Abstract
Background
Biofilms are communities of bacteria that are characterized by specific phenotypes, including an increased resistance towards anti-microbials and the host immune system. This calls for the development of novel biofilm prevention and treatment options to combat infectious disease. In Escherichia coli, numerous global regulators have been implicated in the control of biofilm associated cell surface organelles. These include the flagellar regulator FlhD/FlhC, the osmoregulator EnvZ/OmpR, and the colanic acid activator RcsCDB. Using flow cell technology and fluorescence microscopy, we determined the temporal expression from flhD::gfp, ompR::gfp, and rcsB::gfp in E. coli biofilm, as well as the impact of the negative regulation of flhD by OmpR and RcsB. Spatial gene expression was investigated from flhD::gfp.
Results
The temporal gene expression profile for flhD yielded an early peak at 12 h, a minimum of expression at 35 h, and a second increase in expression towards 51 h of biofilm development. In contrast, the ompR profile showed a peak at 35 h. A mutation in ompR abolished time dependence of flhD expression after the initial growth period of 12 h. Intriguingly, rcsB expression did not correlate inversely with flhD expression, yet a mutation in rcsB abolished time dependence of flhD expression as well. Spatially, expression of flhD was highest in the outermost layer of the biofilm in the parent strain. In ompR and rcsB mutants, flhD was expressed throughout the biofilm. Mutations in both, ompR and rcsB increased flhD expression throughout all temporal and spatial experiments. This increase was paralleled by reductions in biofilm amounts at four tested time points.
Conclusion
Our data lead to the conclusion that FlhD/FlhC and its regulation by OmpR and RcsB may be our first target mechanism for the development of novel biofilm prevention and treatment techniques.
Keywords: Escherichia coli, Biofilm, Reporter gene fusion, Fluorescence microscopy
Background
Bacterial biofilms are defined as sessile communities of bacteria that form on air-liquid or liquid–solid interfaces, or even intracellularly [1]. Due to their high resistance to any attempts of removing them, biofilms have a profound impact in many clinical settings, including catheter-associated urinary tract infections [2], periodontitis [3], and otitis [4], as well as Pseudomonas aeruginosa infections of cystic fibrosis patients [5]. Much research has been done on disease mechanisms relating to the biofilm lifestyle. Yet, many of the early studies do not consider that growth conditions for the bacteria differ across the biofilm and also change with time. As one example, bacteria residing within the fully matured biofilm have limited access to nutrients and oxygen, but are also well protected from anti-microbials, as well as the host immune system. In contrast, bacteria that grow at the surface of the three-dimensional structure or are still in the early phases of biofilm formation would have better access to nutrients and oxygen, but are also more exposed to anti-microbials. Some temporal studies of gene expression in biofilms were done years ago [6]. Spatial studies have been done more recently. These were facilitated by advances in microscopy techniques, as well as the development of fluorescent probes [7-9].
Fusions of gene promoters to the structural genes of fluorescence proteins were used to study heterogeneity in biofilms of several bacterial species. This was done to measure: i) spatial gene regulation in biofilm of Bacillus subtilis[10], ii) real-time spatial gene expression in Geobacter sulfurreducens electricity producing biofilm [11], iii) quantitative gene expression in biofilm of Salmonella[12], iv) single cell gene expression in B. subtilis biofilm [13], and v) the effect of inhibitors on Pseudomonas aeruginosa biofilm [14]. To reduce complexity and facilitate genetics experiments, flow cell technology was developed to grow the biofilm [8,15]. This allows the biofilm to form under continuous hydrodynamic conditions at a controlled and reproducible flow rate. In this study, we used promoter fusions to green fluorescence protein (GFP), flow cell biofilms, and fluorescence microscopy to measure temporal and spatial expression of selected biofilm associated genes in Escherichia coli biofilms.
The genetic system that is used for this study consists of the flagellar [16] and global regulator [17-19] complex FlhD4/FlhC2[20] and the two-component systems for osmoregulation EnvZ/OmpR [21] and colanic acid activation RcsCDB [22]. These three regulatory systems are part of a partial transcriptional network that centered around FlhD/FlhC and regulated all the biofilm associated cell surface organelles [23]. In particular, OmpR and RcsB in their phosphorylated form are inhibitors of flhD expression [24]. RcsB and OmpR are regulators of type I fimbriae [25,26], as well as expression of many other genes [27,28]. In planktonic E. coli, growth phase dependent expression of flhD required OmpR. Additionally, flhD expression in the ompR mutant was much higher [29]. This was also true for flhD expression and swarming of Xenorhabdus nematophila[30].
While all the above research involving OmpR, RcsB, and FlhD/FlhC was done with planktonic bacteria, this study investigates the impact of this regulation on biofilm formation. In particular, we wanted to accomplished three goals: i) provide proof of concept that the study of temporal and spatial expression of biofilm associated genes can lead to the identification of novel targets or target mechanisms for the development of biofilm prevention techniques (gene is expressed early in biofilm development) and treatment options (gene is expressed late and at the edge of the biofilm); ii) attempt to identify FlhD/FlhC as the first such targets, because it is a transmitter between numerous environmental conditions and many cellular responses, and iii) establish OmpR and RcsB as control mechanisms that can be taken advantage of to increase flhD expression and reduce biofilm amounts.
Results
Temporal gene expression of flhD, ompR, and rcsB in E. coli biofilm
Expression of flhD peaked at 12 h and increased again towards 51 h of biofilm formation
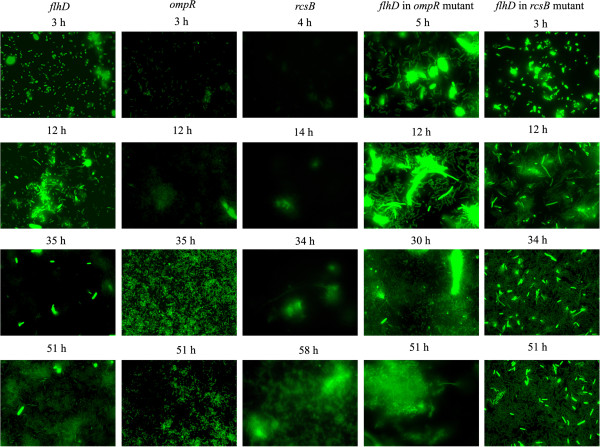
Fluorescence microscopy images were produced from flow cell grown biofilm of the E. coli genetic parent strain AJW678 that contained the flhD::gfp fusion plasmid, called pPS71. Fluorescence signals obtained from these biofilms were highest at 12 h, lowest at 35 h, and then increased again towards 51 h of biofilm formation. This was seen in all four time series of images that had been taken from four independently formed biofilms. A selection of images from one of these experiments is shown in the left column of Figure 1. Occasionally, we observed high signals in individual bacteria of the 3 h sample, but the number of bacteria on the slides was not indicative of a biofilm at that point in time.
