Abstract
During development of the mouse forebrain interneurons, the Dlx genes play a key role in a gene regulatory network (GRN) that leads to the GABAergic phenotype. Here, we have examined the regulatory relationships between the ascl1a, dlx, and gad1b genes in the zebrafish forebrain. Expression of ascl1a overlaps with dlx1a in the telencephalon and diencephalon during early forebrain development. The loss of Ascl1a function results in a loss of dlx expression, and subsequent losses of dlx5a and gad1b expression in the diencephalic prethalamus and hypothalamus. Loss of Dlx1a and Dlx2a function, and, to a lesser extent, of Dlx5a and Dlx6a, impairs gad1b expression in the prethalamus and hypothalamus. We conclude that dlx1a/2a act downstream of ascl1a but upstream of dlx5a/dlx6a and gad1b to activate GABAergic specification. This pathway is conserved in the diencephalon, but has diverged between mammals and teleosts in the telencephalon.
Keywords: ascl1a, dlx, gad1b, GABAergic interneuron, Telencephalon, Diencephalon
Introduction
Gene regulatory networks (GRNs) are made up of dynamic interactions between transcription factors and cis-regulatory elements (CREs) found within the genome (for reviews see: Levine and Davidson (2005) and Davidson and Levine (2008)). CREs are classically thought to be non-coding regulatory sequences, comprised of clustered transcription factor binding sites; the binding of these transcription factors are able to affect the transcription of specific genes (for reviews see: Kulkarni and Arnosti (2003), Kadonaga (2004) and Panne (2008)). The overall levels and timing of gene expression are conferred by the cumulative contributions of multiple transcription factors on a myriad of regulatory regions. The genes regulated by this process during development often encode transcription factors that will play a role in the regulation of other transcription factor genes located downstream in the GRN, eventually resulting in the expression of terminal differentiation genes leading to a specified cell type.
Ascl1 (Mash1) is one of the basic helix–loop–helix (bHLH) transcription factors thought to play important roles in GRNs controlling neurogenesis (for reviews see: Bertrand et al. (2002) and Allan and Thor (2003)). Ascl1 is expressed in proliferating neural precursors in the subpallial telencephalon and prethalamus of the mouse (Lo et al., 1991; Guillemot and Joyner, 1993; Porteus et al., 1994; Yun et al., 2002; Andrews et al., 2003). Ascl1 mutants have defects in neural specification and in the timing of differentiation in the ventral forebrain, including altered telencephalic expression of the Dlx genes and Gad1 (Gad67), which encodes glutamic acid decarboxylase, the enzyme responsible for the production of γ-amino butyric acid (GABA) (Casarosa et al., 1999; Horton et al., 1999: Yun et al., 2002; Long et al., 2009a). Ectopic expression of Ascl1 leads to Gad1 expression in the mouse dorsal telencephalon, further supporting a role for Ascl1 in GABAergic interneuron development (Fode et al., 2000). In zebrafish there are two Ascl1 orthologs, ascl1a and ascl1b. These genes are expressed in the embryonic forebrain, including the subpallial telencephalon and prethalamus, reminiscent of Ascl1 expression in the mouse (Allende and Weinberg, 1994; Wullimann and Mueller, 2002).
In the mouse forebrain, expression of Ascl1 and Dlx genes overlap suggesting these genes may genetically interact during mouse forebrain development (Porteus et al., 1994; Yun et al., 2002; Andrews et al., 2003). Consistent with this hypothesis, Ascl1−/− mutant mice have mis-expression of Dlx in the ganglionic eminences (Casarosa et al., 1999; Horton et al., 1999; Yun et al., 2002; Long et al., 2009a) and ASCL1 proteins have been shown to activate and directly bind to a Dlx1/Dlx2 regulatory element (Poitras et al., 2007).
The Dlx genes encode homeodomain transcription factors expressed in the ganglionic eminences of the telencephalon and diencephalon in the mouse. More specifically, four Dlx genes are expressed in the forebrain of the mouse: Dlx1, Dlx2, Dlx5, and Dlx6 (Liu et al., 1997; Yang et al., 1998; Anderson et al., 1997a; Eisenstat et al., 1999), while five orthologous dlx genes are expressed in the forebrain of the zebrafish: dlx1a, dlx2a, dlx5a, dlx6a, and dlx2b (Akimenko et al., 1994; Ellies et al., 1997; Hauptmann and Gerster, 2000). The Dlx genes are expressed in highly overlapping but also distinct domains within the forebrain of mice and zebrafish, often correlating with neuronal differentiation and Gad expression (Liu et al., 1997; Eisenstat et al., 1999; MacDonald et al., 2010a; Stühmer et al., 2002a,b; Yun et al., 2002). Functional studies have shown that the Dlx genes are required for the differentiation and migration of most GABAergic neurons in the telencephalon and diencephalon (Anderson et al., 1997a,b; Stühmer et al., 2002a,b; Long et al., 2007; Long et al., 2009a,b; Wang et al., 2012). Additionally, DLX1 and DLX2 are involved in the suppression of neurite growth and branching, thus enabling the proper tangential migration of GABAergic neurons (Cobos et al., 2007).
The zebrafish dlx genes are involved in branchial arch and sensory placode development (Solomon and Fritz, 2002; Kaji and Artinger, 2004; Walker et al., 2006; Jackman and Stock, 2006; Sperber et al., 2008; Talbot et al., 2010), as are the mouse Dlx genes (Qiu et al., 1995; Depew et al., 2002; Jeong et al., 2008). However, despite their common use as forebrain markers, there has been little functional analysis of the dlx genes in the zebrafish brain. To characterize the role of ascl1a and dlx in the GRN(s) controlling GABAergic interneuron differentiation in the zebrafish forebrain, we have knocked down their function and assayed the effects on downstream targets. Our results show that the ascl1a gene regulates dlx genes necessary for proper gad1b expression in the diencephalon of the zebrafish. Thus, these genes are key part of a GRN involved in early forebrain development that is conserved among bony vertebrates.
Materials and methods
Zebrafish strains and staging
Embryos were obtained and housed using standard procedures described in Westerfield (2000). The following transgenic zebrafish lines were used in this study: Tg(dlx1a/2aIG:GFP) (MacDonald et al., 2010a), Tg(dlx1URE2:GFP) (MacDonald et al., 2010b), and Tg(dlx5a/6a: GFP) (Ghanem et al., 2003). All developmental stages are reported as hours post-fertilization (hpf) All experiments were performed in accordance with the Canadian Council on Animal Care guidelines and approved by institutional animal care committees.
Morpholino and mRNA injections
Morpholino oligonucleotides (MO) were injected into one-cell stage wild type or transgenic zebrafish embryos at concentrations ranging from 2 to 4 ng/μl. The following translation blocking morpholinos were used: dlx1a (Sperber et al., 2008), dlx2a (Sperber et al., 2008), dlx2b (Jackman and Stock, 2006), dlx5a (Walker et al., 2006), dlx6a (5′TGGTCATCATCAAATTTTCTGCTTT3′). The ascl1a5′UTR MO (Cau and Wilson, 2003) was kindly provided by Dr. S. Wilson. Splice blocking MOs for dlx5a (Talbot et al., 2010) were kindly provided by Dr. C. B. Kimmel, and were used to confirm the translation blocking MO phenotypes. The dlx6a splice-blocking morpholino binds to the end of the second exon and inhibits the splicing of the second intron (5′AAATGAGTTCA-CATCTCACCTGCGT3′).
