Abstract
T-cell acute lymphoblastic leukaemia (T-ALL) is a blood malignancy afflicting mainly children and adolescents1. T-ALL patients present at diagnosis with increased white cell counts and hepatosplenomegaly, and are at an increased risk of central nervous system (CNS) relapse2,3. For that reason, T-ALL patients usually receive cranial irradiation in addition to intensified intrathecal chemotherapy. The marked increase in survival is thought to be worth the considerable side-effects associated with this therapy. Such complications include secondary tumours, neurocognitive deficits, endocrine disorders and growth impairment3. Little is known about the mechanism of leukaemic cell infiltration of the CNS, despite its clinical importance4. Here we show, using T-ALL animal modelling and gene-expression profiling, that the chemokine receptor CCR7 (ref. 5) is the essential adhesion signal required for the targeting of leukaemic T-cells into the CNS. Ccr7 gene expression is controlled by the activity of the T-ALL oncogene Notch1 and is expressed in human tumours carrying Notch1-activating mutations. Silencing of either CCR7 or its chemokine ligand CCL19 (ref. 6) in an animal model of T-ALL specifically inhibits CNS infiltration. Furthermore, murine CNS-targeting by human T-ALL cells depends on their ability to express CCR7. These studies identify a single chemokine–receptor interaction as a CNS ‘entry’ signal, and open the way for future pharmacological targeting. Targeted inhibition of CNS involvement in T-ALL could potentially decrease the intensity of CNS-targeted therapy, thus reducing its associated short- and long-term complications.
Recent studies have shown that mutations of the developmental regulator Notch1 can be identified in most T-ALL patients7. It is estimated that activation of the Notch1 signalling pathway occurs in at least 80% of all T-ALL cases7-10. To investigate the mechanisms of T-ALL CNS infiltration and derive information that could be useful for treatment, we have attempted to establish animal models involving expression of oncogenic Notch1 (intracellular Notch1 fragment, Notch1-IC). The first model entails the transplantation of wild-type haematopoietic progenitors carrying Notch1-IC introduced by retroviral transfer (WTNotch-IC)11. The second model is on the basis of Cre/loxP recombination and involves Mx-Cre mice crossed with partners carrying dormant transgenic Notch1-IC, which was knocked-in into the ubiquitously expressed Eef1a1 locus12. The dormant Notch1-IC exerts oncogenic action after excision of a DNA segment blocking its expression, when Cre is expressed in haematopoietic progenitor cells by the IFN-α-inducible Mx1 promoter after polyinosinic:polycytidylic acid (poly(I:C)) injection. Both models developed T-ALL, presented atypical CD4+ CD8+ T cells in the peripheral blood as well as characteristic pathological features of T-ALL (Fig. 1 and Supplementary Figs 1 and 2). Immunohistochemical analysis demonstrated that in both models Notch1-IC–EGFP+ (enhanced green fluorescent protein) and CD3+ leukaemic cells efficiently infiltrated the leptomeningeal spaces of the brain (Fig. 1b, c and Supplementary Fig. 1). Further studies showed that the CNS infiltration was progressive, and was initially detected in mice in which leukaemic blasts were readily detected in their peripheral blood (Supplementary Fig. 3) and secondary lymphoid tissue (data not shown). We were thus able to show that oncogenic Notch1-IC expression was able to induce T-ALL and target the transformed cells to the CNS.
Figure 1. Notch1 activation induces T-ALL and targets leukaemic cells into the CNS.