Figure 1.
Fluorescence images of flhD::gfp, ompR::gfp, rcsB::gfp in AJW678 and flhD in the ompR and rcsB mutant strains. Biofilms of BP1470, BP1432, BP1462, BP1531, and BP1532 were grown in flow cells and subjected to fluorescence microscopy. Four time points were selected for each strain; these are printed on top of the respective images. At the very top of each column, promoter names are printed. Images were taken at 1,000 fold magnification.
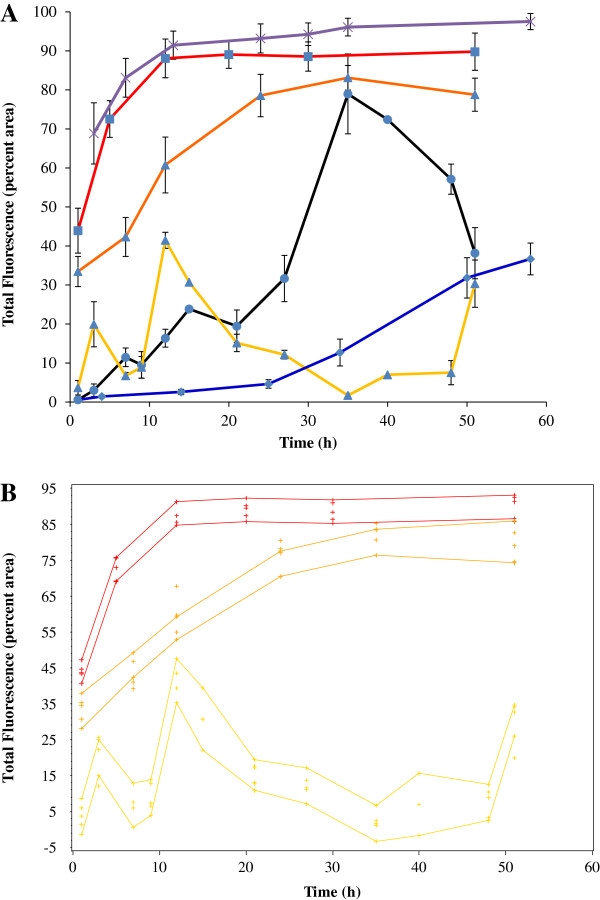
The images from Figure 1 were converted into quantitative data by calculating the percent area of the images that were fluorescent. The resulting expression profile for flhD showed a peak at 12 h (Figure 2A, yellow line, blue triangles). Fluorescence was lowest at 35 h and increased again towards 51 h. We also noticed a small single point peak at 3 h, which is in agreement with the occasional high fluorescence of small numbers of individual bacteria that was visualized on the images (Figure 1). Since fluorescence from the green fluorescence protein reporter is indicative of flhD expression, we conclude that flhD expression was highest at 12 h, lowest at 35 h, and increased again towards 51 h.
Figure 2.
Temporal expression of flhD, ompR, rcsB in AJW678 and flhD in the ompR and rcsB mutant strains. A. Fluorescence was quantified as percent area of the images that were fluorescent, averages and standard deviations were determined. The x-axis indicates the time (hours) of biofilm formation. The y-axis indicates the total fluorescence intensity in percent area for the different strains at the different time points. The yellow, black, and blue lines are showing the gene expression profile of BP1470 (AJW678 flhD::gfp), BP1432 (AJW678 ompR::gfp), and BP1462 (AJW678 rcsB::gfp), respectively. The red line is the temporal expression profile of BP1531 (flhD::gfp ompR::Tn10), the orange line that of BP1532 (flhD::gfp rcsB::Tn5). The purple line is our housekeeping strain BP1437 which contains the aceK::gfp fusion plasmid. B. Confidence bands were calculated using the loess procedure. Upper and lower lines of each colors are indicating the highest and the lowest level of the total fluorescence intensity. The color code is identical to A.
The temporal expression of ompR, but not rcsB, correlated inversely with that of flhD
Expression of the negative regulator of flhD expression, OmpR, exhibited a temporal profile (Figure 1, second column from the left and Figure 2A, black line, blue circles) that was almost the inverse of flhD expression between 21 h and 51 h of biofilm formation. Specifically, ompR expression increased between 21 h and 34 h, while flhD expression decreased. Between 34 and 51 h, ompR expression decreased, while flhD expression increased. Expression of another negative regulator of flhD expression, RcsB, did not correlate with the temporal expression profile for flhD (Figure 1, center column and Figure 2A, blue line, blue diamond’s). Until 25 h, the fluorescence signal from the rcsB::gfp plasmid containing strain was very weak, but increased steadily after this point in time.
Mutations in ompR and rcsB abolished temporal differences in flhD expression
The fluorescence signals from flhD::gfp in the ompR and rcsB mutant strains were higher than those from the other strains at all times. Expression of flhD in the ompR mutant increased over the first 12 h and reached a steady state level after that (Figure 2A, red line, blue squares). Between 12 h and 24 h, expression of flhD in the rcsB mutant (Figure 2A, orange line, blue triangles) increased more slowly than in the ompR mutant, but was reasonably growth phase independent after 24 h as well. This slower increase in flhD expression in the rcsB mutant (relative to the ompR mutant) correlates with the reduced increase in rcsB expression (blue line) during the same time period, relative to the increase in ompR expression (black line). Statistical analysis of the data with the Loess procedure yielded confidence bands for the ompR and rcsB mutant strains that did not overlap with that of the parent (Figure 2B).This indicates that there is indeed a statistically significant difference between the parent strain and either of the two mutants.
