Abstract
Firefly bioluminescence has attracted great interest because of its high quantum yield and intriguing modifiable colours. Modifications to the structure of the enzyme luciferase can change the emission colour of firefly bioluminescence, and the mechanism of the colour change has been intensively studied by biochemists, structural biologists, optical physicists, and quantum-chemistry theorists. Here, we report on the quantitative spectra of firefly bioluminescence catalysed by wild-type and four site-directed mutant luciferases. While the mutation caused different emission spectra, the spectra differed only in the intensity of the green component (λmax ~ 560 nm). In contrast, the orange (λmax ~ 610 nm) and red (λmax ~ 650 nm) components present in all the spectra were almost unaffected by the modifications to the luciferases and changes in pH. Our results reveal that the intensity of the green component is the unique factor that is influenced by the luciferase structure and other reaction conditions.
Natural terrestrial bioluminescent beetles utilize a common substrate molecule d-luciferin (LH2) to emit colourful light ranging from green to red through variation in the structure of the enzyme luciferase1,2. The reaction of the LH2 with adenosine-5′-triphosphate (ATP) and oxygen (O2) in the active pocket of luciferase produces oxyluciferin (OL) in the excited state. The bioluminescence of the beetles originates from the photon emission process accompanying the relaxation of OL* to the ground state. A magnesium ion is the cofactor of the enzymatic reaction.
In vitro bioluminescent emission colours of certain click beetles and railroad worms do not depend on the reaction conditions. In contrast, the natural yellow-green colour of firefly bioluminescence changes to red when the reaction solutions are altered, such as by decreasing the pH value3,4, increasing the temperature, or adding bivalent transition metal ions1,5.
The colour changes in firefly bioluminescence are considered to be due to a pH-dependent equilibrium between two chemical forms or states of the emitter OL. White et al. proposed a pH-dependent equilibrium between keto anionic and dianionic forms of OL as a way to explain the colour change6. McCapra and coworkers considered the coplanar structure and twisting structure between the benzothiazole and thiazole parts of anionic keto OL to be the sources of the yellow-green and red emissions, respectively7. Branchini et al. suggested that delocalization of π-electrons in the green-emitting anionic keto OL produces the red-emitting form with lower energy. Most of the previous reports based their analyses on the shift in the peak wavelength of the normalized spectra1,8,9,10.
We have quantitatively investigated the spectra of bioluminescence from the North-American firefly (Photinus pyralis, Ppy) at various pH values4 and in the presence of bivalent metal ions5. We analysed the experimental data by assuming three components of Gaussian functions4,5. This analysis has shown that only the intensity of a green Gaussian component peaking at 560 nm depended on pH or metal-ion additives, and that its intensity determined the spectral change, whereas the intensities of the other two Gaussian components did not depend on the pH value or metal-ion additives.
Colour changes in bioluminescence caused by luciferase mutations have attracted a great deal of interest from researchers. Many mutant luciferases have been derived from bioluminescent beetles, including fireflies, in order to investigate the influence of microenvironmental changes in luciferase on bioluminescence9,10,11,12,13. The size of the active pocket or hydrogen-bond network and certain amino acid residues have been cited as having an effect on the emission colour. Most of the previous reports on bioluminescence catalysed by mutant luciferases only dealt with normalized spectra and summarized the influence of the mutation in the form of the induced shift in the peak wavelength. Niwa et al. measured the quantum yield of bioluminescent reactions using various mutant luciferases8, and Ugarova et al. analysed the normalized spectra with three Gaussian functions9. However, the absolute intensities of the three Gaussian components and the dependence of these components on mutagenesis have never been quantitatively investigated.
