Abstract
Mass spectrometry imaging (MSI) methods and protocols have become widely adapted to a variety of tissues and species. However, the MSI literature contains minimal information on whole-body cryosection preparation for the zebrafish (ZF; Danio rerio), a model organism routinely used in developmental, toxicity, and carcinogenicity studies. The optimal medium for embedding and cryosectioning a whole organism or soft-tissue specimen for histological examination is a synthetic polymer mixture that is incompatible with MSI as a result of ion suppression. We describe the optimal methods and results for embedding and cryosectioning whole-body ZF for MALDI-MSI. We evaluated 13 distinct embedding media formulations and found a supportive hydrogel with the consistency of cartilage to be the optimal embedding medium. The hydrogel medium does not interfere with MSI data collection, aids in tissue stability, is readily available for purchase, and is easy to prepare and handle during cryosectioning. Additionally, we decreased the matrix cluster interference commonly caused by α-cyano-4-hydroxycinnamic acid by adding ammonium phosphate to the solvent spray solution. The optimized methods developed in our laboratory produced high-quality cryosections, as well as high-quality mass spectral images of sectioned ZF.
Keywords: α-cyano-4-hydroxycinnamic acid, ammonium phosphate, cryosectioning, hydrogel, MALDI-MS, whole-body imaging, zebrafish proteins
INTRODUCTION
MALDI-mass spectrometry imaging (MSI) provides the opportunity to examine the spatial context of both small molecules and biological macromolecules in a whole organism or specific tissue. The clinical, pharmacological, and toxicological research communities have adopted MALDI-MSI as a research tool and have speculated that the technique has the potential to become as revolutionary as the microscope.1 Recently, MSI sample preparation and handling were emphasized as the most important and challenging first step in the process.2 Among the many critical preparation factors to consider are the embedding medium and matrix application, which greatly affect the quality of the data and detection of the molecules of interest. The cryosectioning of a number of tissues and sample types, from human tumor biopsies3 to sea cucumbers,4 has been described in the literature, and a variety of embedding and sectioning methods exist for whole-body rat and mouse model species, including ice5 and carboxymethyl cellulose (CMC).6 However, the MSI literature contains little information for processing small fish species that are increasingly used as replacements for more expensive, higher vertebrate models.
The zebrafish (ZF; Danio rerio) is an established model species with a fully sequenced and well-annotated genome that is used routinely in molecular, reproductive, developmental, and chemical toxicity studies.7 Accordingly, the literature demonstrates the usefulness of ZF for studies concerned with proteomic endpoints,8 including MSI, using an ambient ionization MS approach,9 although there is a paucity of experimental or descriptive peer-reviewed studies that make reference to ZF preparation for MALDI-MSI. In a single technology note, the use of MALDI-MSI, using ZF, was demonstrated, although embedding medium methods were not discussed.10 A personal communication to an author of the tech note revealed that two treatments were tried: no-embedding medium (frozen, whole-body only) or the common tissue-embedding polymer, optimal cutting temperature (OCT; Sakura Finetek, Torrance, CA, USA). The OCT caused ion suppression during MALDI-MSI and therefore, was an unsuitable ZF-embedding medium (U. Binkle, University of Konstanz, Germany, personal communication). Most fish species contain an air-filled swim bladder that once frozen, creates a very delicate structure that requires support for cryosectioning, can be crushed easily during cryosectioning and dislocate additional organ structures, and is therefore not amenable to the no-embedding or ice-embedding methods established for whole-body mice or rat frozen sections.5 The objective of this study was to determine the optimal embedding medium, sectioning considerations, and matrix application conditions specifically suited for whole-body ZF peptide and protein detection using MALDI-MSI.
MATERIALS AND METHODS
Chemicals
The Tissue-Tek OCT compound was purchased from Sakura Finetek; TBS medium was provided by Triangle Biomedical Sciences, (Durham, NC, USA); Richard-Allen Scientific H&E Y stains were purchased from Cole Parmer (Vernon Hills, IL, USA); chloroform was purchased from J. T. Baker (Phillipsburg, NJ, USA); ethanol (EtOH) was purchased from AAPER Alcohol and Chemical (Shelbyville, KY, USA); and acetonitrile (ACN) was purchased from Burdick and Jackson (Honeywell International, Morristown, NJ, USA). The following chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA): tricaine methanesulfonate (MS-222), CMC, gelatin, α-cyano-4-hydroxycinnamic acid (CHCA), sinapinic acid (SPA), sucrose, trifluoroacetic acid (TFA), agarose, ammonium phosphate monobasic (AmP), and acetic acid.
ZF Production and Tissue Preparation
The use of fish in the present study was approved by the Gulf Ecology Division's Animal Care and Use Committee (Gulf Breeze, FL, USA). Adult WT ZF were obtained from the Gulf Coast Research Laboratory (Ocean Springs, MS, USA). Male and female adult fish were maintained in 60-liter tanks receiving continuous flow of aerated, filtered freshwater at 26 ± 2°C under a 14-h light:10-h dark photoperiod. Diet consisted of Artemia spp. nauplii (Brine Shrimp Direct, Ogden, UT, USA), fed twice daily and Tetramin flake food (Tetra Holding, Blacksburg, VA, USA) to satiation once daily. On weekends, fish were fed Tetramin and Artemia to satiation once daily. Adults were maintained and spawned in the laboratory for over 1 year prior to specimen selection. Only adult ZF with clearly differentiated gonads, 3 months–2 years of age, were used for experimentation.