In situ hybridization and imaging
Whole mount mRNA in situ hybridization was carried out as described in Thisse and Thisse (1998). The antisense mRNA probes were labeled with digoxygenin-11-UTP (Roche, 11277073910) and synthesized from the following cDNA clones: dlx1a (Ellies et al., 1997), dlx2a (Akimenko et al., 1994), dlx5a (Akimenko et al., 1994), dlx6a (Ellies et al., 1997), dlx2b (Ellies et al., 1997), gad2 (Martin et al., 1998), gad1b (Mueller et al., 2008), ascl1a (Cau et al., 2000), nkx2.1a (Rohr and Concha, 2000), emx2 (Morita et al., 1995), lhx5a (Toyama et al., 1995), gfp (Dorsky et al., 2002). After the procedure, embryos were post fixed in 4% PFA and cleared overnight in glycerol.
Fluorescent RNA in situ hybridization was carried out with a protocol modified from those described previously (Jowett and Yan, 1996; Welten et al., 2006; Talbot et al., 2010). The DNP-labeled probe was revealed with tyr-Cy5, whereas dig-labeled probes were revealed using tyr-Cy3. Fluorescein-labeled probes were revealed with tyrfluorescein (available from Perkin-Elmer). The full tissue labeling protocols can be found online: http://wiki.zfin.org/display/prot/Triple+Fluorescent+In+Situ.
For confocal imaging, embryos were placed in mounting media on glass slides and positioned under coverslips. Confocal z-stacks were obtained by using a Zeiss LSM5 PASCAL (Carl Zeiss, Germany) with an excitation laser at 488 (Fluorescein), 543 nm (Cy3), and 633 nm (Cy5).
Rescue experiments and morphant phenotype scoring
For exogenous expression of dlx genes, capped full-length mRNA was synthesized in vitro using linearized PCS2+ plasmids (mMessageMachine; Ambion) and purified. The following plasmids as templates: mutdlx2a (mutagenized at MO binding site) and mutdlx5a (Supplementary Table 1). A solution containing 40 ng/μl of mRNA, along with MO, was co-injected into single cell embryos. Individuals were classified and scored in two groups: either as having reduced or normal prethalamic expression. Embryos from each treatment were scored in a double-blind manner and plotted with standard error from three individual experiments. One way ANOVA was used to compare data.
Results
The ascl1a, dlx, and gad1b genes are co-expressed in the forebrain
We utilized triple fluorescent in situ hybridizations to determine if the zebrafish ascl1a, dlx and gad genes show overlapping expression in the forebrain as they do in the mouse. The expression of ascl1a begins in the prospective forebrain at 10 hpf and lasts until at least 72 hpf (Allende and Weinberg, 1994). The dlx1a and dlx2a (hereafter called dlx1a/2a) genes are expressed starting at 13 hpf in the prospective forebrain (Akimenko et al., 1994; Ellies et al., 1997). At 24 hpf, dlx1a is expressed in the telencephalon and two domains of the diencephalon, the prethalamus (or ventral thalamus) and the hypothalamus (Fig. 1A). The dlx2a expression domains are identical to dlx1a (MacDonald et al., 2010a), so we consider dlx1a expression as representative of the two genes. At 24 hpf, expression of ascl1a is detected in the telencephalon and prethalamus, and partially overlaps with the dlx1a expression domain (Fig. 1A). The ascl1b gene is paralogous to ascl1a, but each has unique and overlapping expression domains with the central nervous system, including the forebrain (Allende and Weinberg, 1994). At 24 hpf, expression of ascl1b was detected in the forebrain but had very little co-expression with dlx1a or ascl1a positive cells (Fig. 1A), indicating that ascl1b cannot activate dlx gene expression at this stage in the forebrain.
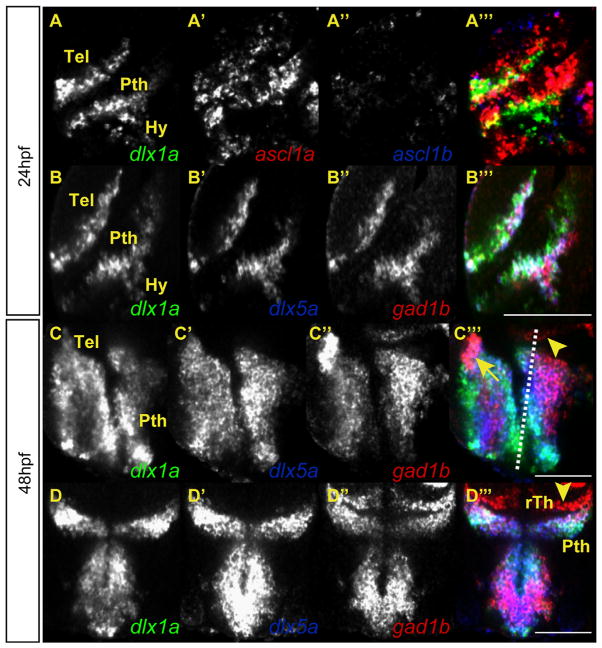
Fig. 1.
The expression domains of the ascl1a, dlx, and gad1b genes overlap in the forebrain of the embryonic zebrafish. Single z sections on triple fluorescent in situ hybridizations. (A) In the forebrain lateral view of a WT embryo, the dlx1a and ascl1a are co-expressed throughout the telencephalon and prethalamus at 24 hpf, while ascl1b is not co-expressed at this stage. (B) The dlx1a, dlx5a, and gad1b genes are co-expressed in the telencephalon, prethalamus, and hypothalamus at 24 hpf. (C) The dlx1a, dlx5a, and gad1b genes continue to be co-expressed in the telencephalon and prethalamus at 48 hpf. The gad1b expression is increased in the dorsal tip of the telencephalon (arrow). The plane of section for (D) is shown as a dotted line in C‴. (D) Cross-section showing the co-expression of dlx1a, dlx5a, and gad1b in the prethalamus (arrow). The rostral thalamus (arrowhead) is positive for gad1b expression but not dlx1 or dlx5a. Tel=telencephalon, Pth=prethalamus, Hy=hypothalamus, rTh=rostral thalamus. Scale bar in A–D=100 μm and in E=40 μm.