Here, we present the quantitative characteristics of the spectral change caused by substituting an amino acid residue on firefly luciferase. We prepared four site-directed mutant luciferases of Luciola cruciata (Lcr, Japanese Genji firefly) by substituting the amino acid residue tyrosine at position 257 with phenylalanine, alanine, arginine, and glutamate. We quantitatively measured the bioluminescent spectra catalyzed by the WT luciferase and the four mutants. The spectra were fitted with Gaussian functions. The spectral peak wavelength, intensity, and their sensitivity to pH changes were compared and the influence of the substitution on each emission component was analyzed. We found that mutagenesis mainly changed the intensity of the emission component peaking around 560 nm and thus regulated the pH sensitivity of the bioluminescence spectrum.
Results
Quantitative spectra of wild-type and mutant Lcr luciferases
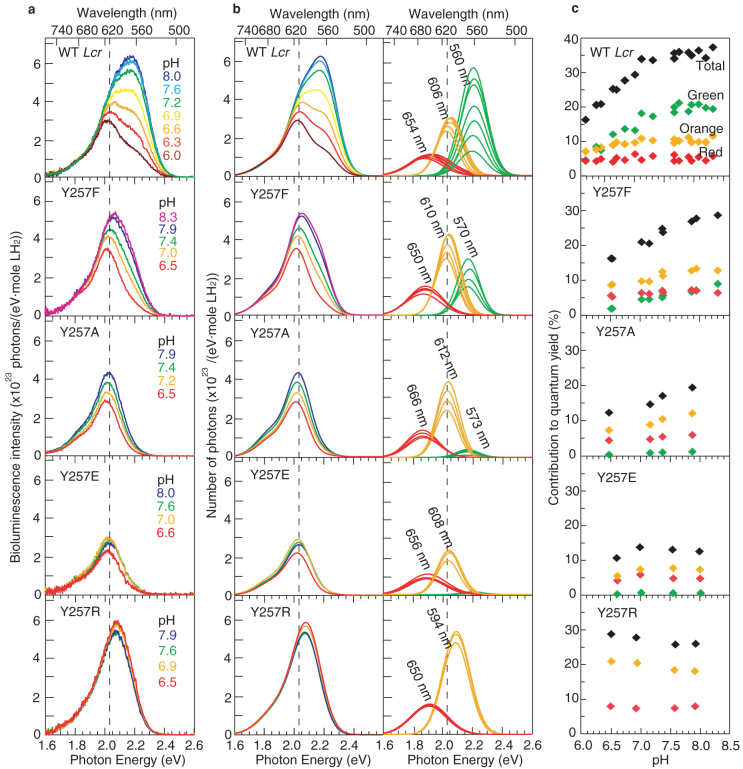
Figure 1 shows photographs of the bioluminescence catalyzed by the wild-type (WT), Y257F, Y257A, Y257E, and Y257R Lcr luciferases at pH 8.0, the colours of which are yellow-green, yellow, orange, red, and yellow, respectively. Figure 2 (a) shows the quantitative spectra of bioluminescence catalyzed by wild-type (WT) Lcr luciferase and the four mutants Y257F/A/E/R at various pH values. The absolute spectra show all the photons emitted by consuming one mole of LH2. The quantum yields and spectral peak wavelengths (λmax) at pH 8.0 are summarized in Table 1.
Figure 1. Photographs of bioluminescence catalyzed by wild-type (WT), Y257F, Y257A, Y257E, and Y257R mutant of Lcr luciferase at pH 8.0.
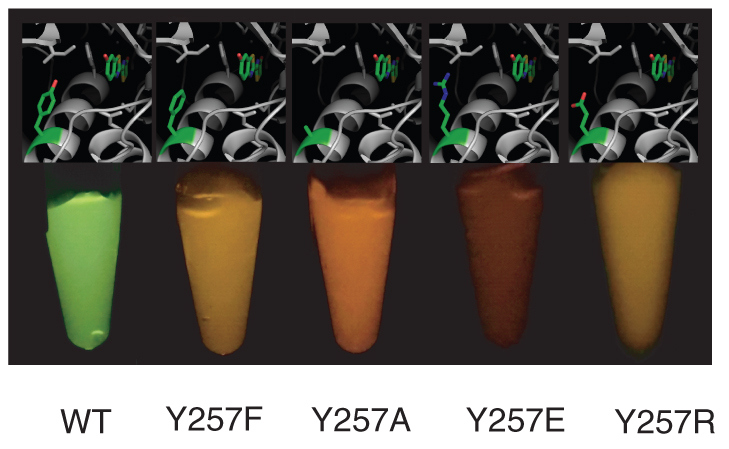
The corresponding structures were generated from the crystal structure of Lcr luciferase according to ref. 12 (code 2D1R in Protein Data Bank) with PyMOL14.
