Abstract
Cell-based therapies using pluripotent stem cells hold great promise as regenerative approaches to treat many types of diseases. Nevertheless many challenges remain and, perhaps foremost, is the issue of how to direct and enhance the specification and differentiation of a desired cell type for potential therapeutics. We have examined the molecular basis for the inverse correlation of cardiac and skeletal myogenesis in small molecule-enhanced stem cell differentiation. Our study shows that activation of premyogenic factor Pax3 coincides with inhibiting gene expression of early cardiac factor GATA4. Interestingly, the inhibitory effect of small molecules on cardiac differentiation depends on the function of Pax3, but not the mesoderm factor Meox1. Thus Pax3 is an inhibitor of cardiac differentiation in lineage specification. Our studies reveal the dual roles of Pax3 in stem cell fate determinations and provide new molecular insights into small molecule-enhanced lineage specification.
Cell-based therapies using embryonic stem (ES) cells or induced pluripotent stem cells hold great promise as regenerative approaches for treating many types of diseases. Nevertheless many challenges remain and, perhaps foremost, is how to direct and enhance the specification and differentiation of a desired cell type for potential therapeutics. Deciphering the molecular mechanisms underpinning lineage commitment is critical to developing the best strategies to direct lineage specific differentiation of pluirpotent stem cells.
Retinoic acid (RA) affects the lineage specification of ES cells in a time- and concentration-dependent manner1,2. Treatment of ES cells with low concentrations of RA (<10−7 M) at the stage of lineage specification results in an enhancement of skeletal myogenesis, but an inhibition of cardiomyogenesis2. However, when the low concentration of RA is administered at the late stage of differentiation, skeletal myogenesis is inhibited, whereas cardiomyogenesis is enhanced2.
On a cellular level, the process of cardiomyogenesis involves the specialization of mesoderm cells into pre-cardiac mesoderm cells, and the differentiation of these precursor cells into cardiomyoblasts and eventually working cardiac muscle3,4. This development is controlled by the cardiac transcription factors such as GATA binding protein 4 (GATA4), NK2 transcription factor related, locus 5 (Nkx2.5), and T-box 5 (Tbx5) which are first expressed in cardiac muscle precursor cells and then proceed in concert to activate the expression of other cardiac muscle genes5,6,7,8,9. The GATA4 is an evolutionarily conserved zinc finger protein and a key regulator of endoderm and mesoderm development in the post gastrula embryo10,11. It controls the expression of genes critical for cardiac differentiation, and interacts with many transcription factors to regulate cardiomyogenesis12. While loss of function mutations in GATA4 or Nkx2.5 causes congenital heart diseases, over-expression of any of these factors can initiate cardiomyogenesis7,13,14,15. Finally, specific over-expression of GATA4 and Tbx5 is sufficient to generate functional cardiomyocytes from cardiac fibroblasts16,17.
Skeletal muscle development is a complex process that requires combined signaling of many myogenic regulatory factors such as Myf5, MyoD and Myogenin18. The premyogenic factor Pax3 is important for embryonic skeletal muscle development by acting upstream of muscle-specific program19,20,21. Ectopic expression of Pax3 at the early stage of differentiation enhances mesoderm formation and increases the muscle potential of Pax3-induced ES cells22. On the other hand, overexpression of mesoderm factor Meox1 per se does not increase myogenic differentiation, but a dominant negative Meox1 downregulates Pax3 expression and decreases the differentiation of pluripotent P19 embryonal carcinoma (EC) cells into skeletal myocytes23.
In this study, we focus on the molecular basis for the inverse correlation of cardiac and skeletal myogenesis in small molecule-enhanced stem cell differentiation. Our studies revealed that the progenitor factor Pax3 plays dual roles in lineage specification.
