). The SLIDE domain appears to be important for DNA binding and ATPase activity and together, the SANT and SLIDE domains are likely important for nucleosome interaction. The HAPR domain dictates ATP-dependent annealing helicase activity (Ghosal et al., 2011). The BRK domain is of unknown function.
The nucleosome is the basic unit of chromatin structure, which is composed of approximately 150 base pairs of DNA wrapped around an octamer of histone proteins. Depending upon the particular chromatin remodeling activity, these enzymes can alter nucleosome position, composition or conformation (Figure 2) (Clapier and Cairns, 2009; Fan et al., 2004). By remodeling nucleosomes, ATP-dependent chromatin remodelers can create or prevent DNA access for factor binding, incorporate histone variants to mark nucleosomes, or space nucleosomes to organize chromatin domains and generate higher order chromatin structures. The activities of SNF2/SWI2 proteins are, however, not restricted to nucleosomes. For instance, the modifier of transcription 1 protein (MOT1) uses energy from ATP hydrolysis to displace the TATA-binding protein, TBP, from DNA (Auble et al., 1994; Madison and Winston, 1997; Wollmann et al., 2011). The HepA-related protein, HARP, has been shown to anneal single-stranded DNA (Yusufzai and Kadonaga, 2008). RAD54, a DNA repair and recombination protein, not only alters nucleosome positions but can increase the efficiency of pairing Rad51-coated single-stranded DNA filaments with duplex DNA (Alexiadis and Kadonaga, 2002; Zhang et al., 2007). The activities of the SNF2/SWI2 family members, therefore, can be quite diverse, but the characteristic feature of these proteins is to use energy from ATP hydrolysis to regulate chromatin fluidity.
Figure 2.
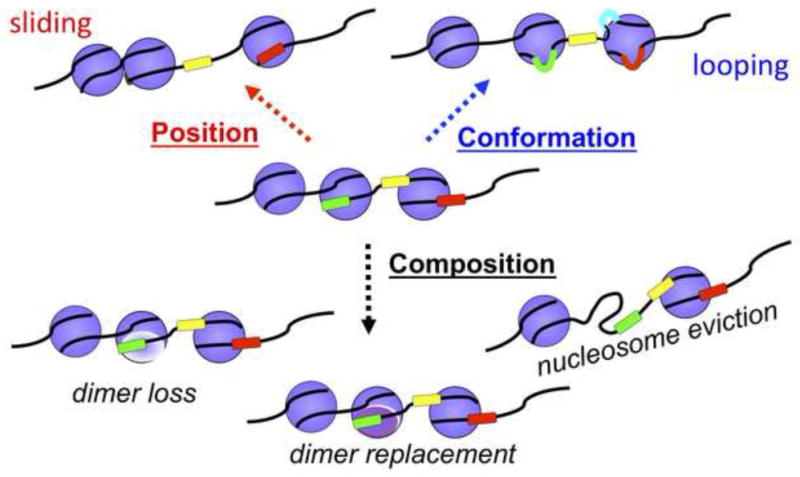
ATP-dependent chromatin remodelers alter chromatin structure by a variety of mechanisms. These mechanisms include changing the translational position of a nucleosome (nucleosome sliding), evicting an entire histone octamer, removing an H2A-H2B dimer, exchanging an H2A-H2B dimer (which can include exchanges between canonical histones or histone variants), and altering the conformation of a nucleosome.
ATP-dependent chromatin remodelers can function within the context of large macromolecular assemblies, such as BRG1 or BRM in the context of the SWI/SNF complexes. They can also function in isolation, such as the CHD1 remodeler (Clapier and Cairns, 2009). In general, ATP–dependent chromatin remodelers are active for remodeling on their own in vitro. The additional, non-catalytic subunits of a remodeling complex can increase the specific activity of the remodeler, regulate the remodeling outcomes or target the remodeling activity to specific genomic loci (He et al., 2008; He et al., 2006; Ho and Crabtree, 2010; Ito et al., 1999; Phelan et al., 1999). What then are the structural and biochemical characteristics of CSB that equip this remodeler to carry out its functions in transcription and DNA repair?
2. FUNCTIONAL INSIGHTS FROM CSB MUTATIONS
CSB can be roughly divided into three parts: a central ATPase domain flanked by N-terminal and C-terminal regions (Figure 1 and 3). Within the N-terminal region there is an acidic-rich region of unknown function (Brosh et al., 1999; Troelstra et al., 1992), and the C-terminal region harbors a ubiquitin-binding domain (UBD) (Anindya et al., 2010). There are also two predicted nuclear localization sequences that lie on either side of the ATPase domain.
Figure 3.

Summary of Cockayne syndrome-associated CSB mutations (Colella et al., 2000; Colella et al., 1999; Jaakkola et al., 2010; Laugel et al., 2010; Mallery et al., 1998).
Human genetic studies have cataloged a number of CSB mutations that are linked to Cockayne syndrome (for review, see this issue), and these mutations have revealed the functional importance of different CSB domains (Figure 3) (Colella et al., 2000; Colella et al., 1999; Jaakkola et al., 2010; Laugel et al., 2010; Mallery et al., 1998). As shown in Figure 3, there is a collection of missense mutations that lie within the central catalytic core. DNA readily stimulates the ATPase activity of CSB, and biochemical analyses of mutant CSB proteins have demonstrated that CSB proteins harboring the R670W or W851R mutations are defective for DNA-stimulated ATP hydrolysis (Lake et al., 2010). A CSB protein harboring the V957G mutation can hydrolyze ATP but with a substantially decreased efficiency (Lake et al., 2010). Each of these missense mutations, as well as the P1042L mutation, were also shown to be inactivating by virtue of their inability to restore resistance to UV irradiation that results from loss of CSB activity (Mallery et al., 1998). In addition to missense mutations, there are a handful of disease mutations that generate small deletions within the catalytic region, and these deletions also likely disrupt ATP hydrolysis. The biochemical defects that are coupled to Cockayne syndrome-associated CSB mutations highlights the importance of CSB’s enzymatic activity to its biological functions.
In addition to the disease-associated CSB mutations described above, several studies examined the consequences of mutating conserved residues that lie in the helicase motifs (Figure 1) (Brosh et al., 1999; Christiansen et al., 2003; Citterio et al., 2000; Muftuoglu et al., 2002; Selzer et al., 2002). These engineered mutations targeted residues that are likely important for coordinating the binding and hydrolysis of triphosphates (I, II and VI), coordinating ATP and nucleic acid binding (III) or contacting nucleic acid (Ia) (Fairman-Williams et al., 2010). As had been shown for other SNF2/SWI2 family members (Richmond and Peterson, 1996), the results of these studies revealed that the integrity of these motifs is important for the biochemical and biological functions of CSB.