Figure 2.
(a) Quantitative spectra of wild-type and Y257F/A/R/E mutant Lcr luciferases at various pH values. (b) Results of the Gaussian curve fitting to the spectra in (a) and the Gaussian components that reconstruct the fitted spectra. (c) The contributions of the integrated areas of each Gaussian component in (b) to the quantum yield are shown with the corresponding colour. The summations are shown as black diamonds. The dashed lines are drawn at 610 nm (2.03 eV) to guide the eye.
Table 1. Summary of spectra catalyzed by wild type and mutant Lcr luciferases.
| Luciferase | λmax (nm) at pH 8.0 (quantum yield) | λmax (nm) of the Gaussian components (ΦGC of each component at pH 8.0*) | ||
|---|---|---|---|---|
| Ppy WT | 558 (40.8%) | 560 (25.5%) | 605 (9.8%) | 649 (5.4%) |
| Lcr WT | 566 (37.2%) | 560 (22.6%) | 608 (9.9%) | 654 (5.2%) |
| Y257F | 598 (27.6%) | 570 (7.2%) | 610 (13.3%) | 650 (7.0%) |
| Y257A | 610 (18.6%) | 573 (0.9%) | 612 (12.0%) | 666 (5.9%) |
| Y257E | 608 (12.4%) | — (~0%) | 608 (7.7%) | 656 (4.5%) |
| Y257R | 594 (25.5%) | — (0%) | 594 (17.8%) | 650 (7.8%) |
*The values were calculated from the fitting data so that the summation of the three ΦGC deviated a little from the experimental value.
The peak wavelength of the spectrum of bioluminescence catalyzed by WT Lcr luciferase gradually shifted from 566 nm to 612 nm, and the quantum yield decreased from 37.2% to 16.3% as the pH value of the reaction solution decreased from 8.0 to 6.0. The shift in the peak wavelength is consistent with the results of the previous report15. The bioluminescent intensity in the green region (~560 nm) changed significantly with pH, whereas it did not in the red region (~610 nm). The intensity of the green peak decreased to a very low level, and the red peak gradually became dominant in acidic solutions.
As the pH value decreased from 8.3 to 6.5, the spectral shape of the bioluminescence catalyzed by Y257F mutant luciferase gradually changed, and the peak wavelength shifted from 598 nm to 613 nm and the intensity decreased. Moreover, there was a significant change in the intensity in the high-energy region, similar to those of WT Lcr luciferase. The changes in the peak wavelength and intensity were less than those of WT luciferase.
The spectral shapes of bioluminescence catalyzed by Y257A, Y257E, and Y257R mutant luciferases did not change in the pH range of 6.5–8.0, as shown in Fig. 2 (a). Their peak wavelengths were 612 nm, 608 nm, and 594 nm, respectively. Only the intensity of the spectra of Y257A decreased moderately in an acidic solution; the other two remained almost constant.
