Abstract
Tuberous sclerosis (TS) is characterized by the development of hamartomas in various organs and is caused by a germ-line mutation in either TSC1 or TSC2 tumor suppressor genes. From the symptomatic resemblance among TS patients, involvement of TSC1 and TSC2 products in a common pathway has been suggested. Here, to analyze the function of the Tsc1 product, we established a line of Tsc1 (TSC1 homologue) knockout mouse by gene targeting. Heterozygous Tsc1 mutant (Tsc1+/−) mice developed renal and extra-renal tumors such as hepatic hemangiomas. In these tumors, loss of wild-type Tsc1 allele was observed. Homozygous Tsc1 mutants died around embryonic days 10.5–11.5, frequently associated with neural tube unclosure. As a whole, phenotypes of Tsc1 knockout mice resembled those of Tsc2 knockout mice previously reported, suggesting that the presumptive common pathway for Tsc1 and Tsc2 products may also exist in mice. Notably, however, development of renal tumors in Tsc1+/− mice was apparently slower than that in Tsc2+/− mice. The Tsc1 knockout mouse described here will be a useful model to elucidate the function of Tsc1 and Tsc2 products as well as pathogenesis of TS.
Tuberous sclerosis (TS) is an autosomal dominantly inherited disease characterized by the development of hamartomas and benign tumors in various organs such as brain, kidney, and heart (1). A germ-line mutation in either TSC1 or TSC2 genes (2, 3), both of which act as tumor suppressors (4, 5), is a genetic factor responsible for pathogenesis of TS. The similar symptoms of TS patients associated with TSC1 or TSC2 mutations suggest that the products of TSC1 and TSC2 are involved in a common physiological pathway (1, 6). TSC1 encodes a protein with a molecular mass of ≈130 kDa, hamartin, which contains a coiled-coil domain in the carboxyl-terminal half (3). TSC2 encodes tuberin, a rap1-GTPase activating protein homology domain-containing protein with a molecular mass of ≈180 kDa (2). Although several studies concerned with functions of these products have been reported, in vivo functions of them remain to be elucidated (1, 7, 8).
The tumor suppressor function of TSC2 became evident by studies of rodents with a germ-line Tsc2 mutation such as the Eker rat (9–13) and Tsc2 knockout mice (14, 15). Both heterozygous Tsc2 mutant rats and mice develop hereditary renal tumors and extra-renal tumors carrying a second hit of Tsc2 gene (14–17). Homozygosity of Tsc2 mutation leads to the embryonic lethality both in rats (9, 18) and mice (15, 16), indicating that the function of tuberin is essential for mammalian development.
We also isolated a rat homologue of TSC1 (Tsc1) and analyzed its mutation in chemically induced renal tumors in wild-type rats, in which Tsc2 mutations were found with high frequency (≈50%) (19). In those tumors, we found Tsc1 mutations in a case with no Tsc2 mutation (19). These results suggest that mutations of Tsc1 and Tsc2 are involved in the development of chemically induced renal tumors in rats, although the latter is more common. These systems of renal tumorigenesis could be an experimental approach to analyze the function of Tsc1 and its relationship with function of Tsc2 in vivo. However, the rat or mouse model carrying the germ-line Tsc1 mutation, which should be essential for precise analysis to elucidate the molecular mechanism of pathogenesis of TS, is not yet available. Therefore, in this study, we generated Tsc1 mutant mice by gene targeting and initially characterized its phenotypes about tumor development and embryonic lethality.
Materials and Methods
Genomic DNA Cloning and cDNA Amplification.
A mouse embryonic stem (ES) cell (clone J1, 129/Sv background) genomic DNA library was screened with rat Tsc1 cDNA (19) as a probe, and a positive clone (λMTSC1) was isolated and analyzed by restriction enzyme digestion and sequencing. The numbering of exons in this study followed those of human and rat Tsc1 genes (3, 19). Mouse Tsc1 cDNA fragments corresponding to the coding region of rat and human counterparts were amplified by reverse transcription-PCR using total RNA from ES cells as a template. The 5′ half was amplified with a primer set, MTRTF1 (5′-AGCATAATTGAGTGAGAGAATG-3′) and MTRTR2 (CCATGCACACAGTCATCTTG-3′). The 3′ half was amplified with a primer set, MTRTF2 (5′-CAAGATGACTGTGTGCATGG-3′) and MTRTR1 (5′-ACACTGACAGACCCTCTTCA-3′). MTRTF1 and MTRTR1 were designed from the rat Tsc1 cDNA sequence in expectation of high homology between rat and mouse. In the above sequences, the positions of putative initiation (in MTRTF1) and termination (in MTRTR1, complementary strand) codons for translation are underlined. MTRTF2 and MTRTR2 were sequences in exon 12 found in λMTSC1. Amplified products were directly sequenced for both strands by primer walking.