The Dlx genes are expressed in very similar domains within the developing forebrain of mice and zebrafish (Liu et al., 1997; Anderson et al., 1997a; Eisenstat et al., 1999; Akimenko et al., 1994; Ellies et al., 1997; Zerucha et al., 2000; Mueller et al., 2008; MacDonald et al., 2010a). At 24 hpf, the expression of dlx5a and gad1b are both highly overlapping with dlx1a expression in the telencephalon, prethalamus, and hypothalamus (Fig. 1B; MacDonald et al., 2010a). Expression of dlx1a, dlx5a, and gad1b remains highly overlapping at 48 hpf in the telencephalon (Fig. 1C), and prethalamus (Fig. 1C and D, arrows). However, there is an area of intense staining for gad1b in the dorsal region of the telencephalon potentially corresponding to GABAergic interneurons that will migrate into the pallium starting at approximately 72 hpf (Fig. 1C′′′, asterisk) (Mione et al., 2008). Additionally, there is an area dorsal to the dlx1a and dlx5a expression domains of the prethalamus that is gad1b positive and dlx negative (Fig. 1D, arrowhead) that may correspond to the rostral thalamus (Peukert et al., 2011; Lauter et al., 2013).
Knockdown of ascl1a reveals a role in the regulation of the Dlx and gad1b genes in the prethalamus and hypothalamus
To determine the role of ascl1a in regulating dlx and gad1b gene expression in the zebrafish forebrain, we knocked down Ascl1a activity with a translation blocking morpholino (Cau and Wilson, 2003). Morpholino knock down of Ascl1a function in the zebrafish affects the development of the pituitary, neurogenesis in the epiphysis, and regeneration in the retina (Cau and Wilson, 2003; Herzog et al., 2004; Pogoda et al., 2006; Fausett et al., 2008), here we examine the consequences of its knockdown on GABAergic fate specification in the forebrain. Injected embryos were examined at 24 and 48 hpf, stages when the genes are co-expressed and the distinct regions of the forebrain are evident. In ascl1a morphants, there is a loss of dlx1a, dlx2a and dlx5a expression in the domain of the prethalamus and hypothalamus at 24 and 48 hpf (Fig. 2A–C, Supplementary Fig. 1, asterisks). A slight reduction in dlx1a, dlx2a and dlx5a expression in the telencephalon is also possible but difficult to assess by in situ hybridization. Expression of gad1b is also impaired in the ventral prethalamus and hypothalamus of ascl1a morphants at 24 and 48 hpf (about 65% of injected embryos; Fig. 2D, Fig. 3A, B and Supplementary Fig. 2A, B, G, H), suggesting the loss of either ascl1a or dlx gene function may play a role in the differentiation of gad1b expressing cells.
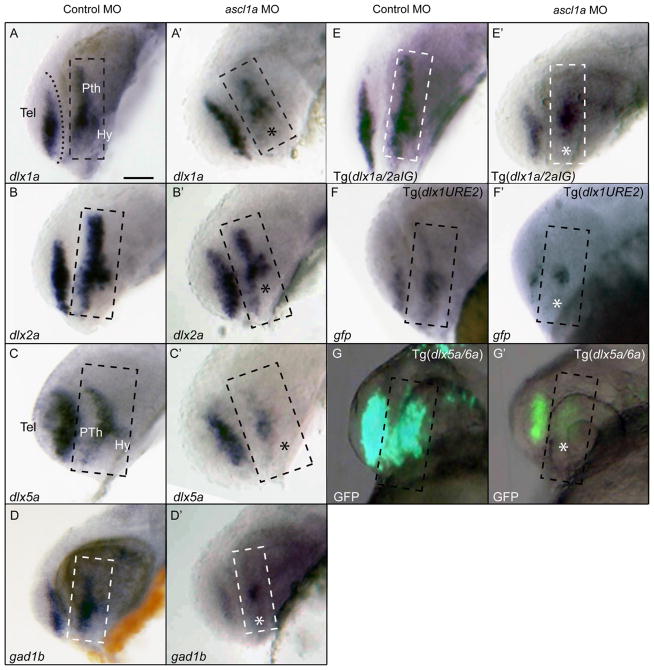
Fig. 2.
Ascl1a function is required for proper expression of the dlx and gad1b genes in the forebrain at 24 hpf. (A, B) Expression of dlx1a and dlx2a is reduced in the prethalamus and hypothalamus (asterisk), but not affected in the telencephalon of ascl1a morphants compared to control embryos. (C) The expression of dlx5a is particularly reduced in the prethalamus and hypothalamus in ascl1a morphants. (D) gad1b expression is unaffected in the telencephalon, but reduced in the prethalamus and hypothalamus. (E) Tg(dlx1a/2aIG:GFP) embryos reduced gfp mRNA expression in the prethalamus of ascl1a morphants but telencephalic expression appears unaffected. (F) Tg (dlx1URE2:GFP) embryos show reduced gfp mRNA expression in the telencephalon and prethalamus. (G) The loss of GFP expression in the Tg(dlx5a/6a:GFP) transgenic line is consistent with a loss of dlx5a expression. Dashed box represents prethalamus region. Tel, telencephalon; Pth, prethalamus; Hy, hypothalamus. Scale bar=50 μm.
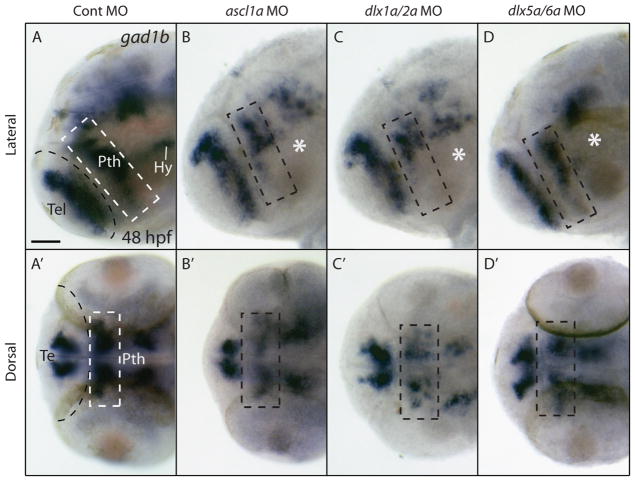
Fig. 3.
The expression of gad1b is impaired in the prethalamus but not in the telencephalon of ascl1a morphants and of dlx1a/2a double morphants at 48 hpf. Lateral (A–D) and dorsal views (A′–D′) of the forebrain of control and morphant embryos. (A) Expression of gad1b in embryos injected with the control morpholino is seen in the telencephalon (Tel), prethalamic (Pth) and hypothalamic (Hy) diencephalon. (B) Morpholino knockdown of ascl1a results in a loss of ventral prethalamic (dashed box) and hypothalamic (asterisk) gad1b expression. (C) Double morpholino knockdown of dlx1a/2a results in decreased prethalamic and hypothalamic gad1b expression. (D) In dlx5a/dlx6a morphants there is mild reduction of prethalamic and a decrease in hypothalamic gad1b expression similar to that observed in ascl1a and dlx1a/2a morphants. Dashed line indicates telencephalon-diencephalon boundary. Scale bar=50 μm.
