Abstract
The functions of gene regulatory networks that control embryonic cell diversification occur on a background of constitutively active molecular machinery necessary for the elaboration of genetic interactions. The essential roles of subsets of such “housekeeping” genes in the regulation of specific aspects of development have become increasingly clear. Pre-mRNA processing is essential for production of functional transcripts by, for example, excision of introns. We have cloned the zebrafish toastb460 locus and found that it encodes splicing factor 3b, subunit 1 (sf3b1). The sf3b1b460 mutation causes aberrant splicing of sf3b1 resulting in functional and predicted nonfunctional transcripts and a 90% reduction in full-length Sf3b1 protein. The sf3b1b460 mutation was isolated in a mutagenesis screen based on the absence of neural crest-derived melanophores. Further analysis revealed specific earlier defects in neural crest development, whereas the early development of other ectodermal populations appears unaffected. The expression of essential transcriptional regulators of neural crest development are severely disrupted in sf3b1b460 mutants, due in part to defects in pre-mRNA processing of a subset of these factors, leading to defects in neural crest sublineage specification, survival and migration. Misexpression of a subset of these factors rescues aspects of neural crest development in mutant embryos. Our results indicate that although sf3b1 is a ubiquitously essential gene, the degree to which it is required exhibits tissue-type specificity during early embryogenesis. Further, the developmental defects caused by the sf3b1b460 mutation provide insights into genetic interactions among members of the gene regulatory network controlling neural crest development.
Keywords: neural crest, pre-mRNA, splicing, zebrafish
Introduction
The removal of introns from pre-mRNA is essential for the ultimate generation of functional proteins. The precise excision of introns is catalyzed by a dynamic protein complex called the spliceosome (Burge et al., 1999). Major elements of the spliceosome include the U1, U2, U5 and U4/6 small nuclear ribonucleoprote in particles (snRNPs) that contain small nuclear RNAs, common Sm proteins and proteins unique to each snRNP (Kambach et al., 1999). The U2 snRNP exists in an inactive 12S form and an active 17S form generated by interaction of the 12S snRNP with the splicing factors Sf3b and Sf3a that are themselves comprised of several subunits (Brosi et al., 1993a, 1993b). Sf3b1 is the largest subunit comprising the Sf3b complex (25 exons encoding a 1315 amino acid protein in zebrafish) and is required for prespliceosome assembly and splicing catalysis and is thus essential for pre-mRNA processing (Gozani et al., 1996; Gozani et al., 1998).Sf3b1 is also a component of the minor U12-dependent spliceosome (Will et al., 1999). sf3b1 is ubiquitously expressed in mouse during early embryogenesis (Isono et al., 2001) and sf3b1-/- mouse embryos arrest at the 16–32 cell stage (Isono et al., 2005) demonstrating its essential function. The ubiquitous expression and function of sf3b1 in eukaryotic cells defines sf3b1 as one of a number of so-called “housekeeping” genes. However, more nuanced functional requirements for a number of housekeeping genes have been documented recently (for example, see Coutinho et al., 2004; Nambiar and Henion, 2004; Nambiar et al., 2007).
The possibility that sf3b1 may also serve more graded functions in different tissues at different times has been suggested by several recent reports. For example, Isono and colleagues, employing sf3b1+/− mice, demonstrate the requirement for sf3b1 in Polycomb group-mediated repression of Hox genes that regulate skeletal growth and patterning (Isono et al., 2005). A prominent differential role in neurogenesis by stem cells in neurogenic compared to non-neurogenic regions of the adult mammalian brain for RNA processing genes, including sf3b1, has been suggested (Lim et al., 2005). Also of interest is the observation that sf3b1 levels can determine the alternative splicing pattern derived from the BCL-x gene, potentially regulating pro- or antiapoptotic responses of cells to external stimuli (Massiello et al., 2006). Together, these studies suggest that in certain contexts sf3b1 may differentially regulate developmentally relevant processes.
In a screen for mutations affecting neural crest development (Henion et al., 1996), we isolated the toastb460 (tst) mutant based on the absence of neural crest-derived pigment cells but presence of the non-neural crest-derived pigmented retinal epithelium, suggesting possible cell type-specific functions of the tst mutant locus during development. Later in development, tst mutant embryos die at approximately 40 hours postfertilization (hpf), presumably due to prominent dorsal CNS cell death and the lack of circulating blood.
