Abstract
Phloem sap contains a large repertoire of macromolecules in addition to sugars, amino acids, growth substances and ions. The transcription profile of melon phloem sap contains over 1000 mRNA molecules, most of them associated with signal transduction, transcriptional control, and stress and defense responses. Heterografting experiments have established the long-distance trafficking of numerous mRNA molecules. Interestingly, several trafficking transcripts are involved in the auxin response, including two molecules coding for auxin/indole acetic acid (Aux/IAA). To further explore the biological role of the melon Aux/IAA transcript CmF-308 in the vascular tissue, a cassette containing the coding sequence of this gene under a phloem-specific promoter was introduced into tomato plants. The number of lateral roots was significantly higher in transgenic plants expressing CmF-308 under the AtSUC2 promoter than in controls. A similar effect on root development was obtained after transient expression of CmF-308 in source leaves of N. benthamiana plants. An auxin-response assay showed that CmF-308-transgenic roots are more sensitive to auxin than control roots. In addition to the altered root development, phloem-specific expression of CmF-308 resulted in shorter plants, a higher number of lateral shoots and delayed flowering, a phenotype resembling reduced apical dominance. In contrast to the root response, cotyledons of the transgenic plants were less sensitive to auxin than control cotyledons. The reduced auxin sensitivity in the shoot tissue was confirmed by lower relative expression of several Aux/IAA genes in leaves and an increase in the relative expression of a cytokinin-response regulator, TRR8/9b. The accumulated data suggest that expression of Aux/IAA in the phloem modifies auxin sensitivity in a tissue-specific manner, thereby altering plant development.
Keywords: Aux/IAA, auxin response, phloem, root development, Solanum lycopersicum
Introduction
It is now evident that phloem sap contains a wide range of mRNA molecules (Vilaine et al., 2003; Omid et al., 2007; Buhtz et al., 2008; Zhang et al., 2009). Transcription profiling of the phloem sap of cucurbit plants has enabled the identification of thousands of mRNA molecules, as well as tRNAs, and small and microRNAs. Intuitively, one would think that all of these phloem-sap molecules are destined for long-distance movement. However, heterografting experiments indicate that only a small proportion of these molecules are capable of long-distance movement (Omid et al., 2007). Interestingly, numerous long-distance trafficking mRNA molecules have been characterized as coding for proteins involved in signal transduction mediated by gibberellin (Haywood et al., 2005), gibberellins and cytokinin (Banerjee et al., 2006) or auxin (Omid et al., 2007; Notaguchi et al., 2012).
We previously examined the ability of 43 melon phloem sap mRNA molecules to traffic long distances using melon–pumpkin heterografting experiments. Interestingly, only six of the examined melon transcripts were identified in the pumpkin scion (Omid et al., 2007). Annotation of these transcripts revealed that two of them were auxin/indole acetic acid (Aux/IAA) and one was small auxin-up RNA (SAUR), while the other three encoded “hypothetical proteins” (Omid et al., 2007).
Aux/IAA is a large family of early auxin response genes with 29 and 26 members in Arabidopsis and tomato, respectively (Overvoorde et al., 2005; Audran-Delalande et al., 2012). These genes encode transcriptional repressors of auxin response factor (ARF), thereby preventing transcription of genes controlled by these ARFs. Interaction of auxin with transport inhibitor response 1 (TIR1) and auxin F-box protein (AFB) forms part of the SCF ubiquitin-ligase (SCFTIR1) complex which catalyzes ubiquitin-mediated degradation of Aux/IAA (Teale et al., 2006). It has recently been shown that efficient auxin binding requires assembly of Aux/IAA and TIR1 proteins. The various combinations of TIR1–Aux/IAA complexes interact with auxin with a wide range of affinities (Calderon Villalobos et al., 2012).
The encoded Aux/IAA proteins are highly conserved in four distinct domains (Woodward and Bartel, 2005). Domain I is a transcriptional repressor, domain II is critical for Aux/IAA instability, and domains III and IV are involved in homodimerization and heterodimerization with other Aux/IAA proteins or with ARF (Reed, 2001). Genetic screens have identified Arabidopsis plants with mutations in various Aux/IAA genes that result in changed morphology. Most of these primary mutations were located in the highly conserved domain II, which is responsible for protein degradation. Such mutations result in stable proteins and gain-of-function phenotypes (e.g., insensitivity to auxin). An excellent example is the solitary root (slr) mutant which has reduced sensitivity to auxin (Fukaki et al., 2002, 2005; Vanneste et al., 2005). This dominant mutant completely lacks lateral roots and is also defective in root-hair formation and in gravitropic responses of its roots and hypocotyls. Map-based cloning and isolation of an intragenic suppressor mutant revealed that SLR encodes IAA14, a member of the Aux/IAA protein family (Fukaki et al., 2002).
Substantial phenotypic changes were also observed in transgenic tomato plants into which an antisense form of SlIAA9, another member of the Aux/IAA protein family, was inserted under the control of the CaMV-35S promoter (Wang et al., 2005). Significant reduction in the accumulation of IAA9 transcript was associated with altered leaf morphology, increased number of lateral shoots, parthenocarpic fruit development, enhanced hypocotyl/stem elongation and increased leaf vascularization. Auxin dose-response assay of cotyledon explants confirmed that SlIAA9 antisense plants are more sensitive to exogenous auxin than control plants (Wang et al., 2005). Interestingly, expression of SlIAA3 was higher in SlIAA9-antisense than control plants. Consistent with these results, roots of tomato plants in which SlIAA3 was silenced by expression of its antisense form were less sensitive to auxin than control roots (Chaabouni et al., 2009). Nevertheless, these antisense plants were also characterized by a higher number of lateral shoots.
The effect of Aux/IAA overexpression on plant development has been only scarcely examined. Transgenic Arabidopsis plants expressing the Vitis vinifera IAA9 (Fujita et al., 2012) or IAA19 (Kohno et al., 2012) grew somewhat faster but were similar to control plants in terms of morphological characteristics. On the other hand, overexpression of AtIAA20, AtIAA30 or AtIAA31 in transgenic Arabidopsis plants caused auxin-related aberrant phenotypes including semi-dwarfism, malformed vasculature and inhibition of primary root growth (Sato and Yamamoto, 2008). Significant inhibition of primary root length and increased number of adventitious roots were also observed when AtIAA17 was overexpressed in transgenic Arabidopsis plants (Worley et al., 2000).