Fluorescence in Situ Hybridization.
For fluorescence in situ hybridization, the insert DNA of λMTSC1 was used as a probe. Mouse pro-metaphase chromosomes were prepared from embryonic fibroblasts in early passages by standard technique. The procedures of fluorescence in situ hybridization and microscopic observation were carried out as described (20).
Construction of a Targeting Vector and Gene Targeting.
For the construction of a targeting vector, pBmTsc1-KO, a 1.8-kb genomic DNA fragment from a BamHI site in intron 4 to the splicing acceptor site in intron 5 and an 8-kb fragment from an ApaI site in intron 8 to a BamHI site in intron 12 were used as 5′ and 3′ homologous regions, respectively. A DNA fragment containing internal ribosome entry site (IRES) of the encephalomyocarditis virus (21), enhanced green fluorescent protein (EGFP)-coding sequence (CLONTECH) followed by a poly(A) addition signal (pA), was introduced between 5′ and 3′ homologous regions to delete exons 6 to 8. IRES-EGFP-pA was inserted just after the splicing acceptor of intron 5, allowing a splicing event between exon 5 and IRES and the expression of EGFP to occur. At the 3′ end of this IRES-EGFP-pA unit, an expression cassette of neomycin resistance gene (neo) was introduced in opposite orientation. The diphtheria toxin A-chain expression cassette was used for negative selection (22). For gene targeting, linearized pBmTsc1-KO was introduced into J1 ES cells by electroporation. Subsequent cloning of homologous recombinant clones and production of chimera mice were carried out as described (23). F1 mice were obtained by crossing male chimera mice with female C57BL/6J mice.
Southern and Northern Blot Analyses and PCR Genotyping.
For genotyping by Southern blot analysis, XbaI-digested DNAs were hybridized with 5′ outside probe (0.5-kb ClaI–BamHI fragment), and EcoRI- or EcoRV-digested DNAs were hybridized with 3′ outside probe (0.3-kb BamHI–EcoRI fragment). Genotyping by PCR was performed with the following primers: INT7F1 (5′-TCTGAGTCCGAGTCAAGAGT-3′ in intron 7) and INT7R1 (5′-TAGATGTGAAGTTGTGTGGCA-3′ in intron 7) for wild-type allele; and EGFPF2 (5′-CATGGACGAGCTGTACAAGT-3′ in EGFP coding sequence) and EGFPR2 (5′-GGACAAACCACAACTAGAATG-3′ in EGFP coding sequence) for mutant allele. Conditions of PCR were essentially the same as described (13). Northern blot analysis was carried out as described (11).
N-Ethyl-N-Nitrosourea (ENU) Treatment.
F1 male Tsc1+/− mice were crossed with female C57BL/6J mice, and ENU solution was i.p. injected (50 mg/kg body weight) into the pregnant mother mice at embryonic day (E) 15.0 (noon of the day on which a vaginal plug was detected was defined as E0.5). Offspring were killed at 11 weeks of age for histological analyses.
Histological Analyses.
Tissues from mice or embryos were fixed in either 10% buffered formaline or Bouin's fixative. Dehydration, embedding in paraffin, sectioning, and staining with hematoxylin and eosin (HE) were performed by standard protocols. Immunostaining with anti-β-tubulin III (clone TuJ1; Babco, Richmond, CA) was performed as described by using VECTASTAIN ABC horseradish peroxidase kit (Vector Laboratories) (13).
Loss of Heterozygosity (LOH) Analysis.
DNA samples from formaline-fixed, paraffin-embedded tissues were prepared as described (13). Primers used for LOH analysis were INT7F1 and INT7R1 for wild-type allele and NEOF1 (5′-GGCTGCTATTGGGCGAAGT-3′ in neo cassette) and NEOR1 (5′-CCTGATGCTCTTCGTCCAG-3′ in neo cassette) for mutated allele. Conditions of PCR were essentially the same as described (13).
Results and Discussion
Cloning and Gene Targeting of Mouse Tsc1.