In comparison, the expression profile for our housekeeping strain that contains the aceK::gfp fusion plasmid was high at all times (Figure 2A, purple line, cross symbols). Expression increases in any strain during the first 12 h can be explained by the increase in bacterial cell numbers during the early development of the biofilm.
Spatial gene expression of flhD in E. coli biofilm
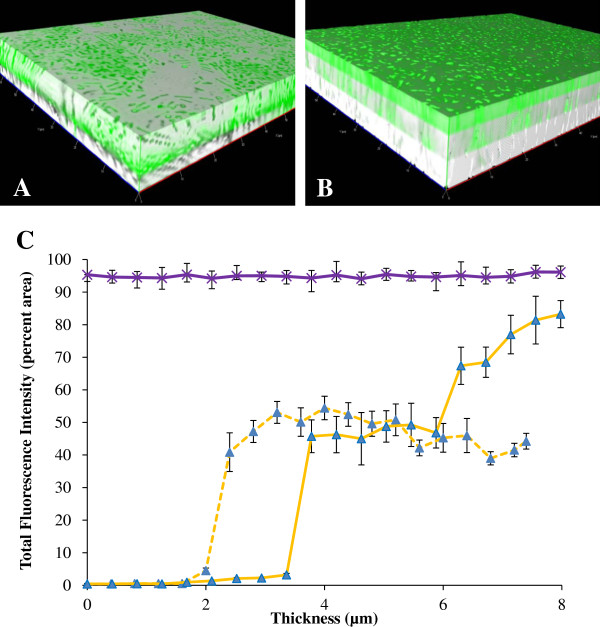
From the temporal gene expression experiment, we knew that the highest expression of flhD was at 12 h and 51 h of biofilm formation. As a consequence, we performed the spatial gene expression experiment for flhD at those two time points. In both the 12 h (Figure 3A) and 51 h (Figure 3B) biofilms, the expression of flhD was highest at the outer layer of the biofilm. Fluorescence calculated from the individual images of the z-stacks showed that at 12 h, there was little or no expression of flhD within the first 2 μm from the surface that the biofilm had formed on (dotted yellow lines). Expression increased rapidly at 2 μm to approximately 50% coverage. In 51 h biofilms, there were three distinct intensity levels (solid yellow lines). Until 3 μm, the expression of flhD was very low; at 3.5 μm, the expression jumped to 50% and maintained this level until 6 μm; across the upper 2 μm of our biofilm, flhD expression increased to approximately 75% of the total area of the images. Our housekeeping gene in comparison was highly expressed all throughout the biofilm (purple lines).
Figure 3.
Spatial gene expression of flhD in the parent strain. (A) and (B) are the 3D images constructed from the z-stacked images (bright field and fluorescence) at 12 hours (A) and 51 hours (B), using BP1470 (AJW678 pPS71). (C) is the quantitative representation of the spatial gene expression of flhD at 12 hours (dashed yellow line) and 51 hours (solid yellow line) of biofilm formation. The purple line is the spatial expression profile from the aceK::gfp fusion at 34 h.
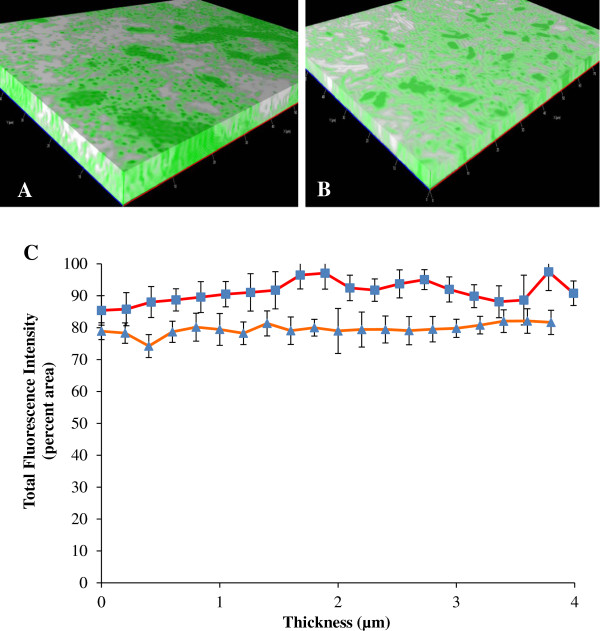
The temporal gene expression study had determined that the expression of flhD in the ompR and rcsB mutant strains was constitutively high throughout the experiment after a primary increase during the initial time period of biofilm formation. As time points for the spatial experiment, we selected 33 h for the ompR mutant (Figure 4A) and 51 h for the rcsB mutant (Figure 4B). Interestingly, expression of flhD in both mutants was high across all layers of the biofilm. Fluorescence was between 80 and 95% coverage across the entire biofilm of both mutants (Figure 4C). By all appearances, both OmpR and RcsB abolished spatial differences in flhD expression together with temporal ones, while increasing overall expression.
Figure 4.
Spatial gene expression of flhD in the ompR and rcsB mutant strains. (A) is the 3D image of the 33 h biofilm from BP1531 (ompR::Tn10 pPS71), (B) is the respective image from the 51 h biofilm from BP1532 (rcsB::Tn5 pKK12). (C) is the quantitative representation of the spatial gene expression of flhD in the ompR mutant (red line) and the rcsB mutant (orange line) at the times points represented in A and B.