Fish were killed using MS-222 and then frozen immediately in a dry ice (DI)/EtOH bath (1:1 ratio of DI:95% EtOH) in a cryobowl (Cole-Parmer Instrument, Chicago, IL, USA), using a foil cup shaped inside of a Peel-A-Way mold (Polysciences, Warrington, PA, USA). A color change from fleshy to pale visually indicated that the specimen was frozen completely. For all trials, the frozen tissue posterior to the anal vent was removed, and only the anterior portion was used for embedding.
To investigate methods for enhanced tissue stabilization during sectioning, a subset of frozen ZF was injected with a 2% CMC medium just below the lateral line, in the area of the dual-chambered swim bladder. The frozen ZF was oriented on its side for injections and a 25-gauge needle was filled with prewarmed 2% CMC solution. The tip of the needle was heated with an alcohol lamp to ensure transfer of the warm 2% CMC into the frozen body cavity; the ZF was injected until the swim-bladder chamber had reached filling capacity, as indicated by minute backflow. ZF were injected twice, once into each swim bladder chamber.
Media Preparation
The following nine pure media were evaluated as suitable substrates for embedding whole ZF: OCT compound, TBS medium, ice [frozen Milli-Q water (MQH2O)], 0.6 M sucrose, 1% agarose, 5% and 10% gelatin, and 2% and 5% CMC. Four additional media mixtures consisting of gelatin with CMC, agarose, or sucrose were investigated as well (Table 1). Each pure medium and medium mixture was prepared by adding the solid(s) to a 50-ml polypropylene tube, followed by MQH2O at room temperature to thoroughly hydrate the media. The uncapped tube was gently heated in a standard microwave oven without boiling over (<1 min; time will vary per microwave unit), and the mixture was stirred to dissolve all solids. Each tube was placed in a warm water bath to maintain a fluid consistency.
TABLE 1.
Observational Comparison of Embedding Media Performance for MALDI-MSI
| Embedding media | Physical property | Stability | Ion suppression | Media acceptability |
|---|---|---|---|---|
| OCT | Pliable | Stable | Yes | Poor |
| TBS | Pliable | Stable | Yes | Poor |
| Gelatin (10%, 5%) | Hard | Partial | No | Acceptable |
| Agarose (1%) | Hard | Disintegrated | N.T. | Poor |
| CMC (5%, 2%) | Hard, semihard | Disintegrated | N.T. | Poor |
| Ice (pure MQH2O) | Hard | Disintegrated | N.T. | Poor |
| Sucrose (0.6 M) | Soft | No integrity | N.T. | Poor |
| 2% CMC + 5% gelatin | Semihard | Stable | No | Acceptable |
| 5% CMC + 10% gelatin | Pliable | Stable | No | Optimal |
| 1% Agarose + 5% gelatin | Semihard | Partial | No | Acceptable |
| 0.6 M Sucrose + 5% gelatin | Semihard | Partial | No | Acceptable |
N.T., Medium not tested for ion suppression effects as a result of poor stability and physical properties.
Embedding and Cryosectioning
Warm medium was poured into a Peel-A-Way mold (Polysciences) and allowed to cool slightly at room temperature. Each frozen fish specimen was placed horizontally into the mold containing the medium using forceps, with the lateral side of the ZF parallel to the bottom of the mold. The tissue placement described herein produced sagittal whole-body sections. The entire mold, containing the embedded specimen, was submerged partially on top of the DI/EtOH bath to quickly freeze the medium without allowing the DI or EtOH to enter the mold. The embedded specimen block inside of the mold was equilibrated on the cryostat freeze plate at −40°C for at least 30 min. The specimen block was then removed from the mold and shaped to fit on a cryostat specimen holder. The back of the block was fastened to the specimen holder with a small amount of OCT while ensuring that the side of the specimen to be sectioned did not contact the OCT. Sectioning was performed using a Minotome Plus Cryostat (Triangle Biomedical Sciences) set to 16 μm thickness and a −20°C chamber temperature. Sections were collected such that the cut began on the ventral surface of the fish.