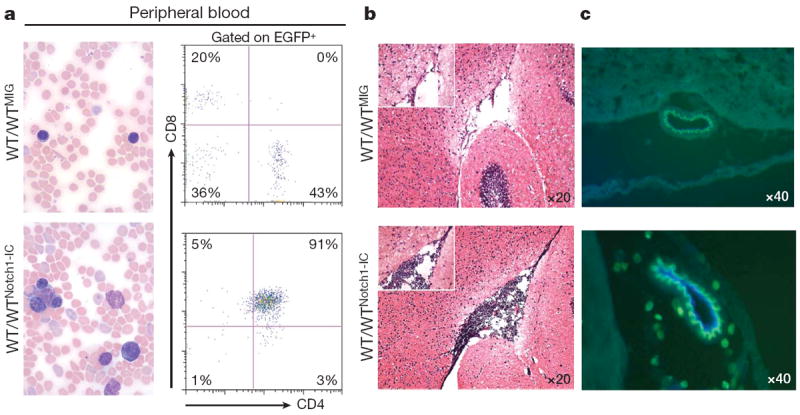
a, Induction of T-ALL in a transplantation model (WT/WTNotch1-IC). Peripheral blood smears (left), and fluorescence-activated cell sorting (FACS, right) analysis using CD4 and CD8 antibodies are shown. ‘WTMIG’ denotes wild-type bone marrow infected with a control MIG retrovirus. b, Notch1-IC+ EGFP + cells in the brain meningeal spaces of transplanted mice. c, Infiltrating lymphocytes surrounding a brain vessel in leukaemic (bottom panel) but not in healthy (control, top panel) recipients. Co-staining with CD31 antibodies (blue) indicates endothelial cells within the infiltrating lymphocytes.
We used a genome-wide transcriptome approach to identify Notch1-induced adhesion regulators that could be essential for CNS infiltration. Uncommitted haematopoietic progenitors were infected with Notch1-IC–EGFP+ retroviruses and gene expression was recorded 48 h later11. Detailed data mining demonstrated that a considerable fraction of Notch-controlled genes are potential regulators of cell adhesion, migration and metastasis (Fig. 2a and Supplementary Table 1). The expression of a specific gene, the chemokine receptor Ccr7, was significantly upregulated (Fig. 2a, b), and its expression remained constant after several days of culture (data not shown). CCR7 overexpression and function was also confirmed by real-time PCR, flow cytometry analysis and in vitro chemotaxis assays towards its known chemokine ligands CCL19 and CCL21 (Fig. 2b–d). CCR7 is an attractive candidate because it is a known regulator of lymphocyte migration6 and has been suggested to be important for the trafficking of lymphocytes participating in CNS immunosurveillance13,14. CCR7 functions through its interactions with CCL19 and CCL21 (ref. 6), and the expression and function of all three have been shown to be involved in the directional metastasis of several types of solid tumours, including melanomas and breast cancers15,16.
Figure 2. CCR7 expression and response to CCL19/CCL21 is induced by Notch1 activation.
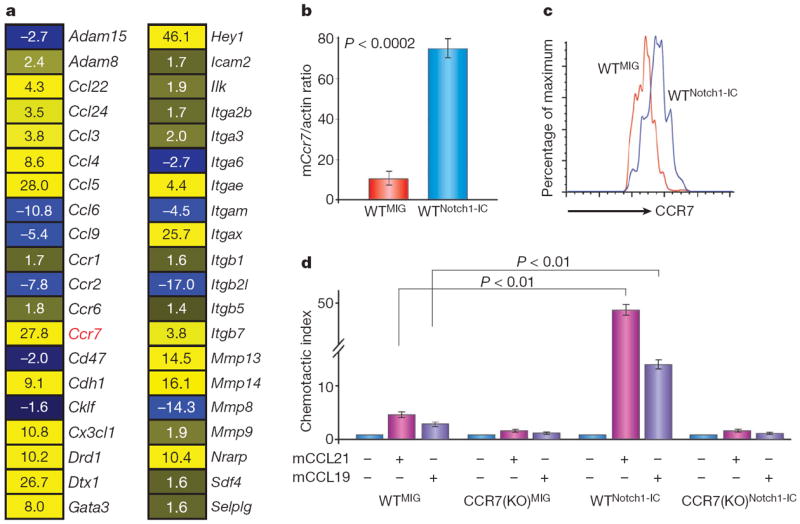
a, Heat diagram of selected adhesion/migration regulators that are controlled by Notch1-IC. A few classical Notch targets (Dtx1, Gata3, Hey1 and Nrarp) are also included. For all genes, P < 0.001. Yellow and blue denote increased and decreased mRNA abundance, respectively. b, c, Real-time PCR (b) and FACS (c) analysis showing the induction of CCR7 gene and protein expression in haematopoietic progenitors in response to Notch1-IC expression; n = 4. d, Notch1-IC expression induces the chemotaxis of wild-type (WTNotch1-IC), but not CCR7(KO)Notch1-IC progenitors towards both CCL19 and CCL21; n = 3. Error bars define s.d. for all experiments.
