Abstract
Neurofibromatosis type 1 (NF1), a neuroectodermal disorder, is caused by germline mutations in the NF1 gene. NF1 affects approximately 1/3,000 individuals worldwide, with about 50% of cases representing de novo mutations. Although the NF1 gene was identified in 1990, the underlying gene mutations still remain undetected in a small but obdurate minority of NF1 patients. We postulated that in these patients, hitherto undetected pathogenic mutations might occur in regulatory elements far upstream of the NF1 gene. In an attempt to identify such remotely acting regulatory elements, we reasoned that some of them might reside within DNA sequences that (1) have the potential to interact at distance with the NF1 gene and (2) lie within a histone H3K27ac-enriched region, a characteristic of active enhancers. Combining Hi-C data, obtained by means of the chromosome conformation capture technique, with data on the location and level of histone H3K27ac enrichment upstream of the NF1 gene, we predicted in silico the presence of two remotely acting regulatory regions, located, respectively, approximately 600 kb and approximately 42 kb upstream of the NF1 gene. These regions were then sequenced in 47 NF1 patients in whom no mutations had been found in either the NF1 or SPRED1 gene regions. Five patients were found to harbour DNA sequence variants in the distal H3K27ac-enriched region. Although these variants are of uncertain pathological significance and still remain to be functionally characterized, this approach promises to be of general utility for the detection of mutations underlying other inherited disorders that may be caused by mutations in remotely acting regulatory elements.
Keywords: NF1, Neurofibromatosis type 1, Gene mutation, Hi-C data, Histone H3K27ac, Enhancer
Background
Germline mutations of the NF1 gene cause the tumour predisposition syndrome, neurofibromatosis type 1 (NF1), which affects 1/3,000–4,000 individuals worldwide. The NF1 gene, spanning 283 kb on chromosome 17q11.2, contains 61 exons that give rise to a 12-kb mRNA transcript encoding neurofibromin [1,2]. The 327-kDa (2,818-amino acid) protein product, neurofibromin, is expressed in most tissues.
Neurofibromin is a tumour suppressor protein, reflecting its role as a key negative regulator of the cellular Ras signalling pathway (reviewed by Bennett et al. [3]). More specifically, it is a Ras-specific GTPase-activating protein (GAP), with strong structural and sequence homologies to the GAP superprotein family [4]. It functions by downregulating Ras, thereby resulting in an overall reduction in cellular mitogenic signalling via the Ras pathway. Thus, any NF1 gene mutation that serves to inactivate neurofibromin function may be expected to increase cellular levels of active Ras-GTP significantly, leading to uncontrolled cell growth and, potentially, tumorigenesis [5]. Germline mutations in the SPRED1 gene have previously been detected in individuals with clinical features overlapping those of NF1 patients [6].
Consistent with Knudson's two-hit hypothesis, NF1 patients harbouring a heterozygous germline NF1 mutation develop neurofibromas upon somatic mutation of the second wild-type NF1 allele. The somatic loss of the second NF1 allele in the progenitor cell (either the Schwann cell or its precursor), combined with haploinsufficiency in a variety of supporting cells [2,7], is then required for tumour formation. The mutation rate at the NF1 locus is one of the highest reported in any human disorder [8]; almost 50% of all NF1 patients exhibit a de novo NF1 mutation. The NF1 mutational spectrum is shaped in large part, and often in remarkably predictable ways, by the local DNA sequence environment [9]. Nearly 1,300 different inherited mutations of the NF1 gene have been reported as a cause of NF1; these vary in size from deletions spanning more than a megabase to subtle single base-pair substitutions that alter an encoded amino acid or the function of a splice junction [9]. In classical NF1 patients, NF1 gene mutations are detectable in 50% to 95% of individuals depending upon the mutation detection techniques employed and the source of tissue used for analysis [2,10]. In our own study, the underlying pathogenic NF1 mutation was detected in approximately 92% of NF1 patients despite sequencing the exons, splice junctions, untranslated regions and proximal promoter region of the NF1 gene, and screening for gross gene deletions and rearrangements as well as subtle lesions [11-13]. The undetected mutations in such patients could in principle reside within a remote regulatory element at some distance from the NF1 gene.