Expression of Dlx1/Dlx2 genes in the forebrain is conferred by at least two conserved CREs, I12b and URE2 whereas that of Dlx5/Dlx6 is conferred at least by CREs located in the intergenic region (Zerucha et al., 2000; Ghanem et al., 2003; Ghanem et al., 2007; Potter et al., 2009; MacDonald et al., 2010a,b). To test if the loss of ascl1a function affects dlx gene regulation by altering the activity of dlx regulatory elements, we knocked down Ascl1a function in the following transgenic lines: Tg(dlx1a/2aIG:GFP) (MacDonald et al., 2010a), Tg(dlx1aURE2:GFP) (MacDonald et al., 2010b), and Tg(dlx5a/6a:GFP) (Ghanem et al., 2003). Both the Tg(dlx1a/2aIG: GFP) (Fig. 2E) and Tg(dlx1URE2:GFP) (Fig. 2F) embryos injected with the ascl1a MO show a reduced reporter gene expression in the prethalamus consistent with the loss of dlx1a/2a expression in this domain. The expression of gfp in Tg(dlx1URE2:GFP) is also reduced in the telencephalon indicating ascl1a is also necessary for proper regulation of dlx1a and/or dlx2a in this part of the forebrain (Fig. 2F). The Tg(dlx5a/6a:GFP) ascl1a morphants showed a severe loss of GFP expression in the prethalamus (Fig. 2G), while expression in the telencephalon may be reduced but is still detectable. Overall, our data support the hypothesis that ascl1a acts as an upstream regulator of the dlx1a, dlx2a, dlx5a and gad1b genes in the embryonic prethalamus and hypothalamus.
Knockdown of dlx paralogs results in the loss of gad1b expression in the forebrain
In the mouse, Gad1 is downstream of Dlx1 and Dlx2 (Stühmer et al., 2002a; Long et al., 2007; Long et al., 2009a), prompting us to test this relationship in zebrafish. To assay a possible role for dlx genes in gad1b expression (orthologous to mouse Gad1) in the zebrafish forebrain at 24 and 48 hpf, we used translation and splice blocking MOs against dlx1a, dlx2a, dlx5a, and dlx6a. Single dlx morphants show no discernible changes in gad1b expression in the telencephalon, prethalamus or hypothalamus at 24 and 48 hpf (Supplementary Figs. 2D and 3A–C, E, F). At 48 hpf, gad1b expression in the telencephalon of dlx1a/2a double morphants is similar to controls, but there is a reduction in the ventral prethalamus and in the hypothalamus (Fig. 3C and Supplementary Fig. 2C). The combinatorial loss of dlx5a/6a results in a mild reduction in prethalamic gadb1 signal compared to dlx1a/2a morphants, and a reduction in the hypothalamic gad1b expression that is comparable to that observed in ascl1a and dlx1a/2a morphants. (Fig. 3D and Supplementary Fig. 2E). Finally, triple knockdown of the paralogs dlx1a, dlx2a and dlx2b does not increase the severity of gad1b loss in the prethalamus or result in any noticeable loss in the telencephalon, prethalamus or hypothalamus (Supplementary Fig. 3D). Therefore, dlx1a/2a and, possibly, dlx5a/6a are necessary for proper expression of gad1b in the prethalamus and hypothalamus. In Dlx1/2−/− mutant mice the expression of Ascl1 was altered in regions of the telencephalon (Yun et al., 2002; Long et al., 2009a); we examined ascl1a expression in dlx1a/2a and dlx5a/6a double morphants. We did not observe any noticeable changes in ascl1a expression in the forebrain in these morphants (Supplementary Fig. 4).
The mouse Dlx genes are involved in auto- and cross-regulatory interactions in the telencephalon (Zerucha et al., 2000; Zhou et al., 2004; Poitras et al., 2007; Bond et al., 2009; Potter et al., 2009) and diencephalon (Long et al., 2009a). To test if the zebrafish dlx1a/2a genes play a role in the regulation of the dlx5a/6a bigene cluster, we examined dlx5a expression and the activity of Tg(dlx5a/6a:GFP) following MO-mediated knock down of dlx1a and/or dlx2a. There is no loss of dlx5a expression in single dlx1a or dlx2a morphant embryos (data not shown). In embryos injected with both MOs, there is a severe reduction of both telencephalic, prethalamus and hypothalamic expression of dlx5a, consistent with the loss of GFP expression in Tg(dlx5a/6a:GFP)(Fig. 4A–B). To confirm the observed differences in expression are not due to alteration of the anatomy of the telencephalon or diencephalon, we verified the expression of marker genes such as nkx2.1a (Fig. 4C), lhx5a (Fig. 4D), and emx2 (Fig. 4E) and their expression domains are not markedly changed after the knockdown of dlx1a/2a. The gad2 gene is also expressed in the telencephalon and prethalamus, in a pattern very similar to gad1b (Martin et al., 1998). However, dlx1a/2a morphants show no change in the expression of gad2 at 24 hpf, indicating the gad1b and gad2 genes may be regulated differently (Fig. 4F). To verify that changes in gad1b expression pattern in morphants is not due to cell death in the forebrain we stained double morphants with acridine orange and did not observe any overt increase in cell death (Supplementary Fig. 5).
Fig. 4.
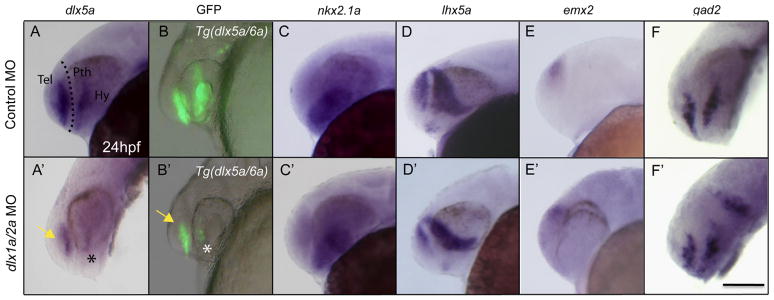
Knockdown of dlx1a/2a identifies cross-regulatory interactions between the dlx1a/dlx2a and dlx5a/dlx6a dlx bigene clusters. (A) Double knockdown of dlx1a/2a results in reduced dlx5a expression in the telencephalon (arrow) and diencephalon (asterisk). (B) The loss of dlx5a/6a regulatory element activity in Tg(dlx5a/6a:GFP) in dlx1a/2a morphants is consistent with the loss of dlx5a expression. The defects in the prethalamus are not due to loss of mis-patterning of the structures as (C) nkx2.1a, (D) emx2, and (E) lhx5a genes are unaffected in dlx1a/2a morphants. (F) The dlx1a/2a morphants do not show a loss of gad2 expression in the forebrain. Scale bar=100 μm.