Results
Bexarotene inhibits the commitment of ES cells into cardiomyocytes
Since the early events of embryonic myogenesis is recapitulated closely by differentiation of ES cells into cardiac and skeletal muscle lineages24,25, we used an embryoid body (EB) formation approach to study the impact of small molecules on myogenic conversion of ES cells. The ES cells were cultivated in the hanging drops for 2 days to form the EBs which were then cultured in suspension for 5 days and maintained as adhering culture for additional 10 days (Fig. 1A). For immunofluorescence microscopy analysis, the cells were co-stained for myosin heavy chain and MyoD to quantify the development of cardiac and skeletal myocytes. Developed cardiac and skeletal myocytes are distinguished by their staining pattern and morphological characteristics. Skeletal myocytes were recognized as elongated bipolar myocytes positively stained for myosin heavy chain and MyoD, while cardiac myocytes are rounded myosin heavy chain positive but MyoD negative cells.
Figure 1. Bexarotene inhibits the differentiation of ES cells into cardiac myocytes.
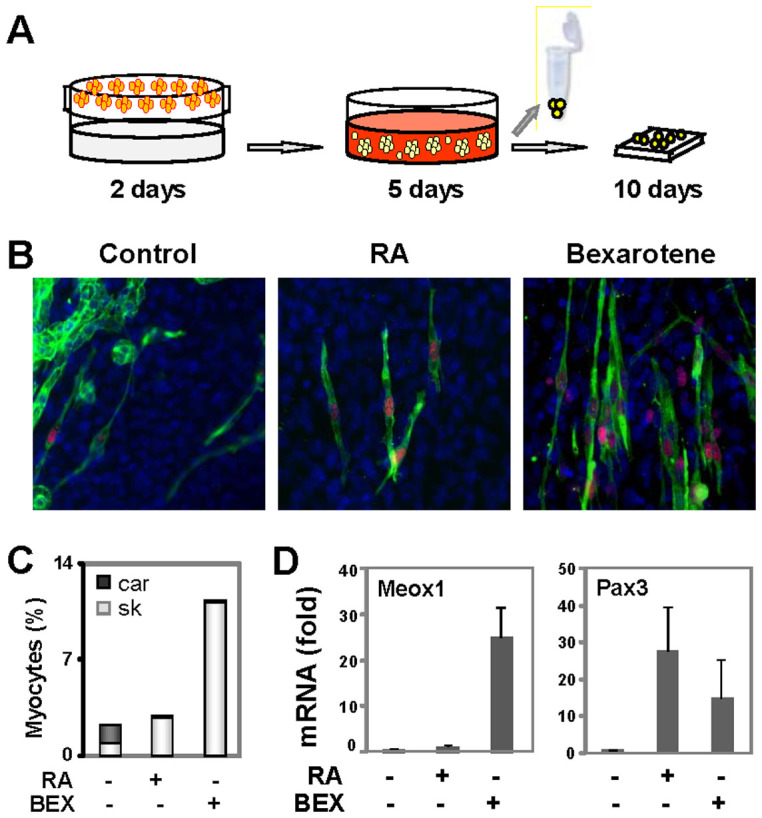
(A) ES cells were grown in hanging drops for 2 days, in suspension with RA (5 nM) or bexarotene (BEX, 50 nM) for 5 days, and maintained on coverslips without any treatments for an additional 10 days. The cells were then harvested for RT-PCR analysis, or costained with specific antibodies for microscopic analysis of MyoD (red), myosin heavy chain (MyHC, green), and with Hoechst for the nuclei (blue). (B) Shown are the representative images of microscopy. (C) Quantification of differentiation is presented as the percentage of cardiac myocytes (car, dark grey) and skeletal myocytes (sk, light grey) in relation to the total cell population (n = 3). (D) Real-time RT-PCR was used to determine the transcript levels of Meox1 and Pax3 on day 7 of differentiation in the same batch of cDNA. Quantification is presented as fold changes in reference to the untreated EBs (n = 3).
Cultured in media favorable for differentiation of skeletal myocytes but in the absence of small molecule inducers, ES cells were able to spontaneously differentiate into cardiac and skeletal myocytes but at low frequencies. Only about 1.5% of cardiac and 1% of skeletal myocytes were generated as determined by the quantitative immunofluorescence microscopy (Fig. 1B and C). In line with the literature2, treatment of the ES cells with RA during day 2–5 of EB formation, attenuated the generation of cardiac myocytes while enhancing skeletal myogenesis moderately to about 3% (Fig. 1B and C).