The N- and C-terminal regions of chromatin remodelers often contain domains that are important for targeting these proteins to specific regions of chromatin (Figure 1). However, such domains are noticeably absent from CSB. Nonetheless, human genetic studies have revealed the importance of these regions to CSB function.
Within the C-terminal region of CSB, there are two disease-associated missense mutations, P1042L and P1095R. It is not yet clear how these mutations disrupt CSB’s functions, but the P1042L mutation does not interfere with ATP hydrolysis. In addition, even though the P1042L mutation lies within a predicted nuclear localization signal, it does not disrupt the ability of CSB to enter the nucleus (Lake et al., 2010). The P1095R mutation has not yet been biochemically examined. The importance of the C-terminal region to CSB function is further revealed by a number of other Cockayne syndrome-associated mutations that generate termination codons, frame-shifts and internal deletions within this region.
Interestingly, no disease-associated, missense mutations that lie within the N-terminal region of CSB have yet been described. There are, however, a number of mutations that introduce frame-shifts and stop codons in this region, indicating that the N-terminal region of CSB is not less prone to mutagenesis. This noticeable lack of disease-associated missense mutations might indicate that single residues in the N-terminal region do not play prominent functional roles, but rather groups of amino acids with similar properties, such as charge or hydrophobicity, may be critical to the function of the N-terminal region.
3. POST-TRANSLATION MODIFICATION OF THE CSB PROTEIN
Post-translational protein modification is a fundamental mechanism that is used to regulate protein function. One of the most common modifications is phosphorylation, which occurs on serine, threonine and tyrosine residues. Proteomic approaches have revealed that CSB is phosphorylated on multiple serine residues (Dephoure et al., 2008; Matsuoka et al., 2007; Nousiainen et al., 2006; Yu et al., 2007). Five of these residues lie in the N-terminal region of CSB (serines 158, 429, 430, 486 and 489) and three lie in the C-terminal region (serines 1142, 1348 and 1461), suggesting that the N- and C-terminal regions of CSB are critical targets for the posttranslational regulation of CSB activity (Dephoure et al., 2008; Matsuoka et al., 2007; Nousiainen et al., 2006; Yu et al., 2007). With the exception of serine 1461, all of the other serine residues have been found phosphorylated in mitotic cells. During mitosis, many DNA-binding proteins are excluded from the highly condensed chromatin, and this exclusion is coincident with increased protein phosphorylation. It has been hypothesized that protein phosphorylation may be a mechanism that is used to exclude DNA-binding proteins from mitotic chromatin. For example, BRG1, the ATPase motor of the SWI/SNF remodeling complex, is phosphorylated during mitosis and disengaged from chromatin (Sif et al., 1998). Although the cell cycle dynamics of CSB phosphorylation has not been examined, it is likely that at least some of these phosphorylation events are important to regulate CSB-chromatin association. Interestingly, Christiansen et al. observed that CSB is dephosphorylated upon UV irradiation, and that dephosphorylated CSB has increased ATPase activity (Christiansen et al., 2003); therefore, in addition to regulating chromatin association, phosphorylation likely plays an important role in modulating the enzymatic properties of CSB.
Among the phosphorylated serine residues, serine 1461 is unique as it is inducibly phosphorylated in response to ionizing radiation, and this residue lies within a putative ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) phosphorylation motif (Matsuoka et al., 2007). ATM senses double-strand DNA breaks and ATR senses single-stranded DNA, typically generated by the processing of double-strand breaks or stalled replication. Both of these kinases phosphorylate proteins to initiate a signaling cascade that results in cell cycle arrest. Together, these observations raise the possibility that phosphorylation of CSB at serine 1461 may be important to the mechanism of DNA-damage checkpoint control.
In addition to serine phosphorylation, CSB is phosphorylated at a single tyrosine residue (tyrosine 932) by the c-Abl kinase (Imam et al., 2007). The c-Abl kinase plays roles in a wide variety of cellular processes, which include cell survival, apoptosis and the DNA damage response. c-Abl-mediated phosphorylation of CSB was shown to impact the localization of CSB in the nucleus and nucleolus and was increased in cells challenged with the free radical generator hydrogen peroxide. Phosphorylation of CSB by c-Abl, therefore, may be important to regulate the subcellular localization of CSB in response to oxidative stress. In this study, it was also shown that c-Abl interacts with the N-terminal region of CSB through its SH3 domain, a region of c-Abl that negatively regulates its own kinase activity. These results suggested that CSB and c-Abl might reciprocally regulate each other’s activity. Given that tyrosine 932 lies within the ATPase domain of CSB, it is tempting to speculate that c-Abl phosphorylation might also play a role in modulating the ATPase activity of CSB, which could have consequences on both CSB-chromatin association and its enzymatic activity (Lake et al. 2010, Christiansen et al., 2003). This study further revealed the importance of the N-terminal region of CSB as a surface for protein-protein interactions.
Together, the results from these studies have demonstrated that the CSB protein is phosphorylated at multiple residues. By generating serine to alanine substitutions, we have found that phosphorylation of serine 158, 486 or 1461 does not impact the UV-induced association of CSB with chromatin (Lake and Fan, unpublished observations; see below). Additional systematic amino acid-substitution studies coupled with biochemical and cell biological assays are needed to understand the full impact of CSB phosphorylation on CSB function.
In addition to phosphorylation, CSB has been shown modified by the attachment of ADP-ribose moieties through the enzyme PARP-1 (poly(ADP-ribose) polymerase-1). This modification appears to occur at both the N- and C-terminal regions of CSB, with the N-terminal region being preferentially modified (Thorslund et al., 2005). Biochemical assays suggest that ADP-ribosylation inhibits the ATP hydrolysis activity of CSB, and evidence suggests that ADP-ribose addition may play a role in modulating the processing of a variety of DNA lesions by CSB (Flohr et al., 2003; Sakai et al., 2012; Thorslund et al., 2005)
4. CSB OLIGOMERIZATION
The oligomeric state of a chromatin remodeler can have a profound impact on the remodeling outcome. For instance, the SWI/SNF remodeler, a large, multiple-subunit remodeling complex, functions as a monomer. SWI/SNF regulates DNA access by opening or occluding sites for factor binding and has an essential role in transcription regulation. In contrast, the ACF remodeling complex functions as a dimer (Racki et al., 2009). ACF plays an important role in creating equally spaced nucleosomes for the formation of silent chromatin and higher order chromatin structures (Racki et al., 2009). By functioning as a dimer, it is believed that ACF can sense the length of linker DNA that lies on either side of a nucleosome to regulate the rate of nucleosome movement.