Analysis via curve fitting with Gaussian functions
To further clarify the quantitative characteristics of the above spectra, we conducted curve fittings assuming Gaussian energy distribution functions (see details of the curve fitting in Methods). The left and right panels of Fig. 2 (b) show the fitting results and the Gaussian functions used to reconstruct the experimental spectra. The summation spectra (left panel of Fig. 2 (b)) match the experimental spectra Fig. 2 (a); most of the fitting correlation coefficients were above 0.999. The three Gaussian functions (right panel of Fig. 2 (b)) had peaks (averaged) at ~560–570 nm, ~610 nm, and ~650 nm (see also Table 1) and are denoted hereafter as the green, orange, and red components, respectively. Note that the total bioluminescence spectrum with a peak wavelength ~610 nm is conventionally called a ‘red' emission. Hereafter, we denote the Gaussian component peaked at ~610 nm and that at ~650 nm as the ‘orange' and ‘red' components, respectively. The peak wavelength and spectral width of each Gaussian component changed only a little with pH in each set of fittings for each species of luciferase. Figure 2 (c) plots the contributions of the three Gaussian components to the total quantum yield (ΦGC); these were obtained by dividing the respective integrated areas by the total molecular number of the substrate LH2. The ΦGC values of each component at pH 8.0 are listed in Table 1. The summations shown in Fig. 2 (c) are very close to the corresponding experimental quantum yields.
The quantitative spectra of the WT Lcr luciferase were reproduced with three Gaussian functions peaking at 560 nm, 606 nm, and 654 nm (see Fig. 2 (b)). The intensity of the green component (λmax ~ 560 nm) was sensitive to the pH value of the reaction solution, whereas those of the orange (λmax ~ 606 nm) and red (λmax ~ 654 nm) components were not. Consistent with the pH dependence of the intensity illustrated in Fig. 2 (b), the integrated area of the green component shown in Fig. 2 (c) decreased rapidly as the pH decreased and dominantly contributed to the change in the total quantum yield; in contrast, the areas of the orange and red components were almost flat. Therefore, change in pH of reaction solution influenced the intensity of the green emission component, and thus changed the quantum yield and spectrum. In other words, the pH dependence of the intensity of the green emission component determined the pH sensitivity of the quantum yield and spectrum.
The average peak wavelengths of the green, orange, and red Gaussian components required to fit the spectra of Y257F mutant luciferase were 570 nm, 610 nm, and 650 nm, respectively. These peak wavelengths were close to those in the fitting of WT luciferase, which were 560 nm, 606 nm, and 654 nm (see Table 1). As shown in the second row of Fig. 2 (b), the intensity of the green component of Y257F depended on pH, and it accounted partly for the spectral change. The orange spectra was moderately sensitive to pH, and it also contributed to the spectral change.
The intensity of the green component peaking at 573 nm was very weak in the fitting for the spectra of Y257A mutant luciferase. In this case, the change in the spectral intensity was mostly due to moderate changes in the intensities of the orange (~612 nm) and red (~666 nm) Gaussian components.
In contrast, the green component was not necessary for reproducing the spectra of Y257R and Y257E (the 4th and 5th rows in 2 (b)); only two components, the orange and red ones, fit their spectra. The peak energies, as well as the intensities, were insensitive to the changes in pH. The intensities and peak wavelengths of the two Gaussian components in the spectra of Y257E were very close to those of WT, but the intensity of the orange peak of Y257R was stronger and its peak was blue shifted by 14 nm compared with WT.
Comparison of Lcr and Ppy luciferase spectra
The pH-dependent spectra of Lcr luciferase shared many similarities with those of Ppy luciferase4. In addition to the well-known red shift in peak wavelength and intensity decrease in acid solution3, both spectra were reproduced with three Gaussian functions peaking at similar wavelengths (see Table 1 and Supplementary Fig. S1 online). The spectral shift and quantum-yield change with pH were due to the pH dependence of the intensity of the green component.
On the other hand, the Lcr luciferase spectra were less sensitive to pH changes than the Ppy luciferase spectra. Although their spectra around pH 8.0 were similar, the Lcr luciferase spectrum at pH 6.3 had a strong green shoulder, whereas the Ppy luciferase spectrum had regressed to a symmetric red one. A plot of the integrated areas of the Gaussian components (see Supplementary Fig. S1 online) revealed the difference more clearly: the green component of Lcr luciferase was lower in intensity at pH 8.0 and decreased more slowly as the pH changed. The lower sensitivity of Lcr luciferase may result from the substitution of Thr 217 with Ile for thermo-stability15,16.