Bexarotene, a selective ligand of retinoid X receptor (RXR), is a more efficient inducer for ES cells to differentiate into skeletal myocytes than RA26. To delineate the mechanisms of bexarotene action, we examined the impact of bexarotene on the specification of cardiac lineage in ES cell differentiation. As shown in figure 1B and C, bexarotene inhibited the differentiation of ES cells into cardiac myocytes, while markedly increased the specification of skeletal muscle lineage to about 11%. Moreover, the quantitative real-time RT-PCR analyses revealed that bexarotene increased the transcripts of Meox1 more potently than RA in the EBs, whereas it was less efficient at increasing the transcripts of Pax3 (Fig. 1D). Thus, our findings establish that bexarotene, is also an inhibitor of cardiac differentiation, similar to RA in ES system, if administered during EB formation.
Inhibition of GATA4 early expression by bexarotene
Pluripotent P19 stem cells can be directed into differentiation to form cell lineages of all three germ layers27,28. They have served as a valuable model system to study myogenesis and to identify small molecule inducers for lineage specification26,28. Specifically, the differentiation of P19 cells into cardiac and skeletal myocytes reflects the cellular and molecular processes occurring in early embryogenesis and ES cell differentiation26,29. In tissue cultures, the P19 cells can be induced into differentiation with an aggregation protocol that involves the formation of early embryo-like cell aggregates also known as EBs (Fig. 2A). Formation of the EBs in the absence of exogenous stimuli results in the expression of markers of mesoderm, but not of cardiac or skeletal myocytes30. Cells treated with DMSO differentiate into small percentages of cardiac and skeletal myocytes along with other mesodermal and endodermal cell types31.
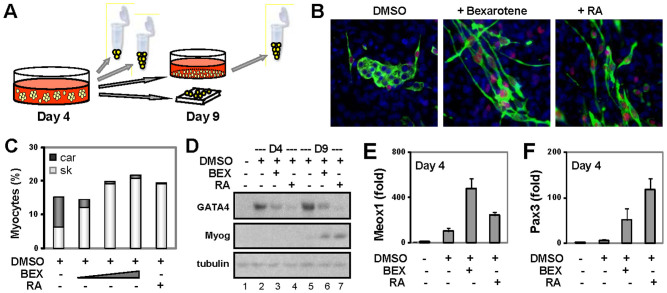
Figure 2. Bexarotene inhibits cardiac differentiation of P19 pluripotent stem cells.
(A) P19 cells were differentiated with DMSO in the absence or presence of increasing concentrations of bexarotene (BEX, 10 nM, 100 nM, 1 μM) or RA (10 nM) during EB formation. The cells were then maintained on coverslips without any treatments for an additional 5 days and costained with specific antibodies for MyoD (red), myosin heavy chain (MyHC, green) and Hoechst for the nuclei (blue). The cells were also harvested at different time points for RT-PCR and Western analyses. (B) Shown are the representative images of microscopy. (C) Quantification of differentiation is presented as the fractions of cardiac myocytes (car, dark grey) and skeletal myocytes (sk, light grey) in relation to the total cell population (n = 5). (D) Gene expression of myogenin and GATA4 and protein on day 4 and day 9 of differentiation was determined by Western analysis in parallel with the use of the same batch of cell extracts. The blots were then stripped and reprobed for β-tubulin as loading controls. Shown are the cropped blot images representing indicated protein. (E) and (F) Real-time RT-PCR was used to determine the transcripts levels of Pax3 and Meox1 on day 4 of differentiation in the same batch of cDNA. Quantification is presented as fold changes in reference to the untreated EBs (n = 3).
As shown in figure 2B and C, DMSO induced about 8% of cardiac and 6% of skeletal myocytes in the P19 cells, respectively, as quantified by quantitative immunofluorescence microscopy, consistent with previous studies. Cotreatment of the EBs with RA enhanced the generation of skeletal myocytes, while attenuating cardiac differentiation (Fig. 2B and C), as previously reported32. Bexarotene enhances the specification of muscle lineage in a concentration dependent manner and the efficacies of bexarotene on the differentiation of P19 cells into skeletal myocytes is comparable to RA26. More interestingly, bexarotene also inhibited the generation of cardiac myocytes by P19 cells in a concentration dependent manner (Fig. 2B and C). While higher concentrations of bexarotene enhanced the specification of skeletal muscle lineage to about 20%, it markedly impeded cardiac differentiation (Fig. 2B and C). Nevertheless, this negative effect of bexarotene on P19 cardiac differentiation appears to be less potent than RA (Fig. 2C).