Scanning force microscopy size measurements indicated that CSB bound DNA as a dimer, as a consistent 1.6-fold increase in CSB volume was observed when CSB was bound to DNA, as compared to unbound CSB (Beerens et al., 2005). As had been proposed, however, an apparent increase in measured volume could also result from a change in CSB conformation upon binding to DNA (Beerens et al., 2005). Co-immunoprecipitation experiments using two differentially tagged forms of CSB also led to a similar conclusion, suggesting that CSB exists as a dimer (Beerens et al., 2005; Christiansen et al., 2005). It is important to note that co-immunoprecipitation experiments cannot directly address the precise stoichiometry of CSB oligomerization; therefore, a dimer would be indistinguishable from a trimer or another multimeric complex, as well as an unstructured aggregate. In yet another study, in vivo cross-linking in conjunction with gel filtration chromatography was used to address the question of CSB oligomerization (Christiansen et al., 2005). It was found that CSB migrated as two forms: as a large molecular mass form that eluted in the void volume, likely representing CSB-containing protein aggregates, and a well defined peak that migrated with a molecular mass of approximately 360 kD, consistent with CSB being a dimer. Molecular mass determinations from gel filtration chromatography, however, rely upon the assumption that a protein exists as a hydrated sphere, which may or may not be the case for CSB. Taken together, these studies suggest that CSB can oligomerize, but additional biophysical techniques, such as analytical ultracentrifugation, are needed to obtain a precise understanding of the CSB oligomeric state, both in isolation and in complex with DNA or nucleosomes.
5. ATP-DEPENDENT CSB ACTIVITIES
5.1. Alteration of chromatin structure
Biochemical studies have demonstrated that the ATP hydrolysis activity of CSB can be stimulated by a variety of DNA substrates, which include double-stranded DNA fragments, stem-looped DNA, DNA fragments with splayed arms, plasmid DNA and nucleosomal DNA. Although there appears to be a large degree of degeneracy in the types of substrates that stimulate CSB’s ATPase activity, the common feature of all these substrates is duplex DNA, and double-stranded DNA appears to be essential, as single stranded DNA oligonucleotides, DNA/RNA hybrids or RNA/RNA duplexes fail to stimulate ATP hydrolysis (Berquist and Wilson, 2009; Citterio et al., 1998; Selby and Sancar, 1997). What then are the biochemical processes that are coupled to DNA-stimulated ATP hydrolysis by CSB?
Using DNase I to monitor DNA accessibility, it was found that CSB could alter the DNase I sensitivity profile of a core mononucleosome in an ATP-dependent manner. Additionally, it was shown in this study that CSB could randomize an array of regularly spaced nucleosomes on plasmid DNA. These observations support the notion that CSB can function as an ATP-dependent chromatin remodeler (Citterio et al., 2000). The chromatin-remodeling activity of CSB, however, appeared to be very weak relative to other well-characterized chromatin remodelers, and the lack of robust remodeling activity has hindered the elucidation of its remodeling mechanism in vitro (Figure 2) and its biological functions in vivo.
In this regard, it is important to note that ATP-dependent chromatin remodelers are often components of macromolecular assemblies, and lessons learned from other ATP-dependent chromatin remodeling complexes indicate that non-catalytic subunits often enhance the specific activity of the ATPase motor. For example, the non-catalytic subunits of the human SWI/SNF remodeling complex, BAF155, BAF170 and INI1, enhance the enzymatic activity of the SWI/SNF2 motor protein BRG1 (Phelan et al., 1999). In fact, not all characterized ATP-dependent chromatin remodelers demonstrate robust in vitro remodeling activity in the absence of ancillary proteins. For instance, the Drosophila ISWI remodeler, the motor protein of the ACF complex that consists of ISWI and Acf1, has a very weak activity on its own, ~3% of the ACF complex (Ito et al., 1999). CSB has been found to interact with multiple proteins and to be a component of a large macromolecular complex (van Gool et al., 1997); therefore, it is likely that other proteins that interact with CSB will enhance its chromatin remodeling activity. In addition, it was found that histone tails, the N-terminal, unstructured regions of histones that protrude from the nucleosome surface, are important for mediating the interaction of CSB with nucleosomes, as removal of histone tails severely reduced the ability of CSB to render nucleosomal DNA accessible to DNase I digestion (Citterio et al., 2000). Given that histone tails are extensively modified, it is likely that histone modifications will also play an important role in modulating the chromatin remodeling activity of CSB.
5.2. Alteration of DNA conformation
Using topological assays with naked plasmid DNA, Beerens et al. found that CSB can change the conformation of DNA by introducing negative supercoils (Beerens et al., 2005). Moreover, it was found that a DNA conformation change occurred more efficiently when a non-hydrolysable ATP analogue was included in the assay. Using scanning force microscopy to study CSB-DNA complex structure, a shortening of DNA-contour length was detected upon CSB binding. As in the topological assays, shorter DNA molecules were observed more frequently in the presence of a non-hydrolysable ATP analogue. From the results of this study, a model was proposed whereby CSB alters the conformation of the DNA duplex by wrapping DNA. Given that the conformational change in DNA occurred more efficiently in the presence of an ATP analogue, it was suggested that the DNA wrapping activity of CSB is dependent upon ATP binding, and ATP hydrolysis would result in DNA unwrapping. What then would be the biological consequences of CSB-mediated DNA wrapping. It was proposed that DNA wrapping by CSB could be a mechanism that is used by CSB to regulate DNA-protein associations by altering DNA conformation. Although intriguing, this hypothesis awaits testing.
6. ATP-INDEPENDENT CSB ACTIVITIES
6.1. ssDNA annealing activity
In vitro biochemical studies have revealed that CSB facilities the annealing of single-stranded DNA (ssDNA), at a rate that is 25-fold faster than spontaneous annealing, and promotes strand exchange (Muftuoglu et al., 2006). CSB has also been shown to promote the annealing of DNA/RNA hybrids and RNA/RNA duplexes as well (Berquist and Wilson, 2009). Interestingly, the ssDNA annealing and strand exchange reactions do not require ATP. In fact, ATP was found to inhibit the strand annealing activity of CSB, as revealed by using CSB proteins that are defective for ATP hydrolysis as well as a non-hydrolysable ATP analog. Indeed, such ATP-independent strand annealing activity has also been observed for other members of the SF2 helicase superfamily, such as RECQ helicases (Wu and Brosh, 2010). Taken together, these results are consistent with a model whereby ATP binding induces a conformational change in CSB that inhibits its strand annealing activity. Consequently, in addition to being a source of energy for catalysis, ATP may also function as a molecular switch to regulate distinct activities of CSB. This molecular switch may, in turn, be regulated by post-translational modification, as in vitro studies suggested that dephosphorylated CSB has higher ssDNA annealing activity (Muftuoglu et al., 2006).