To obtain the structural information about mouse hamartin, mouse Tsc1 cDNA fragments were amplified by reverse transcription-PCR (see Materials and Methods). The mouse Tsc1 cDNA sequence corresponding to the coding region of rat and human counterparts encoded 1,160 aa residues showing ≈87% and ≈95% identities with human and rat hamartins, respectively (data not shown, GenBank accession no. AB047561) (3, 16). By Northern blot analysis, a major ≈8.0-kb mRNA of Tsc1 was detected in brain, heart, kidney, heart, liver, and embryos at E9.5 to E13.5 (data not shown). Also, we isolated a phage genomic DNA clone (λMTSC1) covering exons 4–12 of mouse Tsc1 gene (Fig. 1A). By fluorescence in situ hybridization, Tsc1 gene appeared to be localized on mouse chromosome 2, band B-C1.1 (data not shown). Recently, Cheadle et al. (24) reported the genomic organization and comparative analysis of the mouse Tsc1. An alanine residue at position 381 of predicted mouse hamartin sequence in their report was missed in our sequence, probably because of a difference in the usage of splicing acceptor at the 3′ end of intron 11 (data not shown).
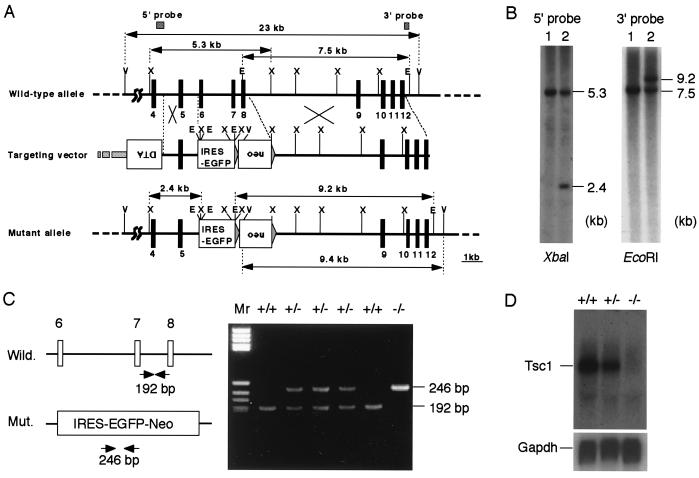
Figure 1.
Inactivation of mouse Tsc1 gene. (A) Structure of targeting vector and wild-type and mutant alleles. Exons are denoted with filled boxes with numbers, and restriction enzyme sites (E, EcoRI; V, EcoRV; X, XbaI) are shown. Expression cassettes for the neomycin resistance gene (neo) and diphtheria toxin A-chain gene (DTA), and the coding sequence for EGFP preceded with an IRES (IRES-EGFP) are shown as open boxes. Length (kb) of restriction fragments and positions of probes (5′ and 3′ probes) used for genotyping are denoted by arrows and hatched boxes, respectively. (B) Southern blot analysis of ES cells. XbaI- or EcoRI-digested DNAs from homologous recombinant (lane 2) and control (lane 1) ES cells were probed with probes shown in A. Sizes (kb) of bands are shown. (C) PCR genotyping of F2 embryos. (Left) Schematic representation of primers and product size (bp). (Right) A representative result of PCR genotyping of E10.0 embryos obtained by double heterozygous breeding. Wild-type (+/+), heterozygous (+/−), and homozygous mutant (−/−) embryos were present in this litter. (D) Northern blot analysis of E11.0 embryos. (Upper) The expression profile of Tsc1 mRNA (≈8.0 kb) in wild-type (+/+), heterozygous (+/−), and homozygous mutant (−/−) embryos. Probe used was a Tsc1 cDNA fragment covering the 3′ region downstream from exon 8. (Lower) The result with a GAPDH cDNA probe using the same blot.
To disrupt Tsc1, a targeting vector (pBmTsc1-KO), in which exons 6–8 were replaced by IRES-EGFP-pA expression unit and a neo-expression cassette, was constructed and used for gene targeting in ES cells (Fig. 1A). These deleted exons encode a part of the highly conserved region between mammalian and fly Tsc1 homologues that may be functionally important (7, 8). Skipping these three exons, splicing from exon 5 to exon 9 should result in frameshift. Male germ-line chimeras were obtained using a correctly targeted ES cell clone, and F1 mice were obtained by crossing of those chimeras with C57BL/6J female mice (Fig. 1B). F1 mice heterozygous for Tsc1 mutation (Tsc1+/−) were born and grew with normal appearance. Northern blot analysis of E11.0 embryos obtained by double heterozygous breeding revealed that ≈8.0-kb mRNA of Tsc1 disappeared in homozygous mutant (Tsc1−/−) embryos (Fig. 1 C and D).
Embryonic Lethality of Homozygous Tsc1 Mutants.