Western analysis revealed that the expression of GATA4, an early cardiac factor, was induced by day 4 of differentiation following treatment of the EBs with DMSO (Fig. 2D). This early expression of GATA4 gene was notably impaired by the administration of bexarotene or RA (Fig. 2D). Again, RA appears to be more effective than bexarotene on the inhibition of GATA4 gene expression. However, the expression of myogenin protein, a specific skeletal muscle factor was enhanced by bexarotene or RA to a similar degree on day 9 of differentiation, correlating with their efficacies on the specification of skeletal muscle lineage (Fig. 2B–D). The expression of Meox1 and Pax3 was increased by bexarotene and RA by day 4 of differentiation. Similar to ES cell differentiation, bexarotene increased the level of Meox1 gene expression more than RA, whereas RA had a higher impact than bexarotene on the expression of Pax3 gene, by about 2-fold (Fig. 2E and F). Thus, the temporal gene expression pattern induced by bexarotene during P19 myogenic differentiation is similar to skeletal myogenesis in vivo and in ES cell differentiation. In addition, the inhibitory effects of bexarotene and RA on cardiac differentiation are exerted through the inhibition of GATA4 gene expression.
Role of Pax3 in myogeneic conversion
One valuable feature of P19 cells is that they are amenable for genetic manipulation to incorporate and express ectopic genes, and can be used to select stably transfected clones which retain their ability to differentiate33. Since the expression of premyogenic factor Meox1 and Pax3, similar to cardiac factor GATA4, was detected by day 4 of differentiation (Fig. 2D–F), we next examined the roles of Meox1 and Pax3 in RA- and bexarotene-inhibited cardiac differentiation. To this end, we employed previously established P19 stable lines expressing a dominant negative Meox1 or Pax323,26,34. In these cells, the transcriptional activation domain of either Meox1 or Pax3 is replaced by an engrailed repressor domain (EN-2) which silences transcription through its interaction with members of the Groucho/TLE family of transcriptional repressors35.
Similar to the wild-type cells, the control cells harboring the empty vector were converted into cardiac and skeletal myocytes, about 7% and 6% respectively, following treatment with DMSO, but failed to produce cardiac myocytes after treatment with bexarotene or RA (Fig. 3A). On the other hand, the dominant negative Meox1 cells generated only about 2% and 1% of cardiac and skeletal myocytes after DMSO treatments, and cotreatment with bexarotene and RA enhanced the generation of skeletal myocytes only to about 5% and 6% respectively (Fig. 3A). This is consistent with previous studies, because Meox1 is important for skeletal myogenesis23,26. Regardless, bexarotene and RA impeded cardiac differentiation in the dominant Meox1 cells (Fig. 3A), suggesting that the inhibitory effects of bexarotene and RA on cardiac differentiation are not dependent on the function of Meox1.
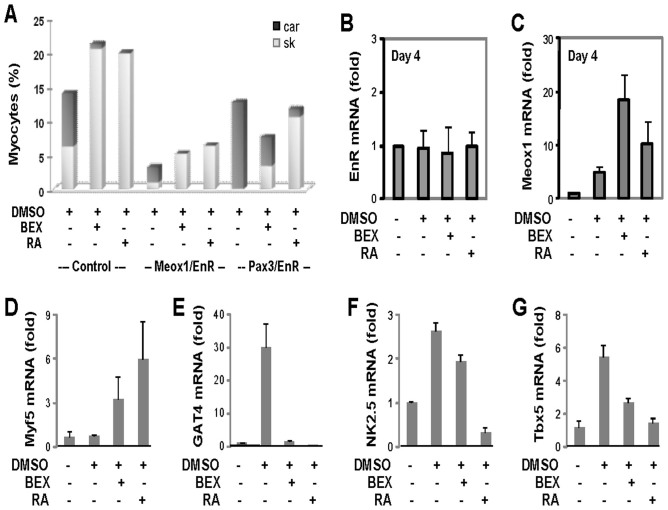
Figure 3. Role of Pax3 in cardiac differentiation.