Immediately upon cryosectioning, tissue slices or media slices without tissue were placed on indium tin oxide (ITO) slides (HTX Technologies, Carrboro, NC, USA) by gently placing the slide (maintained at room temperature) over the frozen section. The slides were desiccated for 5 min and rinsed using chloroform11 or the multistep method similar to the method described by Yang and Caprioli.12 Briefly, slides were dipped in 70% EtOH for 30 s, 100% EtOH for 30 s, Carnoy's solution for 2 min, 100% EtOH for 30 s, water for 30 s, 100% EtOH for 30 s, and desiccated for 3-min to ensure solvent evaporation.12 Serial sections were also collected between each section taken for MSI analysis, placed on glass slides, and stained with H&E using the staining method described by Deutskens et al.13 A low picomolar mix of peptide and protein mass standards (AB SCIEX, Foster City, CA, USA) was mixed with matrix and placed on each media section (described below) or placed in a single 0.5-μl droplet on the posterior end of each MSI tissue section prior to spraying with matrix: des-arg-bradykinin, 0.5 pmol, 904.4681 Da; angiotensin I, 1.0 pmol, 1296.6853 Da; glu-fibrinopeptide β, 0.65 pmol, 1570.6774 Da; ACTH (1–17), 1.0 pmol, 2093.0867 Da; ACTH (18–39), 0.75 pmol, 2465.1989 Da; ACTH (7–38), 1.5 pmol, 3657.9294 Da; insulin, 0.25 pmol, 5734.59+1, 2867.80+2 Da avg; thioredoxin, Escherichia coli, 1.38 pmol, 11,674.48+1, 5837.74+2 Da avg; apomyoglobin, horse, 2.0 pmol, 16,952.56+1, 8476.78+2 Da avg. Photographic images of all sections were taken with a high-resolution digital camera (Sony, New York, NY, USA; Olympus, Center Valley, PA, USA) through a dissecting microscope (Olympus SZ-ST).
Matrix Solution Preparation and Deposition
Three matrix formulations were investigated to assess MALDI-MSI image enhancement: SPA, CHCA, and CHCA + 6 mM AmP. The matrix solutions were prepared at 5 mg/ml by adding SPA or CHCA to 50% ACN/0.1% TFA (v/v; with or without 6 mM AmP additive for CHCA) and mixing vigorously. Media-only sections were hand-spotted with mass standards mixed with SPA and CHCA only, while the tissue sections were sprayed with the three matrix solutions that were applied separately to each slide using an automated TM-Sprayer, (HTX Technologies), at 140°C with a 250 μl/min flow rate. The movable stage velocity was set to obtain a drier spray at 500 mm/min, with five passes made and turning 90° each time, with a 1-mm offset to ensure even spraying. Once sprayed, the ITO slides were secured immediately by both double-sided and conductive copper tape in a MS-specific slide holder adapter plate (LaserBio Labs, Sophia-Antipolis Cedex, France). To aid in locating the exact tissue-imaging area underneath the matrix coating within the mass spectrometer, four corners were etched carefully around the tissue with the rim of a small glass test tube; the ITO slides were then analyzed by MALDI-MSI.
MALDI-MSI Data Collection and Analysis
A 4800 MALDI-TOF/TOF MS (AB SCIEX) with a 355-nm wavelength neodymium-doped yttrium aluminum garnet laser (3–7 ns pulse at 200 Hz with laser-pulse energy >17 μJ), at 100 laser shots/spectrum/pixel, was used to collect standard MALDI-MS data and MALDI-MSI data; MS/MS data were not collected in this study. Data were collected in a positive ion-reflector mode in the range of mass-to-charge ratio (m/z) 800–4000 for media and standards only, and m/z 600–1925 for MSI; in linear mode, data were collected in the range of 4–20 kDa. The whole-body tissue-section imaging data were collected at a spatial resolution of 100 × 100 μm using a manual acquisition method in the 4000 Series Explorer Software (AB SCIEX), directed by the 4800 Imaging Tool v. 3.2 (Novartis, Basel, Switzerland), available through the MALDI-MSI website (www.maldi-msi.org). All MS modes were calibrated immediately prior to sample data collection using mass range-appropriate peptide and protein standards (AB SCIEX). Spectra of media types were processed using baseline correction only for linear-mode data, whereas reflector-mode data required baseline correction with Gaussian smoothing (filter width of five points) and signal-to-noise ratio >20 using Data Explorer Software v.3.7 (AB SCIEX). Image analysis was performed with TissueView Software v. 1.0 (AB SCIEX), wherein linear data were baseline-corrected. Individual .img files were opened and binned conservatively as needed to reduce file size upon import. Image files were manually inspected for masses present in each region of interest (ROI), representing mass standards or tissue type. H&E-stained serial sections were compared directly with the MS images to correlate tissue and organ type to mass spectra present within a ROI (see Supplemental Fig. 3).