It is important to note that all of these studies included transgenic plants expressing the Aux/IAA gene under the control of the CaMV-35S promoter. To better understand the biological function of Aux/IAA in the phloem, CmF-308—the long-distance-trafficking melon Aux/IAA—was expressed in the phloem of tomato plants under control of the AtSUC2 promoter. The phenotype of these plants indicated a modified auxin response while assays established tissue-specific alterations in auxin sensitivity. It is therefore concluded that the Aux/IAA gene product can exert its influence over plant developmental processes while being expressed in the phloem.
Materials and methods
Plant material
Tomato (Solanum lycopersicum), melon (Cucumis melo) and Nicotiana benthamiana plants were grown in a temperature-controlled greenhouse at 25–28/18–20°C (day/night, respectively), under natural sunlight. For hydroponic experiments, N. benthamiana was grown in trays containing coconut mixture. Two-week-old seedlings were transferred to containers (390 × 330 × 163 mm) with a nutrient solution containing 6 mM KNO3, 4 mM Ca(NO3)2, 2 mM KH2PO4, 0.03 mM EDFS [ethylenediamine tetraacetic acid iron (III) sodium salt], 0.5 μM CuSO4, 0.5 μM H2MoO4, 2 μM MnSO4, 50 μM KCl, 2 μM ZnSO4. The seedlings were transplanted into 5-cm plastic tubes that were fitted into holes drilled into the cover of the container such that the roots were inside the solution and the shoots above the cover. The nutrient solution was replaced twice weekly and was continuously aerated with an aquarium pump.
Transgenic tomato plants containing the pAtSUC2:GFP insert were employed to verify promoter activity. GFP fluorescent was visualized using confocal microscopy (CLSM510, Zeiss, Jena GmBH).
RNA isolation and RT-PCR
Total RNA was extracted from different tissues of tomato and melon plants using Tri-reagent (Sigma, http://www.sigmaaldrich.com/) according to the manufacturer's protocol. cDNA was prepared from the same amounts of RNA (1 μg) per sample pretreated with 1 unit μg−1 of RQ1 DNAse (Promega, http://www.promega.com), using the Verso cDNA synthesis kit (Thermo Scientific, http://www.thermoscientific.com). A 2-μl aliquot of cDNA was taken for PCR amplification. Real-time RT-PCR was carried out using 0.5 μl of 2.5 pmol of each primer (Table A1), 4 μl cDNA and 5 μl ABsolute™ Blue QPCR SYBR® Green ROX Mix. PCR conditions were as follows: 95°C for 10 s, 59°C for 15 s and 70°C for 25 s, repeated 45 times. The obtained cycle temperature (CT) values were analyzed with Rotor-Gene 6000 Series software by averaging the three independently calculated normalized expression values of the triplicates. The calculated numerical values were divided by the values obtained for the housekeeping gene tubulin in each respective sample.
Laser-capture microdissection
Expression of CmF-308 in specific cells of melon plants was determined using a laser-capture microdissection system according to Gil et al. (2011). In short, trimmed leaf discs were fixed in Farmer's fixative (3:1 v/v ethanol:acetic acid). Fixed tissue was dehydrated in a graded series of ethanols, after which it was incubated in isopropyl alcohol inside a microwave histoprocessor. Wax impregnation was performed under vacuum.
Cross sections (12 μm) were cut on a rotary microtome (Leica RM2245), floated in water at 42°C to stretch ribbons and incubated on membrane microscope slides. Prior to laser microdissection, slides were deparaffinized twice for 10 min each in 100% Histoclear (Gadot), followed by one wash in 100% ethanol (2 min) and then air-drying in the hood for 5 min. For microdissection, a PALM Laser Microbeam Instrument (Zeiss) was employed. Specific mesophyll and vascular bundle cells were isolated separately by the laser microbeam and collected into the lid of a 0.5-mL reaction tube (Zeiss) filled with 30 μ L ethanol, and placed in a holder located just above the slides.
RNA was extracted and isolated from each reaction tube using the PicoPure RNA isolation kit according to the manufacturer's protocol (Arcturus, http://www.moleculardevices.com/). Isolated samples were treated with the RNase-free DNase set kit (Qiagene, http://www.qiagen.com/). RNA was amplified twice and reverse-transcribed using MessageBOOSTER™ whole transcriptome cDNA synthesis kit (Epicentre, http://www.EpiBio.com/).
Cloning and plant transformation
The coding sequence plus 18bp of the 5' untranslated region (UTR), without the stop codon and the 3' UTR, of the CmF-308 mRNA was amplified from melon (Cucumis melo) cDNA by PCR and then cloned into the pTZ57R vector (Fermentas, 2886 bp) using the T-A ligation protocol. The gene was further restricted by Xho1 and Kpn1 and cloned into the pART7 vector upstream of three HA-tag repeats. The fused CmF-308-3xHA fragment was amplified by PCR, restricted with Sma1 and Hind3, and then cloned into the pART27 binary vector (Gleave, 1992) downstream of the AtSuc2 promoter (Truernit and Sauer, 1995) and upstream of the Ocs terminator. Cotyledon transformation was performed according to McCormick (1991).
Agro-infiltration
Agrobacterium tumefaciens (strain GV3101) containing the pART27 vector was grown overnight at 28°C in Luria-Bertani medium containing 50 mg L−1 gentamycin and spectinomycin. The culture was precipitated by centrifugation for 10 min at 3000 g and then resuspended in inoculation buffer containing 50 mM MES, 0.5% (w/v) glucose, 2 mM Na3PO4, and 200 μM acetosyringone to an OD600 of 0.5. The bacteria were then infiltrated into N. benthamiana leaves using a syringe without a needle. Shoots and roots of the infiltrated plants were collected 14 days after infiltration and dry weight was determined after 4 days at 65°C.
Auxin dose-response assay
Auxin-response assay was performed according to Wang et al. (2005). Tomato seeds were sown in sterile ½ MS medium containing 2.2 g L−1 MS and 30 g L−1 sucrose. Cotyledons (9 days old) were dissected and placed on MS medium containing 4.4 g L−1 MS and 30 g L−1 sucrose with varying concentrations of NAA. Root development from the cut cotyledons was determined 12 days after dissection.