To correlate Notch1 activation and CCR7 expression further, we analysed T-ALL lines containing Notch1-activating mutations and primary T-ALL samples. Notably, surface CCR7 was expressed in 80% (4 out of 5) of T-ALL lines and in 73% (8 out of 11) of peripheral blood from primary T-ALL samples (Supplementary Fig. 4). CCR7 expression in the studied lines was dependent on Notch1 activation, as the repression of Notch1 processing due to the addition of γ-secretase inhibitors (DBZ or compound E) led to significant downregulation of CCR7 messenger RNA and protein expression (Supplementary Fig. 5 and not shown). To obtain a preliminary estimate of the importance of CCR7 expression in CNS infiltration, we selected two human T-ALL lines (Supplementary Fig. 4a), one that expressed (CEM/CCR7+) and one that did not express (DND41/CCR7−) the chemokine receptor, and infected them using a luciferase-expressing lentivirus. To study disease induction and progression, we transplanted the cells into alymphoid Rag2−/− Il2rg−/− hosts. We were able to demonstrate that the transplantation of an identical number of leukaemia cells led to distinct survival patterns, as hosts that received CEM/CCR7+ cells succumbed to the disease earlier than those receiving the DND41/CCR7− cells (Fig. 3a). Using live animal bioluminescent imaging, we were able to demonstrate a higher tumour load in mice that received the CEM/CCR7+ cell line (Fig. 3b). Most importantly, the brain and spinal cord of hosts transplanted with CEM/CCR7+ (but not with DND41/CCR7−) were infiltrated by T-ALL cells. Further histopathological analysis confirmed the localization of the infiltrating cells in leptomeningeal spaces (Fig. 3c, d).
Figure 3. CCR7 expression is sufficient for CNS infiltration of human T-ALL cells.
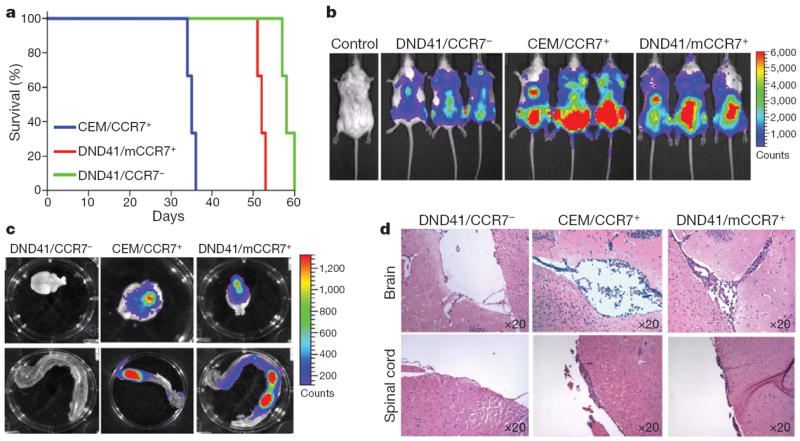
a, Kaplan–Meyer analysis of recipients that received identical numbers of CEM, DND41 or DND41/mCCR7+ cells; n =5.
b, Bioluminescent imaging of mice 2 weeks after transplantation with the indicated cell lines. c–d, Infiltration of T-ALL cells into the CNS of recipient mice as shown using bioluminescence and histochemistry.