Discussion
The spectra of the WT and the four mutant Lcr luciferases showed three patterns of pH sensitivity: the intensities and peak wavelengths of the WT Lcr and Y257F spectra were very sensitive to the change in pH, and their spectra included the green component whose intensity was sensitive to pH; those of the Y257A spectra were less sensitive to pH, and the spectra had a weak green component; in contrast, those of the Y257E and Y257R spectra were insensitive to pH, and the green components were negligible or absent from their spectra. Therefore, we conclude that the intensity of the green Gaussian component dominantly determines the shape and peak wavelength of the spectrum and thus regulates the pH sensitivity of the spectrum and quantum yield. To further illustrate this feature, in Fig. 3, we picked out the green Gaussian components (~560 nm) in the spectra of the WT and Y257F/A/E mutant luciferases at pH 8.0. This figure clearly shows that the substitution for the amino acid residue at position 257 principally modifies the emission intensity of the green Gaussian component and thus regulates the pH sensitivity of the bioluminescent spectra. Only the intensity of the green component was sensitive to pH changes. Since the green component was present in the spectra of the Y257F mutant and WT luciferase, their spectra were pH dependent. However, the green component was absent in the spectra of Y257E and Y257R, and hence their spectra and quantum yields were insensitive to pH.
Figure 3. Green (~560 nm) Gaussian components in the spectra of wild-type and Y257F/A/E mutant Lcr luciferases at pH 8.0.
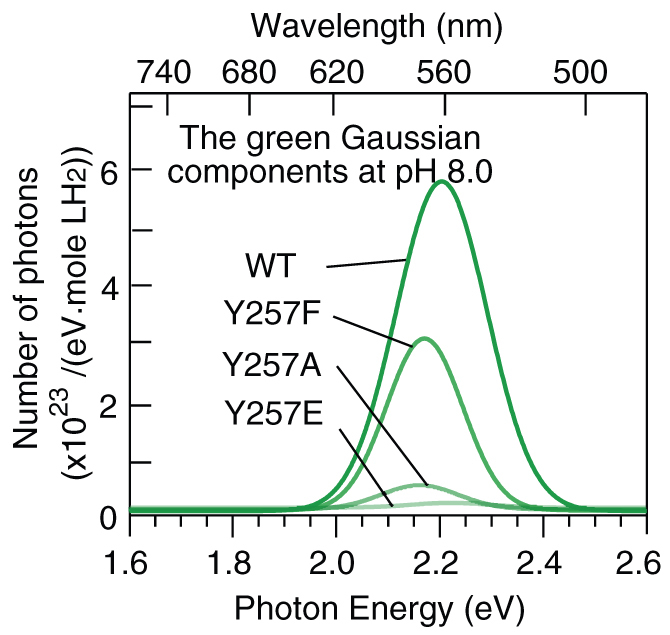
In the spectra of the bioluminescence catalyzed by WT and Y257F/A mutant luciferases, the moderate intensity changes in the orange (~610 nm) and red (~650 nm) Gaussian components also contributed to the spectral change to some extent. But compared with the green component, their intensities were less influenced by the mutagenesis. The intensities of the orange (~610 nm) and red (~650 nm) components of the Y257E/R mutants were insensitive to pH changes, and resulted in their spectra insensitive to pH.
In our previous reports, we measured the total-photon-flux spectra of firefly bioluminescence at various pH values4 and in the presence of bivalent metal ions5. We also reproduced the spectra with three Gaussian functions with various peak wavelengths, among which only the intensity of a green Gaussian component peaking at 560 nm was changed by pH and metal-ion additives, whereas the intensities of the other two peaking at 610 nm and 650 nm was not changed. On the basis of these quantitative measurements and analysis on the spectra of firefly bioluminescence in this and our previous reports4,5, we conclude that the colour change is mainly due to the intensity change in the green (~560 nm) emission component. Moreover, the intensity of the green emission is mainly influenced by the pH of the reaction solution, addition of bivalent metal ions, and mutagenesis. The mechanism of this intensity change is the key to understanding the mechanism of colour changes in firefly bioluminescence.