RESULTS AND DISCUSSION
Embedding, Cryosectioning, and Rinsing Assessment
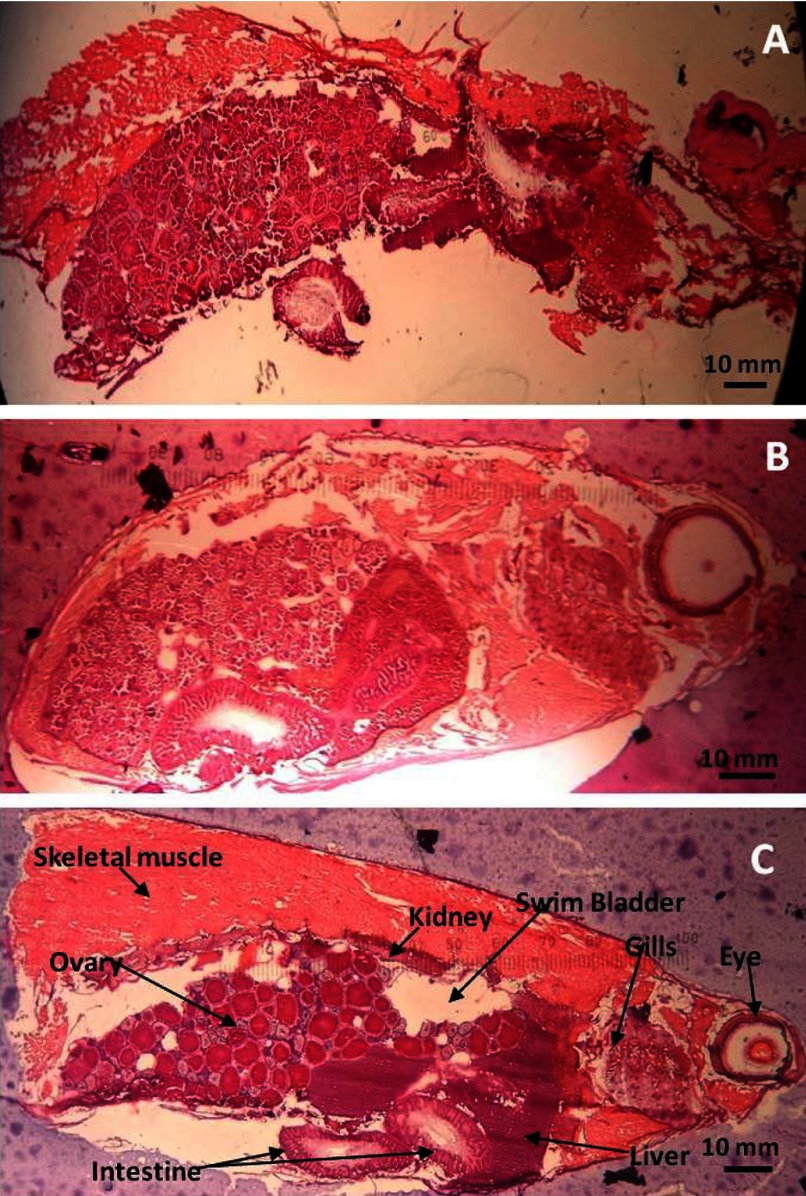
In general, a firm support surrounding any tissue will improve the quality of cryosections. When cryosections are taken from a nonembedded, frozen ZF, the air-filled swim bladder collapses, and skeletal muscles shred (Fig. 1A). Similarly, Strohalm et al.14 described studies concerning the MALDI-MSI preparation of the bumble bee and emphasized filling the body cavity with embedding medium to prevent crushing the insect's delicate structure during cryosectioning. Preserving the structural integrity of cryosections allows researchers to perform histology-defined molecular profiling and data analysis by preserving molecular localization within discrete organs and tissues.15 In our study, 2% CMC injections were made into the swim-bladder chambers in an attempt to support the structural integrity of the whole-body ZF cryosections. This process did not noticeably improve the quality of the cryosections obtained (Fig. 1B and C). Overall, the injection results were not reproducible in the small bodies of ZF (generally, <3 cm total body length), as the injections had the potential for organ and tissue disturbance and required longer specimen handling times, which could increase protein or peptide degradation.16 Therefore, the 2% CMC injection process was not performed on the subsequent evaluations of embedding media using whole-body ZF. The presence of eggs inside of the female ZF body produces a dense abdomen, thus reducing the area occupied by the air-filled swim bladder on most sagittal sections (Fig. 1C). Therefore, we observed that the best cryosections were produced by gravid adult, female ZF.
Figure 1.
Representative H&E-stained sections of a whole-body, female ZF: (A) without embedding medium; (B) with 5% CMC + 10% gelatin hydrogel-embedding medium; (C) with 5% CMC + 10% gelatin hydrogel-embedding medium, injected with 2% CMC. ZF anatomy labeled.
To find an optimal ZF embedding medium for use with MALDI-MSI, we compared the cryosectioning characteristics of a variety of readily available media with OCT, the positive control. OCT is the embedding medium used in routine cryosectioning for light microscopy as a result of its ability to freeze quickly, its ease of use, and its ability to maintain the structural integrity of the tissue sample.17 Table 1 describes our observations of the physical qualities of 13 distinct frozen embedding media for cryosectioning at a −20°C chamber temperature. The sectioning efficiency of the medium alone is listed as “physical property” and qualitatively ranked as soft, pliable, semihard, and hard, where a pliable medium is optimal, as it maintains shape well and sections easily without shredding. The structural integrity of the medium section is listed as “stability” and qualitatively ranked in four categories: disintegrated, no integrity, partial integrity, and stable. The sections ranged from fragmented pieces, lacking integrity during handling, or were partially able to be handled and transfered with minimal damage. The stable sections have optimal handling and transfer characteristics. “Ion suppression” was not determined for each of the 13 media; rather, the eight media that produced acceptable physical sections were tested further for ion-suppression effects to determine if the medium caused direct ion suppression or peak interference with the mass standards placed on the medium (see Media Ion Suppression and Matrix Effects below). “Media acceptability” describes medium selection for sectioning of ZF for MALDI-MSI based on the qualitative rankings of physical properties, stability, and ion suppression with a rating of poor, acceptable, or optimal medium.