As a next step, we addressed the sufficiency of CCR7 overexpression for the recruitment of leukaemic T cells in the CNS. We expressed mouse CCR7 (mCCR7) in the DND41/CCR7− human T-ALL cell line using a bicistronic retroviral vector expressing CCR7 in conjunction with expression of the mCherry fluorochrome (Supplementary Fig. 6a). We verified that retroviral transduction induced mCherry and mCCR7 expression, and that mCherry+ DND41 (DND41/mCCR7+) cells acquired the ability to efficiently respond to both CCL19 and CCL21 ligands (Supplementary Fig. 6b). Infected cells and control (mCherry-only expressing counterparts) were injected into Rag2−/− Il2rg−/− hosts. All recipients developed T-ALL (Fig. 3a, b) and no CNS infiltration was detected in the mice transplanted with the control DND41/CCR7− cells. On the other hand, we were able to demonstrate that CCR7 ectopic expression was sufficient to allow these cells to infiltrate both the brain and the spinal cord (Fig. 3c, d).
Although these results indicate that CCR7 expression is sufficient to recruit leukaemic T-cells into the CNS, they do not address whether CCR7 alone is necessary for this function. We infected haematopoietic progenitors from Ccr7−/− (CCR7(KO))17 and Ccr7+/+ (wild-type) mice with Notch1-IC-expressing retroviruses (CCR7(KO)Notch1-IC and WTNotch1-IC), and infected EGFP+ cells were transplanted into wild-type hosts (WT/CCR7(KO)Notch1-IC and WT/WTNotch1-IC). All recipient mice developed T-cell leukaemia, characterized by the detection of atypical peripheral blood CD4+ CD8+ cells as early as 2 weeks after transplant. As before, histopathological analysis showed notable tissue infiltration in all recipients (Supplementary Figs 2, 7 and data not shown). Hosts receiving leukaemic CCR7(KO) cells survived significantly longer than the hosts that received leukaemic wild-type cells (Supplementary Fig. 8). Although both hosts had similar leukaemic infiltration of most tissues, histopathological analysis of the CNS demonstrated that leukaemic CCR7(KO) cells were not found in the brain at 5 weeks after bone marrow transplantation, but leukaemic wild-type cells were (Fig. 4a). This was also true even at later time points just before the hosts succumbed, at 9–11 weeks after transplant (data not shown), uncoupling the tumour load to CNS infiltration. These results indicate that CCR7 is necessary for this process, and that the elimination of a single chemokine receptor in vivo is able to abrogate CNS involvement in T-ALL. CCR7 function seems to be specific for Notch1-induced T-cell malignancy, because deletion of this chemokine in two models of B-cell ALL failed to suppress CNS infiltration (Supplementary Fig. 9).
Figure 4. CCR7–CCL19 interactions are essential for CNS infiltration in an animal model of T-ALL.
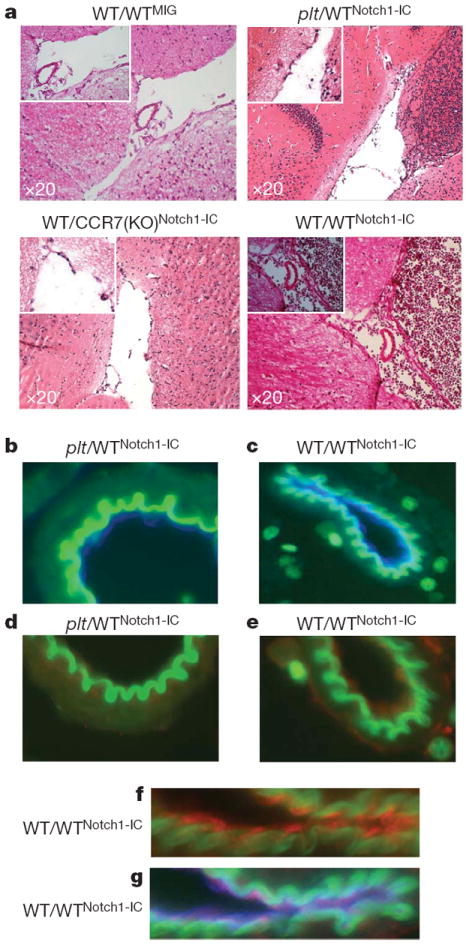
a, Leukaemic cells in the brain meningeal spaces of the indicated transplanted recipients. A positive control (WT/WTNotch1-IC) is also included. b–g, Immunofluorescent staining of brain sections: magnification of a brain microvessel in plt/WTNotch1-IC (b) and WT/WTNotch1-IC (c) mice. Endothelial, CD31+ cells are shown in blue. d–g, CCL19 (red) expression in WT/WTNotch1-IC (e, f, g) but not plt/WTNotch1-IC (d) microvessels. f, g, Magnification of a WT/WTNotch1-IC microvessel and costaining with CCL19 (red) and CD31 (blue).