Here we present the molecular identification and characterization of the tstb460 locus and a detailed phenotypic analysis of the consequences of this mutation. Specifically, we identify tstb460 as a hypomorphic mutant allele of sf3b1 that results in a dramatic reduction in wild-type transcripts and protein. As a result of a T>G point mutation in the 5’ splice site of intron 4 of sf3b1, pre-mRNA splicing errors in tst mutant sf3b1 result in the production of 3 aberrant, presumably non-functional transcripts as well as a minority proportion of wild-type transcripts. The primary consequences of the tstb460 mutation in the ectoderm are selective defects in neural crest development during early embryogenesis. These defects initially present as defects in the levels of expression of key transcriptional regulators of early neural crest development. In specific cases, reduced expression levels result from aberrant pre-mRNA splicing producing severely truncated transcripts. Partial restoration of a subset of these transcriptional regulators by misexpression results in differential degrees of neural crest phenotype rescue. Our results demonstrate differential sensitivities to the levels of expression of the essential RNA processing gene sf3b1 in different subdivisions of the ectoderm and among genes that control the early development of the neural crest. These results indicate that the status of specific elements of essential cellular machinery is an additional regulatory component in the control of neural crest cell diversification during embryogenesis.
Results
Visible phenotype of live tstb460 mutant embryos
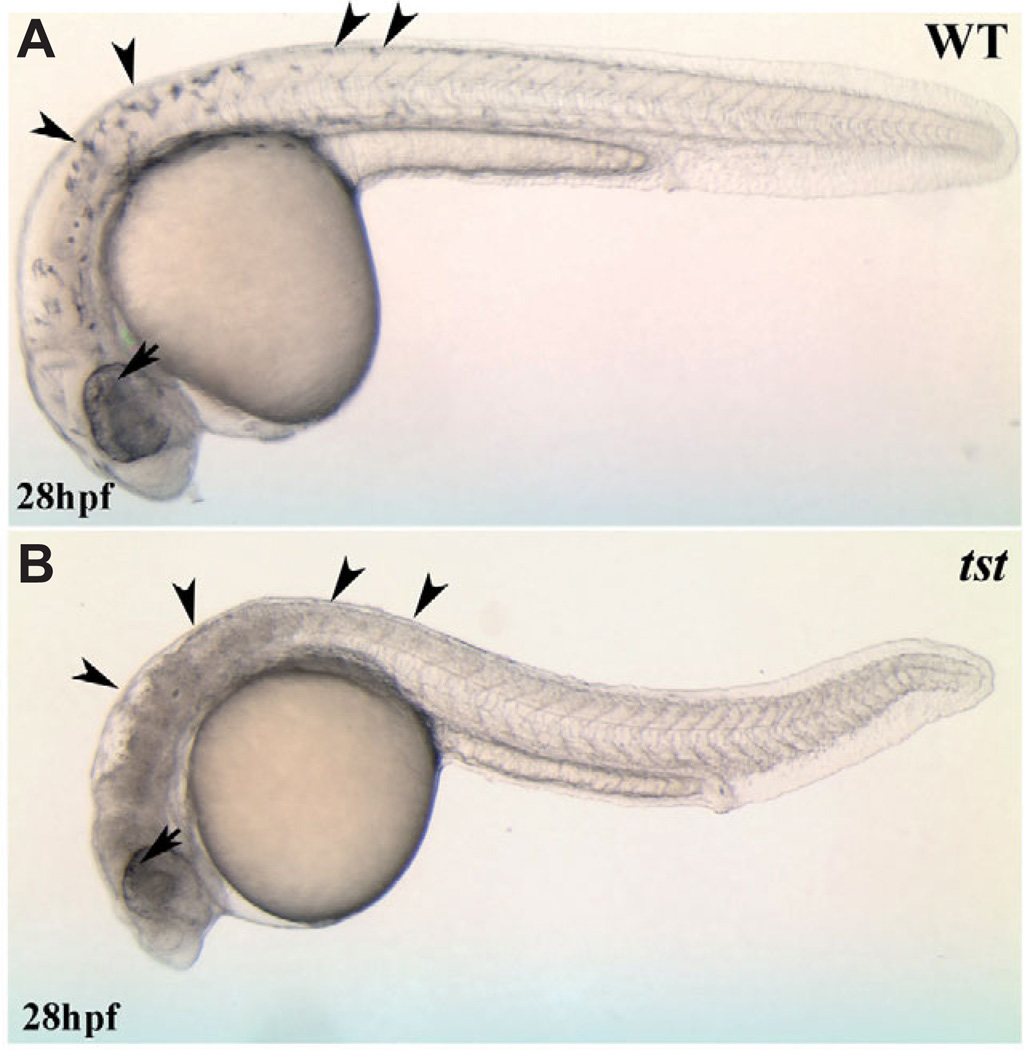
The tstb460 mutant was identified in an EN U-based mutagenesis screen for mutations that affect neural crest development (Henion et al., 1996). Differentiated neural crest-derived melanophores are completely absent in tstb460 embryos (Fig. 1). In contrast, melanized pigmented retinal epithelial cells are present suggesting that, among pigment cells, the mutant phenotype is neural crest-specific. The tstb460 mutation is recessive and tstb460 mutants die by approximately 40 hpf most likely due to progressive dorsal CNS cell death and the absence of circulation.