We cannot directly correlate the spectral change with a specific parameter of the amino acid residue introduced at position 257. In the crystal structure of the luciferase-ligand complex, tyrosine 257 participates in the hydrogen-bond network related to the movement of a key amino acid residue Ile288 in the active pocket12. Decreasing the size of the key amino acid residue at position 288 changes the emission colour to red12. In our experiments, decreasing the size of the side chain at position 257 from Tyr to Phe to Ala decreases the intensity of the green emission component and results in a red shift in the bioluminescence at pH 8.0. However, the size of the amino acid residue is not the only factor that determines the intensity of the green emission component because such green component was disappeared in Y257E and Y257R mutants, which have the long side chain of glutamate and arginine. This system is so complicated that we cannot exclude the influence of other alterations such as disturbances in the hydrogen-bond network or changes in the rigidity of the active pocket due to substitution.
The orange and red Gaussian components of Y257F/A/E had similar intensities and peak wavelengths to those of WT luciferase. However, the orange components of the Y257R mutant had stronger intensities and higher peak energies than those of the other luciferases. The reason may be that the positive charge of arginine exerts a large electrostatic potential on the benzothiazole ring of the oxyluciferin. Theoretical studies have indicated that the electrostatic interaction of positively charged amino acid residues near the benzothiazole ring of oxyluciferin may greatly shift the emission peak of phenolate-keto-OL− to the high energy side and alter the emission oscillation strength17,18.
The quantitative analysis that we performed on the spectra of the Y257R mutant demonstrated that there is a shift in the emission peak of the orange component. However, we would like to point out that the shift in the orange (~610 nm) emission peak does not play a critical role in the pH-dependent colour change in the other mutant luciferases. Despite the blue shift, the intensity of the orange component in the Y257R mutant is still insensitive to changes in pH, just like the other luciferases. Rather than the peak shift in the orange component, it is the variation in the intensity of the green emission component that is the crucial determinant of the pH-sensitive emission colour of firefly bioluminescence.
In summary, we quantitatively measured the spectra of bioluminescence catalyzed by WT Lcr luciferase and four site-directed mutants at position 257 at various pH values. We analysed their characteristics via curve fitting with Gaussian functions. All spectra were well reproduced with two or three Gaussian emission components, and the Gaussian components had similar peak wavelengths around 560 nm, 610 nm, and 650 nm. The spectra of WT and Y257F luciferases with emissions having the green component at 560 nm were sensitive to changes in the pH value, whereas those of Y257R and Y257E without the green component were insensitive to pH. This means the mutagenesis mainly influenced the intensity of the green emission component and hence altered the pH sensitivity of the bioluminescence spectrum. The emission intensities of the orange (~610 nm) and red (~650 nm) components were less sensitive or insensitive to changes in the reaction conditions such as pH, metal-ion additives, or mutated luciferase. Although the introduction of specific amino acids such as arginine changed the peak wavelength and intensity of the orange (~610 nm) emission component, it remained insensitive to changes in pH.
The intensity of the green emission component sensitively depends on the pH value4, addition of metal ions5, and mutagenesis, determining the pH sensitivity of the firefly bioluminescence emission. Clarifying the mechanism of the change in intensity of the green emission components is the key to solving the mystery of colour changes in firefly bioluminescence. We suggest that studying the influence of microenvironmental changes on the green emitters, such as in the process of generating its excited state and its emission efficiency, in addition to studying the shift in peak wavelength will lead to a fuller understanding of the colour change of firefly bioluminescence.