We then sought to exclude the possibility that CCR7-deficient cells do not infiltrate the CNS because of a more general defect in their ability to migrate. As discussed before, CCR7(KO)Notch1-IC cells infiltrate as well as leukaemic wild-type cells into several tissues. Also, the peripheral blood of hosts reconstituted with CCR7(KO)Notch1-IC contained a similar (or even an increased) number of leukaemic blasts (Supplementary Figs 2 and 7). To attain a more dynamic view of the ability of the migratory properties of CCR7(KO)Notch1-IC and WTNotch1-IC leukaemic cells, we used two-photon imaging microscopy and traced EGFP+ cells within the lymph nodes of living leukaemic hosts. As illustrated in Supplementary Movies 1 and 2, we were unable to demonstrate any differences in the ability of these cells to migrate within the lymph nodes. When individual cells were traced, no statistically significant differences were found between the velocity, turning angle, arrest coefficient, confinement and random walk18 of CCR7(KO)Notch1-IC and WTNotch1-IC cells (Supplementary Fig. 10). These results underlined the specificity of CCR7 function in the targeting of T-ALL cells to the CNS.
Although these studies clearly demonstrate the importance of CCR7-mediated leukaemic cell recruitment to the CNS, they do not identify the ligand involved. It was previously shown that both chemokines are expressed in brain–blood barrier endothelium in an animal model of experimental autoimmune encephalomyelitis19,20. Moreover, CCL21 expression was detected previously in the choroid plexus—a possible site of lymphocyte entry into the subarachnoid space21. Thus, we performed immunohistochemical analyses to compare CCL19 and CCL21 expression in brain sections of control and leukaemic mice. CCL21 expression was undetectable in either sample (data not shown). However, CCL19 was detectable mainly in brain venules in the vicinity of infiltrating lymphocytes (Fig. 4b–g). Using immunofluorescence costaining, we confirmed that CD31+ endothelial cells produced CCL19 (Fig. 4f, g). In addition, we purified brain CD31+ endothelial cells and show a significant (8–10-fold when compared to CD31− brain cells) induction of Ccl19 message (Supplementary Fig. 11). Furthermore, endothelial cells from leukaemic animals expressed moderately higher (~twofold) levels of Ccl19 (Supplementary Fig. 12) when compared to endothelial brain cells from recipients that received EGFP-only infected cells (did not show any CNS infiltration). To prove that this slight overexpression is not, by itself, sufficient to attract CCR7+ T cells in the CNS, we have transplanted wild-type CD4+ T cells into leukaemic recipients. Analysis of recipient brain sections 12–16 h after transplantation failed to show any non-Notch1-IC–EGFP T-cell accumulation (Supplementary Fig. 12).
To prove the essential role of CCL19 expression, we modified our transplantation protocol and used plt mice as hosts, which lack CCL19 expression owing to a naturally occurring mutation22. We transplanted these mice (or background- and age-matched controls) with Notch1-IC-expressing haematopoietic progenitors (plt/WTNotch1-IC). In agreement with our previous transplantations, plt mice survived longer than their wild-type counterparts (Supplementary Fig. 8). As before, there was an identical T-ALL induction and disease manifestation between different hosts, suggesting that the plt mutation does not affect leukaemic cell infiltration in peripheral lymphoid tissues (Supplementary Figs 2 and 7). However, leukaemic cells were unable to infiltrate the brain of plt hosts, further strengthening our argument that CCR7–CCL19 interactions are essential for CNS infiltration in T-ALL (Fig. 4a, b).