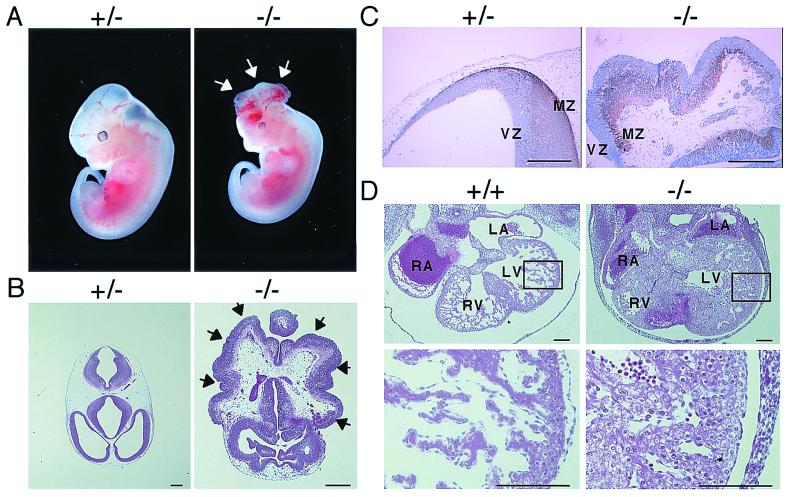
Tsc1−/− mice were not obtained by F1 double heterozygous breedings (32 wild-type and 62 Tsc1+/− mice among 94 F2 mice at birth). Analysis of embryos revealed that most of Tsc1−/− embryos died between E10.5 and E11.5 (Table 1). This stage for lethality is similar to that of Tsc2-deficient embryos, although more embryos alive at E11.0 to E12.5 were found in the Tsc1 case (30% of Tsc1−/− embryos versus 13% of Tsc2−/− ones; refs. 14 and 15). Typically, Tsc1−/− embryos alive at E9.0-E12.5 were smaller than control embryos, and about one-third (6 of 19) exhibited neural tube unclosure at the head region (Fig. 2 A and B). Histologically, the neural tube of those embryos exhibited a disorganized architecture, although they were positive for β-tubulin III, a neuronal differentiation marker, suggesting the differentiation of neural lineage in them (Fig. 2C). Hearts of some Tsc1−/− embryos exhibited abnormal morphology of myocardial cells (Fig. 2D). Although hypoplastic appearance of livers was noticed in Tsc1−/− embryos as reported in Tsc2−/− embryos (data not shown, ref. 15), the Tsc1−/− embryos were much smaller than the control ones; therefore, developmental delay and hypoplasia of livers in those embryos should be carefully analyzed further. Nonetheless, phenotypic similarities found between Tsc1- and Tsc2-deficient embryos suggest that, during the mouse embryonic development, hamartin and tuberin may function in a common pathway.
Table 1.
Genotype analysis of embryos obtained by double heterozygous breedings
| Embryonic day | Litters | Total number | Genotype of Tsc1
|
Tsc2−/−‡ embryos | |||
|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | Resorbed† | ||||
| E9.0∼9.5 | 2 | 21 | 5 | 10 | 6 (1)* | 0 | 19 (7)* |
| E10.0∼10.5 | 6 | 52 | 13 | 25 | 12 (4) | 2 | 10 (5) |
| E11.0∼11.5 | 7 | 63 | 17 | 32 | 12 (7)§ | 2 | 17 (15)§ |
| E12.0∼12.5 | 5 | 43 | 12 | 20 | 8 (7) | 3 | 6 (5) |
| E13.0∼13.5 | 4 | 38 | 9 | 18 | 7 (7) | 4 | 1 (1) |
The number in parentheses indicates the number of dead embryos among total −/− embryos.
Number of resorbed embryos could not be genotyped.
Data from ref. 14 for comparison.
Survival rates did not show the significant difference (P = 0.092) by Fisher's exact test.
Figure 2.
Analysis of homozygous Tsc1 mutant embryos. (A) Macroscopic appearance of heterozygous (+/−) and homozygous Tsc1 mutant (−/−) embryos at E12.0 with heartbeat. Arrows point to the unclosed neural tube exhibiting exencephaly. (B) Histology of head region of heterozygous (+/−) and homozygous (−/−) mutant embryos at E12.0 with heartbeat shown by HE staining. Field of homozygous mutant is enlarged with twice the magnitude of that of the heterozygous mutant. Arrows point to the unclosed region of the neural fold exposed to the outside. (C) Immunostaining of neural tube with anti-class III β-tubulin antibody (TuJ1). Sections of the head region from heterozygous (+/−) and homozygous (−/−) mutant embryos at E12.0 with heartbeat were stained with TuJ1. After immunostaining, a counterstain with HE was performed. Positive signals were seen as brown staining. VZ, ventricular zone; MZ, marginal zone. (D) Histological analysis of heart from wild-type (+/+) and homozygous Tsc1 mutant (−/−) embryos at E12.0 with heartbeat shown by HE staining. (Lower) Enlarged view of the region enclosed with a rectangle in the upper panel. LA, left atrium; LV, left ventricle; RA, right atrium; RV, right ventricle. (Scale bars: B and C = 200 μm; D = 100 μm.)