In the human genome, not all regulatory elements occur immediately 5′ to the genes that they regulate; indeed, some are located at considerable distances from their cognate genes. A variety of micro-lesions causing human inherited disease have been found to occur >10 kb 5′ upstream of the corresponding disease genes [14]. These include a total of eight mutations within a 1-kb region (termed the long-range or limb-specific enhancer, ZRS) approximately 979 kb 5′ to the transcription initiation site of the sonic hedgehog (SHH) gene [15]. Far upstream polymorphic variants that influence gene expression and impact on disease have also been documented. Thus, for example, the C>T functional single nucleotide polymorphism (SNP) 14.5 kb upstream of the interferon regulatory factor 6 (IRF6) gene, which is associated with cleft palate, alters the binding of transcription factor AP-2α [16]. Similarly, a T>C functional SNP approximately 6 kb upstream of the α-globin-like HBM gene serves to create a binding site for the erythroid-specific transcription factor GATA1 and interferes with the activation of the downstream α-globin genes [17]. Finally, a T>G functional SNP approximately 335 kb upstream of the MYC gene increases the risk of colorectal and prostate cancer by altering the binding strength of transcription factors TCF4 and/or TCF7L2 within a transcriptional enhancer [18]. On the basis of these findings in a number of different genes, we postulated that remotely acting regulatory elements might be present far upstream of the NF1 gene and that these could be mutated in NF1 patients in whom no pathogenic mutations had been detected.
The NF1 gene contains a TATA-less promoter within a classic CpG island that extends from the proximal promoter into exon 1, and a 454-bp 5′ untranslated region [19]. The CpG island is normally unmethylated, and the gene is actively transcribed in all tissues so far examined. Despite the high degree of conservation of the proximal regulatory sequences in rodent orthologs [20,21], the distal upstream sequence, potentially harbouring additional regulatory elements, diverges quite significantly between different mammalian species.
Histone H3K27ac serves to distinguish active enhancers from inactive ones [22]. Information on various functional elements, including regions enriched in histone H3K27ac, encoded by the human genome, have recently become available through the ENCODE project resources (ENCODE Project Consortium 2011 [23]; http://genome.ucsc.edu/index.html), a comprehensive collection of experimental data produced using biochemical assays mapped to human genome data. Using a variety of high-throughput techniques for identifying features characteristic to enhancers (e.g. specific histone modifications), up to 1.4 million putative enhancers have been identified in the human genome [23,24].
It has long been hypothesized that communication between widely spaced genomic elements is facilitated by the spatial organization of chromosomes that brings genes and their cognate regulatory elements into close spatial proximity [25]. The development of ‘chromosome conformation capture’ techniques has led to the discovery of many long-range interactions, both intra- and inter-chromosomal (reviewed in [26]). Recently, Lieberman-Aiden et al. [27] employed a novel approach, based on chromosomal conformation capture techniques, and termed Hi-C, to probe the 3D architecture of the entire human genome by coupling proximity-based ligation with massively parallel sequencing to produce a catalogue of approximately 8.4 million inter- and intra-chromosomal interacting fragments. Approximately 6.7 million of these fragments were found to be of long range (>20 kb apart). A genome-wide matrix of DNA-DNA interactions (also known as a contact map), created by dividing the genome into 1-Mb regions and counting the number of interactions between these 1-Mb regions [27], was subsequently analyzed by Yaffe and Tanay [28] to eliminate various biases in experimental procedure.
In this study, we postulated that remotely acting regulatory elements might occur within DNA fragments that interact with the DNA fragment containing the NF1 gene, and further that these elements might be located within an H3K27ac-enriched region. We first combined long-range DNA-DNA interaction data and ENCODE resources to predict in silico the location of novel remotely acting regulatory regions upstream of the NF1 gene. Finally, we screened these regions for mutation in those NF1 patients in whom no mutations had been found in either the NF1 or SPRED1 genes. Germline mutations in the SPRED1 gene have previously been detected in individuals with clinical features overlapping those of NF1 patients [6].