The root-development response to auxin was analyzed in developing seedlings. Seeds were sown on germination papers placed in plastic bags supplemented with tap water. Germinated seeds were transferred to new germination papers soaked in solutions with varying concentrations of NAA, in plastic bags. The bags were kept in the dark at a temperature of 25 ± 2/18 ± 2°C for 10 days, after which the length of the primary root was measured.
Results
CmF-308 transcript accumulates in the vasculature of melon plants
Following our finding that the melon phloem-sap transcript CmF-308 is capable of long-distance trafficking, our initial set of experiments was aimed at identifying its level of accumulation in various tissues of melon plants. Quantitative (q) RT-PCR analyses revealed highest relative expression of this transcript in the hypocotyls and stems of young melon seedlings, with significantly lower relative expression in roots, leaves and shoot apices (Figure 1A). To further verify that CmF-308 accumulates in tissues enriched with vascular bundles, veins were separated from young mature leaves. Here again, relative expression of CmF-308 in the veins was three times higher than that in the rest of the leaf (Figure 1B). Predominant accumulation of CmF-308 in vascular cells was confirmed by collection of specific cell types using the laser-capture microdissection system (Figure 1C).
Figure 1.
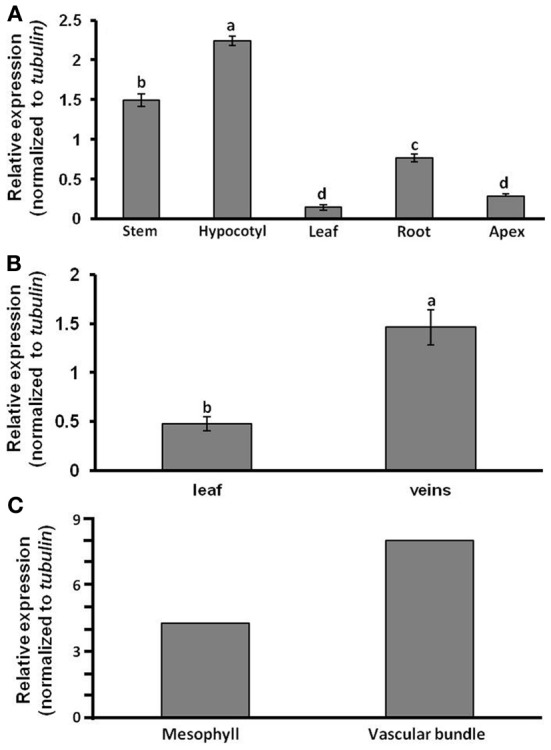
Relative expression levels of CmF-308, as determined by real-time RT-PCR. Relative expression was compared in various melon plant organs (A), in veins and leaf lamina (B) and in mesophyll and vascular bundle cells isolated by laser-capture microdissection system (C). Leaf discs and single cells were collected from the youngest mature source leaf (no. 4). Data represent means of three to six replications (±SE). Different letters indicate significant differences between plant organs or cell types at p < 0.05 by Tukey's HSD-test.
Transient expression of CmF-308 modifies root development in N. benthamiana plants
As indicated, F-308 codes for Aux/IAA, one of the auxin-response regulators. The next set of experiments was aimed at studying the potential functioning of this gene product in the phloem, as a component of the auxin response pathway. A cassette harboring the coding sequence of CmF-308 under the AtSUC2 promoter was agroinfiltrated into leaves of N. benthamiana plants (Figure 2D). Presence of the melon transcript in the apices of these N. benthamiana plants 48 h post-infiltration established that this transcript is indeed capable of long-distance movement (Figure 2A). It is important to note that the gfp transcript that served as a control was absent from the stem and shoot apices, indicating that long-distance movement of RNA molecules is not a general phenomenon (Figure 2B). PCR analysis failed to detect segments of the binary plasmid, inserted into the Agrobacterium, outside the infiltrated leaf, indicating that the bacteria were restricted to this tissue during the first 48 h post-infiltration (Figure 2C).
Figure 2.
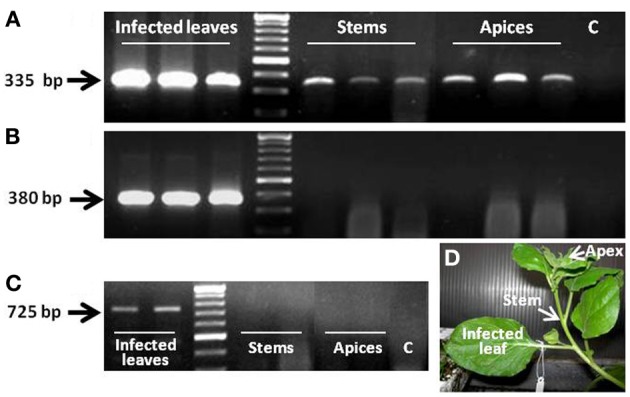
Presence of CmF-308 transcript in the stems and apices of N. benthamiana plants 48 h after agroinfiltration of source leaves with plasmid containing the coding sequence of this transcript, indicating long-distance movement of the transcript (A). Absence of the gfp transcript from the apices and stems and presence of the transcript in the agroinfiltrated leaf (B). (C) Presence of the agrobacterium in the infected leaf 48 h after agroinfiltration. C, negative control RT-PCR with no cDNA template. (D) Picture of the infiltrated N. benthamiana plant.
Interestingly, root mass of plants infected with the plasmid containing CmF-308 was significantly higher than that of plants into which the control CaMV35S:GFP cassette was inserted (Figure 3). The enhanced root growth in plants transiently expressing the CmF-308 gene product was evident both in pot-grown (Figures 3A vs. 3B) and hydroponically grown (Figure 3C) plants. The higher root weight of plants expressing the CmF-308 gene product was mainly due to extensive lateral root development (Figure 3A).
Figure 3.