HARP, a member of the SWI2/SNF2 ATPase family, also displays strand-annealing activity; however, HARP requires ATP to carryout the annealing reaction (Betous et al., 2012; Yusufzai and Kadonaga, 2008). HARP has been shown to play a role in replication fork regression, a process important for maintaining genome stability, possibly by aiding DNA excision repair through the relocation of single-stranded DNA lesions back into duplex DNA. Similar to HARP, which binds selectively to forked DNA, CSB also interacts with forked DNA, but this interaction is not selective, as CSB associates with other forms of duplex nucleic acid (Berquist and Wilson, 2009; Citterio et al., 1998). Although the biological importance of the strand annealing activity of CSB has not been directly examined, it is tempting to speculate that it plays a role in the re-annealing of DNA to promote the repair of transcription-stalling lesions.
6.2. Enhancement of p53-chromatin association
The p53 tumor suppressor is known to interact with CSB both in vivo and in vitro (Wang et al., 1995), and both proteins are implicated in overlapping biological processes, such as DNA repair and aging. p53 binds to DNA in both sequence-dependent and sequence-independent manners. In vitro binding assays demonstrated that CSB facilitates the sequence-independent association of p53 with chromatin when p53 concentrations are low, and that this is achieved by the direct interaction of CSB with the C-terminal region of p53 (Lake et al., 2011). By sequestering the C-terminal region of p53, CSB would expose the p53 core domain and, thus, enhance DNA binding. This mechanism of enhanced p53-chromatin association appears similar to that resulting from the binding of the monoclonal antibody PAb421, which recognizes the C-terminal region of p53, and by deletion or covalent modification of the C-terminal 30 amino acids of p53 (Hupp et al., 1992). The facilitation of p53 binding to chromatin occurs independently of CSB’s ATPase activity. These in vitro observations are supported by results from in vivo chromatin immunoprecipitation experiments, which demonstrated that increased p53-chromatin association occurs upon reintroduction of CSB into a CSB-deficient cell line (Lake et al., 2011). The results of this study suggest that CSB may facilitate the sequence-independent binding of p53 to chromatin to assist p53 in scanning the genome for damaged DNA or in finding its target genes.
6.3. Enhancement of p53 ubiquitination
In addition to impacting the association of p53 with chromatin, evidence indicates that CSB also stimulates the ubiquitination of p53, mediated by the E3 ubiquitin ligase, MDM2 (Latini et al., 2011). It was suggested that the stimulation of p53 ubiquitination is part of a mechanism by which CSB refines p53 activity. Given that ATP was not included in the ubiquitination assays, it is likely that this stimulation occurs in an ATP-independent manner. Taken together, current data suggests that the functional relationship between CSB and p53 is complex and operates at several different cellular levels.
6.4. Enhancement of NEIL1 function
CSB has been found to enhance the in vitro incision activity of the DNA glycosylase NEIL1: a protein that cleaves the DNA backbone during base-excision repair, having a preference for oxidized pyrimidines (Muftuoglu et al., 2009). By analyzing CSB mutants, it was revealed that the enhancement of NEIL1 activity occurs independently of CSB’s enzymatic activity, and that this enhancement is mediated by the N-terminal 341 amino acids of CSB. Although the mechanism by which CSB enhances the incision activity of NEIL1 is unknown, the results of this study suggest that one function of the N-terminal region of CSB may be to stimulate either NEIL1 binding to DNA or NEIL1-mediated cleavage of DNA lesions.
6.5. Enhancement of APE1 function
APE1 is an apurinic/apyrimidinic (AP) endodeoxyribonuclease that functions in DNA base excision repair. APE1 initiates the repair of AP sites by catalyzing incision of the phosphodiester backbone immediately adjacent to a damaged base. Biochemical assays revealed that CSB associates with APE1 and stimulates its incision activity, and that this stimulation occurs in an ATP-independent manner. Interestingly, it was found that the incision activity of APE1 was more pronounced when the AP site was located in a transcription bubble, suggesting that CSB may preferentially stimulate APE1 activity in regions of active transcription (Wong et al., 2007). It will be interesting to determine which domain of CSB is responsible for the stimulatory effect.
6.6. DNA-protein complex dissociation
Two studies have examined the impact that CSB might have on the stability of a DNA-protein complex. In one study, it was shown that CSB stimulated the removal of an inactive restriction enzyme from a DNA template harboring a binding site for that restriction enzyme (Berquist and Wilson, 2009). In another study, it was shown that CSB stimulated the dissociation of TFAM (Transcription factor A, mitochondrial) bound to DNA (Berquist et al., 2012). In each instance, stimulation of protein dissociation occurred independently of ATP and, therefore, was different than that observed for other chromatin remodelers, such as the dissociation of TBP from DNA by MOT1, which is ATP-dependent (Auble et al., 1994; Madison and Winston, 1997; Wollmann et al., 2011). The mechanisms of DNA-protein dissociation, however, were suggested to be different. In the case of the restriction enzyme, it was hypothesized that protein dissociation was likely induced by CSB-mediated DNA distortion and, thus, the mechanism would lack specificity. In the case of TFAM, it was suggested that a direct CSB-TFAM interaction might promote the dissociation of TFAM from DNA. Such a mechanism would rely on competition between DNA and CSB for TFAM binding and, consequently, be specific and dictated by binding equilibria. These in vitro results suggest that CSB can promote the dissociation of a DNA-protein complex by different mechanisms. Additional studies are needed to understand further the mechanisms that CSB might use to promote the dissociation of DNA-protein complexes in vivo as well as the mechanisms by which specificity might be achieved.
Interestingly, TFAM along with two other mitochondrial proteins, POLMRT, a mitochondrial RNA polymerase and TBF2M, the mitochondrial transcription factor B2, were shown to enhance the ATPase activity of CSB in vitro (Berquist et al., 2012). These results led to the hypothesis that communication between these proteins might play a role in the control of mitochondrial transcription.
7. REGULATION OF CSB-CHROMATIN ASSOCIATION
ATP-dependent chromatin remodelers interact with DNA in a sequence-independent manner; however, they do not impact chromatin structure randomly but function in spatially regulated manners. How do ATP-dependent chromatin remodelers know where to go? ATP-dependent chromatin remodelers can be recruited to their sites of action by different mechanisms, which include interactions with specifically modified histones, with histone variants or with sequence-specific transcription factors. These interactions can be mediated directly by a remodeler (Figure 1) or indirectly through non-catalytic subunits of a remodeling complex (Hogan and Varga-Weisz, 2007; Horn and Peterson, 2001; Wu et al., 2007). Protein-protein interactions, therefore, play important roles in targeting specific chromatin-remodeling activities.