Fig. 1. Visible live phenotype of tstb460 mutants.
Neural crest-derived melanophores; see arrowheads in (A) are absent in tstb460 mutants; see arrowheads in (B). In contrast, melanized pigmented retinal epithelial cells are present in the mutants (arrow).
The tstb460 locus encodes sf3b1 and is a hypomorphic sf3b1 mutant allele
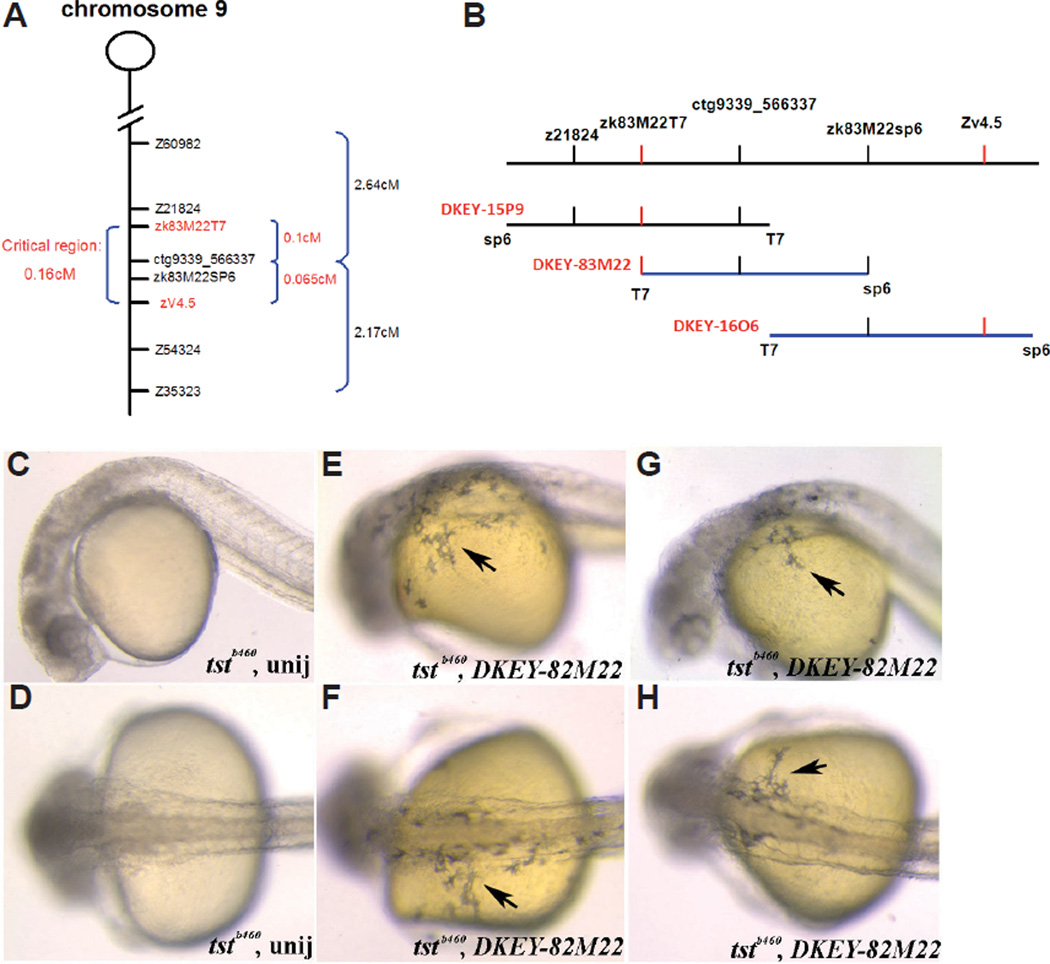
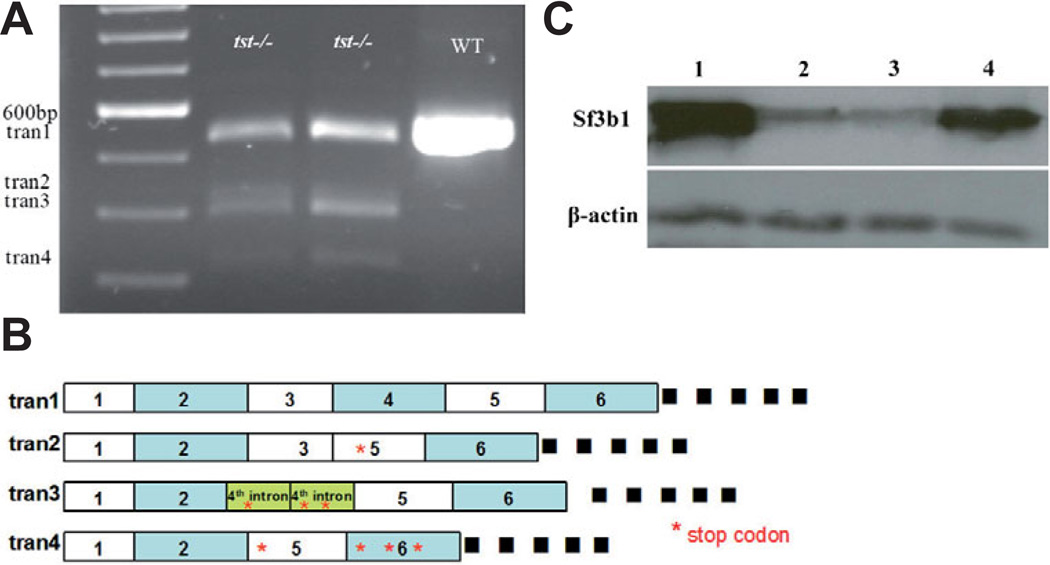
We mapped the tstb460 mutation to chromosome 9 using standard linkage analysis. We then generated a mapping panel comprised of 2018 meioses to conduct recombination frequency analysis using several closely linked SSLP (z21824, Z54324, and Z35323) and SSCP (zk83M22T7, ctg9339_566337, ZK83M22SP6 and zv4.5) markers (see Materials and Methods). The result of this analysis defined a 0.164cM critical region containing the tstb460 locus (Fig. 2A). Three BACs (DKEY-83M22, DKEY-1606, DKEY-15P9) were found to map to this region (Fig. 2B; http://www.sanger.ac.uk/cgi-bin/ Projects/D_rerio/mapsearch). Because tstb460 mutant embryos completely lack easily visualized neural crest-derived melanophores, we performed phenotype rescue experiments by misexpressing the DKEY-83M22 BAC in embryos from tstb460 hetrozygote crosses. All injected embryos were subsequently genotyped with tightly linked SSLP markers (z21824, z54324). We found that misexpression of BAC DKEY-83M22 resulted in robust melanophore phenotype rescue in tstb460 mutants at efficiencies ranging from 57%- 100% per experiment (Fig. 2 C–H and not