(A) Clones of P19 cells expressing either a dominant negative Meox1 (Meox1/EnR) or Pax3 (Pax3/EnR) were treated with bexarotene (BEX, 100 nM) or RA (10 nM) during EB formation. The cells were then maintained on coverslips without any treatments for an additional 5 days and stained with specific antibodies for MyoD, myosin heavy chain and Hoechst for the nuclei. Cells harbouring the empty vector were used as controls. Quantification is plotted as the fraction of cardiac myocytes (car, dark grey) and skeletal myocytes (sk, light grey) in relation to the total cell population (n = 5). (B)–(G) Real-time RT-PCR was used to determine the transcript levels of EnR, Meox1, Myf5, GATA4, Nkx2.5 and Tbx5 on day 4 differentiation in the same batch of cDNA from the cells harboring the dominant negative Pax3. Quantification is presented as fold changes in reference to the untreated EBs (n = 3).
In agreement with previous reports26,34, DMSO failed to convert the dominant negative Pax3 cells into skeletal myocytes (Fig. 3A). However, the capacity of these cells to generate cardiac myocytes increased by about 2-fold following DMSO treatment compared with the control cells (Fig. 3A). Moreover, while treatment of the EBs with bexarotene or RA moderately induced the differentiation of these dominant negative Pax3 cells into skeletal myocytes, they were still able to generate cardiac myocytes, albeit with a lesser degree in comparison to DMSO treated cells (Fig. 3A). Thus, the function of Pax3 is important for bexarotene- and RA-inhibited cardiac differentiation.
Interestingly, while the transcript levels of EnR remained constant following different treatments (Fig. 3B), the negative effect of bexarotene on cardiac differentiation was less potent than RA just as in the wild-type cells (Fig. 3A). Similar to the wild-type cells, bexarotene had a larger effect than RA on Meox1 gene expression, about 2-fold, in the Pax3/EnR cells, whereas RA was more effective at inducing Myf5 gene expression (Fig. 3C–D). In addition, DMSO effectively induced the gene expression of cardiac factors, GATA4, Nkx2.5 and Tbx5 in the Pax3/EnR cells (Fig. 3E–G). Importantly, the inhibitory effect of bexarotene on the transcript levels of GATA4, Nkx2.5 and Tbx5 was less potent than RA, correlating with their efficacy on the inhibition of cardiac differentiation (Fig. 3A and E–G). Taken together, our data suggest that bexarotene and RA inhibits cardiac differentiation in part through the activation of Pax3 gene expression which appears to play dual roles in stem cell fate determinations.
Discussion
We have examined the molecular basis on the inverse correlation of cardiac and skeletal myogenesis in small molecule-enhanced stem cell differentiation. Small molecule inducers such as RA and bexarotene are able to enhance the generation of skeletal myocytes by activating premyogenic transcription factors at the stage of lineage specification. Our findings show that the activation of premyogenic factor Pax3 coincides with inhibiting gene expression of early cardiac factors such as GATA4, Tbx5 and Nkx2.5. Interestingly, the inhibitory effect of bexarotene and RA on cardiac differentiation requires the function of Pax3, but not Meox1. Thus Pax3 is an inhibitor of cardiac differentiation in lineage specification. Our studies reveal the dual roles of Pax3 in stem cell fate determinations and provide new molecular insights into small molecule-enhanced lineage specification.
Pluripotent stem cells, regardless of their origin, possess the potential of developing into many types of cell lineages. The central issue is how differentiation cues commit the cells into a particular lineage and what impact this would have on the specification of other cell lineages. Understanding the molecular mechanisms of lineage specification is central to devise the best strategies for preferentially enhancing lineage specific differentiation in an efficacy suitable for potential therapeutics. Consistent with the literature, RA is able to enhance skeletal myogenesis in pluripotent stem cells, but inhibits cardiac differentiation if used at the stage of lineage specification (Fig. 1 and 2). We also found that bexarotene has similar effects as RA on cardiac differentiation (Fig. 1 and 2). This is particularly interesting as we have previously established that bexarotene activates the skeletal muscle program through a mechanism distinct from RA-mediated pathway26.