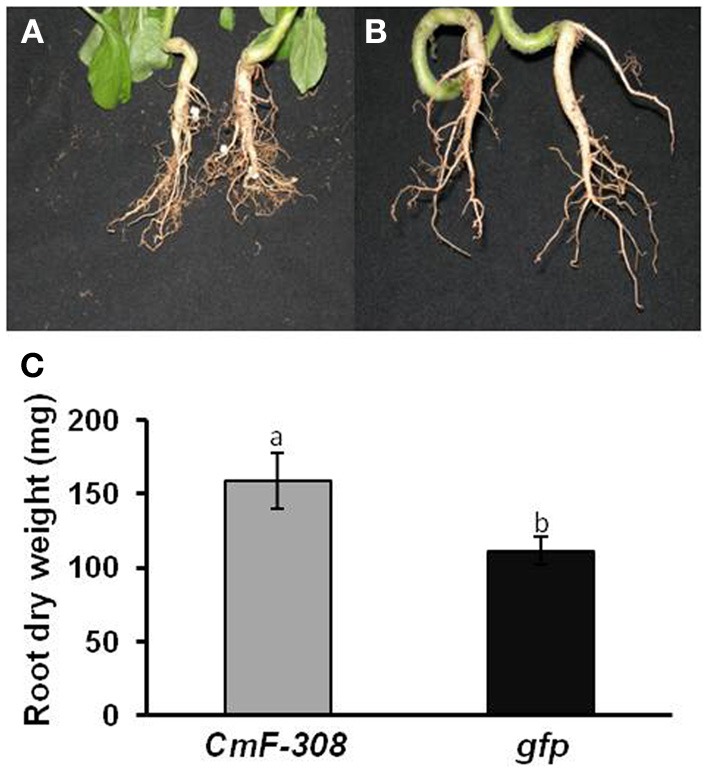
Effect of transient expression of CmF-308 on root mass of N. benthamiana plants. Roots of pot-grown plants agroinfiltrated with plamids containing the coding sequence of CmF-308 (A) or gfp (B). Pictures were taken 14 days after infiltration of the respective plasmids into the source leaves. (C) Root mass of hydroponically grown N. benthamiana plants 14 days after infiltration of the CmF-308 or gfp coding sequence into the source leaves. Data represent means of eight replications ± SE. Different letters indicate significant differences between plant lines at p < 0.05 by Student's t-test.
Overexpression of CmF-308 in the phloem alters root and shoot development of tomato plants
To further explore the effect of phloem-limited expression of CmF-308 on plant development, the gene's coding sequence was inserted into transgenic tomato plants under the control of the phloem specific AtSUC2 promoter (Shalit et al., 2009). Activity of the AtSUC2 promoter, as phloem specific promoter in tomatoes was verified using GFP as a reporter protein (Figure 4A). In agreement with the activity of AtSUC2 promoter along the vasculature, CmF-308 was expressed in the shoot apices and roots of the transgenic tomato plants (Figure 4B). Over ten independent homozygous CmF-308-transgenic tomato plants were generated. Most of them had similar phenotype and two representative lines were selected for further study. Similar to the effect observed after transient expression, constitutive expression of CmF-308 in the phloem of tomato plants caused a significant increase in the number of lateral roots and root weight (Figures 5A–C). Note that shoot weight was similar in CmF-308 and control tomato plants (Figure 5D).
Figure 4.
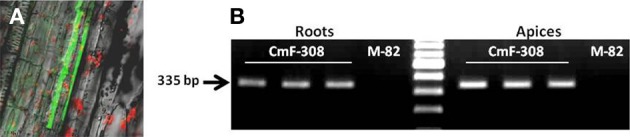
(A) Activity of AtSUC2 promoter in transgenic tomato plants. GFP is visualized the vasculature (companion cells) of tomato stem. (B) Presence of CmF-308 transcript in shoot apices and roots of three independent transgenic tomato plants expressing the gene under the AtSUC2 promoter. The commercial variety M-82 served as a control for the RT-PCR assay.
Figure 5.
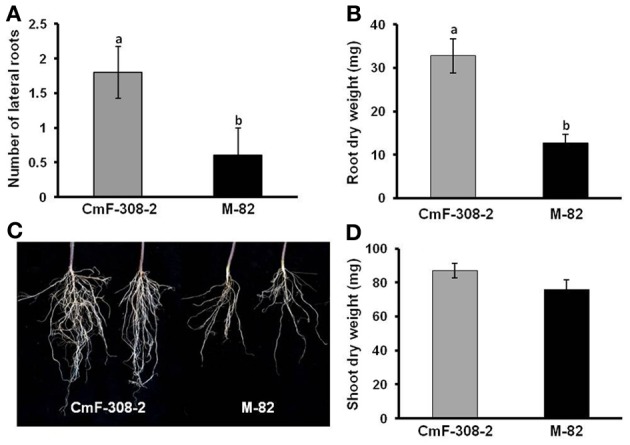
Effect of CmF-308 expression in the phloem of tomato plants on root development. Number of lateral roots (A), root dry weight (B), and a picture (C) presenting the differences between roots of transgenic tomato plants expressing CmF-308 under the AtSUC2 promoter (CmF-308-2) as compared with the control variety M-82. (D) Shoot dry weight of transgenic tomato plants expressing CmF-308 under the AtSUC2 promoter (CmF-308-2) as compared with the control variety M-82. Lateral roots were counted in 9-days old seedlings germination on germination papers (A). Shoot and root weight was measured in 3-week old pot-grown plants (B–D). Data represent means of six replications ± SE. Different letters indicate significant differences between the plant lines at p < 0.05 by Student's t-test.
In addition to the influence on root development, phloem-specific expression of CmF-308 significantly affected shoot development (Figure 6). Transgenic plants expressing CmF-308 in the phloem were significantly shorter (Figure 6F) and had a higher number of axillary shoots than the control tomato variety. Whereas in the control variety, lateral shoots developed in about 50% of the nodes, the percent of lateral shoots in the transgenic plants was between 70 and 80% (Figure 6E). The axillary shoots were longer, exhibiting a typical phenotype of reduced apical dominance (Figures 6A–D). In addition, these transgenic plants were characterized by delayed flowering.
Figure 6.
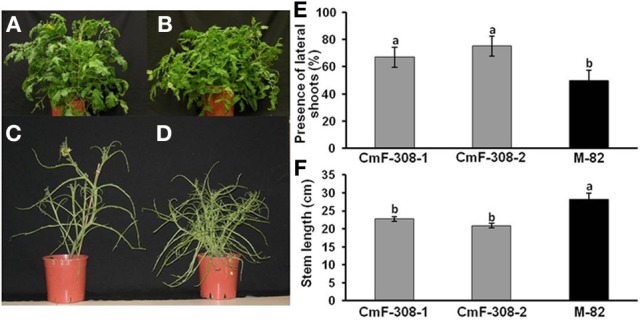
Effect of CmF-308 expression in the phloem of tomato plants on shoot development. Control variety M-82 (A,C) as compared with transgenic plants expressing CmF-308 under the AtSUC2 promoter (B,D). Pictures were taken 60 days after germination. Plants were stripped of all their leaflets (C,D) to assist in visualizing the branching phenotype. Percentage of lateral shoots (E) and stem length (F) of the control tomato variety M-82 and two transgenic lines expressing CmF-308 under the AtSUC2 promoter, 30 days after germination. Data represent means of 12 replications ± SE. Different letters indicate significant differences between the plant lines at p < 0.05 by Student's t-test.