Mutations in ompR and rcsB reduced biofilm biomass
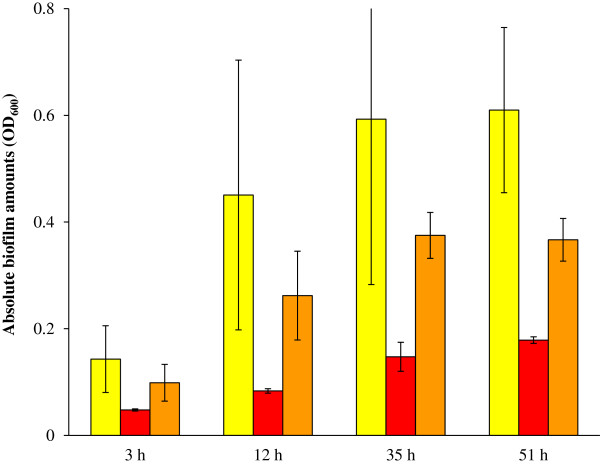
The 3D reconstructions of the biofilms showed that the biofilm from the ompR and rcsB mutants was much thinner than that of the parent strain. The mutant biofilms were no more than 4 μm, as opposed to >8 μm for biofilm from the parent strain (notice x-axis of Figure 4C versus that of Figure 3C). This observation indicates that the elevation of flhD expression levels in the two mutants does indeed have the predicted outcome of reducing biofilm amounts. However, we were unable to quantify thickness of the parental biofilm with the fluorescence microscopy beyond 8 μm due to optical limitations of the objective used for these experiments. To quantify biofilm biomass, the crystal violet (CV) assay was performed with parent bacteria, and ompR and rcsB mutants (Figure 5). Both mutants produced a considerably smaller amount of biofilm than the parent. This difference was more pronounced for the ompR mutant (red bars) than for the rcsB mutant (orange bars).
Figure 5.
CV assay to quantify the biofilm amounts of the ompR and rcsB mutants in comparison to the parent strain. The biofilm biomass was determined for BP1470 (AJW678 pPS71), BP1531 (ompR::Tn10 pPS71) and BP1532 (rcsB::Tn5 pKK12). This was done at four different time points, which are indicated on the x-axis. The yellow bars are the biofilm biomass of the parent strain, the red bars are for the ompR mutant, and the orange bars are for the rcsB mutant. Averages and standard deviations were calculated across three replicate experiments.
Discussion
In the Introduction, we postulated that a biofilm prevention target would be characterized by its expression early in biofilm development. This was the case for flhD whose expression peaked at 12 h. A biofilm treatment target was postulated to be characterized by expression late in biofilm development and at the outermost edge of the biofilm. This, too, was true for FlhD/FlhC. Expression of flhD increased again towards 51 h, the highest expression of flhD was in the outer layer of the biofilm. Based upon these results, we come to the conclusion that the flagella master regulator complex FlhD/FlhC may be our first target for both, biofilm prevention and treatment techniques. This would fulfill our first two goals: i) provide proof of concept that our approach can identify targets for biofilm prevention and treatment techniques and ii) establish FlhD/FlhC as the first such target. In fulfillment of the final goal of this study, we identified two mechanisms to increase flhD expression and reduce biofilm amounts. Mutations in the two-component response regulator genes ompR and rcsB increased flhD expression to the point where temporal and spatial differences in expression were abolished. These expression increases where paralleled by decreases in biofilm amounts, relative to the parent strain.
The expression profiles of flhD, ompR, and rcsB can be related to Biofilm phases
Originally described in Pseudomonas aeruginosa, it is now widely accepted that biofilm development in many bacteria involves reversible attachment, irreversible attachment, maturation, and dispersion [31]. These phases are characterized by cell surface organelles such as flagella, type I fimbriae and curli, as well as numerous exopolysaccharides. The following three paragraphs relate the temporal expression profiles of flhD (positive regulator of flagella), ompR (negative regulator of flagella and positive regulator of curli), and rcsB (negative regulator of flagella and positive regulator of type I fimbriae and colanic acid capsule) to current literature on biofilm developmental phases. According to our previous review [23], the hypothesis for the temporal expression profiles was that flhD expression may peak during reversible attachment, ompR expression during irreversible attachment, and rcsB expression may increase towards maturation.
A recent review article summarized the regulation of motility during biofilm formation [32]. The authors believe that flagella are important in the motility-to-biofilm transition in a way that inhibition of motility encourages biofilm formation by means of several functional (e.g. YcgR) and regulatory (e.g. RcsB) mechanisms [22,33,34]. Our temporal expression profile of flhD is partially in agreement with this postulate. We saw a peak in expression at 12 hours (Figure 2), which may resemble reversible attachment, and a time period of low flhD expression around 34 h, possibly resembling irreversible attachment. However, expression of flhD increased again towards 51 h (Figure 2). This late increase is not necessarily in agreement with current biofilm models. However, Guttenplan and Kearns [32] leave room for flagella regulators that may still be discovered. Also, the role for flagella in dispersal is controversial.
The hypothesis [23] that ompR expression may be highest during irreversible attachment was built upon the fact that phospho-OmpR was a negative regulator of flhD expression [24] and a positive regulator of curli [28,35]. Our temporal expression profile of ompR is in agreement with this hypothesis. The peak for ompR was at 34 h, where flhD expression was minimal (Figure 2). The production of curli has previously been recognized as a control mechanism for biofilm formation [36], an adherence tool to human uroepithelical cells [37], and part of the motility-to-biofilm transition. CsgD contributes to this transition by activating the expression of curli and inhibiting flagella biosynthesis [38]. The expression peak of the positive curli regulator, OmpR, at 34 h could be our marker for irreversible attachment.
Maturation of a biofilm typically requires the synthesis of an exopolysaccharide capsule that serves as a ‘glue’ to keep the microcolony together and contributes to adherence to the surface. This capsule can consist of many different substances, among them the K-capsule polysaccharide that is a contributor to the intracellular lifestyle of uropathogenic E. coli[1] and colanic acid, which has been recognized early as an important factor in forming the three dimensional structures that constitute the biofilm [39]. The phosphorelay system RcsCDB is an activator of colanic acid production [40], while also activating the synthesis of type I fimbriae [25]. These multiple functions of RcsB may explain the slow and steady increase of rcsB expression during biofilm formation (Figure 2) that cannot be correlated with a single phase of biofilm development. With the exception of the late increase in flhD expression, our temporal expression profiles are in agreement with our hypothesis from the review article [23], as well as current literature.
Regulation of flhD by multiple response regulators offers ample opportunity to control biofilm amounts and cell division
Since the goal of our research was to modulate signal transduction pathways and reduce biofilm amounts, the next step after the identification of FlhD/FlhC as our first target would be the attempt to increase flhD expression levels, ultimately causing a reduction in biofilm amounts.