shown). BAC DKEY-83M22 has been completely sequenced by the Sanger Center (http://www.ebi.ac.uk/embl.; http://www.ncbi.nlm.nih.gov). Based on this sequence and the regions of all three BACs that overlap, we identified several genes within this region using GENESCAN (http://genes.mit.edu/GENSCAN.html). We designed primers to amplify these candidate genes from tstb460mutant and wildtype embryos for RT-PCR and sequence analysis. We found that for one of these candidate genes, sf3b1, there were 4 different transcripts in tstb460 mutants as compared to AB* wild-type embryos that express a single transcript (Fig. 3A) revealed by amplification using RT-PCR and the Sf3b1aa (46) F and Sf3b1aa (579) R primers. Of the transcripts expressed by tstb460 embryos (Fig. 3B), sequence comparisons revealed that transcript 1 is the wild-type sf3b1 mRNA. Transcript 2 is an aberrant transcript in which the 4th exon is missing. The 3rd transcript is missing the 3rd and 4th exons with the 4th intron aberrantly included between the 2nd and 5th exons. Transcript 4 is missing part of exon 2nd and all of exons 3 and 4. The splicing errors that result in the three aberrant transcripts all introduce premature stop codons that predict highly truncated proteins that lack the essential TP and RWD repeats that are believed to be essential functional domains for phosphorylation and protein-protein interactions, respectively (Seghezzi et al., 1998;Gozani et al., 1998), and thus non-functional or severely compromised products.