Exogenous administration of dlx2a and dlx5a mRNA in ascl1a morphants partially rescues diencephalic gad1b expression
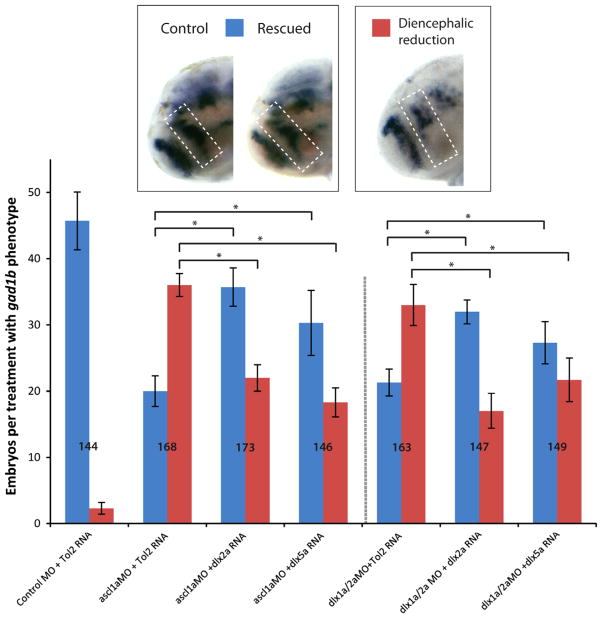
Rescue experiments were carried out to determine whether ascl1a and dlx1a/2a act sequentially or in parallel cascades to regulate prethalamic gad1b expression. We used exogenous expression of dlx2a or dlx5a mRNA to rescue the gad1b phenotype in the forebrain of ascl1a morphants. Rescue experiments were carried out by co-injecting ascl1a MO with dlx2a or dlx5a mRNA or co-injecting the dlx1a/2a MOs with mismatched dlx2a or dlx5a mRNA, and performing in situ hybridization for gad1b at 48 hpf. As both genes in a dlx bigene pair have highly redundant expression patterns and, possibly, biochemical function, we injected mRNA from only one gene of each pair. The gad1b phenotype of a given individual was classified as being “normal” (resembling the wild type expression, blue bars in Fig. 5), or “reduced” in the prethalamus and hypothalamus (red bars in Fig. 5). A cohort of injected embryos for a given treatment would have varying proportions of these two phenotypes. Tol2 mRNA was injected as a control because this RNA is not expected to affect the ascl1a-dlx-gad1b pathway. Sixty-four percent of ascl1a morphants show reduced gad1b expression. However upon co-injection with exogenous dlx2a or dlx5a mRNA this phenotype was significantly rescued in the diencephalon of ascl1a morphants (Fig. 5). Similarly, 61% of dlx1a/2a morphants show reduced gad1b but exogenous expression of dlx2a or dlx5a mRNA significantly rescues this phenotype in the diencephalon (Fig. 5).
Fig. 5.
Exogenous dlx2a and dlx5a mRNA expression in ascl1a and dlx morphants rescues gad1b expression in the diencephalon (prethalamus and hypothalamus). Average number of embryos per injected treatment with normal (blue), and reduced (red) diencephalic gad1b expression. A clear difference in expression is observed between the embryos injected with the control MO+Tol2 mRNA (left insert) and the embryos injected with the ascl1aMO+Tol2 RNA (right insert). The latter is thereafter treated as the baseline for comparison with ascl1aMO+dlx2a (middle insert). Furthermore, exogenous expression of dlx5a, and particularly of dlx2a mRNA, is able to significantly decrease the proportion of embryos with reduced gad1b expression in the diencephalon of dlx1a/2a morphants. Data from 3 experimental replicates, with total individuals per treatment shown within graph ranging from n=144–173, p<0.05.
Discussion
ascl1a expression is necessary for the proper regulation of the dlx and gad1b genes
The ascl1a gene is co-expressed with dlx1a, which is later co-expressed with dlx5a and gad1b in the forebrain, reminiscent of the expression of their mouse orthologs (Porteus et al., 1994; Yun et al., 2002; Andrews et al., 2003; Stühmer et al., 2002a,b; Long et al., 2009a,b). Overlap in ascl1a, dlx and gad1b gene expression suggests that the GRNs necessary for GABAergic interneuron development in mammals and teleosts may be similar. In the mouse, Ascl1 is required for the generation of early born neurons in the subcortical telencephalon, whereas Dlx genes play a role in late neurogenesis (Casarosa et al., 1999; Horton et al., 1999; Anderson et al., 1997a, Yun et al., 2002; Long et al., 2009a,b). The gene expression data, coupled with their relationship during neurogenesis in the forebrain suggest that Ascl1 is upstream of Dlx. Knockdown of the zebrafish Ascl1 ortholog, ascl1a, results in the loss of dlx1a/2a and dlx5a expression in the prethalamus and hypothalamus, and possibly lower expression in the telencephalon, showing that ascl1a is involved in the regulation of dlx genes in diencephalic territories. The loss of Ascl1a function also results in altered gad1b expression in the same region (Fig. 6). Although Ascl1 mutant mice show increased Dlx expression due to premature differentiation of the subventricular zone (Long et al., 2009a, b), we do not see increased dlx expression in the telencephalon of zebrafish ascl1a morphants. This may also be explained by a potentially redundant function of the gsx gene family in the zebrafish forebrain. Whereas mouse Ascl1 is a downstream effector of Gsx function in the telencephalon (Wang et al., 2009), gsx1 is not expressed in the zebrafish telencephalon at early developmental stages (Scholpp et al., 2007), and thus should not control dlx expression. It remains possible that gsx2 may play a role in the telencephalon, however this has yet to be tested.
Fig. 6.
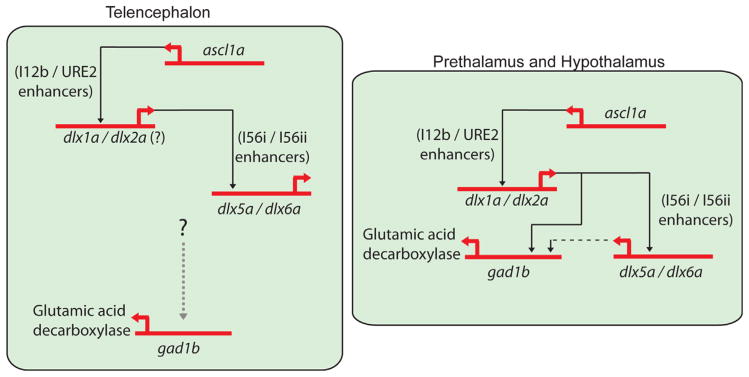
Model showing the regulation of the dlx and gad1b genes in the zebrafish forebrain. In the telencephalon, ascl1a controls Tg(dlx1URE2:GFP) activity and may modulate dlx1a/2a expression. The dlx1a/2a genes will regulate dlx5a expression via its CREs. A currently unknown genetic pathway controls the expression of gad1b in the telencephalon as knockdown of ascl1a, dlx1a/2a or dlx5a/6a has little or no effect on transcription. In the diencephalon, specifically the ventral part of the prethalamus and the hypothalamus, ascl1a controls the expression of dlx1a/2a which then regulates dlx5a/6a and gad1b in this tissue. The dlx5a/6a genes may play a minor role in gad1b regulation in the diencephalon (as shown by dashed black line).