In addition, our data also revealed a role of Pax3 in cardiac differentiation. Pax3 is important for skeletal myogenesis, but overexpression of Pax3 leads to a loss of cardiac differentiation19,23. Our studies establish Pax3 as a molecular pathway of small molecule-enhanced skeletal myogenesis, but -inhibited cardiac differentiation (Fig. 3). Although the specification events differ between skeletal and cardiac lineages, they both are from mesoderm origin. During in vitro stem cell differentiation, lineage specification factors apparently not just exert their functions through the activation of the lineage they command, but also through the inhibition of other lineages to compete for the pool of progenitor cells, when spatial partition is nonexistent. While skeletal and cardiac myogenesis may be segregated spatially during embryogenesis, small molecules are effective means to employ the dual roles of lineage specific factors for generating specific lineage from pluripotent stem cells for potential therapeutics.
Our studies have not examined the genomic targets of Pax3 in relation to cardiac inhibition. Systems studies are required to uncover additional targets of Pax3 involved in the inhibition of cardiac differentiation. Understanding the molecular mechanisms of myogenic conversion is central to direct and enhance lineage specification in cell-based therapies. We have identified a potential mechanism for lineage competition, which suggests early lineage factors regulate lineage commitment by regulating and integrating different signaling pathways. The dual roles of lineage specific factors in stem cell fate determination provide interesting avenue in identifying effective molecular targets to convert pluripotent stem cells into desired cell lineage.
Methods
Cell culture and differentiation
ES Cells (ATCC) were maintained in Dulbecco's modified Eagle medium (D-MEM) (Gibco-Invitrogen) supplemented with 15% fetal bovine serum (PAA), 1% non essential amino acids (Gibco-Invitrogen) and 1.18 mM β-mercaptoethanol (OmniPur) at 37°C with 5% of CO2. Maintenance cultures were supplemented with 1,000 units/ml of Leukemia Inhibitory Factor (Chemicon). For differentiation, ES cells were cultivated in hanging drops on inverted lids for 2 days and in Petri dishes for additional 5 days to form EBs which were then transferred into tissue culture dishes or onto coverslips coated with 0.1% gelatin, and maintained for another 10 days26. P19 cells (ATCC) were maintained in minimum essential medium α (α-MEM) (Gibco-Invitrogen) supplemented with 5% fetal bovine serum (PAA) and 5% bovine calf serum (PAA) at 37°C with 5% of CO2. For differentiation, cells were aggregated in Petrie dishes for 4 days to form EBs. The EBs was then transferred into tissue culture dishes or onto coverslips coated with 0.1% gelatin and grown for a further 5 days36. All-trans retinoic acid (RA) was purchased from the Sigma-Aldrich, and bexarotene was from the LC Laboratories.
Immunofluorescence microscopy
Following differentiation, cells were fixed on the coverslips with cold methanol, rehydrated with PBS, and incubated with the appropriate primary antibodies overnight at 4°C, and then with the corresponding fluorescent secondary antibodies at room temperature for 2 hours. After washing with PBS, the cells were incubated with Hoechst (Molecular Probes) for 3 minutes to stain the DNA. Finally, the coverslips were mounted on slides with 50% glycerol. Microscopy analysis was performed with the Zeiss Axiovert 200 M microscope. Cells were observed through a Zeiss 20X objective and image acquisition was carried out with the AxioCam HRm monochrome camera37. Myogenesis was assessed as the percentage of cells stained positively for skeletal or cardiac markers in relation to total cell populations. Images captured through different fluorescence filters were processed and merged by the Zeiss AxioVision Rel 4.6 software. The primary antibodies used were myosin heavy chain (MF20) and MyoD (Santa Cruz).