Results and discussion
The Hi-C data indicated that a 1-Mb fragment of chromosome 17, starting at position 29,000,000 and ending at position 29,999,999, termed the NF1-containing fragment since it contained the entire NF1 gene (positions 29,421,945–29,709,134), was found to interact most strongly with the adjacent 1-Mb fragment on chromosome 17 (703 and 255 intra-chromosomal interactions with the NF1-containing fragment recorded in HindIII- and NcoI-maps, respectively) which occupies positions 28,000,000–28,999,999 upstream of the NF1-containing fragment (Figure 1). The ENCODE data indicated the presence of an H3K27ac-enriched region somewhere between positions 28,846,790 and 28,847,790 in the interacting fragment (region A in Figure 1). This region is distal (approximately 600 kb upstream) to the NF1 gene. Another peak in H3K27ac enrichment was observed within the NF1-containing fragment (region B between positions 29,378,421 and 29,379,750) which is proximal (approximately 42 kb) to the NF1 gene. The level of enrichment was higher in the distal region A than in the proximal region B. ENCODE data also revealed the presence in region A of three overlapping binding sites for the transcription factors, p300 (positions 28,846,840–28,847,158), c-Fos (28,846,815–28,847,158) and c-Jun (28,846,819–28,847,094), all of which were identified by in vitro ChIP-seq assay (http://genome.ucsc.edu/index.html).
Figure 1.
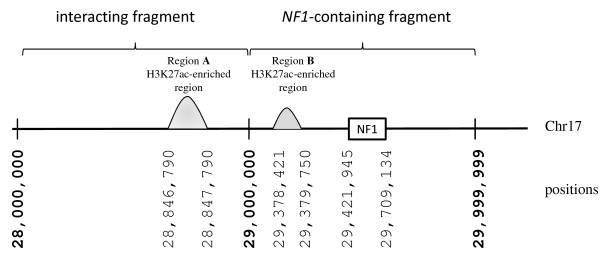
Schematic of the occurrence and level of H3K27ac enrichment within interacting and NF1-containing DNA fragments. The level of enrichment is shown by the height of the curves (not to scale).
Two sets of primers (Table 1) were designed using Primer3 software [29]. These were used for the polymerase chain reaction (PCR) amplification and sequencing of the two regions of interest: region A (positions 28,846,790–28,847,790) and region B (positions 29,378,421–29,379,750), from the genomic DNA of 47 NF1 patients as described in the ‘Methods’ section.
Table 1.
List of primers used to amplify and sequence DNA regions A (28,846,790–28,847,790) and B (29,378,421–29,379,750)
| Primer ID | Sequence (5′>3′) |
|---|---|
| chr17_28846841_1F |
GCAAACATAGGAAGTGGCTTAC |
| chr17_28846841_2F |
GAGGGGAGAAGTGTCTGTGTG |
| chr17_28846841_3F |
TTTTCCCATTTAGAGATAAGTCAGTG |
| chr17_28846841_1R |
TTCTGAGGTGCAGTCTTTTAGG |
| chr17_28846841_2R |
GGTTACAACAAACAAGTGGAGAATG |
| chr17_28846841_3R |
TACTTAGGGTGACAAGGCAAG |
| chr17_29387421_1F |
TCTCCCTATGATGTCCAGGC |
| chr17_29387421_2F |
TGCAGCAATAGGTGACTAAAACAC |
| chr17_29387421_3F |
AGGTCCAAATGCAGGTGTG |
| chr17_29387421_4F |
TCTGGAAGCTCTGCTTGG |
| chr17_29387421_1R |
AATTGTTGGCATGAAAACTGG |
| chr17_29387421_2R |
AGATGTGTACCCTGCGGAAG |
| chr17_29387421_3R |
ACGAAGAGATGTGGTTTCCC |
| chr17_29387421_4R | AACATCCCCTTCCAATTTCC |
Three sequence variants were identified in the distal region, the region that corresponds to the highest level of histone H3K27ac enrichment. DNA sequencing revealed the presence of a heterozygous novel C>T variant at position 28,846,793 in two NF1 patients. This variant is not present in the dbSNP database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp). Two other variants, rs78190160 (position 28,847,605) and rs71372224 (position 28,846,883), were detected each in three different patients. Both of these variants are listed in dbSNP as being of unknown clinical significance; minor allele frequency/minor allele count based on 1000 Genomes Project data was 0.007/16, indicating that they are rare variants. Unfortunately, no linkage disequilibrium data (HapMap Project; http://hapmap.ncbi.nlm.nih.gov/) were available for this region. The rs71372224 SNP was however found to occur within the three overlapping binding sites for transcription factors p300, c-Fos and c-Jun. Recent studies [30] have shown that enhancer elements have a high propensity to be bound by p300, an enhancer-binding transcription factor. One may therefore speculate that a mutation in this putative enhancer element could influence the expression of the NF1 gene in a spatially specific manner. No variants were found in the H3K27ac-enriched region (region B) lying proximal to the NF1 gene region and corresponding to the lower level of H3K27ac enrichment.