The expression of flhD is regulated by many environmental and genetic factors. Environmental factors include temperature [41], osmolarity [24], and the nutritional state of the cell [42]. Genetic factors are similarly diverse and include the Catabolite Repressor Protein CRP and the nucleoid associated protein H-NS [43], the transcriptional regulator LrhA [44], the LysR family protein HdfR [33], and the insertion of IS elements into the flhD promoter [45-47]. Post transcriptional regulation involves the carbon storage regulator CsrA [48] and a negative regulator of cell motility, YdiV [49]. At the transcriptional level, regulation of flhD expression can be accomplished by several of the response regulators of two-component systems, such as RcsB [50], OmpR [24], and QseC [51].
In this study, knock-out mutations in rcsB and ompR yielded an impressive increase in flhD expression in the ompR and rcsB mutants (Figures 2 and 4). Additionally, expression of flhD was not anymore dependent upon the biofilm phase, after the biofilm had formed (Figure 2) or the location of the individual bacterium within the biofilm (Figure 4). The temporal expression profile of flhD in the ompR mutant is similar to the one that was observed previously in planktonic bacteria [29]. However, in planktonic bacteria, we never observed more than 2 or 3 fold increases in flhD expression in the ompR mutant, relative to the parent. Considering the fact that the images for flhD in the ompR mutant had been obtained at a much reduced excitation intensity (10% versus 90% in the parent strain), the difference in flhD expression between the two strains must be much higher in biofilm than in planktonic bacteria.
Intriguingly, the ompR and rcsB mutants are also our first two mechanisms to reduce biofilm amounts by elevating the expression levels of FlhD/FlhC. This observation provides confidence in our conclusion that impacting the signal transduction cascade, consisting of multiple two-component response regulators and FlhD/FlhC can be used to control biofilm amounts. Since the number of two-component systems in E. coli is rather large [28] and response regulators respond to a broad range of environmental signals, the two-component signal transduction mechanism offers ample opportunity at controlling bacterial phenotypes and behaviors by deliberately changing the bacterial environment.
Conclusions
The bacterial species E. coli includes many pathogens, in particular biofilm formation [52,53] and prevention [54] in uropathogenic E. coli (UPEC) have been researched intensively over the past few years. The goal of this study was to use an E. coli K-12 strain as a model to show that the study of temporal and spatial gene expression can lead to the identification of targets for the development of novel biofilm prevention and treatment options. We propose FlhD/FlhC as the first of such targets and OmpR and RcsB as two mechanisms to control this target. Our intention is to identify more of these targets/target mechanisms, using the temporal/spatial gene expression approach on a selection of biofilm associated genes. With respect to FlhD/FlhC, we believe that a gene that is this highly regulated by so many environmental and genetic factors is ideally suited to be controlled by deliberate changes to the environment, through a signal transduction cascade that may involve additional two-component response regulators beyond OmpR and RcsB, ultimately impacting biofilm amounts. The two-component control mechanism may be particularly important for UPEC strains where two-component signaling plays a large role in motility, quorum sensing, biofilm formation, and virulence [55,56].
Methods
Bacterial strains, plasmids, and growth conditions
All the bacterial strains and plasmids that are used for this study are listed in Table 1. Throughout the study, we use the E. coli K-12 strain AJW678 as a parental strain because it is a good biofilm former [57] and wild-type for the biogenesis of flagella and type I fimbriae and curli. AJW678 is lacking the IS element [42] in the flhD promoter that makes bacteria highly motile. MC1000 is another K-12 strain [58,59]. It contains an IS5 in the flhD promoter [47], is highly motile, but produces much reduced biofilm amounts. To assure maximal expression of flhD, we use this promoter to construct the flhD::gfp fusion plasmid pPS71.
Table 1.
Bacterial strains and plasmids used for this study
| Strains | Relevant genotypes | Reference |
|---|---|---|
| AJW678 |
thi-1 thr-1(am) leuB6 metF159(Am) rpsL136 ΔlaxX74 |
[57] |
| AJW2050 |
AJW678 ompR::Tn10 |
[42] |
| AJW2143 |
AJW678 rcsB::Tn 5 |
[60] |
| MC1000 |
F-, araD139 Δ(araAB leu)7696 Δ(lacX74) galU galK strA prsL thi |
[59] |
| BP1470 |
AJW678 pPS71 |
This study |
| BP1531 |
AJW2050 pPS71 |
This study |
| BP1532 |
AJW2143 pKK12 |
This study |
| BP1432 |
AJW678 ompR::gfp |
This study |
| BP1462 |
AJW678 pEC2 |
This study |
| BP1437 |
AJW678 aceK::gfp |
This study |
|
Plasmids | ||
| pPS71 |
pUA66 flhD::gfp |
This study |
| pKK12 |
pPS71 CmR |
This study |
| pOmpR::gfp |
pUA66 ompR::gfp |
[62] |
| pEC2 |
pAcGFP rcsB::gfp |
This study |
| pAceK::gfp | pUA66 aceK::gfp | [62] |
The Tn10 and Tn5 transposons confer resistance towards tetracycline and kanamycin, respectively. Δ constitutes a deletion of the respective gene. CmR indicates chloramphenicol resistance. gfp encodes green fluorescence protein.
AJW2050 is an ompR mutant strain due to the insertion of a Tn10 transposon [42], AJW2143 is an rcsB mutant strain due to Tn5 insertion [60]. AJW678, AJW2050, and AJW2143 were kindly provided by Dr. Alan J. Wolfe (Loyola University Chicago, Maywood IL) and used in several of our previous studies [42,61]. Plasmids pPS71 (flhD::gfp), pKK12 (pPS71 CmR) and pEC2 (rcsB::gfp) were constructed for this study. The ompR::gfp plasmid was obtained from the Open Biosystems promoter collection [62] (Thermo Scientific, Huntsville, AL). As a housekeeping gene, we used aceK which encodes isocitrate dehydrogenase. This gene was selected because genes encoding enzymes of the tricarboxylic acid cycle have previously been shown to be uniformly expressed in biofilms of Geobacter sulfurreducens[11]. In addition, expression from the aceK::gfp fusion was reasonably steady in a temporal expression experiment with planktonic bacteria (Wilson T., and B.M. Prüß, unpublished data). The aceK::gfp fusion plasmid was also part of the Open Biosystems promoter collection.