Development of Renal Tumors in Heterozygous Tsc1 Mutant Mice.
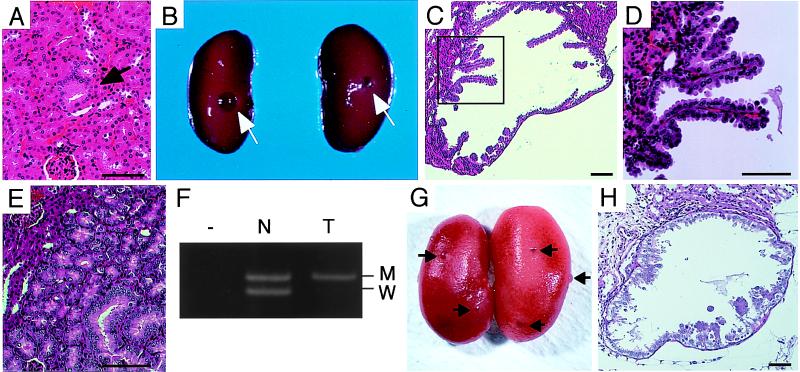
Tsc2+/− mice develop macroscopically visible renal carcinomas and/or renal cystadenomas by the age of 10 months in all cases (14, 15). In addition, hepatic hemangiomas and other tumors in extremities developed in Tsc2+/− mice with a high frequency (14, 15). We also observed tumor development in F1 Tsc1+/− mice. At 9–12 months of age, although macroscopic tumors were not observed, phenotypically altered tubules, the preneoplastic lesions of renal carcinogenesis, were found in kidney sections from Tsc1+/− mice (n = 5) (Table 2 and Fig. 3A). At least one phenotypically altered tubule was observed in the largest dorsoventral section from each kidney. By the age of 15–18 months, small cysts with a diameter of 0.5–2.0 mm appeared on the surface of kidneys in Tsc1+/− mice (9 of 14 = 64%) (Table 2 and Fig. 3B). Histologically, these cysts were similar to those developed in Tsc2+/− mice, showing papillary neoplastic outgrowth into the lumen (Fig. 3 C and D) (14, 15). In addition to cystic lesions, there were tumors exhibiting tubular structure (Fig. 3E). Using DNA samples from microdissected specimens, we examined the LOH at the Tsc1 locus in renal tumors by PCR (Fig. 3F and data not shown). Of six available cases examined, two cases clearly showed the loss of wild-type Tsc1 allele, which suggests that a second hit of Tsc1 may be a critical event for the development of renal tumors in Tsc1+/− mice. As an attempt to accelerate renal carcinogenesis in Tsc1+/− mice, we performed transplacental administration of ENU (25) as in our previous study for Tsc2 knockout mice (14). Of 13 Tsc1+/− mice treated with ENU, six mice developed bilateral macroscopic cysts in kidneys by 11 weeks of age (Fig. 3G). These cysts had a diameter less than 1.5 mm and showed similar histological features to those developed in ENU-treated Tsc2+/− mice (Fig. 3H) (14). Although kidneys from the other seven ENU-treated Tsc1+/− mice did not show macroscopic cysts, microscopic phenotypically altered tubules and adenomatous lesions were observed in the cortex region (data not shown). None of the kidney sections from ENU-treated wild-type mice (n = 6) showed such renal lesions. Thus, transplacental ENU treatment accelerated renal tumorigenesis in Tsc1+/− mice. To confirm that a second hit in the wild-type Tsc1 allele is induced by ENU, mutational analysis of Tsc1 in these ENU-accelerated renal tumors is to be desired. Such analysis will be useful to analyze the mutation spectrum of Tsc1 for comparison with that of human TSC1 (1).
Table 2.
Development of macroscopic renal tumors in Tsc1+/− mice
| Age (months) | Tsc1+/−* | Tsc2+/−*† |
|---|---|---|
| 9∼12 | n = 5 | n = 9 |
| Mice with unilateral tumors | 0 | 2 |
| Mice with bilateral tumors | 0 | 7 |
| Tumor multiplicity (/kidney)‡ | — | 2.3 ± 0.3 |
| Diameter of tumor (mm)‡ | — | 0.8 ± 0.1 |
| 15∼18 | n = 14 | n = 8 |
| Mice with unilateral tumors | 4 | 0 |
| Mice with bilateral tumors | 5 | 8 |
| Tumor multiplicity (/kidney)‡ | 1.7 ± 0.3§ | 5.5 ± 0.6§ |
| Diameter of tumor (mm)‡ | 1.0 ± 0.1 | 1.1 ± 0.2 |
All of examined mice were F1 on a hybrid 129/SvJ-C57BL6/J background.