Fig. 2. Fine mapping of the tst locus and BAC-mediated tstb460 mutant phenotype rescue.
(A) Critical region containing the tstb460 locus between closely linked SSLPs (z21824, z54324, z60982 and z35323) and SSCPs (zk83M22T7, ctg9339_566337, zk83M22SP6 and zv4.5) markers (B) BACs (DKEY-83M22, DKEY-16O6, and DKEY-15P9) that map to the critical region containing tstb460 locus (C–H) Injection of DKEY-83M22 partially rescues the tstb460 mutant melanophore phenotype (C,D) Uninjected tstb460 homozygous mutant embryos; (E–H) DKEY-83M22 injected tstb460 homozygous mutant embryos, showing partial rescue of the melanophore phenotype (arrows).
Fig. 3. Molecular consequences of sf3b1 mutations (A,B).
Variant transcripts of sf3b1 in tstb460 mutants. (A) RT-PCR products were amplified between exon1 and exon6 of sf3b1 with 20–24 hpf AB wildtype and tstb460 homozygous mutant total RNA as template using Sf3b1aa (46)F and Sf3b1aa (579)R primers. The single AB wildtype RT-PCR product was 534 bp (tran1). tstb460 homozygous mutants contained 4 different transcripts; tran1(534 bp), and variant transcripts tran2(412 bp), tran3(390 bp), and tran4(312 bp). (B) Diagram of variant transcripts in tstb460 homozygous mutants. (C) Western blot analysis of Sf3b1 protein levels in wildtype, tstb460 mutant and hi3394a mutant embryos at 24 hpf. Lane 1: wildtype embryos; Lane 2: tstb460 homozygous mutants; Lane 3: hi3394a homozygous mutants; Lane 4: hsp>sf3b1 injected tstb460 mutants with melanophore phenotype rescue.
We employed a Sf3b1 monoclonal antibody that recognizes a central portion of the Sf3b1 protein (kindly provided by Dr. K. Isono, Chiba University; Horie et al., 2003) and densitometer measurements of chemiluminescence detection in Western blots to obtain a quantitative determination of wildtype Sf3b1 protein levels in tstb460 mutant embryos compared to wildtype siblings. Actin protein expression was used as a control. We found that presumptive full length functional Sf3b1 protein levels in pooled tstb460 mutant embryos was dramatically reduced to 9.6 +/− 4.2% of wildtype levels at 24hpf (Fig. 3C; n=10 different protein pools, each containing 20–30 embryos per pool). In addition, we found that the Sf3b1 antibody employed did not recognize any products derived from the aberrant 5’ truncated transcripts produced in tstb460 mutants, as predicted by the antigen sequence used to produce the antibody (Horie et al., 2003). These results suggest that the tstb460 locus is likely to be sf3b1 and that the allele is hypomorphic.
To further assess whether the tstb460 mutant locus is sf3b1, we constructed the full-length wildtype cDNA for misexpression and phenotype rescue analysis (GenBank accession number pending). Because of the large size of the predicted full-length cDNA (4393 bp), we constructed the full-length cDNA from two fragments, one derived from EST clone fb99f09 and the other a RT-PCR fragment of the 5’ region of the transcript generated from RT-PCR amplification from 18–24hpf wildtype total RNA(see Methods). The presumptive full-length wildtype sf3b1 cDNA was sequenced to confirm frame and sequence. The full-length cDNA was then ligated to the heatshock vector pCSHSP (Halloran et al., 2000) to drive expression.