Real-Time RT-PCR
Total RNA was isolated using RNeasy Mini kit (Qiagen) and reverse transcribed with High Capacity cDNA Archive Kit (Applied Biosystems)38. Quantitative real-time PCR was conducted by using the Applied Biosystems 7500 Fast Real-Time PCR System. The amount of targets, normalized to the GAPDH endogenous reference and relative to calibrator control is calculated using the arithmetic formula 2−ΔΔCT. The primers used as the following:
Meox1 fwd-GAGAGGTCAGACAACCAGGAG
Meox1 rev-CGTAGCTGCTCCTTGGTGAAG
Pax3 fwd-GCTGCAGTCAGAGACTGGAAC
Pax3 rev-GAAAGGCACTTTGTCCATACTGC
EnR fwd-TCGGTGTGAACGGATTTG
EnR rev-GGTCTCGCTCCTGGAAGA
Myf5 fwd- GGCATGCCTGAATGTAACAGC
Mfy5 rev- CAATCCAAGCTGGACACGGA
GATA4 fwd- GAAGACACCCCAATCTCGATATG;
GATA4 rev- GGCATTGCACAGGTA GTGTCC
Nkx2.5 fwd- AGTGCTCTCCTGCTTTCCCA;
Nkx2.5 rev- ATCCGTCTCGGCTTTGTC C
Tbx5 fwd- AACAGTAGCAGCTAGCTTGG
Tbx5 rev- GAGGTTCTATTCTCGCTCTGT
Western analysis
Cells were lysed by incubation with whole cell extract buffer (10% glycerol, 50 mM Tris-HCl pH 7.6, 400 mM NaCl, 5 mM EDTA, 1 mM DTT, 1 mM PMSF, 1% NP-40) for 30 minutes on ice and centrifuged at 12,000 g for 15 minutes to collect the supernatant39. Protein concentrations were assessed by Bradford Method following the manufacture's recommendation using Bio-Rad Protein Assay Reagent (Biorad). Equal amounts of protein were separated on SDS-PAGE and transferred onto Immun-Blot PVDF membrane (Bio-Rad). Proteins were then visualized using Western Lightning Chemiluminescence reagents (Perkin Elmer). The primary antibodies used were GATA4 (Santa Cruz), myogenin (F5D) and β-tubulin (E7).
Author Contributions
Q.L. and J.C. designed the research, interpreted the data and prepared the manuscript. M.L. executed microscopy and Western analyses. N.L. participated in the RT-PCR analysis. All authors reviewed the manuscript.
Acknowledgments
This work was sponsored by an operating grant from the Canadian Institutes of Health Research (to Q.L.). We thank Mr. H. Mach for excellent technical assistance.
References
- Wobus A. M. et al. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J Mol Cell Cardiol 29, 1525–39 (1997). [DOI] [PubMed] [Google Scholar]
- Wobus A. M., Rohwedel J., Maltsev V. & Hescheler J. In vitro differentiation of embryonic stem cells into cardiomyocytes or skeletal muscle cells is specifically modulated by retinoic acid. Roux's Arch Dev Biol 204, 36–45 (1994). [DOI] [PubMed] [Google Scholar]
- Buckingham M., Meilhac S. & Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 6, 826–35 (2005). [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell 126, 1037–48 (2006). [DOI] [PubMed] [Google Scholar]
- Grepin C., Robitaille L., Antakly T. & Nemer M. Inhibition of transcription factor GATA-4 expression blocks in vitro cardiac muscle differentiation. Mol Cell Biol 15, 4095–102 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints T. J., Parsons L. M., Hartley L., Lyons I. & Harvey R. P. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 969 (1993). [DOI] [PubMed] [Google Scholar]
- Garg V. et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–7 (2003). [DOI] [PubMed] [Google Scholar]
- Watt A. J., Battle M. A., Li J. & Duncan S. A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A 101, 12573–8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I. et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev 9, 1654–66 (1995). [DOI] [PubMed] [Google Scholar]
- Heikinheimo M., Scandrett J. M. & Wilson D. B. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev Biol 164, 361–73 (1994). [DOI] [PubMed] [Google Scholar]
- Rossi J. M., Dunn N. R., Hogan B. L. & Zaret K. S. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev 15, 1998–2009 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Charron F., Warren R., Schwartz R. J. & Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. Embo J 16, 5687–96 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D. A. et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol 41, 2072–6 (2003). [DOI] [PubMed] [Google Scholar]
- Grepin C., Nemer G. & Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development 124, 2387–95 (1997). [DOI] [PubMed] [Google Scholar]