Overexpression of CmF-308 in the phloem affects auxin sensitivity
The changes in lateral root and axillary shoot development suggested altered sensitivity to auxin in plants expressing CmF-308 under a phloem-specific promoter. We therefore examined the sensitivity of roots and shoots of transgenic and control plants to auxin. A dose-response assay revealed significant inhibition of primary root lengthening in CmF-308 plants at a concentration of 0.5 μM NAA, with no significant effect of this concentration on root length of control tomato plants (Figure 7A). This indicated that the roots of plants expressing the CmF-308 gene product are more sensitive to exogenous auxin than control roots. Primary root elongation of both control and CmF-308 plants was significantly inhibited by 1 μM NAA.
Figure 7.
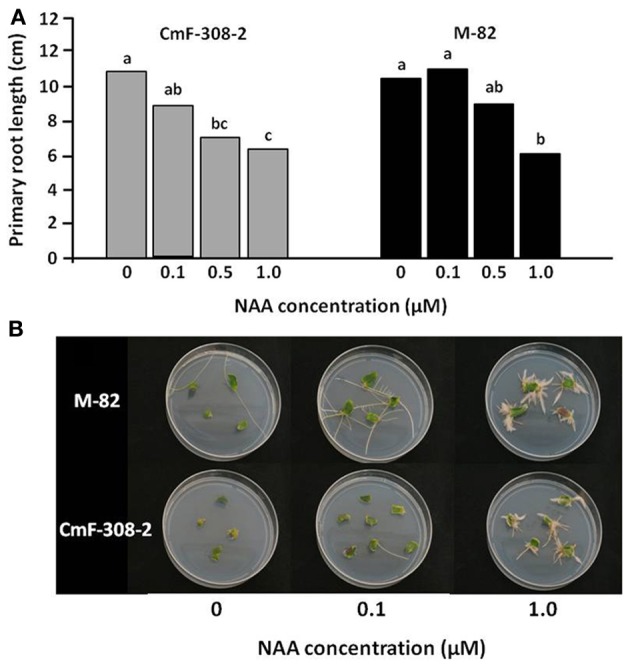
Auxin response of CmF-308-transgenic tomato plants. (A) Primary root length of CmF-308-2 and control M-82 tomato seedlings after incubation in various concentrations of NAA. Seedlings were germinated on filter paper soaked in the auxin solutions in the dark. Lengths of primary roots were measured 10 days after germination. Data represent means of eight replications ± SE. Different letters indicate significant differences between auxin concentration treatments in each plant line Axillary at p < 0.05 by Tukey's HSD-test. (B) Auxin dose-response assay of cotyledon explants showing reduced auxin sensitivity in CmF308-2 cotyledons as compared to controls. Root regeneration is promoted by 0.1 μM NAA in the control variety and by 10 times higher concentration (1.0 μM NAA) in the transgenic line CmF-308-2. Pictures of representative plates were taken 12 days after placing the cut cotyledons on the various auxin media.
Additional assay was aimed to examine the auxin response in shoot tissue. Cotyledon segments were subjected to various NAA concentrations and the number of developing roots was monitored. Interestingly, substantial rooting was evident in control cotyledons subjected to 0.1 μM NAA, whereas only a negligible number of roots developed from cotyledons of CmF-308-transgenic plants (Figure 7B). Similar differences in rooting level could be observed when the cotyledons were subjected to 1 μM NAA.
Collectively, these results indicated that overexpression of CmF-308 in the phloem of tomato plants causes a decrease in the shoot segments' sensitivity to auxin and an increase in roots' sensitivity.
Overexpression of CmF-308 in the phloem affects hormone-related gene expression
It is logical to assume that tissue-specific alteration in auxin sensitivity due to overexpression of CmF-308 in the phloem is associated with changes in related genes' expression. To further explore the mode by which CmF-308 expression affects auxin sensitivity, relative expression of various Aux/IAA transcripts was analyzed in leaves and roots of CmF-308-transgenic and control tomato plants. Consistent with reduced sensitivity to auxin, relative expression of the tomato IAA7, IAA10 and IAA14 was significantly lower in the leaves of CmF-308-transgenic vs. control plants (Figure 8A). Interestingly, relative expression of IAA3 was higher in CmF-308 vs. control leaves, while no significant differences were observed in the relative expression of IAA8 and IAA9 between leaves of the two tomato lines.
Figure 8.
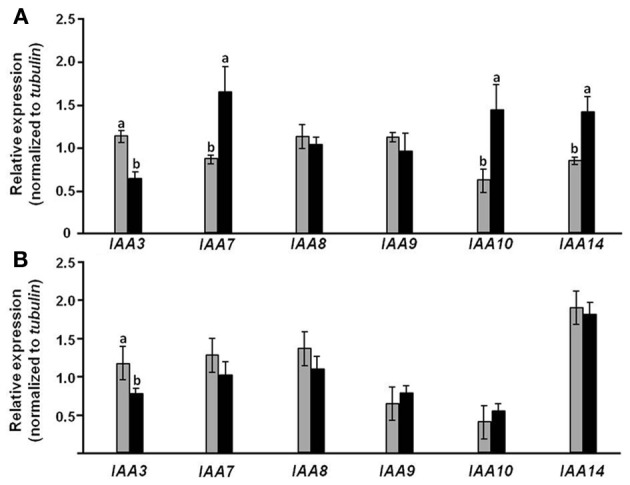
Effect of CmF-308 expression in the phloem of tomato plants on the expression of auxin-response genes. Expression of members of the Aux/IAA gene family was determined by qRT-PCR in leaves (A) and roots (B) of 21-day-old CmF-308-transgenic (gray columns) and control M-82 (black columns) plants. Data represent means of four replications ± SE. Different letters indicate significant differences in relative expression of specific genes between the two plant lines at p < 0.05 by Student's t-test.