Whereas OCT and the alternative polymer medium TBS had optimal sectioning qualities, the majority of the embedding media provided suboptimal frozen sections at a −20°C chamber temperature. Semihard and very hard media sections curled or crumbled during the slicing process, while soft media sections did not maintain shape. Gelatin mixtures and gelatin alone were able to maintain their physical properties and structural integrity during sectioning, whereas agarose, CMC, and sucrose alone disintegrated easily and lacked structural integrity. Aside from the ability to maintain structural integrity of the ZF tissues, we chose media based on cost, ease of preparation, and commercial availability to appeal to potential MSI researchers interested in multiple target tissues within whole-body ZF sections. A recent study described a MALDI-MSI, polymer-based embedding medium, poly[N-(2-hydroxypropyl)methacrylamide] (pHPMA), that is capable of supporting delicate bumble-bee tissues but does not display the ion-suppression qualities of OCT.14 Although ideal, the pHPMA medium synthesized by the authors was not commercially available for testing with ZF during our studies.
The embedding medium that produced optimal results in Table 1 was a CMC + gelatin mixture. When mixed properly, the CMC+gelatin mixture forms a hydrogel that is similar in molecular structure and function to cartilage18 and is therefore capable of providing the structural support necessary for optimal frozen sections. It is very important that the dry CMC and gelatin media are combined prior to the addition of water and heat, as the complexation and hydrogel formation of the CMC and gelatin are temperature- and concentration-dependent.18,19 Although all gelatin mixtures maintained structural integrity upon handling and were capable of producing acceptable media sections, the 5% CMC + 10% gelatin mixture was highly pliable and stable.
The 5% CMC + 10% gelatin mixture, having optimal physical properties and stability, reproducibly provided quality sections at 16 μm thickness. Although Eberlin et al.9 have had success with CMC alone, as a ZF-embedding medium, their cryosections were thicker (20 μm), and their desorption and ionization technique (desorption electrospray ionization) did not require matrix or placement inside of a vacuum. At the beginning of the study, we observed a range of cryosections between 10 μm and 20 μm and found that media-only and ZF cryosections thicker than 16 μm did not adhere to an ITO slide or a MALDI plate (data not shown). Additionally, media-only and ZF sections <16 μm were not consistent in terms of physical properties or stability and therefore, not used.