Methods
Materials
Recombinant wild-type Lcr luciferase16 produced from E. coli was provided by Kikkoman Corporation, Japan. The luciferase mutants Y257F/R/A/E were generated using a QuikChange site-directed mutagenesis kit (Stratagene). The expression and purification methods of the Y257F/R/A/E were only slightly modified from those described previously12. Cold-shock vector, pColdI (TaKaRa), was used for expression. Histidine tag affinity purification was carried out instead of cation exchange chromatography before the hydroxyapatite chromatography. All the reactants were diluted to the expected concentration with a GTA buffer solution (0.15 M, containing 0.05 M 3,3-dimethylglutaric acid, 0.05 M 2-amino-2-hydroxymethyl-1,3-protanediol, and 0.05 M 2-amino-2-methyl-1,3-propanediol (Wako), the pH of which was adjusted to the expected value by adding NaOH or HCl aqueous solution). The assays for quantitative bioluminescence measurement were performed by adding 50 μL of ATP aqueous solution (50 nmol) to a 50 μL mixture containing d-LH2 (0.5 pmol), luciferase (0.1–0.9 nmol), and MgSO4 (0.5 μmol) at 25 ± 1°C. The assays were similar to those used by Seliger et al.3 and Ando et al.4. The concentration of the substrate LH2 was minimized to enable its complete consumption for obtaining the quantum yield of the bioluminescence. The concentrations of the luciferases were made two-fold higher than that of LH2 in order to provide adequate binding sites for the substrate LH2 and to ensure a quick reaction. Over 99% of the LH2 was confirmed to have been consumed.
Setup
The homemade setup described in reference19 was used to measure a quantitative spectrum. The spectrum was scaled by the integrated number of photons in the bioluminescence flux. A low-noise charge-coupled device (CCD, Spec10:400BR, Princeton Instruments) cooled with liquid nitrogen to −120°C was utilized as a light detector. The absolute sensitivity of the optical system and the detector, the transmissivity of the system, and the light-collection efficiency of the sample cell were calibrated in order to convert the raw CCD data into quantitative spectra in units of photons/nm. The quantum yield was obtained by dividing the integrated area of the above quantitative spectrum by the molecular number of luciferin.
Measurements
The CCD was set to exposure mode from the start of the bioluminescent reaction. The spectra were recorded until no signal was detectable. Simultaneous monitoring of the bioluminescence with a luminometer (ATTO, AB2200) provided the reference reaction time. The pH values of the reaction solution were measured after the spectrum measurement.
Curve fitting
A program developed by our group was used in the curve fitting with Gaussian functions. The spectra were processed to have a horizontal axis in units of photon energy (eV). Two or three Gaussian functions were used to reproduce the experimental spectrum (see Supplementary Fig. S2 online). Three parameters, i.e., peak intensity, peak energy, and full width at half maximum (FWHM) of each peak, were adjusted so that the best coincidence was obtained between their summation curve and the experimental spectrum. The degree of coincidence was statistically judged by correlation coefficients. Most fittings gave correlation coefficients above 0.999, except those of Y257E, which were in the range of 0.997–0.998 due to the relatively low S/N ratio. The areas of the Gaussian functions were then integrated, and the corresponding contribution of each Gaussian component to the total quantum yield, ΦGC, was calculated by dividing the integral area by the molecule number of the substrate LH2.
Author Contributions
All authors designed the experiments. Y.W. performed the spectral measurements and analyses. K.T. and T.N. prepared the mutant enzymes. Y.W. and H.A. wrote the first version of the manuscript. All discussed the data and revised the manuscript.
Supplementary Material
Impact of Site-Directed Mutant Luciferase on Quantitative Green and Orange/Red Emission Intensities in Firefly Bioluminescence
Acknowledgments
We are grateful to Dr. Gomi and Dr. Fukuda (Kikkoman Co., Japan) for providing the wild-type Luciola cruciata luciferase and for their helpful discussions. This research was partly supported by the Japan Society for the Promotion of Science (Kakenhi 23360135), the Ministry of Education, Culture, Sports, Science & Technology (MEXT; Kakenhi 20108016), and the Photon Frontier Network Program of MEXT, Japan. One of us (Y. W.) is also grateful for the support given by the International Priority Graduate Programs of MEXT, Japan.