7.1. CSB targeting by sequence-specific DNA binding proteins
7.1.1. Targeting of CSB by TTF1
CSB has been found to be in a complex with RNA polymerase I and to be important for efficient ribosomal RNA (rRNA) expression (Bradsher et al., 2002; Xie et al., 2012; Yuan et al., 2007) (see this issue for a review article). Moreover, CSB has been shown to interact with TTF-1 (transcription termination factor I), a sequence-specific DNA-binding protein important for regulating rRNA transcription. Evidence indicates that the binding of TTF-1 to promoter proximal sites can recruit NoRC, the SNF2h-containing nucleolar remodeling complex, and NoRC recruitment leads to transcriptional repression at rDNA locus (Guetg et al., 2010). Given the interaction of CSB with RNA Polymerase I and TTF-1, as well as the impact of CSB on rRNA expression, it was hypothesized that TTF-1 might also recruit CSB to rDNA but, in this case, to activate rDNA expression (Yuan et al., 2007). Indeed, TTF-1 binding to rDNA was found to be important for CSB recruitment and enhanced rRNA expression. Together, these studies revealed that a sequence-specific DNA binding protein is important for targeting and regulating the activity of CSB in RNA polymerase I-mediated gene expression. However, it is still unclear how the enzymatic activity of CSB is used to modulate RNA polymerase I-mediated transcription.
7.1.2. Targeting of CSB by c-Jun
Evidence indicates that CSB participates in both transcription initiation and elongation mediated by RNA polymerase II, and CSB has been found in a large RNA polymerase II-containing complex (van Gool et al., 1997) (see this issue for a review article on CSB in transcription regulation). Interestingly, gene expression profiling has revealed that CSB does not impact transcription globally, but instead, CSB appears to have specific impacts on the genetic program of a cell (Kyng et al., 2003; Newman et al., 2006). How then is specificity achieved?
In a recent study, ChIP-seq was used to identify genomic sites that are occupied by the CSB-PGBD3 fusion protein, produced by alternative splicing and containing the N-terminal 465 amino acids of CSB fused to a PGBD3 transposase (Gray et al., 2012; Newman et al., 2008) (see this issue for a review article on CSB-PGBD3). In addition to shedding light on the causes and mechanisms of Cockayne syndrome, understanding the functions and activities of the CSB-PGBD3 fusion protein can also offer novel insights into the functions of the CSB N-terminal region. Gray et al. (2012) found that CSB-PGBD3 associates with the tumor promoting antigen response element TRE, which is best known as the binding site for activator protein 1 (AP-1) complexes (Eferl and Wagner, 2003; Gray et al., 2012). This result suggested that CSB-PGBD3 might interact with a TRE-binding protein. Indeed, it was found that the c-Jun transcription factor interacts with CSB-PGBD3 and that this interaction is mediated through the N-terminal CSB domain. Interestingly, isolated, full-length CSB did not interact with c-Jun, suggesting that an interaction between c-Jun and the N-terminal region of full-length CSB would likely be conformation dependent (Gray et al., 2012), possibly occurring on chromatin or in a transcription or repair complex. Importantly, these results suggest that the activity of CSB in RNA polymerase II-mediated gene expression might also be regulated by recruitment through sequence-specific DNA-binding proteins, such as c-Jun, similar to the recruitment of CSB to RNA polymerase I-responsive genes. This study also uncovered an additional role of the N-terminal region of CSB in protein-protein interaction and targeting. It will be of great interest to determine the extent to which c-Jun recruits CSB to sites containing TRE elements and if chromatin remodeling by CSB facilitates the association of c-Jun with DNA to regulate transcription.
7.1.3. Targeting of CSB by TRF2
Telomere shortening has been hypothesized to be an important component of the aging mechanism. Given that patients with Cockayne syndrome have features of premature aging, it was suggested that CSB may play a role in the maintenance of telomere length and integrity (Batenburg et al., 2012). It was found that CSB colocalizes with telomeres, albeit to a small subset at any given time. Additionally, it was found that CSB-deficient cells display defects in telomere maintenance. These observations raised the possibility that CSB plays a role in telomere biology. How then is CSB recruited to telomeres?
The telomeric repeat binding factor 2 (TRF2) was found to directly interact with CSB. TRF2 binds to telomeric DNA and plays a key role in telomere maintenance (van Steensel et al., 1998), Interestingly, this interaction did not appear to be confined to a single region, as the N-terminal, C-terminal and central catalytic regions of CSB could independently interact with TRF2 in vitro (Batenburg et al., 2012). Although the role of TRF2 in the recruitment of CSB to telomeres was not directly addressed, the results of this study suggest that TRF2 may be important for the sequence-specific targeting of CSB to telomeres to regulate telomere function or integrity.
7.2. An autoregulatory mechanism ensures the specific targeting of CSB to DNA lesion-stalled transcription
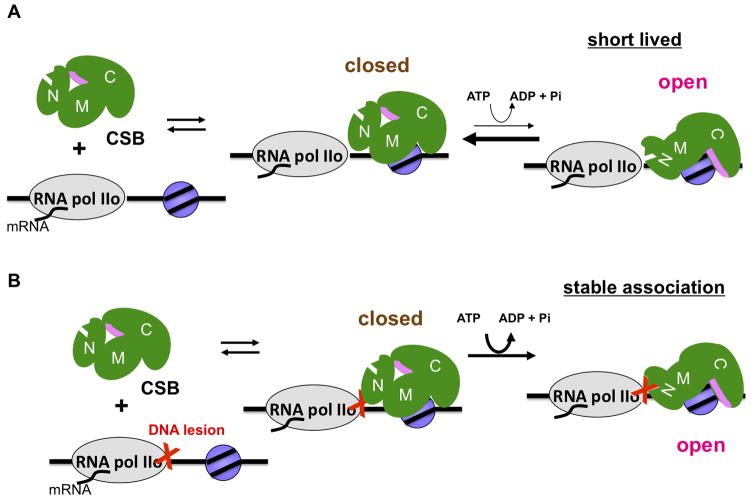
In contrast to most other well-characterized ATP-dependent chromatin remodelers, the vast majority of CSB is not stably bound to chromatin during normal cell growth (Figure 4) (Lake et al., 2010; van den Boom et al., 2004). By expressing a GFP-tagged CSB protein in CSB-deficient cells, van den Boom et al. found that CSB was evenly dispersed throughout the nucleoplasm, with additional accumulation in the nucleolus and nuclear foci of unknown composition (van den Boom et al., 2004). FRAP (Fluorescence Recovery After Photobleaching) studies revealed that the GFP-CSB protein in the nucleoplasm was freely mobile under normal growth conditions, suggesting that the association of CSB with chromatin is dynamic. The mobility of GFP-CSB decreased when cells expressing GFP-CSB were challenged with transcription stalling agents, such as actinomycin D or UV irradiation, suggesting that the association of CSB with chromatin was stabilized by stalled transcription.