MO-mediated knockdown of dlx1a/2a leads to impaired expression of dlx5a and gad1b in the prethalamus and hypothalamus. We therefore attribute, at least in part, the loss of dlx5a and gad1b expression in the diencephalon of ascl1a morphants to the loss of proper dlx1a/2a function. Our data suggest ascl1a regulates dlx1a/2a expression, which may then regulate dlx5a/6a (Fig. 4) and gad1b expression (Fig. 6). Furthermore, the results of dlx5a/6a morpholino injections show a mild modification of gad1b expression in the ventral prethalamus when compared to dlx1a/2a morphants. This suggests that dlx5a/6a may play a relatively smaller role in the regulation of gad1b in this region. The rescue of the prethalamic gad1b phenotype in ascl1a morphants by exogenous expression of dlx2a also supports the existence of a genetic cascade consisting of ascl1a, dlx1a/2a, dlx5a/6a, leading to expression of the Gad1b enzyme.
Dlx function during prethalamus development in the zebrafish
The Dlx genes have largely redundant functions within the mouse forebrain, as suggested by the synergistic phenotypes see after combinatorial loss of Dlx genes (Qiu et al., 1995; Acampora et al., 1999; Anderson et al., 1997b; Marin et al., 2000; Cobos et al., 2007; Long et al., 2007; Mao et al., 2009; Long et al., 2009a,b; Wang et al., 2012). For instance, although Dlx1−/− and Dlx2−/−mutants have only subtle neural defects, Dlx1−/−/Dlx2−/− double mutants show a major block in neurogenesis and differentiation resulting in a loss of Gad expression in the ventral telencephalon. Similarly, we show that knockdown of individual dlx genes in the zebrafish had no discernible effect on gad1b expression whereas double knockdowns resulted in reduced gad1b expression in the prethalamus and hypothalamus. Impaired gad1b expression is unlikely due to increased cell death or loss of the structures as specific molecular markers for this forebrain region are unaltered in dlx morphants.
In contrast to the prethalamus, the combined knockdown of both dlx1a/2a does not result in discernible loss of gad1b expression in the telencephalon. However, it is possible that the expression level of gad1b is altered but is not detectable with our methods. The presence of dlx2b, a teleost specific paralog of dlx2a that is co-expressed with dlx1a and dlx2a in the forebrain, could explain the absence of gad1b phenotype in the telencephalon. However, MO-mediated knock down of dlx2b did not affect forebrain gad1b expression and the combined administration of MOs for dlx1a, dlx2a and dlx2b did not enhance the phenotype observed with the dlx1a/2a MO combination. The lack of a gad1b phenotype in the zebrafish telencephalon points to differences in the genetic pathways regulating gad1b expression in this tissue between teleosts and mammals.
Dlx1−/−/Dlx2−/− mutant mice show a loss of Gad1 expression in the olfactory bulb and the ventral telencephalon (Anderson et al., 1997a,b; Bulfone et al., 1998, Long et al., 2007; Long et al., 2009a,b; Wang et al., 2012). Furthermore, ectopic expression of the Dlx genes leads to expression of Gad genes, Gad1 and Gad2, in mouse telencephalon slice cultures (Anderson et al., 1999; Stühmer et al., 2002a).The knockdown of dlx5a/6a causes less dramatic gad1b changes in the ventral prethalamus. This suggests that dlx1a/2a function may be sufficient for gad1b expression in this region. Unfortunately, the mouse Dlx5−/−/Dlx6−/− mutant forebrain phenotype cannot be fully studied due to exencephaly (Robledo et al., 2002, Wang et al., 2010). However, our morpholino data may provide some clarity as to the role of Dlx1/2 and/or Dlx5/6 for the GABAergic neuron differentiation and migration in the forebrain. Due to the close genetic interactions between Dlx genes, it has been challenging to uncouple their functions. Our data suggests that Dlx1/2 function is critical for gad1b expression, whereas Dlx5/6 may have a relatively minor role.
Loss of dlx function does not affect the expression of gad2. Distinct genetic pathways may regulate the gad1b and gad2 genes (as suggested by Szabó et al. (1996), Pinal et al. (1997) and Yanagawa et al. (1997)) although some upstream factors such as fgf3/8 and her6 were shown to be necessary for the proper expression of both gad1b and gad2 in the prethalamus (Miyake et al., 2005; Scholpp et al., 2009).
Regulatory interactions involving Dlx genes
In mammals, the Dlx proteins participate in auto- and cross-regulatory interactions by binding regulatory elements both within their own bigene cluster and in paralogous Dlx cluster (Zerucha et al., 2000; Zhou et al., 2004; Bond et al., 2009; Potter et al., 2009). Altered Dlx binding to such regulatory elements results in the loss of reporter gene expression in the forebrain of transgenic mice (Zerucha et al., 2000; Poitras et al., 2007). To explore this issue in the zebrafish, we tested the effect of the ascl1a and dlx MO injections on transgenic lines where the reporter gene (gfp) is under the control of zebrafish CREs from the dlx loci. The Tg(dlx1a/2aIG:GFP) contains the I12b element and is active in telencephalon and prethalamus, similarly to its orthologous mouse sequences (Ghanem et al., 2003; Ghanem et al., 2007; Poitras et al., 2007; MacDonald et al., 2010a). Furthermore this regulatory element contains a highly conserved ASCL1 binding site (E box) that that is essential for enhancer activity (Poitras et al., 2007). Expression of Tg(dlx1a/2aIG:GFP) is impaired in the diencephalon of ascl1a morphants with no severe impact in the telencephalon. The zebrafish URE2 CRE found upstream of dlx1a does not contain a conserved E-box (MacDonald, R.B. and Ekker, M., Unpublished observations) and may not require Ascl1a for its activity. Surprisingly, expression of the URE2-containing Tg(dlx1aURE2:GFP) transgene is impaired in both the telencephalon and diencephalon of ascl1a morphants. This large effect contrasts with changes in dlx1a/2a mRNA expression that were observed only in the diencephalon of morphants. Similarly, Tg(dlx5a/6a:GFP) expression was severely affected in ascl1a morphants without a comparable loss of dlx5a or dlx6a mRNA expression. These apparent discrepancies are likely attributable to the redundancy in gene regulatory controls and/or would suggest influence of factors, in addition to Ascl1a, involved in dlx regulation.
Lineage-specific changes GRNs in the forebrain of mice and zebrafish
The early patterning of the diencephalon seems to have been highly conserved amongst extant vertebrates. The roles of Shh, Fez, Otx, Wnts, Ascl1, Neurog1 and FGFs in prethalamic and thalamic regionalization are similar in the mouse, chick, Xenopus and zebrafish (Scholpp and Lumsden, 2010). Our results suggest that regulatory relationships between ascl1a, dlx1a/2a, dlx5a/6a and gad1b are present in the developing zebrafish prethalamus and hypothalamus, but the position of dlx5a and dlx6a in such a pathway remains ambiguous due to a smaller dlx5a/6a knock down effect on gad1b expression. The loss Dlx5 and Dlx6 expression in the forebrain of Dlx1−/−/Dlx2−/− mice and the regulatory relationships between them (Zerucha et al., 2000; Anderson et al., 1997a) support an implication of Dlx5/Dlx6 in GABAergic neuron development. It is possible that the Dlx5/Dlx6 genes of the last common ancestor of mice and zebrafish were involved in such a developmental pathway, but this role became less important in the lineage leading to zebrafish. If this were the case, dlx5a/6a may have been examples of nodes in a GRN that were more vulnerable to exclusion, perhaps due to regulatory redundancy with other dlx genes.