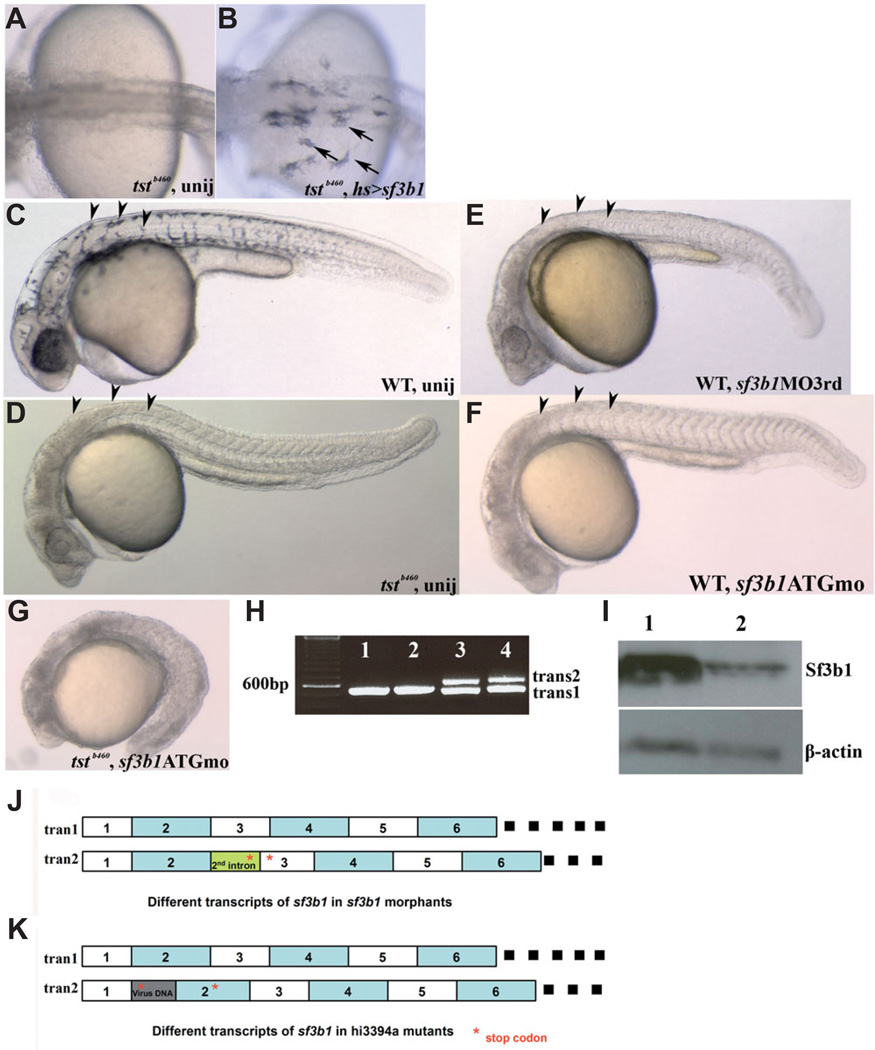
The hsp>sf3b1 expression construct was injected into a single blastomere of 1–2 cell stage embryos derived from tstb460heterozygous parents and injected embryos were heat-shocked at 6–7hpf and 11–12hpf and raised to 27–36hpf. In a subset of experiments, embryos were heat-shocked an additional time at 23–24hpf. No phenotypic differences were observed between those embryos heat-shocked 2 or 3 times. Injected embryos were examined for visible melanophores at 27- 36hpf and genotyped with the closely-linked SSLP markers z21824 and z54324. We found that misexpression of wildtype sf3b1 by injection of hsp>sf3b1 resulted in significant melanophore rescue in 80% of injected homozygous tstb460 mutant embryos (Fig. 4 A,B; n=92 mutant embryos). Together with the efficient rescue exhibited by misexpressing the sf3b1 -containing BACs and the severe reductions of wildtype Sf3b1 protein and full-length transcript levels in mutant embryos, these results provide further evidence that the tstb460 locus is likely to be sf3b1.
Fig. 4. sf3b1 gain- and loss-of-function analyses.