Our data demonstrated that a single chemokine–receptor pair is both necessary and sufficient for T-cell ALL leukaemic cell targeting to the CNS. Although CCR7 expression seems to be an important signal targeting T-ALL cells in the CNS, other factors that are present in transformed leukaemic cells but not in wild-type T cells are needed for CNS infiltration. These factors could include extra adhesion regulators (that is, integrins and metalloprotease, see Fig. 2a) that could potentially interact with CCR7 function. We believe that our studies open the way for the development of different therapeutic protocols, in which specific adhesion antagonists23 can be used together with either current chemotherapy-based protocols or molecularly targeted approaches, for example, the use of antagonists of Notch signalling1. Putative T-ALL CNS infiltration antagonists could include inhibitors of CCR7 expression and function, as well as drugs targeting specific migration regulators, which could be activated by CCR7 signalling.
METHODS
Animals
C57BL/6 and Rag2−/− Il2rg−/− mice were purchased from Jackson Laboratories and Taconic Farms. CCR7(KO) and plt mice have been described previously17,22. All mice were kept in specific pathogen-free animal facilities at the NYU-SOM and Mount Sinai Medical School. All animal procedures were performed in accordance to the guidelines of the Institutional Animal Care and Use Committee of the NYU-SOM.
Recombinant DNA constructs and retrovirus production and infection
The Notch1-IC retroviral plasmid, its parent CMMP-based vector27, the pMXs IRES-mCherry retroviral vector and the pWPI lentivirus (a gift from E. Hernando) were used. Viral supernatants were generated as described previously28. Isolation, retroviral infection and reconstitution experiments were performed as previously described27.
Antibodies and reagents
Mouse CD4-APC, CD8-PE-Cy7, CCR7-PE, B22 and CD31, and human CCR7-PE and CD3-APC primary antibodies were purchased from BD Bioscience. Human CD3 antibody was obtained by DAKO. Mouse CCL19 and CCL21 were procured from R&D. Mouse CD31 was from Pharmingen. Cytokines and chemokines were obtained from Peprotech. Secondary antibodies were purchased from Jackson Laboratories. Immunohistochemical analysis was performed using standard methods as described previously11.
Chemotaxis assays
Chemotaxis assays were performed as previously described29.
Quantitative RT–PCR
RNA was isolated using RNeasy Plus Mini Kit (Qiagen) columns and used to synthesize cDNA with the SuperScript First-Strand Kit (Invitrogen). Real-time PCR was performed using iQ SYBR Green Supermix and an iCycler (Bio-Rad). Relative expression was determined from cycle threshold (CT) values, and normalized using β-actin as an internal control. All primer oligonucleotides are available on request.
Bioluminescent imaging
Imaging of luciferase-tagged leukaemic cells were performed as previously described24.
Intravital microscopy
Two-photon imaging was performed as previously described25. Individual cell tracing and data analysis was performed as previously described26.
Microarray analysis
The accession numbers for the individual array comparisons are Gene Expression Omnibus (GEO) series GSE6396, samples GSM147443, GSM147464 and GSM147508. Sample preparation and processing is detailed previously11. Pathway analysis of the microarray mRNA profiling results was performed using the Gene Ontology and KEGG pathway mapping within the web-based tool Database for Annotation, Visualization and Integrated Discovery. The results of category/pathway enrichment were manually curated for focused contents based on gene number and associated P value, and are summarized in Supplementary Table 1.