Cloning of flhD::gfp (pPS71), pPS71 CmR (pKK12) and rcsB::gfp (pEC2) plasmids
pPS71
To construct the flhD::gfp containing plasmid, the flhD promoter region that starts 1,419 bp upstream of the +1 transcriptional start site and ends 502 bp downstream of the +1 was amplified from MC1000, using 5′-TCCTCGAGTGACTGTGCGCAACATCCCATT-3′ as forward primer and 5′-AGGTACCTGCCAGCTTAACCATTTGCGGA-3′ as reverse primer. This promoter fragment contains the IS5 that increases flhD expression and is located at −1,294 bp to −94 bp [47], making the fragment 1,921 bp in length. The forward and reverse primers were designed with XhoI and BamHI restriction enzyme recognition sites at the 5′ ends. The flhD promoter fragment was then digested with XhoI and BamHI. The vector pUA66 (Open Biosystems, Huntsville, AL), containing gfpmut2 as a reporter gene and a kanamycin resistance cassette, was also digested with these enzymes. To reduce re-ligation of the plasmid, digested pUA66 vector was treated with Calf Intestinal Alkaline Phosphatase (CIAP, Promega, Madison WI) that removes the 5′ phosphate. The double digested flhD promoter region was ligated into the digested and CIAP-treated pUA66 vector. Competent JM109 cells (Promega, Madison WI) were transformed with the resulting plasmid pPS71. The insertion was confirmed by restriction digest and sequencing. Ultimately, pPS71 was transformed into chemically competent AJW678 and AJW2050.
pKK12
The antibiotic resistance of pPS71 was changed from KmR to CmR creating pKK12. This permitted transformation of the flhD::gfp fusion plasmid into KmR mutants. pPS71 was digested with EagI to remove 280 bp from pPS71. This deleted region started upstream of the flhD promoter and extended upstream into the kanamycin resistance gene. This caused inactivation of kanamycin resistance. The digested plasmid was blunt ended with Klenow (Promega, Madison WI), and treated with CIAP. pHP45Ω-Cm was the source of the chloramphenical resistance gene cassette [63] and was digested with EcoRI and blunt ended with Klenow. The CIAP-treated pPS71 and pHP45Ω-Cm DNA fragments were ligated. Competent JM109 were transformed with the resulting plasmid pKK12, transformants were resistant to chloramphenicol, but not to kanamycin. Competent AJW2143 (rcsB::Kn) were then transformed with pKK12.
pEC2
To construct this plasmid, the rcsB promoter region that starts 100 bp upstream of its +1 transcriptional start site and ends 50 bp downstream was PCR-amplified from AJW678, using 5′-GAGAGATCTGCAACCTGTATCACACCCGATGAAAG-3′ as forward primer and 5′-GCAAAGCTTCGGATGGTCATCGGCAATAATTACG-3′ as reverse primer. The PCR-amplified region was then cleaned up and ligated into pGEM-T Easy (Promega, Madison WI). Successful ligations were identified by white color of the transformed colonies. Plasmids were digested using the HindIII and BglII restriction sites that had been added to the 5′ends of the primers. The promoterless pAcGFP1-1 encodes the green fluorescent protein AcGFP1, a derivative of AcGFP from Aequorea coerulescens, and has a kanamycin resistance gene (Clontech, Mountain View, CA). This plasmid was also double digested with the same enzymes. The digested rcsB promoter region was ligated into the digested pAcGFP1-1 vector. Competent JM109 cells were transformed with the resulting plasmid pEC2. The insertion region was confirmed by restriction digest and sequencing. Ultimately, pEC2 was transformed into chemically competent AJW678.
Bacterial strains were stored at −80°C in 10% dimethyl sulfoxide (DMSO). Before use, the bacterial strains were streaked onto LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl) agar plates and incubated overnight at 37°C. From the plates, cultures were inoculated into liquid tryptone broth (TB, 1% tryptone, 0.5% NaCl) and grown overnight at 37°C. For bacterial strains containing pPS71, 25 μg/ml of kanamycin were added to the bacterial growth medium. For pEC2, 50 μg/ml of kanamycin were added. For pKK12, 50 μg/ml of chloramphenicol were added.
Temporal and spatial expression of flhD, ompR, and rcsB
E. coli strains were grown in TB overnight at 37°C. 1 ml of each culture was injected into one channel of a 3 channel flow cell (Stovall, Greensboro NC) with a syringe as described [8]. The flow cell was incubated at room temperature for one hour without any media flow. After that, TB was pumped by an Isma Tec Low Flow High Accuracy Peristaltic Pump (Stovall) into the flow cell at 1 ml/min, equaling 0.33 ml/min per channel. For temporal expression experiments, the flow cell was disconnected after a maximum of 62 h. For spatial expression experiments, the flow cell was disconnected at time points of interest. Each of the investigated bacterial strains was processed at least three times for both temporal and spatial experiments. The flow cell system was kept free of air bubbles by the bubble trap that is part of the Stovall system.
We used a Zeiss Axio Imager M2 upright fluorescence microscope with ApoTome2 (Zeiss Microimaging, Thornwood NY) to detect the fluorescence signals coming from the promoter::gfp fusions. The Zeiss Axio Imager M2 microscope is equipped with a 100×/1.40 oil Paln-Apochromat objective, a Colibri2 higher power LED light source, and a high-resolution monochrome camera for optimal illumination and imaging. For the temporal experiment, fluorescence images were taken at appropriate time points. For the spatial experiments, 20 z-stacking images were taken at one or two time points, separately for fluorescence and bright field. Due to the objective working distance limit, z-sections could be effectively imaged across 8 μm in depth. In cases where biofilms were thicker than 8 μm on some areas of the slides, we selected areas of the biofilm that were consistent with the limitation of the objective.
The intensities of the fluorescence signals from aceK::gfp and from flhD::gfp in the ompR and rcsB mutants turned out to be much higher than those from the remaining strains and fusions. For this reason, we performed microscopy for BP1437 at 5% of the available excitation light and for BP1531 and BP1532 at 10%. For BP1470, BP1432, and BP1462, we used 90% of the available excitation light.