Figure 4.
An auto-regulatory mechanism ensures the specific targeting of CSB to DNA-lesion stalled transcription. (A) Under conditions of normal cell growth, CSB interacts with chromatin dynamically. Most chromatin interactions are non-productive, as the N-terminal region of CSB inhibits DNA-stimulated ATP hydrolysis. Additionally, the N-terminal region occludes a chromatin interaction surface in the C-terminal region. Occasionally, productive chromatin interactions stimulate ATP hydrolysis, promoting a transition of CSB from a closed to an open conformation. In the absence of DNA-lesion stalled transcription, the open conformation is not maintained and readily converts back to a closed conformation. (B) However, in the presence of lesion-stalled transcription, the N-terminal region of CSB interacts with a component of DNA lesion-stalled transcription (red X). The open conformation is now maintained and stable CSB-chromatin association is accomplished by extending the chromatin interaction surface from the ATPase domain to C-terminal region.
Similar observations were obtained using chromatin fractionation approaches (Lake et al., 2010). In this study, it was found that during normal cell growth, approximately 10% of the endogenous CSB pool co-fractionated with chromatin. In response to UV irradiation, the association of CSB with chromatin increased, and this association was found to be directly proportional to the UV dose. The fraction of chromatin-associated CSB reached a plateau at 25 J/m2 (which introduces ~2 DNA lesions per 10 kb DNA), when approximately 90% of endogenous CSB was stably bound to chromatin (Lake et al., 2010; Mathonnet et al., 2003). The results of these studies are consistent with the notion that the association of CSB with chromatin is highly dynamic under conditions of normal cell growth. This dynamic CSB-chromatin association may be part of a mechanism that CSB uses to probe chromatin for lesion-stalled transcription. Once lesion-stalled transcription is encountered, CSB then stably associates with chromatin to promote DNA repair.
By expressing CSB proteins harboring Cockayne syndrome-associated mutations or engineered CSB derivatives, it was found that the stable association of CSB with chromatin in response to UV irradiation was dependent upon the ability of CSB to hydrolyze ATP (Lake et al., 2010). This observation was surprising, as the association of other ATP-dependent chromatin remodelers with chromatin does not rely upon ATP hydrolysis. Indeed, a genome-wide approach to determine the binding sites of the yeast ATP-dependent chromatin remodeler Isw2 reveled that an ATPase-defective Isw2 mutant protein, but not wild-type Isw2, was enriched at genes regulated by Isw2 (Gelbart et al., 2005). This study of CSB, therefore, demonstrated for the first time that an ATP-dependent chromatin remodeler can use its enzymatic activity to ensure its specific targeting to its site of action (Lake et al., 2010). Further mutational analyses revealed that the C-terminal region of CSB positively regulated chromatin association, as CSB proteins lacking the C-terminal region did not stably associate with chromatin in response to UV-induced DNA lesions. The N-terminal region, on the other hand, prevented CSB from stably associating with chromatin in the absence UV treatment (Figure 4A), as CSB proteins lacking the N-terminal region constitutively associated with chromatin during normal cell growth, and this association no longer required ATP hydrolysis. In addition, it was found that the N-terminal region of CSB functioned as an inhibitor of DNA-stimulated ATP hydrolysis (Lake et al., 2010).
Based upon these results, a model was proposed to explain the mechanism by which the association of CSB with chromatin in response to UV-induced DNA lesions is regulated (Figure 4). During normal cell growth, the association of CSB with chromatin is dynamic (Figure 4A). This dynamic association occurs because the N-terminal region of CSB occludes a chromatin-interaction surface that lies within the C-terminal region and promotes non-productive chromatin interactions by inhibiting ATP hydrolysis. At sites of DNA lesion-stalled transcription, the association of CSB with chromatin is stabilized (Figure 4B). Stable CSB-chromatin association is initiated by sequestration of the N-terminal region of CSB with an as yet unidentified component of lesion-stalled transcription, relieving the inhibitory effect imposed by the N-terminal region. Stabilization of CSB-chromatin association is then accomplished by extending the chromatin interaction surface from the ATPase domain to include residues in the C-terminal region. ATP hydrolysis by CSB is necessary to initiate relief of the autoinhibitory effect imposed by the N-terminal region (Lake et al., 2010). Interestingly, Gray et al. (2012) found that the CSB-PGBD3 fusion protein interacts with RNA polymerase II through the N-terminal CSB domain (Gray et al., 2012). This result suggests that relief of CSB autoinhibition at sites of lesion-stalled transcription may occur through a direct interaction between the N-terminal region of CSB and stalled RNA polymerase.
7.3. Dissociation of CSB from chromatin
7.3.1. p53-mediated CSB-chromatin dissociation
CSB interacts with the p53 tumor suppressor, and the carboxy-terminal region of p53 mediates this interaction (Wang et al., 1995; Yu et al., 2000). In a study exploring the mechanisms that can lead to metaphase chromosome fragility, it was found that loss of CSB or activation of p53 results in chromosome fragility at four loci encoding highly structured RNAs: RNU1, RNU2, RN5S and PSU1 (Yu et al., 1998; Yu et al., 2000). In this study, a model was proposed whereby CSB functions as a transcription elongation factor for structured RNAs, and activated p53 can sequester CSB. This sequestration would stall transcription and ultimately interfere with chromatin condensation at those loci (Yu et al., 2000).
In another study examining the relationship between CSB and p53 activities, it was found that high concentrations of p53 could prevent CSB from associating with chromatin in vitro (Lake et al., 2011). By examining the impact of p53 on the enzymatic properties of CSB, it was suggested that p53 can exclude CSB from nucleosomes by occluding a nucleosome interaction surface, composed of residues that lie within the central ATPase domain and C-terminal regions (Lake et al., 2011).
These studies are consistent with the notion that p53 can play a role in modulating the association of CSB with chromatin by sequestering CSB and, thus, promoting CSB-chromatin dissociation.