- Reecy J. M. et al. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development 126, 839–49 (1999). [DOI] [PubMed] [Google Scholar]
- Ieda M. et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K. et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic T. & Li Q. Skeletal myogenesis and Myf5 activation. Transcription 2, 109–14. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Cossu G. & Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89, 127–38 (1997). [DOI] [PubMed] [Google Scholar]
- Maroto M. et al. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell 89, 139–48 (1997). [DOI] [PubMed] [Google Scholar]
- Bajard L. et al. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev 20, 2450–64 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R. et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med 14, 134–43 (2008). [DOI] [PubMed] [Google Scholar]
- Petropoulos H., Gianakopoulos P. J., Ridgeway A. G. & Skerjanc I. S. Disruption of Meox or Gli activity ablates skeletal myogenesis in P19 cells. J Biol Chem 279, 23874–81 (2004). [DOI] [PubMed] [Google Scholar]
- Rohwedel J. et al. Muscle cell differentiation of embryonic stem cells reflects myogenesis in vivo: developmentally regulated expression of myogenic determination genes and functional expression of ionic currents. Dev Biol 164, 87–101 (1994). [DOI] [PubMed] [Google Scholar]
- Weitzer G., Milner D. J., Kim J. U., Bradley A. & Capetanaki Y. Cytoskeletal control of myogenesis: a desmin null mutation blocks the myogenic pathway during embryonic stem cell differentiation. Dev Biol 172, 422–39 (1995). [DOI] [PubMed] [Google Scholar]
- Le May M. et al. Contribution of Retinoid X Receptor Signaling to the Specification of Skeletal Muscle Lineage. J Biol Chem 286, 26806–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heyden M. A. & Defize L. H. Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res 58, 292–302 (2003). [DOI] [PubMed] [Google Scholar]
- Yu J. & Thomson J. A. Pluripotent stem cell lines. Genes Dev 22, 1987–97 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki M. A., Reuhl K. R. & McBurney M. W. Cell lines with developmental potential restricted to mesodermal lineages isolated from differentiating cultures of pluripotential P19 embryonal carcinoma cells. Development 107, 361–72 (1989). [DOI] [PubMed] [Google Scholar]
- Ridgeway A. G., Petropoulos H., Wilton S. & Skerjanc I. S. Wnt signaling regulates the function of MyoD and myogenin. J Biol Chem 275, 32398–405 (2000). [DOI] [PubMed] [Google Scholar]
- Edwards M. K. & McBurney M. W. The concentration of retinoic acid determines the differentiated cell types formed by a teratocarcinoma cell line. Dev Biol 98, 187–91 (1983). [DOI] [PubMed] [Google Scholar]
- Kennedy K. A. et al. Retinoic acid enhances skeletal muscle progenitor formation and bypasses inhibition by bone morphogenetic protein 4 but not dominant negative beta-catenin. BMC Biol 7, 67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney M. W. P19 embryonal carcinoma cells. Int J Dev Biol 37, 135–40 (1993). [PubMed] [Google Scholar]
- Ridgeway A. G. & Skerjanc I. S. Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J Biol Chem 276, 19033–9 (2001). [DOI] [PubMed] [Google Scholar]
- Chen G. & Courey A. J. Groucho/TLE family proteins and transcriptional repression. Gene 249, 1–16 (2000). [DOI] [PubMed] [Google Scholar]
- Francetic T. et al. Regulation of Myf5 early enhancer by histone acetyltransferase p300 during stem cell differentiation. Mol Biol 1, 103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Halappanavar S., Th' ng J. P. & Li Q. Ubiquitin-dependent distribution of the transcriptional coactivator p300 in cytoplasmic inclusion bodies. Epigenetics 2, 92–9 (2007). [DOI] [PubMed] [Google Scholar]
- St-Germain J. R., Chen J. & Li Q. Involvement of PML nuclear bodies in CBP degradation through the ubiquitin-proteasome pathway. Epigenetics 3, 342–9 (2008). [DOI] [PubMed] [Google Scholar]
- Chen J., St-Germain J. R. & Li Q. B56 Regulatory Subunit of Protein Phosphatase 2A Mediates Valproic Acid-Induced p300 Degradation. Mol Cell Biol 25, 525–32 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]