Quantification of the fluorescence signals and statistical analysis
To quantify the fluorescence signals that were visualized in the images, we used Image-Pro Plus software and determined the percent area of the image that produced a fluorescent signal. Specifically, pixels values from each image were divided by the pixel values that represent the total area of an image. Under the settings that were used for our imaging, this was 42,100 pixels. Resulting values were multiplied by 100 to yield percent. Next, we determined the average and standard deviation across all 9 images (3 images per biological replicate) for BP1531, BP1532, BP1462, and BP1437 and across the 4 images (1 image from each biological replicate) for BP1470 and BP1432 that were obtained at each time point. Finally, the average percent area was plotted against time for the temporal experiment.
Statistical analysis of the temporal data was done with local regression via the Loess procedure [64]. At each time point, a weighted least squares regression polynomial was fitted to a subset of the data to yield a Loess curve. Confidence bands were computed at a 95% confidence interval. This was done independently for the pPS71 containing parent strain and its ompR and rcsB mutant strains. To compare temporal expression profiles, overlaps of the confidence bands were determined. A lack of overlap between the confidence bands of any two strains is indicative of a statistically significant difference between the strains. The statistical analysis was done with SAS version 9.2.
For spatial gene expression experiments, 3D reconstructions of the biofilms were done from the z-stacked images with AxioVision v-4.7.1 software from Zeiss, using both fluorescence and bright field images. Quantification of the fluorescence signals from these images was done as described for the temporal experiment.
Crystal violet assay to determine biofilm biomass
Biofilm of BP1470, BP1531, and BP1532 were grown in individual wells of a 24 well plate in TB for 3 h, 12 h, 35 h, and 51 h at room temperature. Liquid bacterial growth medium was removed and biofilms were washed twice with phosphate buffered saline (PBS). Biofilms were stained with crystal violet (CV) as described [65-68]. The OD600 of the extracted CV was determined from a 1:10 dilution with a Synergy H1 plate reader from BioTek (Winooski, VT). Averages and standard deviations were determined across the three replicate experiments.
Abbreviations
CV: crystal violet; GFP: green fluorescence protein.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PS constructed the flhD::gfp plasmid pPS71, performed the fluorescence microscopy, and analyzed the data. He also wrote the first draft of the manuscript. ERC constructed the rcsB::gfp plasmid pEC2, KK changed the kanamycin resistance of pPS71 to chloramphenicol to yield plasmid pKK12. SMH designed the cloning strategies for all plasmids and supervised the undergraduate students. BMP designed the project, helped PS to set up the flow cells and the microscopy, and contributed to the analysis and interpretation of the data. All authors read the manuscript, made suggestions for changes, and approved the final manuscript.
Authors’ information
PS is a Ph.D. student in the Molecular Pathogenesis program and the main student working on this NIH funded project. ERC and KK were undergraduate researchers in the Prüß lab. SMH is the research associate in the lab. BMP is the principal investigator of the lab.
Contributor Information
Priyankar Samanta, Email: priyankar.samanta@my.ndsu.edu.
Emily R Clark, Email: emliyclark2912@gmail.com.
Katie Knutson, Email: katie.knutson.2@my.ndsu.edu.
Shelley M Horne, Email: sm.horne@ndsu.edu.
Birgit M Prüß, Email: birgit.pruess@ndsu.edu.
Acknowledgements
The AJW678 parental strains and its ompR and rcsB mutant strains were kindly provided by Dr. Alan J. Wolfe (Loyola University Chicago, Maywood IL). Pawel Borowicz (Advanced Imaging and Microscopy Core Laboratory, NDSU) helped with the microscopy and imaging conducted in this study, as well as with the quantification of the fluorescence signals. Curt Doetkott and Jamie Kubat (Department of Statistics, NDSU) helped with the statistical analysis of the data. The work was funded by grant 1R15AI089403 from the NIH/NIAID. The Synergy plate reader was purchased from grant 2012-67006-19659 from the USDA/NIFA.
References
- Goller CC, Seed PC. Revisiting the Escherichia coli polysaccharide capsule as a virulence factor during urinary tract infection: contribution to intracellular biofilm development. Virulence. 2010;1:333–337. doi: 10.4161/viru.1.4.12388. [DOI] [PubMed] [Google Scholar]
- Saint S, Chenoweth CE. Biofilms and catheter-associated urinary tract infections. Infect Dis Clin North Am. 2003;17:411–432. doi: 10.1016/S0891-5520(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Schaudinn C, Gorur A, Keller D, Sedghizadeh PP, Costerton JW. Periodontitis: an archetypical biofilm disease. J Am Dent Assoc. 2009;140:978–986. doi: 10.14219/jada.archive.2009.0307. [DOI] [PubMed] [Google Scholar]
- Hoa M, Tomovic S, Nistico L, Hall-Stoodley L, Stoodley P, Sachdeva L, Berk R, Coticchia JM. Identification of adenoid biofilms with middle ear pathogens in otitis-prone children utilizing SEM and FISH. Int J Pediatr Otorhinolaryngol. 2009;73:1242–1248. doi: 10.1016/j.ijporl.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T, Wood TK. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Pamp SJ, Sternberg C, Tolker-Nielsen T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry A. 2009;75:90–103. doi: 10.1002/cyto.a.20685. [DOI] [PubMed] [Google Scholar]
- Villena GK, Fujikawa T, Tsuyumu S, Gutierrez-Correa M. Structural analysis of biofilms and pellets of Aspergillus niger by confocal laser scanning microscopy and cryo scanning electron microscopy. Bioresour Technol. 2010;101:1920–1926. doi: 10.1016/j.biortech.2009.10.036. [DOI] [PubMed] [Google Scholar]
- McLoon AL, Kolodkin-Gal I, Rubinstein SM, Kolter R, Losick R. Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J Bacteriol. 2011;193:679–685. doi: 10.1128/JB.01186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks AE, Glaven RH, Lovley DR. Real-time spatial gene expression analysis within current-producing biofilms. ChemSusChem. 2012;5:1092–1098. doi: 10.1002/cssc.201100714. [DOI] [PubMed] [Google Scholar]
- Grantcharova N, Peters V, Monteiro C, Zakikhany K, Romling U. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J Bacteriol. 2010;192:456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Betancur JC, Yepes A, Schneider J, Lopez D. Single-cell analysis of Bacillus subtilis biofilms using fluorescence microscopy and flow cytometry. J Vis Exp. 2012. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- Jakobsen TH, van GM, Christensen LD, Bjarnsholt T, Givskov M. Qualitative and quantitative determination of quorum sensing inhibition in vitro. Methods Mol Biol. 2011;692:253–263. doi: 10.1007/978-1-60761-971-0_18. [DOI] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Bartlett DH, Frantz BB, Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß BM, Markovic D, Matsumura P. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J Bacteriol. 1997;179:3818–3821. doi: 10.1128/jb.179.11.3818-3821.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß BM, Liu X, Hendrickson W, Matsumura P. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol Lett. 2001;197:91–97. doi: 10.1111/j.1574-6968.2001.tb10588.x. [DOI] [PubMed] [Google Scholar]