Relative expression of most examined Aux/IAA transcripts was similar in the roots of transgenic and control tomato plants. An exception was IAA3, whose relative expression was significantly higher in roots of CmF-308 vs. control plants (Figure 8B).
Further study was aimed at verifying the interaction between expression of auxin- and cytokinin-responsive genes (Figure 9). Relative expression of four cytokinin-induced Type A-tomato response regulators (TRRs) was similar in roots of CmF-308-transgenic and control plants (Figure 9B). However, relative expression of TRR8/9b was almost five times higher in leaves of CmF-308 vs. control plants (Figure 9A), suggesting a higher cytokinin response associated with the observed reduced auxin response.
Figure 9.
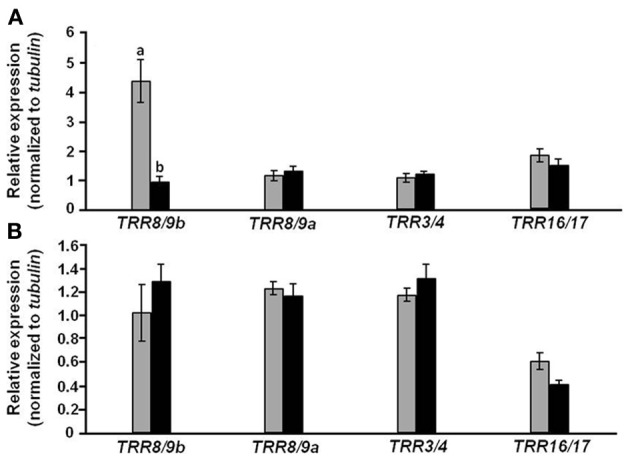
Effect of CmF-308 expression in the phloem of tomato plants on the expression of cytokinin-response genes. The expression of members of the tomato response regulator (TRR) gene family was determined by qRT-PCR in leaves (A) and roots (B) of 21-day-old CmF-308-transgenic (gray columns) and control M-82 (black columns) plants. Data represent means of four replications ± SE. Different letters indicate significant differences in relative expression of specific genes between the two plant lines at p < 0.05 by Student's t-test.
One of the most pronounced characteristics of CmF-308-transgenic plants was massive branching, indicating reduced apical dominance. A major regulator of axillary shoot development is the branching inhibitor strigolactone (Gomez-Roldan et al., 2008). It is generally assumed that strigolactone is synthesized in the roots and move via the xylem into the shoots. We therefore, examined whether expression of genes coding for strigolactone biosynthesis is altered in the roots of CmF-308-transgenic plants. Relative expression of CCD7 (carotenoid cleavage dioxygenase), a gene coding for one of the key enzymes involved in strigolactone biosynthesis, was significantly (about twofold) higher in the roots of CmF-308 plants vs. controls (Data not shown). Relative expression of CCD8 was similar in roots of CmF-308 and control plants. These results suggested that enhanced branching in CmF-308-transgenic plants is not associated with inhibition of strigolactone biosynthesis.
Discussion
A previous study established that two melon Aux/IAA transcripts are capable of long-distance movement from melon rootstock to pumpkin scion (Omid et al., 2007). The ability of one melon Aux/IAA transcript, CmF-308, to move long distances was further established here in N. benthamiana plants (Figure 2). The presence of CmF-308 in the shoot apex 48 h after agroinfiltration indicated that this transcript has the ability to move from cell to cell and enter the companion cell–sieve element (CC–SE) complex. It is logical to assume that such trafficking requires a chaperoning mechanism, probably as a ribonucleoprotein (RNP) complex. This notion is supported by the fact that phloem sap contains numerous RNA-binding proteins (Giavalisco et al., 2006; Lin et al., 2009), and by the demonstrated in vitro interaction between phloem sap-specific proteins and RNA molecules (Yoo et al., 2004; Ham et al., 2009). The absence of gfp transcripts from tissues outside the infiltrated leaf indicates that cell-to-cell and long-distance movement of mRNA are characteristic of specific molecules. Higher accumulation of CmF-308 in the veins and vascular bundles (Figure 1) suggests that this transcript is indeed destined for long-distance movement. The biological role for the long-distance trafficking of Aux/IAA transcript has yet to be explored.
To further explore the significance of CmF-308 expression in the vascular tissue, the gene was expressed in transgenic tomato plants under the AtSUC2 promoter. Various mutations in Arabidopsis Aux/IAA genes result in minor or no phenotypic changes, suggesting functional redundancy among Aux/IAA members (Overvoorde et al., 2005). However, antisense silencing of SlIAA9 (Wang et al., 2005) or SlIAA3 (Chaabouni et al., 2009) affected leaf architecture, root and fruit development. One should remember that the above-described Aux/IAA antisense constructs were inserted into tomato plants under the CaMV-35S promoter. The substantial phenotypic changes in tomato plants expressing CmF-308 predominantly in the phloem indicate that developmental processes are affected by altered auxin response imposed by the CC–SE complex. In this respect, it is important to note that we made numerous attempts to insert an antisense construct of CmF-308 into tomato plants under the AtSUC2 promoter. None of these attempts enabled regeneration of even one transgenic tomato plant, suggesting that silencing Aux/IAA in the phloem might be lethal.
Interestingly, overexpression of CmF-308 under a phloem-specific promoter resulted in significant modification of both root and shoot development (Figures 5, 6). Root development was altered in Arabidopsis and tomato plants in which various Aux/IAA genes were either mutated or silenced. For example, dominant Arabidopsis mutants of IAA19 (Tatematsu et al., 2004) or IAA14 (Fukaki et al., 2002) were characterized by few or complete lack of lateral roots, respectively. Similarly, gain of function mutations in IAA28 and IAA18 also resulted in defected formation of lateral root (Rogg et al., 2001; Uehara et al., 2008). However, a dominant IAA7 Arabidopsis mutant, had more lateral roots then control plants (Nagpal et al., 2000). These results indicate that different Aux/IAA gene products have contrasting effect on root growth and lateral root formation. A comparison of CmF-308 coding sequence and its Arabidopsis and tomato homologs revealed that the closest Arabidopsis homologs are IAA14 (1e-98) IAA7 (2e-89), IAA16 (4e-80) and IAA17 (6e-78) while the closest tomato homologs are IAA14 (5e-128) IAA9 (2e-108) IAA16 (7e -108) and IAA7 (3e-100). Due to the high degree of similarity between the indicated Aux/IAA genes, one cannot determine which one is the CmF-308 ortholog. It is possible that functioning of the CmF-308 gene product resembles the functioning of AtIAA7, namely enhanced formation of lateral roots. In this respect it is important to note that silencing SlIAA9 in transgenic tomato plants, expressing the antisense construct, enhanced auxin sensitivity and resulted in higher number of lateral roots (Wang et al., 2005). Interestingly, this phenotype was associated with upregulation of SlIAA3 expression predominantly in the vasculature (Chaabouni et al., 2009). Expression of SlIAA3 was upregulated in roots of our CmF-308 plants, raising the possibility that overexpression of Cm-F308 in the phloem exerts an effect on lateral root formation via upregulation of SlIAA3.