7.3.2. Ubiquitin-mediated CSB-chromatin dissociation
In addition to being important for chromatin association, the C-terminal region of CSB also plays an important role in regulating the dissociation of CSB from chromatin. A ubiquitin-binding domain (UBD) was identified in the C-terminal region of CSB (Figure 1) (Anindya et al., 2010). Interestingly, when the UBD was mutated, either through amino acid substitution or deletion, the resulting CSB derivatives failed to complement CSB function. However, functional complementation could by restored by attaching a heterologous UBD onto a CSB-deletion mutant. Interestingly, the UBD is dispensable for CSB-mediated assembly of nucleotide excision repair factors at sites of DNA lesion-stalled transcription, but the assembled repair complex failed to repair the DNA lesion (Anindya et al., 2010). By using FRAP analysis, it was found that, in the absence of a functional UBD, CSB became less mobile. The results of this study led to the intriguing hypothesis that the binding of CSB to a ubiquitinylated protein is necessary to trigger the dissociation of CSB from sites of DNA-lesion-stalled transcription and, thus, permit DNA repair (Anindya et al., 2010). Given the impact of p53 on the association of CSB with chromatin, as discussed above, it would be interesting to determine if ubiquitinylated p53 is part of the mechanism that triggers CSB dissociation (Lake et al., 2011). In addition to facilitating CSB-chromatin dissociation, the UBD might also be important for regulating chromatin remodeling by CSB or recruiting other unidentified factors necessary for the completion of DNA repair (Anindya et al., 2010).
8. WHAT DOES CSB DO ONCE RECRUITED TO SITES OF DNA LESION-STALLED TRANSCRIPTION?
Transcription-coupled DNA repair is a multistep process and CSB likely participates in different steps (Hanawalt and Spivak, 2008). The mutation frequency decline protein, Mfd, is a bacterial ATPase that is believed to be the prokaryotic counterpart of CSB. Mfd plays an essential role in displacing DNA lesion-stalled stalled RNA polymerase and initiating repair protein recruitment through direct protein-protein interaction (Selby and Sancar, 1995). In comparison, CSB has not yet been found to be capable of displacing a stalled RNA polymerase (Selby and Sancar, 1997). However, elegant work by Fousteri et al. revealed that CSB does recruit proteins important for carrying out DNA repair, and that this recruitment occurs without displacement of the stalled RNA polymerase (Fousteri et al., 2006). By comparing repair complex composition in different cell lines harboring mutations in several genes critical for transcription-coupled DNA repair, an order of protein recruitment was established. The results of this study revealed that CSB is the first protein recruited to DNA lesion-stalled transcription and is essential for repair protein recruitment. It will be important to determine if protein recruitment occurs through direct interactions with CSB and if it relies upon the ATP-dependent chromatin remodeling activity of CSB. In eukaryotes, the chromatin remodeling activity of CSB might be needed to reposition nucleosomes to create DNA access for repair factor binding or the processing of DNA lesions.
9. FUTURE PERSPECTIVES
From the in vitro and in vivo studies described above, it is clear that CSB performs like a multi-talented gymnast, displaying strength, balance and agility in a variety of competitive cellular events. These events include, but are not limited to, nuclear and mitochondrial transcription regulation, transcription-coupled nucleotide excision repair, base excision repair, autophagy and apoptosis (see other articles in this issue for comprehensive reviews on these topics). Although much progress has been made in understanding the structure, function and regulation of CSB, many important, unanswered questions still remain. For instance, how are the different biochemical activities of CSB used to carry out its biological functions? How is the ATP hydrolysis activity of CSB coupled to transcription regulation and DNA repair? How are the ATP-independent activities that have been observed in vitro utilized in vivo? How does the post-translational modifications of CSB impact its activities? What are the features of CSB that permit such diversity of function in transcription regulation and DNA repair? What are the mechanisms that CSB uses to remodel chromatin, and what are the remodeling outcomes: does CSB alter nucleosome position, composition or conformation (Figure 2)? How does the chromatin remodeling activity of CSB compare to that of other remodelers? In addition to the proteins described above, are there other proteins that might impact the activity of CSB? What is the molecular structure of CSB and how does this structure account for the different activities of this multi-talented gymnast? These and other questions open up a number of exciting and challenging avenues for future investigations.
HIGHLIGHTS.
Mechanisms of CSB-chromatin interaction
ATP-dependent CSB activities
ATP-independent CSB activities
CSB modification and oligomerization
Acknowledgments
Work from the authors’ laboratory is supported by the US National Institutes of Health (GM 084983 to H.-Y. Fan).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexiadis V, Kadonaga JT. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM, Svejstrup JQ. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol Cell. 2010;38:637–648. doi: 10.1016/j.molcel.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auble DT, Hansen KE, Mueller CG, Lane WS, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- Batenburg NL, Mitchell TR, Leach DM, Rainbow AJ, Zhu XD. Cockayne Syndrome group B protein interacts with TRF2 and regulates telomere length and stability. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. J Biol Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- Berquist BR, Canugovi C, Sykora P, Wilson DM, 3rd, Bohr VA. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquist BR, Wilson DM., 3rd Nucleic acid binding activity of human Cockayne syndrome B protein and identification of Ca(2+) as a novel metal cofactor. J Mol Biol. 2009;391:820–832. doi: 10.1016/j.jmb.2009.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L, Bohr VA. The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol Biol Cell. 1999;10:3583–3594. doi: 10.1091/mbc.10.11.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM, Jr, Bohr VA. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen M, Thorslund T, Jochimsen B, Bohr VA, Stevnsner T. The Cockayne syndrome group B protein is a functional dimer. Febs J. 2005;272:4306–4314. doi: 10.1111/j.1742-4658.2005.04844.x. [DOI] [PubMed] [Google Scholar]
- Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Colella S, Nardo T, Botta E, Lehmann AR, Stefanini M. Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-cacchione variant of xeroderma pigmentosum. Hum Mol Genet. 2000;9:1171–1175. doi: 10.1093/hmg/9.8.1171. [DOI] [PubMed] [Google Scholar]