The current study shows the conservation of the diencephalic GRN regulating GABAergic interneuron development and the apparent divergence of the same process in the telencephalon. Our observations reinforce the neuromeric model of brain development and evolution (Rubenstein et al., 1994; Hauptmann and Gerster, 2000; Puelles and Rubenstein, 2003). This model postulates that the early vertebrate forebrain is composed of relatively discrete morphogenetic units termed neuromeres, between which cellular migration is limited, and within each neuromere developmental GRNs may undergo dynamic evolutionary changes. It is possible that during the approximately 300 million since the divergence of lineages leading to mice and zebrafish, the GRN underlying GABAergic interneuron specification in the telencephalic neuromere has changed. Our work suggests that divergence in the activity of GRNs responsible for forebrain neurodevelopment has occurred between mice and zebrafish vertebrate lineages.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Stephen Wilson for his generous gifts of the ascl1a5′UTR MO and ascl1a plasmid. The authors would also like to thank Dr. Charles Kimmel for dlx5a splice blocking MOs and fluorescent in situ reagents. This work was supported by CIHR Grant MOP14460 to ME and NIH Grants RO1DE13834 and PO1HD22486 to Charles Kimmel, University of Oregon.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.05.025.
References
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126 (17):3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distalless: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende ML, Weinberg ES. The expression pattern of two zebrafish achaetescute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev Biol. 1994;166:509–530. doi: 10.1006/dbio.1994.1334. [DOI] [PubMed] [Google Scholar]
- Allan DW, Thor S. Together at last: BHLH and LIM-HD Regulators cooperate to specify motor neurons. Neuron. 2003;38:675–677. doi: 10.1016/s0896-6273(03)00329-5. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JLR. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late-born striatal neurons. Neuron. 1997a;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JLR. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997b;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson S, Mione M, Yun K, Rubenstein JL. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9 (6):646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- Andrews GL, Yun K, Rubenstein JL, Mastick GS. Dlx transcription factors regulate differentiation of dopaminergic neurons of the ventral thalamus. Mol Cell Neurosci. 2003;23:107–120. doi: 10.1016/s1044-7431(03)00016-2. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfone A, Wang F, Hevner R, Anderson S, Cutforth T, Chen S, Meneses J, Pedersen R, Axel R, Rubenstein JL. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron. 1998;21:1273–1282. doi: 10.1016/s0896-6273(00)80647-9. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127:2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- Cau E, Wilson SW. Ash1a and neurogenin1 function downstream of floating head to regulate epiphysial neurogenesis. Development. 2003;130:2455–2466. doi: 10.1242/dev.00452. [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JLR. Dlx Transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc Natl Acad Sci USA. 2008;105:20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298 (5592):381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241 (2):229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Eisenstat DD, Liu J, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JLR. DLX-1, DLX-2 and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Ellies DL, Stock DW, Hatch G, Giroux G, Weiss KM, Ekker M. Relationship between the genomic organization and the overlapping embryonic expression patterns of the zebrafish Dlx genes. Genomics. 1997;45:580–590. doi: 10.1006/geno.1997.4978. [DOI] [PubMed] [Google Scholar]
- Fausett BV, Gumerson JD, Goldman D. The proneural basic helix–loop–helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Jarinova O, Amores A, Hatch G, Park BK, Rubenstein JLR, Ekker M. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long J, Hatch G, Rubenstein JL, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci. 2007;27:5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Joyner AL. Dynamic expression of the murine achaetescute homologue Mash-1 in the developing nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Regulatory gene expression patterns reveal transverse and longitudinal subdivisions of the embryonic zebrafish forebrain. Mech Dev. 2000;91 (1–2):105–118. doi: 10.1016/s0925-4773(99)00277-4. [DOI] [PubMed] [Google Scholar]
- Herzog W, Sonntag C, Walderich B, Odenthal J, Maischein HM, Hammerschmidt M. Genetic analysis of adenohypophysis formation in zebrafish. Mol Endocrinol. 2004;18:1185–1195. doi: 10.1210/me.2003-0376. [DOI] [PubMed] [Google Scholar]
- Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Stock DW. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc Natl Acad Sci USA. 2006;103:19390–19395. doi: 10.1073/pnas.0609575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Li X, McEvilly RJ, Rosenfeld MG, Lufkin T, Rubenstein JL. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development. 2008;135 (17):2905–2916. doi: 10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T, Yan YL. Double fluorescent in situ hybridization to zebrafish embryos. Trends Genet. 1996;12:387–389. doi: 10.1016/s0168-9525(96)90091-8. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Regulation of RNA Polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- Kaji T, Artinger KB. Dlxfib and dlx4b function in the development of Rohon–Beard sensory neurons and trigeminal placode in the zebrafish neurula. Dev Biol. 2004;276:523–540. doi: 10.1016/j.ydbio.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Arnosti DN. Information display by transcriptional enhancers. Development. 2003;130:6569–6575. doi: 10.1242/dev.00890. [DOI] [PubMed] [Google Scholar]
- Lauter G, Söll I, Hauptmann G. Molecular characterization of prosomeric and intraprosomeric subdivisions of the embryonic zebrafish diencephalon. J Comp Neurol. 2013;521 (5):1093–1118. doi: 10.1002/cne.23221. [DOI] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JLR. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Lo LC, Johnson JE, Wuenschell CW, Saito T, Anderson DJ. Mammalian achaete-scute homolog 1 is transiently expressed by spatially restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27 (12):3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009a;512 (4):556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex. 2009b;19:i96–i106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald RB, Debiais-Thibaud M, Talbot JC, Ekker M. The relationship between dlx and gad1 expression indicates highly conserved genetic pathways in the zebrafish forebrain. Dev Dyn. 2010a;239 (8):2298–2306. doi: 10.1002/dvdy.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald RB, Debiais-Thibaud M, Martin K, Tay BH, Venkatesh B, Ekker M. Functional conservation of a forebrain enhancer from the elephant shark (Callorhinchus milii) in zebrafish and mice. BMC Evol Biol. 2010b;10:157. doi: 10.1186/1471-2148-10-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Page DT, Merzlyak I, Kim C, Tecott LH, Janak PH, Rubenstein JL, Sur M. Reduced conditioned fear response in mice that lack Dlx1 and show subtype-specific loss of interneurons. J Neurodev Disord. 2009;1 (3):224–236. doi: 10.1007/s11689-009-9025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SC, Heinrich G, Sandell JH. Sequence and expression of glutamic acid decarboxylase isoforms in the developing zebrafish. J Comp Neurol. 1998;396:253–266. [PubMed] [Google Scholar]