Conclusions
This pilot study has shown that a combination of long-range DNA sequence interaction data and information about the location of functional elements available from ENCODE allows us to predict the possible location of novel regulatory regions that may act at distance from the genes whose expression they might influence. Mutations in these regions may in turn modulate the expression of their cognate target gene(s) that are spatially close to these regions. Although the functional ascertainment of these variants far upstream of the NF1 gene has not yet been performed, the type of in silico analysis described here served to guide the mutation screening procedure, allowing us to detect sequence variants in five unrelated patients in whom no pathogenic mutations had previously been found in either the NF1 gene or the SPRED1 gene [6]. Even if eventually none of the detected sequence variants turn out to be of pathogenic significance, the Hi-C data indicate that several other 1-Mb fragments have a high number of interactions with the NF1-containing fragment and could potentially harbour remotely acting regulatory elements. Among them are two 1-Mb fragments occurring 1 Mb (positions 31,000,000–31,999,999) and 2 Mb (positions 32,000,000–32,999,999) downstream of the NF1-containing fragment that have, respectively, 699 and 150 intra-chromosomal interactions with the NF1-containing fragment in the NcoI-map and, respectively, 222 and 59 interactions in the HindIII-map. Another, albeit weaker, inter-chromosomal interaction (21 interactions in the NcoI-map) was recorded between the NF1-containing fragment and a 1-Mb fragment on chromosome 1 (positions 1–1,999,999). Hence, applicability of this ap-proach is not only confined to cis-acting regulatory elements but could also be employed with trans-acting regulatory elements. The use of additional information that recently became available through the ENCODE resources, i.e. on the occurrence of CTCF binding factors linked to a range of gene regulatory functions including enhancer-blocking activity of insulator elements; DNase I hypersensitive sites characterizing open chromatin conformations, etc., may help to improve the in silico prediction of remotely acting regulatory elements. This approach therefore promises to be of potential utility in investigating those other disorders in which mutations could occur in remotely acting regulatory elements.
Methods
Patient samples
A sample of 47 unrelated NF1 patients (out of a total of 624 NF1 patients studied in our diagnostic centre), who tested negative for NF1 (and SPRED1) point mutations and intra-genic insertions/deletions, was selected for this study. This project was approved by the local Ethics committee. Informed consent for sample collection was obtained according to the protocols approved by this committee.
Bioinformatics analysis
Long-range chromosomal DNA sequence interaction data were downloaded from http://compgenomics.weizmann.ac.il/tanay/?page_id=283. These data comprised the Hi-C contact maps [27], normalized to eliminate various biases associated with the experimental procedure [28]. Two matrices of interactions between 1-Mb fragments, each combining the results obtained using, respectively, the restriction enzymes HindIII and NcoI in the experimental procedures, were available. Henceforth, these contact maps will be termed, respectively, the HindIII- and NcoI-maps. Information on the genome-wide occurrence of one particular functional element, the histone H3K27ac-enriched region, was downloaded from the ENCODE database available at http://genome.ucsc.edu/index.html.
The NF1 gene, spanning positions 29,421,945–29,709,134 from pter of chromosome 17 (NCBI build 37, UCSC HG18 and Ensembl GRCh37), occurs almost in the middle of a 1-Mb DNA fragment that starts at position 29,000,000 and ends at position 29,999,999. This 1-Mb fragment will henceforth be termed the NF1-containing fragment. Two of the contact maps described above were searched for DNA fragments, whether occurring on the same chromosome or on different chromosomes that had the most interactions with the NF1-containing fragment.