Data from our previous study (ref. 14) for comparison.
Multiplicity and diameter of tumor were of tumor-bearing kidneys (mean ± SEM).
P < 0.0001 by Mann–Whitney test.
Figure 3.
Renal tumors in Tsc1+/− mice. (A) A phenotypically altered renal tubule (arrow) shown by HE staining. (B) Macroscopic renal cysts (arrows) developed in a 15-month-old mouse. (C and D) Histology of a renal cystadenoma shown by HE staining. D is an enlarged view of region enclosed with a rectangle in C. (E) Histology of a renal adenoma exhibiting tubular structure shown by HE staining. (F) Loss of wild-type Tsc1 allele in a renal tumor of Tsc1+/− mice. Representative result of LOH analysis is shown. Lanes T, tumor; N, normal counterpart; −, no input DNA sample. Products from mutant allele (M, 220 bp) and from wild-type allele (W, 192 bp) are indicated. (G) Macroscopic renal cysts (arrows) developed in an 11-week-old mouse transplacentally treated with ENU. (H) Histology of a renal cystadenoma developed by ENU treatment shown by HE staining. (Scale bars = 50 μm.)
Results obtained in this study indicate that a germ-line Tsc1 mutation causes renal tumorigenesis in mice as a Tsc2 mutation. However, spontaneous development of macroscopic renal tumors was apparently slower in Tsc1+/− mice, compared with Tsc2+/− mice on the same genetic background (C57BL6/J:129/Sv; Table 2, ref. 14). Although the molecular mechanism underlying this difference is currently unknown, it may be relevant to the difference in frequencies of second hit or in functions of gene products. In the case of human, there is a debate about the symptomatic difference between TS patients associated with TSC1 and TSC2 mutations (1). Dabora et al. (26) provided evidence for the increased severity of TSC2-associated patients than TSC1-associated ones. Elucidation of the molecular basis for the differences of renal tumorigenesis found here may provide some important clues for the understanding of pathogenesis of human TS. Nonetheless, development of renal tumors and extra-renal tumors (see below) in both Tsc1+/− and Tsc2+/− mice suggests that tuberin and hamartin have a functional relationship for tumor suppression.
Development of Extra-Renal Tumors in Heterozygous Tsc1 Mutant Mice.
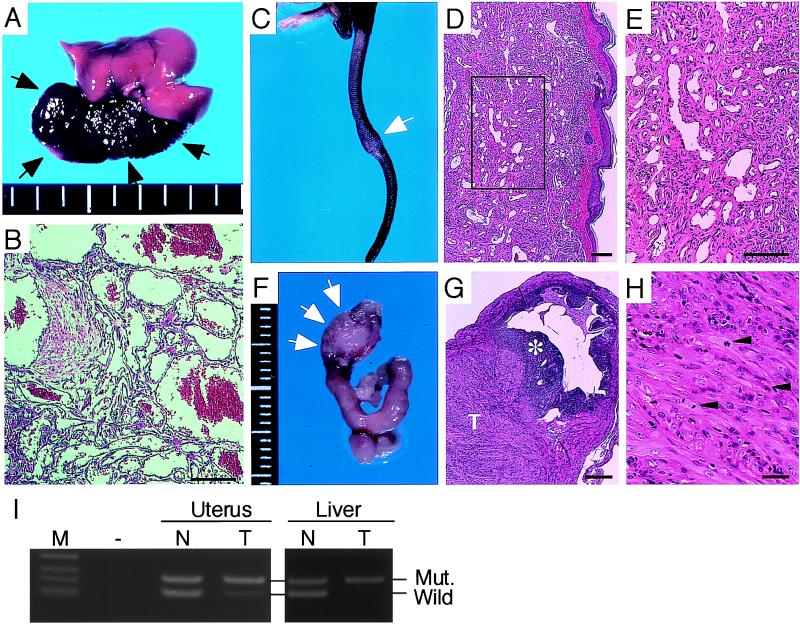
Macroscopic hemangiomas in liver also were developed in Tsc1+/− mice by 15–18 months of age (10 of 14 = 71%) (Fig. 4A). This incidence of hepatic hemangiomas resembled that of Tsc2+/− mice at a similar age (≈80%) (14). Histologically, these hemangiomas could be classified as cavernous hemangiomas and resembled those developed in Tsc2+/− mice (Fig. 4B) (14, 15). Although there was no higher incidence of mortality in Tsc1+/− mice compared with wild-type mice during observation of 18 months, we noticed sudden death of Tsc1+/− mice older than 18 months of age, probably as a result of the rupture of huge hepatic hemangiomas (unpublished observation). In addition, we found a hemangioma in the tail and a leiomyoma/leiomyosarcoma in the uterus (Fig. 4 C–H). Also in Tsc2+/− mice, hemangiomas/hemangiosarcomas in the extremities were often (≈10%) developed by the age of 18 months (ref. 15 and unpublished observation). Although uterus tumors are rare in mice, Tsc2+/− Eker rat females frequently develop leiomyomas/leiomyosarcomas (27). None of these extra-renal tumors was observed in wild-type mice analyzed as a control in this study (n = 15). We examined LOH of Tsc1 in those extra-renal tumors by PCR (Fig. 4I). Of three hepatic hemangiomas examined, one case clearly showed the loss of wild-type Tsc1 allele. The uterine leiomyoma/leiomyosarcoma also showed the loss of wild-type Tsc1 allele. These LOHs suggest that a second hit of Tsc1 may be a critical event for the development of these extra-renal tumors in Tsc1+/− mice. Together, these results indicate that, in mice, a germ-line Tsc1 mutation contributes to the development of extra-renal tumors as a germ-line Tsc2 mutation.