Auxin-response assays confirmed that auxin sensitivity of CmF-308 plants is altered in a tissue-specific manner (Figure 7): roots of CmF-308 plants were indeed more sensitive to auxin than control roots, but auxin sensitivity of CmF-308 shoot tissue (cotyledons) was lower than that of control plants. These findings suggest that CmF-308 can both repress and activate auxin responses in tomato plants.
Auxin responses in Arabidopsis plants expressing either gain-of-function or loss-of-function mutations in IAA3 indicated that this gene's product acts as both a positive and negative regulator of the auxin response (Tian and Reed, 1999). Those authors suggested that weak transient auxin signaling induces a low level of IAA3 in the roots which is sufficient to promote root growth and lateral root formation. However, stronger auxin signaling induces a higher level of IAA3, inhibiting these responses. This dual function of Aux/IAA is supported by previous studies demonstrating that the auxin response is dose-dependent, with stimulation of root growth by low concentrations of exogenous auxin and inhibition of root growth by higher IAA concentrations (Evans et al., 1994). Similarly, one can suggest that the CmF-308 gene product accumulates at higher levels in the shoot apex, resulting in reduced auxin response, inhibition of apical dominance and enhanced development of lateral shoots. In contrast, low levels of CmF-308 in the root cause a slight increase in SlIAA3, thereby enhancing the auxin response and lateral root formation. The finding that light regulates IAA3 expression (Tian et al., 2002) suggests a lower expression level of this transcript in root compared to shoot tissues, resulting in a differential auxin response.
An additional explanation for the effect of CmF-308 on plant development might relate to the expression of other genes mediating responses to growth substances. The expression of three Aux/IAA genes (IAA7, IAA10 and IAA14) was lower in leaves of CmF-308 vs. control plants, in line with the lower auxin response in the former. It is important to note that in parallel to reduced expression of these three Aux/IAA genes, expression level of TRR8/9b was significantly higher in CmF-308 leaves (Figure 8). Expression of TRR8/9b was upregulated by cytokinin (Shani et al., 2010), suggesting that the effect of the CmF-308 gene product on apical dominance and shoot branching is via modulation of the cytokinin response. Reduced expression of IAA7 was recently found in Arabidopsis plants paralleling an increase in the cytokinin response (Brenner and Schmülling, 2012); this supports the notion of cross talk between auxin- and cytokinin-signaling pathways.
The presented results provide support for Aux/IAA functioning in the phloem. The altered developmental program of cells distant from the CC–SE complex suggests an involvement of a phloem borne signal mediating auxin response. Future study should aim to identify potential interacting proteins (molecules) that might be associated with the short- or long-distance trafficking of the Aux/IAA gene product.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by a grant from the United States-Israel Binational Science Foundation (BSF 2007052). This paper is a contribution from the Uri Kinamon Laboratory. Guy Golan and Rotem Betzer were supported by a scholarship from the Uri Kinamon Foundation.
Appendix
Table A1.
List of primers used for amplification of the examined genes.
| Amplified gene | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|
| CmF-308-3XHA | GGG CCAAG AATGATAG AC | CTA CTG AGCAGCGTA ATCTGG |
| TRR8/9b | AGTATGCCGGAAATGACTGG | TGGAACATTTTCCGATGACA |
| TRR16/17 | GGTCTAAGGGCGTTGGAGTA | TCCTGGCATGCAATAATCTG |
| CCD7 | TGGGAAGGTGGTGATCCTTA | TAGCTGAGCAGCAACATCCA |
| CCD8 | CAATCACAGCGGTAACTCTTCCA | GCATCCTGATTCTAAAGCATTT |
| CmF-308 | GACTGGAGTTACCGTCGATCT | CGGAGTCAGGGCTCTTTTGA |
| SlIAA14 | CCTGAAGTTCATCTGCACCA | GTTCACCTTGATGCCGTTCT |
| SlIAA9 | CAAATACGTGAAGGTAGCAGTTGAC | ACACCATTTGTAAGGTCCATAAGCT |
| SlIAA3 | GACTTCTCAAAAGCTTGATCGAGAG | TGAAATCTTTCATTCCTTGGACAA |
| SlIAA7 | AGCCACCAACTAAGGCTCAA | CCATCCATGGAAACCTTCAC |
| SlIAA8 | CAAATACGTGAAGGTAGCAGTTGAC | ACACCATTTGTAAGGTCCATAAGCT |
| SlIAA10 | GACTTCTCAAAAGCTTGATCGAGAG | TGAAATCTTTCATTCCTTGGACAA |
| TRR3/4 | CGTCCCCTAAAGCATTCTCA | CGTCTTGTTGGTGATGTTGG |
| TRR8/9a | TGCTTAGAAGAAGGGGCAGA | GGGGGCTTTTACATTTGGTT |
| SlTubulin | GAAAGCCTACCATGAGCAGC | CTTTGGCACAACATCACCAC |
References
- Audran-Delalande C., Bassa C., Mila I., Regad F., Zouine M., Bouzayen M. (2012). Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 53, 659–672 10.1093/pcp/pcs022 [DOI] [PubMed] [Google Scholar]
- Banerjee A. K., Chatterjee M., Yu Y., Suh S. G., Miller W. A., Hannapel D. J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457 10.1105/tpc.106.042473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner W. G., Schmülling T. (2012). Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol. 12:112 10.1186/1471-2229-12-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A., Springer F., Chappell L., Baulcombe D. C., Kehr J. (2008). Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53, 739–749 10.1111/j.1365-313X.2007.03368.x [DOI] [PubMed] [Google Scholar]
- Calderon Villalobos L. I., Lee S., De Oliveira C., Ivetac A., Brandt W., Armitage L., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S., Jones B., Delalande C., Wang H., Li Z., Mila I., et al. (2009). Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J. Exp. Bot. 60, 1349–1362 10.1093/jxb/erp009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. L., Ishikawa H., Estelle M. A. (1994). Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta 194, 215–222 10.1007/BF01101680 [DOI] [Google Scholar]