References
- Seliger H. H. & McElroy W. D. The colors of firefly bioluminescence: enzyme configuration and species specificity. Proc. Natl. Acad. Sci. U. S. A. 52, 75–81 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviani V. R. The origin, diversity, and structure function relationships of insect luciferases. Cell. Mol. Life Sci. 59, 1833–50 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger H. H. & McElroy W. D. Spectral emission and quantum yield of firefly bioluminescence. Arch. Biochem. Biophys. 88, 136–41 (1960). [DOI] [PubMed] [Google Scholar]
- Ando Y. et al. Firefly bioluminescence quantum yield and colour change by pH-sensitive green emission. Nat. Photonics 2, 44–47 (2007). [Google Scholar]
- Wang Y. et al. Quantum yields and quantitative spectra of firefly bioluminescence with various bivalent metal ions. Photochem. Photobiol. 87, 846–52 (2011). [DOI] [PubMed] [Google Scholar]
- White E. H., Rapaport E., Hopkins T. A. & Seliger H. H. Chemi-and bioluminescence of firefly luciferin. J. Am. Chem. Soc. 91, 2178–80 (1969). [DOI] [PubMed] [Google Scholar]
- McCapra F. Chemical generation of excited states: the basis of chemiluminescence and bioluminescence. Meth. Enzymol. 305, 3–47 (2000). [DOI] [PubMed] [Google Scholar]
- Niwa K. et al. Quantum yields and kinetics of the firefly bioluminescence reaction of beetle luciferases. Photochem. Photobiol. 86, 1046–9 (2010). [DOI] [PubMed] [Google Scholar]
- Ugarova N. N., Maloshenok L. G., Uporov I. V. & Koksharov M. I. Bioluminescence spectra of native and mutant firefly luciferases as a function of pH. Biochemistry (Moscow) 70, 1262–7 (2005). [DOI] [PubMed] [Google Scholar]
- Viviani V. R. et al. The influence of the loop between residues 223-235 in beetle luciferase bioluminescence spectra: A solvent gate for the active site of pH-sensitive luciferases. Photochem. Photobiol. 84, 138–44 (2007). [DOI] [PubMed] [Google Scholar]
- Branchini B. R. et al. An alternative mechanism of bioluminescence color determination in firefly luciferase. Biochemistry 43, 1205–10 (2004). [DOI] [PubMed] [Google Scholar]
- Nakatsu T. et al. Structural basis for the spectral difference in luciferase bioluminescence. Nature 440, 372–6 (2006). [DOI] [PubMed] [Google Scholar]
- Hosseinkhani S. Molecular enigma of multicolor bioluminescence of firefly luciferase. Cell. Mol. Life Sci. 68, 1167–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.
- Hirokawa K., Kajiyama N. & Murakami S. Improved practical usefulness of firefly luciferase by gene chimerization and random mutagenesis. Biochim. Biophys. Acta 1597, 271–9 (2002). [DOI] [PubMed] [Google Scholar]
- Kajiyama N. & Nakano E. Thermostabilization of firefly luciferase by a single amino acid substitution at position 217. Biochemistry 32, 13795–99 (1993). [DOI] [PubMed] [Google Scholar]
- Nakatani N., Hasegawa J. & Nakatsuji H. Red light in chemiluminescence and yellow-green light in bioluminescence: color-tuning mechanism of firefly, photinus pyralis, studied by the symmetry-adapted cluster-configuration interaction method. J. Am. Chem. Soc. 129, 8756–65 (2007). [DOI] [PubMed] [Google Scholar]
- Cai D., Marques M. A. L. & Nogueira F. Accurate color tuning of firefly chromophore by modulation of local polarization electrostatic fields. J. Phys. Chem. B 115, 329–332 (2011). [DOI] [PubMed] [Google Scholar]
- Ando Y. et al. Development of a quantitative bio/chemiluminescence spectrometer determining quantum yields: re-examination of the aqueous luminol chemiluminescence standard. Photochem. Photobiol. 83, 1205–10 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Impact of Site-Directed Mutant Luciferase on Quantitative Green and Orange/Red Emission Intensities in Firefly Bioluminescence