- Colella S, Nardo T, Mallery D, Borrone C, Ricci R, Ruffa G, Lehmann AR, Stefanini M. Alterations in the CSB gene in three Italian patients with the severe form of Cockayne syndrome (CS) but without clinical photosensitivity. Hum Mol Genet. 1999;8:935–941. doi: 10.1093/hmg/8.5.935. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Narlikar GJ, Kingston RE. Noncovalent modification of chromatin: different remodeled products with different ATPase domains. Cold Spring Harb Symp Quant Biol. 2004;69:183–192. doi: 10.1101/sqb.2004.69.183. [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohr C, Burkle A, Radicella JP, Epe B. Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein. Nucleic Acids Res. 2003;31:5332–5337. doi: 10.1093/nar/gkg715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray LT, Fong KK, Pavelitz T, Weiner AM. Tethering of the Conserved piggyBac Transposase Fusion Protein CSB-PGBD3 to Chromosomal AP-1 Proteins Regulates Expression of Nearby Genes in Humans. PLoS Genet. 2012;8:e1002972. doi: 10.1371/journal.pgen.1002972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Yuan J, Chen J. The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1. EMBO Rep. 2011;12:574–580. doi: 10.1038/embor.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüne T, Brzeski J, Eberharter A, Clapier CR, Corona DFV, Becker PB, Müller CW. Crystal Structure and Functional Analysis of a Nucleosome Recognition Module of the Remodeling Factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. Embo J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- He X, Fan HY, Garlick JD, Kingston RE. Diverse Regulation of SNF2h Chromatin Remodeling by Noncatalytic Subunits. Biochemistry. 2008 doi: 10.1021/bi702304p. [DOI] [PubMed] [Google Scholar]
- He X, Fan HY, Narlikar GJ, Kingston RE. Human ACF1 Alters the Remodeling Strategy of SNF2h. Journal of Biological Chemistry. 2006;281:28636–28647. doi: 10.1074/jbc.M603008200. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C, Varga-Weisz P. The regulation of ATP-dependent nucleosome remodelling factors. Mutat Res. 2007;618:41–51. doi: 10.1016/j.mrfmmm.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Gerhold CB, Lakomek K, Wollmann P. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr Opin Struct Biol. 2012;22:225–233. doi: 10.1016/j.sbi.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Peterson CL. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front Biosci. 2001;6:D1019–1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- Imam SZ, Indig FE, Cheng WH, Saxena SP, Stevnsner T, Kufe D, Bohr VA. Cockayne syndrome protein B interacts with and is phosphorylated by c-Abl tyrosine kinase. Nucleic Acids Res. 2007;35:4941–4951. doi: 10.1093/nar/gkm386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola E, Mustonen A, Olsen P, Miettinen S, Savuoja T, Raams A, Jaspers NG, Shao H, Wu BL, Ignatius J. ERCC6 founder mutation identified in Finnish patients with COFS syndrome. Clin Genet. 2010;78:541–547. doi: 10.1111/j.1399-0004.2010.01424.x. [DOI] [PubMed] [Google Scholar]
- Kyng KJ, May A, Brosh RM, Jr, Cheng WH, Chen C, Becker KG, Bohr VA. The transcriptional response after oxidative stress is defective in Cockayne syndrome group B cells. Oncogene. 2003;22:1135–1149. doi: 10.1038/sj.onc.1206187. [DOI] [PubMed] [Google Scholar]
- Lake RJ, Basheer A, Fan HY. Reciprocally regulated chromatin association of Cockayne syndrome protein B and p53 protein. J Biol Chem. 2011;286:34951–34958. doi: 10.1074/jbc.M111.252643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake RJ, Geyko A, Hemashettar G, Zhao Y, Fan HY. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell. 2010;37:235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini P, Frontini M, Caputo M, Gregan J, Cipak L, Filippi S, Kumar V, Velez-Cruz R, Stefanini M, Proietti-De-Santis L. CSA and CSB proteins interact with p53 and regulate its Mdm2-dependent ubiquitination. Cell Cycle. 2011;10:3719–3730. doi: 10.4161/cc.10.21.17905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugel V, Dalloz C, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Cormier-Daire V, Gener B, Tobias ES, Tolmie JL, Martin-Coignard D, Drouin-Garraud V, Heron D, Journel H, Raffo E, Vigneron J, Lyonnet S, Murday V, Gubser-Mercati D, Funalot B, Brueton L, Sanchez Del Pozo J, Munoz E, Gennery AR, Salih M, Noruzinia M, Prescott K, Ramos L, Stark Z, Fieggen K, Chabrol B, Sarda P, Edery P, Bloch-Zupan A, Fawcett H, Pham D, Egly JM, Lehmann AR, Sarasin A, Dollfus H. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- Madison JM, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M, Lehmann AR. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet. 1998;62:77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Leger C, Desnoyers J, Drouin R, Therrien JP, Drobetsky EA. UV wavelength-dependent regulation of transcription-coupled nucleotide excision repair in p53-deficient human cells. Proc Natl Acad Sci U S A. 2003;100:7219–7224. doi: 10.1073/pnas.1232161100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muftuoglu M, Selzer R, Tuo J, Brosh RM, Jr, Bohr VA. Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene. 2002;283:27–40. doi: 10.1016/s0378-1119(01)00870-8. [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, Sharma S, Thorslund T, Stevnsner T, Soerensen MM, Brosh RM, Jr, Bohr VA. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 2006;34:295–304. doi: 10.1093/nar/gkj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Bailey AD, Fan HY, Pavelitz T, Weiner AM. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 2008;4:e1000031. doi: 10.1371/journal.pgen.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proceedings of the National Academy of Sciences. 2006;103:9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Sakasai R, Kakeji Y, Kitao H, Maehara Y. PARP and CSB modulate the processing of transcription-mediated DNA strand breaks. Genes Genet Syst. 2012;87:265–272. doi: 10.1266/ggs.87.265. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM, Jr, Bohr VA. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- Thorslund T, von Kobbe C, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol Cell Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- van den Boom V, Citterio E, Hoogstraten D, Zotter A, Egly JM, van Cappellen WA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J Cell Biol. 2004;166:27–36. doi: 10.1083/jcb.200401056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers JH. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. Embo J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Wang XW, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly JM, Wang Z, Freidberg EC, Evans MK, Taffe BG, et al. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- Wollmann P, Cui S, Viswanathan R, Berninghausen O, Wells MN, Moldt M, Witte G, Butryn A, Wendler P, Beckmann R, Auble DT, Hopfner KP. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011;475:403–407. doi: 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Wu Y, Brosh RM., Jr Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair (Amst) 2010;9:315–324. doi: 10.1016/j.dnarep.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Bailey AD, Weiner AM. Metaphase fragility of the human RNU1 and RNU2 loci is induced by actinomycin D through a p53-dependent pathway. Hum Mol Genet. 1998;7:609–617. doi: 10.1093/hmg/7.4.609. [DOI] [PubMed] [Google Scholar]
- Yu A, Fan HY, Liao D, Bailey AD, Weiner AM. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]
- Yu LR, Zhu Z, Chan KC, Issaq HJ, Dimitrov DS, Veenstra TD. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J Proteome Res. 2007;6:4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Fan HY, Goldman JA, Kingston RE. Homology-driven chromatin remodeling by human RAD54. Nat Struct Mol Biol. 2007;14:397–405. doi: 10.1038/nsmb1223. [DOI] [PubMed] [Google Scholar]