- Fujita K., Horiuchi H., Takato H., Kohno M., Suzuki S. (2012). Auxin-responsive grape Aux/IAA9 regulates transgenic Arabidopsis plant growth. Mol. Biol. Rep. 39, 7823–7829 10.1007/s11033-012-1625-9 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Nakao Y., Okushima Y., Theologis A., Tasaka M. (2005). Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44, 382–395 10.1111/j.1365-313X.2005.02537.x [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tameda S., Masuda H., Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Giavalisco P., Kapitza K., Kolasa A., Buhtz A., Kehr J. (2006). Towards the proteome of Brassica napus phloem sap. Proteomics 6, 896–909 10.1002/pmic.200500155 [DOI] [PubMed] [Google Scholar]
- Gil L., Yaron I., Shalitin D., Sauer N., Turgeon R., Wolf S. (2011). Sucrose transporter plays a role in phloem loading in CMV infected melon plants that are defined as symplastic loaders. Plant J. 66, 366–374 10.1111/j.1365-313X.2011.04498.x [DOI] [PubMed] [Google Scholar]
- Gleave A. P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207 10.1007/BF00028910 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pagès V., Dun E. A., Pillot J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- Ham B. K., Brandom J. L., Xoconostle-Cazares B., Ringgold V., Lough T. J., Lucas W. J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21, 197–215 10.1105/tpc.108.061317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V., Yu T. S., Huang N. C., Lucas W. J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68 10.1111/j.1365-313X.2005.02351.x [DOI] [PubMed] [Google Scholar]
- Kohno M., Takato H., Horiuchi H., Fujita K., Suzuki S. (2012). Auxin-nonresponsive grape Aux/IAA19 is a positive regulator of plant growth. Mol. Biol. Rep. 39, 911–917 10.1007/s11033-011-0816-0 [DOI] [PubMed] [Google Scholar]
- Lin M.-K., Lee Y.-J., Lough T. J., Phinney B. S., Lucas W. J. (2009). Analysis of the pumpkin phloem proteome provides functional insights into angiosperm sieve tube function. Mol. Cell Proteom. 8, 343–356 10.1074/mcp.M800420-MCP200 [DOI] [PubMed] [Google Scholar]
- McCormick S. (1991). Transformation of tomato with Agrobacterium tumefaciens, in Plant Tissue Culture Manual, ed Lindsey K. (Dordrecht: Kluwer Academic Publishers; ), 1–9 [Google Scholar]
- Nagpal P., Walker L. M., Young J. C., Sonawala A., Timpte C., Estelle M., et al. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–574 10.1104/pp.123.2.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M., Wolf S., Lucas W. J. (2012). Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 54, 760–772 10.1111/j.1744-7909.2012.01155.x [DOI] [PubMed] [Google Scholar]
- Omid A., Keilin T., Glass A., Leshkowitz D., Wolf S. (2007). Characterization of phloem-sap transcription profile in melon plants. J. Exp. Bot. 58, 3645–3656 10.1093/jxb/erm214 [DOI] [PubMed] [Google Scholar]
- Overvoorde P. J., Okushima Y., Alonso J. M., Chan A., Chang C., Ecker J. R., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17, 3282–3300 10.1105/tpc.105.036723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J. W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425 10.1016/S1360-1385(01)02042-8 [DOI] [PubMed] [Google Scholar]
- Rogg L. E., Lasswell J., Bartel B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Yamamoto K. T. (2008). Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 133, 397–405 10.1111/j.1399-3054.2008.01055.x [DOI] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J. P., Bowman J. L., Eshed Y., et al. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. U.S.A. 106, 8392–8397 10.1073/pnas.0810810106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Ben-Gera H., Shleizer-Burko S., Burko Y., Weiss D., Ori N. (2010). Cytokinin regulates compound leaf development in tomato. Plant Cell 22, 3206–3217 10.1105/tpc.110.078253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Kumagai S., Muto H., Sato A., Watahiki M. K., Harper R. M., et al. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393 10.1105/tpc.018630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W. D., Paponov I. A., Palme K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell. Biol. 7, 847–859 10.1038/nrm2020 [DOI] [PubMed] [Google Scholar]
- Tian Q., Reed J. W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721 [DOI] [PubMed] [Google Scholar]
- Tian Q., Uhlir N.J., Reed J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14, 301–319 10.1105/tpc.010283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196, 564–570 10.1007/BF00203657 [DOI] [PubMed] [Google Scholar]
- Uehara T., Okushima Y., Mimura T., Tasaka M., Fukaki H. (2008). Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 49, 1025–1038 10.1093/pcp/pcn079 [DOI] [PubMed] [Google Scholar]
- Vanneste S., De Rybel B., Beemster G. T., Ljung K., De Smet I., Van Isterdael G., et al. (2005). Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17, 3035–3050 10.1105/tpc.105.035493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaine F., Palaqui J. C., Amselem J., Kusiak C., Lemoine R., Dinant S. (2003). Towards deciphering the phloem: a transcriptome analysis of the phloem of Apium graveolens. Plant J. 36, 67–81 10.1046/j.1365-313X.2003.01855.x [DOI] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z., Frasse P., Delalande C., Regad F., et al. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17, 2676–2692 10.1105/tpc.105.033415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A. W., Bartel B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley C. K., Zenser N., Ramos J., Rouse D., Leyser O., Theologis A., et al. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562 10.1046/j.1365-313x.2000.00703.x [DOI] [PubMed] [Google Scholar]
- Yoo B. C., Kragler F., Varkonyi-Gasic E., Haywood V., Archer-Evans S., Lee Y. M., et al. (2004). A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000 10.1105/tpc.104.023614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Sun L., Kragler F. (2009). The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150, 378–387 10.1104/pp.108.134767 [DOI] [PMC free article] [PubMed] [Google Scholar]