Rinsing the mounted cryosection with solvents ensures that lipids and salts are removed to decrease ion suppression of peptides and proteins.12 Lemaire et al.11 found that chloroform improves peptide and protein detection and sample reproducibility compared with other solvent rinses for MSI. We tested two rinsing methods at the beginning of our study and observed that the single-step chloroform method was less physically destructive to the delicate ZF cryosections than the multistep method12 (Supplemental Fig. 1). Therefore, each section was rinsed using the chloroform procedure prior to coating with the matrix.
Media Ion Suppression and Matrix Effects
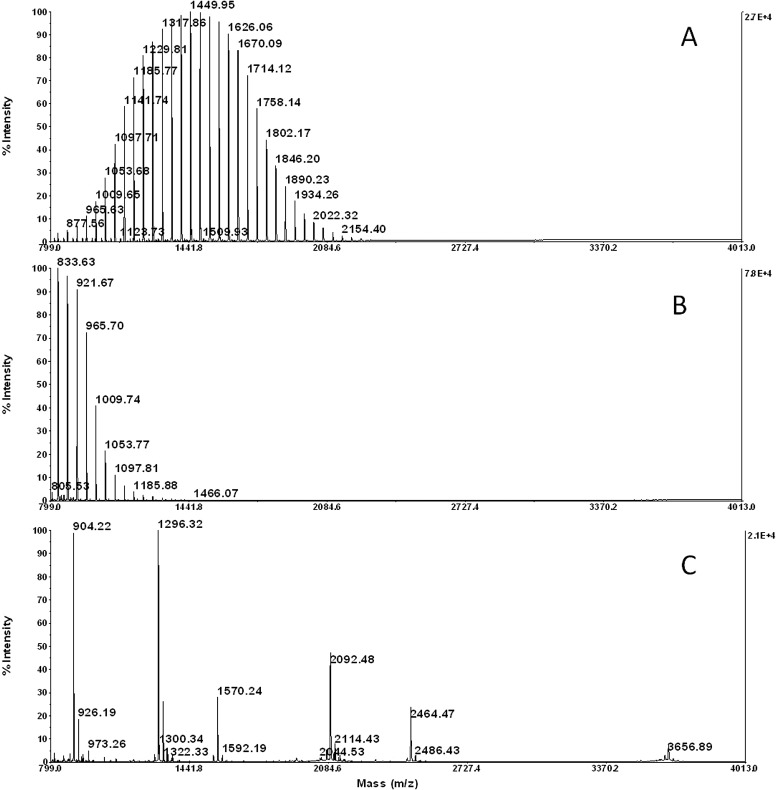
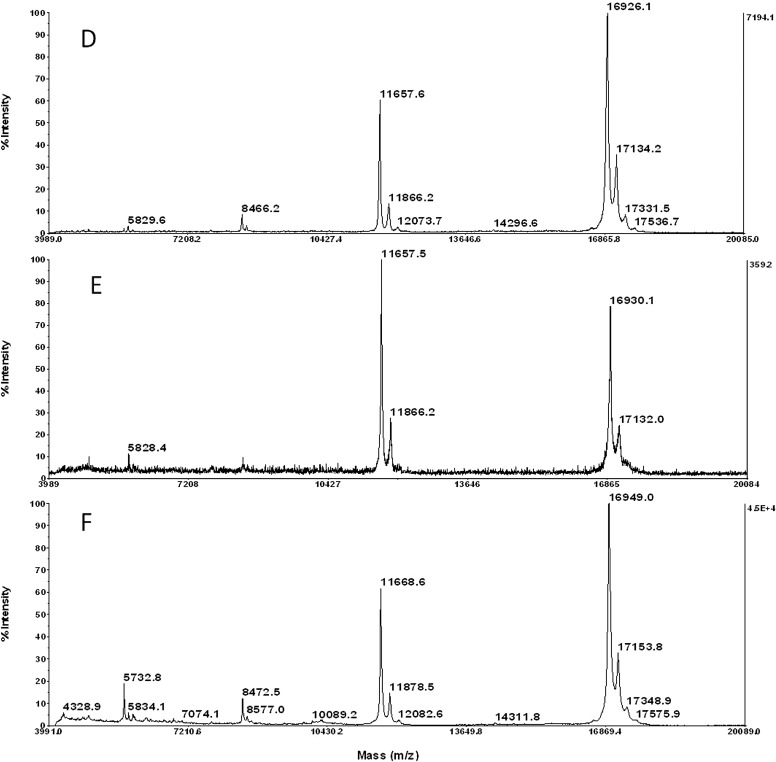
Based on the qualitative results displayed in Table 1, the media that demonstrated suitable physical properties and stability were tested further for ion suppression to determine the overall optimal medium. To test for ion-suppression effects, low pmol/μl peptide and protein mass standards were mixed with SPA or CHCA matrix and spotted on each of eight media types: OCT, TBS, 5% gelatin, 10% gelatin, 2% CMC + 5% gelatin, 5% CMC + 10% gelatin, 0.6 M sucrose + 5% gelatin, and 1% agarose + 5% gelatin. We confirmed that polymer-based OCT and TBS media cause ion suppression via contaminant peaks in the typical peptide mass range of 1–4 KDa (Fig. 2A and B), as well as slight protein ion suppression <10 KDa (Fig. 2D and E), compared with the optimal medium 5% CMC + 10% gelatin (Fig. 2C and F). Similarly, the other gelatin mixtures did not demonstrate ion-suppression effects on peptides or protein mass standards (Supplemental Fig. 2).
Figure 2.
Representative spectra of media sections spotted with peptide/protein standards mixed with saturated matrix in 50% ACN, 0.1% TFA. (A) OCT media with CHCA; (B) TBS media with CHCA; (C) 5% CMC + 10% gelatin media with CHCA; (D) OCT media with SPA; (E) TBS media with SPA; (F) 5% CMC + 10% gelatin media with SPA. See Supplemental Fig. 2 for spectra representing the remaining five media types that also did not suppress peptide or protein ions.