Figure 4.
Extra-renal tumors in Tsc1+/− mice. (A) Macroscopic view of a hepatic hemangioma (arrows) developed in a 17-month-old mouse. (B) Histology of a hepatic hemangioma shown by HE staining. (C) Macroscopic view of a tail of 15-month-old mouse showing a knob (arrow). (D and E) Histology of a hemangioma developed in the knob in C shown by HE staining. E is an enlarged view of the region enclosed with a rectangle in D. (F) Macroscopic view of a uterus leiomyoma/leiomyosarcoma developed in a 16-month-old mouse (arrows). (G and H) Histology of the leiomyoma/leiomyosarcoma in F shown by HE staining. In G, * and T indicate the endometrium and tumor region, respectively. Arrowheads in H point to mitotic cells. (I) Loss of wild-type Tsc1 allele in extra-renal tumor of Tsc1+/− mice. Representative results of LOH analysis are shown. Lanes T, tumor; N, normal counterpart. M, size marker; −, no input DNA sample. Products from mutant allele (Mut., 220 bp) and from wild-type allele (Wild, 192 bp) are indicated. (Scale bars: B, D, E, and G = 100 μm; H = 10 μm.)
In this study, similarities of phenotypes between Tsc1 and Tsc2 knockout mice were revealed for both tumor development and embryonic lethality, although some differences were also observed. As a whole, these phenotypical similarities must reflect the involvement of hamartin and tuberin in a common pathway, as has been suggested from the symptom of TS patients. Although phenotypes caused by Tsc1 and Tsc2 mutations in mice are different from those of human TS patients, involvement of hamartin and tuberin in a common pathway may be conserved in both species. Elucidation of this presumptive common pathway is important for the understanding of tumorigenesis in mice as well as pathogenesis of TS patients. In combination with the use of Tsc2 knockout mouse, our Tsc1 knockout mouse will be a useful experimental model to unravel the functions of hamartin and tuberin and to elucidate the mechanism of tumor development associated with Tsc1 and Tsc2 mutations.
Acknowledgments
We thank H. Yamanaka, M. Miyagawa, T. Fukuda, and Y. Hirayama for technical assistance; H. Akazawa, I. Komuro, and H. Takeshima for helpful discussions; I. Ishikawa for plasmid preparation; and H. Sugano, T. Kitagawa, and A. G. Knudson for encouragement throughout this work. This work was supported in part by grants from the Ministry of Education, Science and Culture, the Ministry of Health and Welfare of Japan, the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research, and the Program of Core Research for Evolutional Science and Technology of the Japan Science and Technology Corporation.
Abbreviations
- TS
tuberous sclerosis
- E
embryonic day
- ES
embryonic stem
- IRES
internal ribosome entry site
- EGFP
enhanced green fluorescent protein
- ENU
N-ethyl-N-nitrosourea
- HE
hematoxylin and eosin
- LOH
loss of heterozygosity
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB047561).