Supplementary Material
Acknowledgments
We are grateful to G. Randolph, E. Kuan and T. Vilimas for technical help and discussions. We would like to thank P. Lopez and G. De La Cruz for cell sorting; D. Littman, W. Carroll, E. Raetz, S. Lira and S. Schwab for advice and illuminating discussions; C. Loomis and the Histology Facility for advice and troubleshooting tips. This work was supported by the National Institutes of Health (RO1CA105129, RO1CA133379, R56AI070310, P30CA016087 to I.A., RO1AI41428, RO1AI072039 to J.S.B.), the American Cancer Society (RSG0806801 to I.A.), the Dana Foundation, The Chemotherapy Foundation, the Alex’s Lemonade Stand Foundation (to I.A.), the Lauri Strauss Leukemia Foundation (to F.S.), the G&P Foundation, NYU Molecular Oncology and Immunology training grant (to S.B.), American Society of Hematology (to S.B.), Juvenile Diabetes Research Foundation (JDRFI1-2008-90 and 5-2008-236 to J.S.B.), the National Cancer Institute (1 P01 CA97403, Project 2 to A.E.) and a gift from the Berrie Foundation (to A.E.). A.K. was supported by a Fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
Author Contributions S.B., B.G.M. and I.A. designed experiments, performed experiments and wrote the manuscript. M.L.D., M.Li and M.G.R. performed the in vivo two-photon microscopy experiments and analysis. D.M. and J.-C.T. performed the bioluminescent imaging experiments. A.K. and A.E. generated the EF1-Notch1-IC mice. J.Z. performed the microarray data mining. Y.L., E.N. and D.Z. performed in the CNS pathology analysis. M.Lipp and J.S.B. assisted with experimental design, provided reagents and advice. T.T., L.R., S.C. and E.G. performed experiments.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nature Rev Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 2.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nature Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Howard SC. Current management and challenges of malignant disease in the CNS in paediatric leukaemia. Lancet Oncol. 2008;9:257–268. doi: 10.1016/S1470-2045(08)70070-6. [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 5.Cyster JG. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 6.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nature Rev Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 7.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 8.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Neil J, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilimas T, et al. Targeting the NF-κB signaling pathway in Notch1-induced T-cell leukemia. Nature Med. 2007;13:70–77. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- 12.Klinakis A, et al. Igf1r as a therapeutic target in a mouse model of basal-like breast cancer. Proc Natl Acad Sci USA. 2009;106:2359–2364. doi: 10.1073/pnas.0810221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 14.Cardona AE, Li M, Liu L, Savarin C, Ransohoff RM. Chemokines in and out of the central nervous system: much more than chemotaxis and inflammation. J Leukoc Biol. 2008;84:587–594. doi: 10.1189/jlb.1107763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 16.Shields JD, et al. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell. 2007;11:526–538. doi: 10.1016/j.ccr.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 17.Forster R, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 18.Mempel TR, Scimone ML, Mora JR, von Andrian UH. In vivo imaging of leukocyte trafficking in blood vessels and tissues. Curr Opin Immunol. 2004;16:406–417. doi: 10.1016/j.coi.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Kivisakk P, et al. Human cerebrospinal fluid central memory CD4+ T cells: evidence for trafficking through choroid plexus and meninges via P-selectin. Proc Natl Acad Sci USA. 2003;100:8389–8394. doi: 10.1073/pnas.1433000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giunti D, et al. Phenotypic and functional analysis of T cells homing into the CSF of subjects with inflammatory diseases of the CNS. J Leukoc Biol. 2003;73:584–590. doi: 10.1189/jlb.1202598. [DOI] [PubMed] [Google Scholar]
- 21.Kivisakk P, et al. Expression of CCR7 in multiple sclerosis: implications for CNS immunity. Ann Neurol. 2004;55:627–638. doi: 10.1002/ana.20049. [DOI] [PubMed] [Google Scholar]
- 22.Gunn MD, et al. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J Exp Med. 1999;189:451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ransohoff RM. Natalizumab for multiple sclerosis. N Engl J Med. 2007;356:2622–2629. doi: 10.1056/NEJMct071462. [DOI] [PubMed] [Google Scholar]
- 24.Tseng JC, et al. Tumor-specific in vivo transfection with HSV-1 thymidine kinase gene using a Sindbis viral vector as a basis for prodrug ganciclovir activation and PET. J Nucl Med. 2006;47:1136–1143. [PubMed] [Google Scholar]
- 25.Shakhar G, et al. Stable T cell-dendritic cell interactions precede the development of both tolerance and immunity in vivo. Nature Immunol. 2005;6:707–714. doi: 10.1038/ni1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 27.Sicinska E, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–461. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 28.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scimone ML, Aifantis I, Apostolou I, von Boehmer H, von Andrian UH. A multistep adhesion cascade for lymphoid progenitor cell homing to the thymus. Proc Natl Acad Sci USA. 2006;103:7006–7011. doi: 10.1073/pnas.0602024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