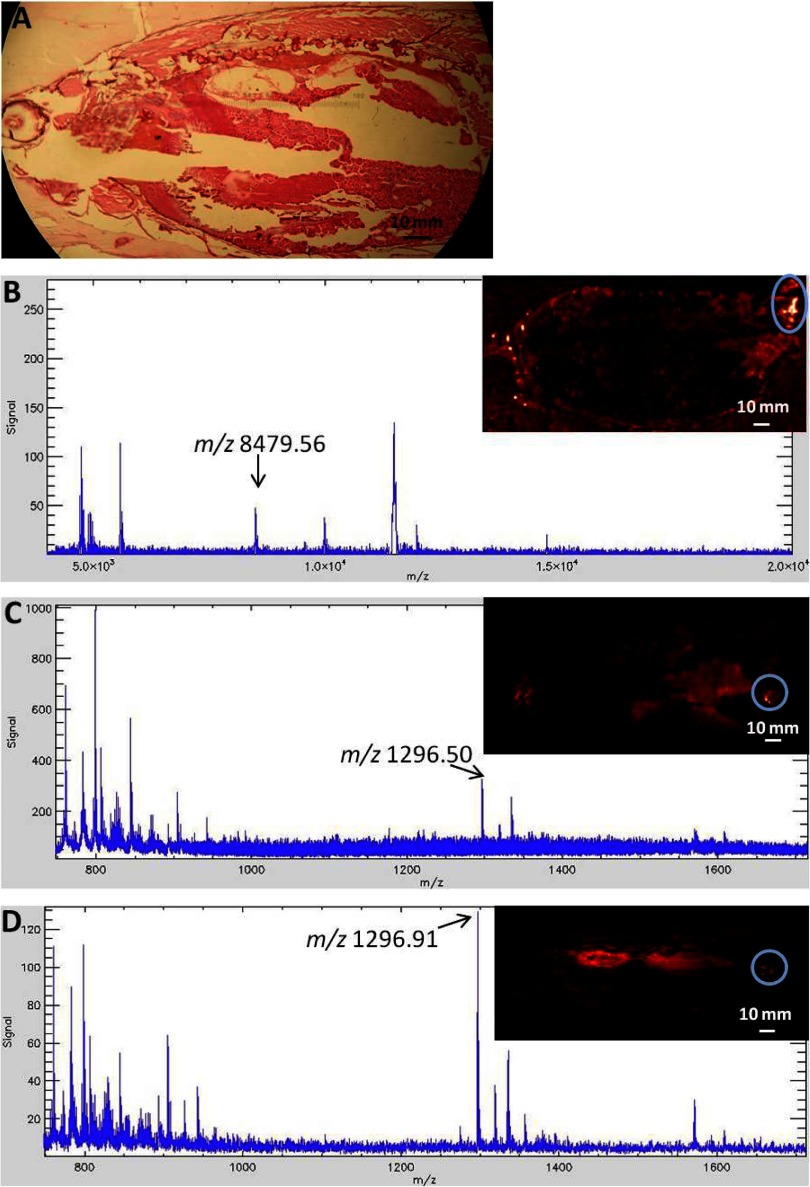
Additionally, ZF cryosections embedded in each of the eight media types described above were sprayed with SPA, CHCA, or CHCA + AmP matrices using an automated sprayer unit. Figure 3 demonstrates MALDI-MS images and representative spectra from an adult, female ZF, embedded in 2% CMC + 5% gelatin and treated with each matrix type. The spectra presented in Fig. 3 clearly demonstrate that the SPA, CHCA, and CHCA + AmP matrices ionized the low-concentration (pmol/μl) peptide and protein standards applied to each ZF cryosection. For MALDI-MS, the addition of AmP to the CHCA matrix reduces alkali metal adducts causing matrix cluster interference20 and has been shown to enhance phosphopeptide signals.21 In our experiments, the addition of 6 mM AmP to 5 mg/ml CHCA aided in decreasing the predominance of CHCA matrix ion clusters that typically interfere in the peptide mass range 650–1300 m/z (Fig. 3B and C and Supplemental Fig. 3).22 Although the addition of AmP to the CHCA matrix solution is not a new concept, we demonstrate the first use of AmP as a CHCA matrix additive applied with an automated sprayer, as opposed to dipping a matrix-coated section into AmP23 prior to MALDI-MSI. The act of dipping the delicate ZF sections into solutions resulted in partial section destruction, as demonstrated in the multistep-rinsing photos (Supplemental Fig. 1). However, it should be noted that the act of physically rinsing a MALDI spot with an ammonium salt solution has proven to be more effective at suppressing CHCA matrix cluster formation than simply adding the ammonium salt to the matrix.22 Therefore, dipping a tissue section in an appropriate ammonium salt solution prior to MALDI-MSI may be beneficial for less delicate tissues.
Figure 3.
A representative H&E-stained section and MS images of peptide and protein standard mass spectra from serial sections of a single adult, female ZF embedded in 2% CMC + 5% gelatin and sprayed with three different matrices in 50% ACN, 0.1% TFA. (A) H&E-stained serial section; (B) 5 mg/ml SPA, mass standard apomyoglobin+2, average mass 8476.78 Da; (C) 5 mg/ml CHCA, mass standard angiotensin I, monoisotopic mass 1296.6853; (D) 5 mg/ml CHCA + 6 mM AmP, mass standard angiotensin I, monoisotopic mass 1296.6853. The standard mass for which the droplet is highlighted in the MSI is labeled on the mass spectrum. Arrows in the spectra denote appearance of expected mass-standard peak; a circle in the MSI denotes mass-standard droplet placement; overall average mass spectrum for the circled area and total ion current is shown for each mass spectrum.
Conclusion
Based on physical properties, stability, and ion-suppression data, we conclude that gelatin hydrogels form acceptable embedding media for ZF cryosectioning, and 5% CMC + 10% gelatin is the optimal embedding medium for whole-body ZF cryosectioning. The addition of AmP to the CHCA matrix spray further optimized the MALDI-MSI preparation methods for the detection of peptide standards in ZF sections. Future efforts will include optimizing MALDI-MSI preparation and handling methods for multiple life stages of this important model species.
ACKNOWLEDGMENTS
The authors thank Peggy Harris for expertise in ZF culturing and production and Rachel Pryor for ZF culture maintenance at the Gulf Ecology Division.
DISCLOSURES
The authors declare no financial conflict of interests. The views expressed in this paper are those of the author(s) and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names, products, or services does not convey and should not be interpreted as conveying official EPA approval, endorsement, or recommendation.