References
- 1.Cheadle J P, Reeve M P, Sampson J R, Kwiatkowski D J. Hum Genet. 2000;107:97–114. doi: 10.1007/s004390000348. [DOI] [PubMed] [Google Scholar]
- 2.The European Chromosome 16 Tuberous Sclerosis Consortium. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 3.van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, et al. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 4.Green A J, Johnson P H, Yates J R W. Hum Mol Genet. 1994;3:1833–1834. doi: 10.1093/hmg/3.10.1833. [DOI] [PubMed] [Google Scholar]
- 5.Green A J, Smith M, Yates J R W. Nat Genet. 1994;6:193–196. doi: 10.1038/ng0294-193. [DOI] [PubMed] [Google Scholar]
- 6.van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Shell R, van den Ouweland A, Reuser A, Sampson J, Halley D, van der Sluijs P. Hum Mol Genet. 1998;7:1053–1058. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- 7.Ito N, Rubin G M. Cell. 1999;96:529–539. doi: 10.1016/s0092-8674(00)80657-1. [DOI] [PubMed] [Google Scholar]
- 8.Lamb R F, Roy C, Diefenbach T J, Vinters H V, Johnson M W, Jay D G, Hall A. Nat Cell Biol. 2000;2:281–287. doi: 10.1038/35010550. [DOI] [PubMed] [Google Scholar]
- 9.Hino O, Klein-Szanto A J P, Freed J J, Testa J R, Brown D Q, Vilensky M, Yeung R S, Tartof K D, Knudson A G. Proc Natl Acad Sci USA. 1993;90:327–331. doi: 10.1073/pnas.90.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hino O, Kobayashi T, Tsuchiya H, Kikuchi Y, Kobayashi E, Mitani H, Hirayama Y. Biochem Biophys Res Commun. 1994;203:1302–1308. doi: 10.1006/bbrc.1994.2324. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T, Hirayama Y, Kobayashi E, Kubo Y, Hino O. Nat Genet. 1995;9:70–74. doi: 10.1038/ng0195-70. [DOI] [PubMed] [Google Scholar]
- 12.Yeung R S, Xiao G-H, Jin F, Lee W-C, Testa J R, Knudson A G. Proc Natl Acad Sci USA. 1994;91:11413–11416. doi: 10.1073/pnas.91.24.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi T, Mitani H, Takahashi R, Hirabayashi M, Ueda M, Tamura H, Hino O. Proc Natl Acad Sci USA. 1997;94:3990–3993. doi: 10.1073/pnas.94.8.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T, Minowa O, Kuno J, Mitani H, Hino O, Noda T. Cancer Res. 1999;59:1206–1211. [PubMed] [Google Scholar]
- 15.Onda H, Lueck A, Marks P W, Warren H B, Kwiatkowski D J. J Clin Invest. 1999;104:687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubo Y, Mitani H, Hino O. Cancer Res. 1994;54:2633–2635. [PubMed] [Google Scholar]
- 17.Yeung R S, Xiao G-H, Everitt J I, Jin F, Walker C L. Mol Carcinog. 1995;14:28–36. doi: 10.1002/mc.2940140107. [DOI] [PubMed] [Google Scholar]
- 18.Rennebeck G, Kleymenova E V, Anderson R, Yeung R S, Artzt K, Walker C L. Proc Natl Acad Sci USA. 1998;95:15629–15634. doi: 10.1073/pnas.95.26.15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satake N, Kobayashi T, Kobayashi E, Izumi K, Hino O. Cancer Res. 1999;59:849–855. [PubMed] [Google Scholar]
- 20.Hagiwara T, Tanaka K, Takai S, Maeno-Hikichi Y, Mukainaka Y, Wada K. Genomics. 1996;33:508–515. doi: 10.1006/geno.1996.0226. [DOI] [PubMed] [Google Scholar]
- 21.Ghattas I R, Sanes J R, Majors J E. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yagi T, Ikawa Y, Yoshida K, Shigetani Y, Takeda N, Mabuchi I, Yamamoto T, Aizawa S. Proc Natl Acad Sci USA. 1990;87:9918–9922. doi: 10.1073/pnas.87.24.9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, et al. Genes Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 24.Cheadle J P, Dobbie L, Idziaszczyk S, Hodges A K, Smith A J H, Sampson J R, Young J. Mamm Genome. 2000;11:1135–1138. doi: 10.1007/s003350010203. [DOI] [PubMed] [Google Scholar]
- 25.Hino O, Mitani H, Knudson A G. Cancer Res. 1993;53:5856–5858. [PubMed] [Google Scholar]
- 26.Dabora S L, Jozwiak S, Franz D N, Roberts P S, Nieto A, Chung J, Choy Y-S, Reeve M P, Thiele E, Egelhoff J C, et al. Am J Hum Genet. 2001;68:64–80. doi: 10.1086/316951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hino O, Mitani H, Katsuyama H, Kubo Y. Cancer Lett. 1994;83:117–121. doi: 10.1016/0304-3835(94)90307-7. [DOI] [PubMed] [Google Scholar]