- Mione M, Baldessari D, Deflorian G, Nappo G, Santoriello C. How neuronal migration contributes to the morphogenesis of the CNS: insights from the zebrafish. Dev Neurosci. 2008;30:65–81. doi: 10.1159/000109853. [DOI] [PubMed] [Google Scholar]
- Miyake A, Nakayama Y, Konishi M, Itoh N. Fgf19 regulated by Hh signaling is required for zebrafish forebrain development. Dev Biol. 2005;288 (1):259–275. doi: 10.1016/j.ydbio.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Morita T, Nitta H, Kiyama Y, Mori H, Mishina M. Differential expression of two zebrafish Emx homeoprotein mRNAs in the developing brain. Neurosci Lett. 1995;198:131–134. doi: 10.1016/0304-3940(95)11988-9. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF, Guo S. Early teleostean basal ganglia development visualized by zebrafish Dlx2a, Lhx6, Lhx7, Tbr2 (Eomesa), and GAD67 gene expression. J Comp Neurol. 2008;507:1245–1257. doi: 10.1002/cne.21604. [DOI] [PubMed] [Google Scholar]
- Panne D. The Enhanceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Pinal CS, Cortessis V, Tobin AJ. Multiple elements regulate GAD65 transcription. Dev Neurosci. 1997;19 (6):465–475. doi: 10.1159/000111244. [DOI] [PubMed] [Google Scholar]
- Peukert D, Weber S, Lumsden A, Scholpp S. Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol. 2011;9 (12):e1001218. doi: 10.1371/journal.pbio.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda HM, von der Hardt S, Herzog W, Kramer C, Schwarz H, Hammerschmidt M. The proneural gene ascl1a is required for endocrine differentiation and cell survival in the zebrafish adenohypophysis. Development. 2006;133:1079–1089. doi: 10.1242/dev.02296. [DOI] [PubMed] [Google Scholar]
- Poitras L, Ghanem N, Hatch G, Ekker M. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development. 2007;134:1755–1765. doi: 10.1242/dev.02845. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Bulfone A, Liu JK, Puelles L, Lo LC, Rubenstein JL. DLX-2, MASH-1, and MAP-2 expression and bromodeoxyuridine incorporation define molecularly distinct cell populations in the embryonic mouse forebrain. J Neurosci. 1994;14:6370–6383. doi: 10.1523/JNEUROSCI.14-11-06370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JL. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci. 2009;40:167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26 (9):469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JL. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9 (20):2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16 (9):1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr KB, Concha ML. Expression of nk2.1a during early development of the thyroid gland in zebrafish. Mech Dev. 2000;95:267–270. doi: 10.1016/s0925-4773(00)00345-2. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266 (5185):578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Foucher I, Staudt N, Peukert D, Lumsden A, Houart C. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development. 2007;134 (17):3167–3176. doi: 10.1242/dev.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S, Delogu A, Gilthorpe J, Peukert D, Schindler S, Lumsden A. Her6 regulates the neurogenetic gradient and neuronal identity in the thalamus. Proc Natl Acad Sci USA. 2009;106 (47):19895–19900. doi: 10.1073/pnas.0910894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 2010;33 (8):373–380. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon KS, Fritz A. Concerted Action of two Dlx paralogs in sensory placode formation. Development. 2002;129:3127–3136. doi: 10.1242/dev.129.13.3127. [DOI] [PubMed] [Google Scholar]
- Sperber SM, Saxena V, Hatch G, Ekker M. Zebrafish dlx2a contributes to hindbrain neural crest survival, is necessary for differentiation of sensory ganglia and functions with dlx1a in maturation of the arch cartilage elements. Dev Biol. 2008;314:59–70. doi: 10.1016/j.ydbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Anderson SA, Ekker M, Rubenstein JLR. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002a;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Puelles L, Ekker M, Rubenstein JL. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb Cortex. 2002b;12:75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- Szabó G, Katarova Z, Körtvély E, Greenspan RJ, Urbán Z. Structure and the promoter region of the mouse gene encoding the 67-kD form of glutamic acid decarboxylase. DNA Cell Biol. 1996;15 (12):1081–1091. doi: 10.1089/dna.1996.15.1081. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137 (15):2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High resolution whole-mount in situ hybridization. Zebrafish Science Monitor. 1998;5:8–9. [Google Scholar]
- Toyama R, Curtiss PE, Otani H, Kimura M, Dawid IB, Taira M. The LIM class homeobox gene lim5: implied role in CNS patterning in xenopus and zebrafish. Dev Biol. 1995;170:583–593. doi: 10.1006/dbio.1995.1238. [DOI] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Talbot JC, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wang B, Waclaw RR, Allen ZJ, Guillemot F, Campbell K. Ascl1 is a required downstream effector of Gsx gene function in the embryonic mouse telencephalon. Neural Dev. 2009;4(5) doi: 10.1186/1749-8104-4-5. http://dx.doi.org/10.1186/1749-8104-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dye CA, Sohal V, Long JE, Estrada RC, Roztocil T, Lufkin T, Deisseroth K, Baraban SC, Rubenstein JLR. Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J Neurosci. 2010;30:5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Long JE, Flandin P, Pla R, Waclaw RR, Campbell K, Rubenstein JL. Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants. J Comp Neurol. 2012;521 (7):1561–1584. doi: 10.1002/cne.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welten MC, de Haan SB, van den Boogert N, Noordermeer JN, Lamers GE, Spaink HP, Meijer AH, Verbeek FJ. ZebraFISH: fluorescent in situ hybridization protocol and three-dimensional imaging of gene expression patterns. Zebrafish. 2006;3:465–476. doi: 10.1089/zeb.2006.3.465. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. A Guide for the Laboratory use of Zebrafish (Danio Rerio) University of Oregon Press; Eugene, Oregon: 2000. [Google Scholar]
- Wullimann MF, Mueller T. Expression of Zash-1a in the postembryonic zebrafish brain allows comparison to mouse Mash1 domains. Brain Res Gene Expr Patterns. 2002;1:187–192. doi: 10.1016/s1567-133x(02)00016-9. [DOI] [PubMed] [Google Scholar]
- Yanagawa Y, Kobayashi T, Kamei T, Ishii K, Nishijima M, Takaku A, Tamura S. Structure and alternative promoters of the mouse glutamic acid decarboxylase 67 gene. Biochem J. 1997;326 (Pt2):573–578. doi: 10.1042/bj3260573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang H, Hu G, Wang H, Abate-Shen C, Shen MM. An early phase of embryonic Dlx5 expression defines the rostral boundary of the neural plate. J Neurosci. 1998;18:8322–8330. doi: 10.1523/JNEUROSCI.18-20-08322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JLR, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between DLx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QP, Le TN, Qiu X, Spencer V, de Melo J, Du G, Plews M, Fonseca M, Sun JM, Davie JR, Eisenstat C. Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res. 2004;32:884–892. doi: 10.1093/nar/gkh233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.