Supplemental Data Section
REFERENCES
- 1. Cameron LC. Mass spectrometry imaging: facts and perspectives from a non-mass spectrometrist point of view. Methods 2012;57:417–422 [DOI] [PubMed] [Google Scholar]
- 2. Goodwin RJ. Sample preparation for mass spectrometry imaging: small mistakes can lead to big consequences. J Prot 2012;75:4893–4911 [DOI] [PubMed] [Google Scholar]
- 3. Gustafsson JO, Oehler MK, Ruzkiewicz A, McColl SR, Hoffmann P. MALDI imaging mass spectrometry (MALDI-IMS)-application of spatial proteomics for ovarian cancer classification and diagnosis. Int J Mol Sci 2011;12:773–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Dyck S, Flammang P, Meriaux C, et al. Localization of secondary metabolites in marine invertebrates: contribution of MALDI MSI for the study of saponins in cuverian tubules of H. forskali. PLoS ONE 2010;5:e13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khatib-Shahidi S, Andersson M, Herman JL, et al. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal Chem 2006;78:6448–6456 [DOI] [PubMed] [Google Scholar]
- 6. Takai N, Tanaka Y, Inazawa K, et al. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun Mass Spectrom 2012;26:1549–1556 [DOI] [PubMed] [Google Scholar]
- 7. Herbert SP, Stainier DY. Molecular control of endothelial cell behavior during blood vessel morphogenesis. Nat Rev Mol Cell Biol 2011;12:551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Souza AG, MacCormack TJ, Wang N, et al. Large-scale proteome profile of the zebrafish (Danio rerio) gill for physiological and biomarker discovery studies. Zebrafish 2009;6:229–238 [DOI] [PubMed] [Google Scholar]
- 9. Eberlin LS, Chramow A, Hamid TS, Girod M, Cooks RG, Ifa DR. Chemical imaging of zebrafish by desorption electrospray ionization-mass spectrometry (DESI-MS). J Am Soc Mass Spectrom 2012;23:Suppl [Google Scholar]
- 10. Willett M, Deininger SO, Ketterlinus R. Protein analysis moves into the fast lane. Biophot Int January 2006 [Google Scholar]
- 11. Lemaire R, Wisztorski M, Desmons A, et al. MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal Chem 2006;78:7145–7153 [DOI] [PubMed] [Google Scholar]
- 12. Yang J, Caprioli RM. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal Chem 2011;83:5728–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deutskens F, Yang J, Capriolo RM. High spatial resolution imaging mass spectrometry and classical histology on a single tissue section. J Mass Spectrom 2011;46:568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strohalm M, Strohalm J, Kaftan F, et al. Poly[N-(2-hydroxypropyl) methacrylamide]-based tissue-embedding medium compatible with MALDI mass spectrometry imaging experiments. Anal Chem 2011;83:5458–5462 [DOI] [PubMed] [Google Scholar]
- 15. Jones EA, Deininger SO, Hogendoorn PC, Deelder AM, McDonnell LA. Imaging mass spectrometry statistical analysis. J Prot 2012;75:4962–4989 [DOI] [PubMed] [Google Scholar]
- 16. Goodwin RJ, Dungworth JC, Cobb SR, Pitt AR. Time-dependent evolution of tissue markers by MALDI-MS imaging. Proteomics 2008;8:3801–3808 [DOI] [PubMed] [Google Scholar]
- 17. Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom 2003;38:699–708 [DOI] [PubMed] [Google Scholar]
- 18. Chatterji PR, Kaur H. Interpenetrating hydrogel networks: 3. Properties of the gelatin-sodium carboxymethylcellulose system. Polymer 1992;33:2388–2391 [Google Scholar]
- 19. Buhus G, Peptu C, Popa M, Desbrières J. Controlled release of water soluble antibiotics by carboxymethylcellulose- and gelatin-based hydrogels crosslinked with epichlorhydrin. Cell Chem Technol 2009;43:141–151 [Google Scholar]
- 20. Zhu X, Papayannopoulos IA. Improvement in the detection of low concentration protein digests on a MALDI TOF/TOF workstation by reducing α-cyano-4-hydroxycinnamic acid adduct ions. J Biomol Tech 2003;14:298–307 [PMC free article] [PubMed] [Google Scholar]
- 21. Asara JM, Allison J. Enhanced detection of phosphopeptides in matrix-assisted laser desorption/ionization mass spectrometry using ammonium salts. J Am Soc Mass Spectrom 1999;10:35–44 [DOI] [PubMed] [Google Scholar]
- 22. Smirnov IP, Zhu X, Taylor T, et al. Suppression of α-cyano-4-hydroxycinnamic acid matrix clusters and reduction of chemical noise in MALDI-TOF mass spectrometry. Anal Chem 2004;76:2958–2965 [DOI] [PubMed] [Google Scholar]
- 23. Lagarrigue M, Becker M, Lavigne R, et al. Revisiting rat spermatogenesis with MALDI imaging at 20-μm resolution. Mol Cell Proteomics 2011;10:M110.005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.