Data on the occurrence and level of H3K27ac enrichment (as measured by the ChIP-seq assay) within the 1-Mb fragment that had the strongest interaction with the NF1-containing fragment were downloaded from the ENCODE database. H3K27ac data were also downloaded for the NF1-containing fragment. For each of these two 1-Mb fragments (i.e. the fragment that had the strongest interaction with the NF1-containing fragment and the NF1-containing fragment itself), relatively short regions, corresponding to the highest level of H3K27ac enrichment, termed regions A and B, respectively, were selected for DNA sequencing in 47 NF1 patients in whom no pathogenic mutations had been found by screening both the NF1 and SPRED1 genes.
PCR and sequencing
Two sets of primers for regions A and B, selected as described above, were designed using Primer3 software [29] and purchased from Eurogentec (Southampton, UK). Specificity was confirmed by BLAST analysis.
The genomic DNA isolated from the peripheral blood cells of the NF1 patients was subjected to PCR and Sanger sequencing as described [31]. PCR reaction mixtures of 25 μl contained 5 μl of genomic DNA (6 ng/μl), 5 pmol each of the forward and reverse primers, 0.25 U AmpliTaq Gold polymerase, 3 mM MgCl2, PCR buffer and 0.125 mM each dNTP. PCR cycling conditions were 95°C for 10 min followed by 40 cycles of 95°C × 30 s, 60°C × 30 s, 72°C × 30 s with a final extension step of 72°C for 10 min. PCR fragments were confirmed by gel electrophoresis in 1.5% agarose gels and visualized with ethidium bromide staining. The sequencing of the PCR products was performed with BigDye Terminator v3.1 Cycle Sequencing Kits and run on an ABI 3730 genetic analyzer. The obtained sequences were analyzed using Sequencher software version 4.7 (Gene Codes Corp., Ann Arbor, MI, USA). All reagents were purchased from Life Technologies Ltd, Paisley, UK.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NC conceived the approach. SEH performed the in silico analysis. MU and PR performed the PCR and sequencing. NC, PR and DNC interpreted the results. NC drafted the manuscript. All authors participated in the preparation of the manuscript, and read and approved the final manuscript.
Contributor Information
Stephen E Hamby, Email: sh529@leicester.ac.uk.
Pablo Reviriego, Email: Pablo.Reviriego@wales.nhs.uk.
David N Cooper, Email: CooperDN@cardiff.ac.uk.
Meena Upadhyaya, Email: Upadhyaya@cardiff.ac.uk.
Nadia Chuzhanova, Email: nadia.chuzhanova@ntu.ac.uk.
References
- Upadhyaya M, Chuzhanova N, Cooper DN. In: Neurofibromatosis Type 1: Molecular and Cellular Biology. Upadhyaya M, Cooper DN, editor. Heidelberg: Springer; 2012. The somatic mutational spectrum of the NF1 gene; pp. 211–233. [Google Scholar]
- Upadhyaya M. Neurofibromatosis type 1: diagnosis and recent advances. Expert Opin Med Diagn. 2010;4(4):307–322. doi: 10.1517/17530059.2010.494660. [DOI] [PubMed] [Google Scholar]
- Bennett E, Thomas N, Upadhyaya M. Neurofibromatosis type 1: its association with the Ras/MAPK pathway syndromes. J Paediatr Neurol. 2009;7:105–115. [Google Scholar]
- Scheffzek K, Ahmadian MR, Wiesmuller L, Kabsch W, Stege P, Schmitz F, Wittinghofer A. Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 1998;17(15):4313–4327. doi: 10.1093/emboj/17.15.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Welti S. In: Neurofibromatosis Type 1: Molecular and Cellular Biology. Upadhyaya M, Cooper DN, editor. Heidelberg: Springer; 2012. Neurofibromin: protein domains and functional characteristics; pp. 305–326. [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, Cools J, Marynen P, Thomas G, Yoshimura A, Legius E. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39(9):1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104(4):593–604. doi: 10.1016/S0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN. Mosaicism in sporadic neurofibromatosis type 1: variations on a theme common to other hereditary cancer syndromes? J Med Genet. 2008;45(10):622–631. doi: 10.1136/jmg.2008.059329. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Upadhyaya M. In: Neurofibromatosis Type 1: Molecular and Cellular Biology. Upadhyaya M, Cooper DN, editor. Heidelberg: Springer; 2012. The germline mutational spectrum in neurofibromatosis type 1 and genotype-phenotype correlations; pp. 115–134. [Google Scholar]