- Prüß BM, Campbell JW, Van Dyk TK, Zhu C, Kogan Y, Matsumura P. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J Bacteriol. 2003;185:534–543. doi: 10.1128/JB.185.2.534-543.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Kato M, Jo YL, Mizushima S. Interaction of OmpR, a positive regulator, with the osmoregulated ompC and ompF genes of Escherichia coli. Studies with wild-type and mutant OmpR proteins. J Biol Chem. 1988;263:1008–1012. [PubMed] [Google Scholar]
- Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß BM, Besemann C, Denton A, Wolfe AJ. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J Bacteriol. 2006;188:3731–3739. doi: 10.1128/JB.01780-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan WR, Shibata S, Aizawa SI, Wolfe AJ. The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli. J Bacteriol. 2007;189:7159–7163. doi: 10.1128/JB.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentschler AE, Lovrich SD, Fitton R, Enos-Berlage J, Schwan WR. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology. 2013;159:316–327. doi: 10.1099/mic.0.059386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J Bacteriol. 2003;185:5735–5746. doi: 10.1128/JB.185.19.5735-5746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- Prüß BM. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch Microbiol. 1998;170:141–146. doi: 10.1007/s002030050626. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Boylan B, George N, Forst S. Inactivation of ompR promotes precocious swarming and flhDC expression in Xenorhabdus nematophila. J Bacteriol. 2003;185:5290–5294. doi: 10.1128/JB.185.17.5290-5294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Li H, Spangler C, Kasper CA, Kaever V, Sourjik V, Roth V, Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel U, Romling U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/JB.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V, Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990;172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Zhou Y, Wild J, Adler J, Gross CA. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992;174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß BM, Verma K, Samanta P, Sule P, Kumar S, Wu J, Horne SM, Christianson DA, Stafslien SJ, Wolfe AJ. et al. Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch Microbiol. 2010;192:715–728. doi: 10.1007/s00203-010-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol. 2002;45:521–532. doi: 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- Lee C, Park C. Mutations upregulating the flhDC operon of Escherichia coli K-12. J Microbiol. 2013;51:140–144. doi: 10.1007/s12275-013-2212-z. [DOI] [PubMed] [Google Scholar]
- Wang X, Wood TK. IS5 inserts upstream of the master motility operon flhDC in a quasi-Lamarckian way. ISME J. 2011;5:1517–1525. doi: 10.1038/ismej.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CS, Prüß BM, Matsumura P. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol. 2004;186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüß BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Li B, Li N, Wang F, Guo L, Huang Y, Liu X, Wei T, Zhu D, Liu C, Pan H. et al. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res. 2012;40:11073–11085. doi: 10.1093/nar/gks869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A, Laugel B, Van GA, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol. 2003;49:823–832. doi: 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Hadjifrangiskou M, Hultgren SJ. What does it take to stick around? Molecular insights into biofilm formation by uropathogenic Escherichia coli. Virulence. 2012;3:231–233. doi: 10.4161/viru.19763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck RR, Tarrien RJ, Mobley HL. Enzymatically active and inactive phosphodiesterases and diguanylate cyclases are involved in regulation of motility or sessility in Escherichia coli CFT073. MBio. 2012;3 doi: 10.1128/mBio.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiton PS, Cusumano CK, Kline KA, Dodson KW, Han Z, Janetka JW, Henderson JP, Caparon MG, Hultgren SJ. Combinatorial small-molecule therapy prevents uropathogenic Escherichia coli catheter-associated urinary tract infections in mice. Antimicrob Agents Chemother. 2012;56:4738–4745. doi: 10.1128/AAC.00447-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Palaniyandi S, Herren CD, Zhu X, Mukhopadhyay S. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS One. 2013;8:e55492. doi: 10.1371/journal.pone.0055492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol. 2009;73:1020–1031. doi: 10.1111/j.1365-2958.2009.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Beatty CM, Browning DF, Busby SJ, Simel EJ, Hovel-Miner G, Wolfe AJ. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182:4173–4179. doi: 10.1128/JB.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura P. Remembering Malcolm J. Casadaban. J Bacteriol. 2010;192:4261–4263. doi: 10.1128/JB.00484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Fredericks CE, Shibata S, Aizawa SI, Reimann SA, Wolfe AJ. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol Microbiol. 2006;61:734–747. doi: 10.1111/j.1365-2958.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- Sule P, Wadhawan T, Carr NJ, Horne SM, Wolfe AJ, Prüß BM. A combination of assays reveals biomass differences in biofilms formed by Escherichia coli mutants. Lett Appl Microbiol. 2009;49:299–304. doi: 10.1111/j.1472-765X.2009.02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Cleveland W. Robust locally weighted regression and smoothing scatter plots. J Americ Statist Assoc. 1979;74:829–836. doi: 10.1080/01621459.1979.10481038. [DOI] [Google Scholar]
- O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Stafslien S, Daniels J, Chisholm B, Christianson D. Combinatorial materials research applied to the development of new surface coatings III. Utilisation of a high-throughput multiwell plate screening method to rapidly assess bacterial biofilm retention on antifouling surfaces. Biofouling. 2007;23:37–44. doi: 10.1080/08927010601127311. [DOI] [PubMed] [Google Scholar]
- Stafslien SJ, Bahr JA, Feser JM, Weisz JC, Chisholm BJ, Ready TE, Boudjouk P. Combinatorial materials research applied to the development of new surface coatings I: a multiwell plate screening method for the high-throughput assessment of bacterial biofilm retention on surfaces. J Comb Chem. 2006;8:156–162. doi: 10.1021/cc050047m. [DOI] [PubMed] [Google Scholar]