- Messiaen L, Wimmer K. In: Monographs in Human Genetics. Volume 16 . Kaufmann D, editor. Basel: Karger; 2008. NF1 mutational spectrum; pp. 63–77. [Google Scholar]
- Cowley GS, Murthy AE, Parry DM, Schneider G, Korf B, Upadhyaya M, Harper P, MacCollin M, Bernards A, Gusella JF. Genetic variation in the 3′ untranslated region of the neurofibromatosis 1 gene: application to unequal allelic expression. Somat Cell Mol Genet. 1998;24(2):107–119. doi: 10.1023/b:scam.0000007113.28381.53. [DOI] [PubMed] [Google Scholar]
- Osborn M, Cooper DN, Upadhyaya M. Molecular analysis of the 5′-flanking region of the neurofibromatosis type 1 (NF1) gene: identification of five sequence variants. Clin Genet. 2000;57(3):221–224. doi: 10.1034/j.1399-0004.2000.570308.x. [DOI] [PubMed] [Google Scholar]
- Horan MP, Osborn M, Cooper DN, Upadhyaya M. Functional analysis of polymorphic variation within the promoter and 5′ untranslated region of the neurofibromatosis type 1 (NF1) gene. Am J Med Genet A. 2004;131(3):227–231. doi: 10.1002/ajmg.a.30358. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Chen JM, Ball EV, Howells K, Mort M, Phillips AD, Chuzhanova N, Krawczak M, Kehrer-Sawatzki H, Stenson PD. Genes, mutations and human inherited diseases at the dawn of the age of personalized genomics. Hum Mutat. 2010;31(6):631–655. doi: 10.1002/humu.21260. [DOI] [PubMed] [Google Scholar]
- Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet. 2009;46(10):649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2α binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40(11):1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, Cheng JF, Rubin EM, Wood WG, Bowden D, Higgs DR. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312(5777):1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, Yao K, Kehoe SM, Lenz HJ, Haiman CA, Yan C, Henderson BE, Frenkel B, Barretina J, Bass A, Tabernero J, Baselga J, Regan MM, Manak JR, Shivdasani R, Coetzee GA, Freedman ML. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41(8):882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wallace MR. In: Neurofibromatosis Type 1: Molecular and Cellular Biology. Upadhyaya M, Cooper DN, editor. Heidelberg: Springer; 2012. NF1 gene: promoter, 5′ UTR, and 3′UTR; pp. 105–134. [Google Scholar]
- Bernards A, Snijders AJ, Hannigan GE, Murthy AE, Gusella JF. Mouse neurofibromatosis type 1 cDNA sequences reveals high degree of conservation of both coding and non-coding mRNA segments. Hum Mol Genet. 1993;2(6):645–650. doi: 10.1093/hmg/2.6.645. [DOI] [PubMed] [Google Scholar]
- Hajra A, Martin-Gallardo A, Tarlé SA, Freedman M, Wilson-Gunn S, Bernards A, Collins FS. DNA sequences in the promoter region of the NF1 gene are highly conserved between human and mouse. Genomics. 1994;21(3):649–652. doi: 10.1006/geno.1994.1328. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I. ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S. et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28(10):1089–1095. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20(7):849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 2011;43(11):1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Hwang YC, Zheng Q, Gregory BD, Wang LS. High-throughput identification of long-range regulatory elements and their target promoters in the human genome. Nucleic Acids Res. 2013;41(9):4835–4846. doi: 10.1093/nar/gkt188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L, Kluwe L, Chuzhanova N, Mautner V, Upadhyaya M. Analysis of NF1 somatic mutations in cutaneous neurofibromas from patients with high tumor burden. Neurogenetics. 2010;11(4):391–400. doi: 10.1007/s10048-010-0240-y. [DOI] [PubMed] [Google Scholar]