(A,B) The tstb460 phenotype is partially rescued by sf3b1 misexpression. (A) Melanophores are absent in uninjected tstb460 mutant embryos. (B) tstb460 homozygous mutant embryos were injected with hs>sf3b1 and heat-shocked 2 or 3 times (6 hpf, 10 or 11 hpf, 22 or 23 hpf), 1h/each time. Several melanophores present in the dorsal trunk region and over the yolk region in injected tstb460 mutant embryos (arrows). (C–F) Phenocopy of tstb460 mutant embryos using morpholino knockdown analysis. (C) Lateral view of a live uninjected wildtype embryo at 30 hpf. (D) Lateral view of a tstb460 homozygous mutant at 30 hpf. (E) Lateral view of a live sf3b1 MO3rd morphant showing the absence of melanophores (arrowheads) and CNS (brain) cell death at 30 hpf. (F)Lateral view of a live sf3b1 MOATG morphant showing phenocopy of the tstb460 mutant at 30 hpf. (G) The phenotype was more severe in the sf3b1 MOATG injected tstb460 mutant as compared with uninjected tstb460 mutant, suggesting that the tstb460 allele is hypomorphic. (H) Variant transcripts in sf3b1 MO3rd morphants. Lane1 and lane2: RT-PCR product from uninjected wildtype total RNA. Lane 3 and lane 4: RT-PCR product from sf3b1 MO3rd morphant total RNA. (I) Western blot analysis of Sf3b1 protein levels in uninjected wildtype embryos and sf3b1 MOATG morphants at 12 hpf Lane 1: uninjected wildtype embryos; Lane 2: sf3b1 MOATG morphants. (J) Schematic diagram of different sf3b1 transcripts in sf3b1 MO3rd morphants. (K) Schematic diagram of different sf3b1 transcripts in hi3394a mutants.
To further test whether the tstb460 locus corresponds to sf3b1, we designed two antisense oligonucleotide morpholinos (MO) to knockdown Sf3b1 in wildtype embryos when injected at the 1–2 cell stage in wildtype embryos. We designed and tested sf3b1 ATG MO, a start-site blocking MO and sf3b13rdMO, a MO targeted to disrupt preMRNA splicing in a manner approximating the splicing errors found in sf3b1 transcripts in tstb460 mutant embryos. We confirmed by Western analysis that sf3b1 ATG MO effectively knocked down Sf3b1 protein levels in injected wildtype embryos (Fig. 4I). Sequence analysis revealed that the sf3b13rd MO causes a frequent failure in the splicing of the 3’ splice site of the 2nd intron of sf3b1 (Fig. 4 H,J) resulting in a premature stop codon in the 2nd intron. The injected sf3b13rd MO phenocopied the melanophore and gross morphology phenotypes of tstb460mutants in injected wildtype embryos at 88–99% efficiency (Fig. 4E). The sf-3b1 ATG MO also resulted in phenocopy in 87–98% of groups of injected embryos (Fig. 4F). Injection of tstb460 mutants with sf3b1ATG MO resulted in the arrest embryonic development during early somitogenesis (Fig. 4G), reminiscent of Sf3b1 null mice and consistent with the suggestion that tstb460 is a hypomorphic allele. The phenocopy of tstb460 mutants by two verified MOs targeting sf3b1 provides further evidence that tstb460 corresponds to sf3b1.
As mentioned previously, we isolated the full-length zebrafish sf3b1 cDNA (Genbank accession number pending). Sequence analysis of the entire sf3b1 coding sequence of wildtype and tstb460 mutants failed to detect any nucleotide mutations in the tstb460 coding sequence. Therefore, because tstb460 mutants produce aberrant transcripts in the 5’ end of the gene upstream of the 5th exon, we sequenced genomic sequences of wildtype and tstb460 mutants upstream of the 6th exon of sf3b1. Our analysis revealed a T>G point mutation in the 5’ splice site of the 4th intron in the tstb460 sf3b1 sequence (Fig. 5). The U1 snRNP recognizes the 5’ splice site to form a complex that commits the premRNA to spliceosome assembly (reviewed by Nilsen, 1997). The recognition of the 5’ splice site involves base-pairing between the conserved nucleotide at the 5’ end of U1 and 5’ intronic splice site. The specific nucleotide mutation at the 5’ splice site of the 4th intron in sf3b1 from tstb460 mutant embryos is consistent with the production of the aberrant transcripts observed based on the current knowledge of the mechanics of pre-mRNA processing. The observed nucleotide mutation in tstb460 mutant sf3b1 combined with the evidence presented above strongly suggest that tstb460is a sf3b1 mutant allele.
Fig. 5. The nucleotide mutation at the 5’ splice site of the 4th intron of sf3b1 genomic DNA identified in tstb460 mutants.
In tstb460, T is changed to G.
Lastly, as our analysis was proceeding, we learned that a viral insertion mutation in sf3b1 was identified in a mutant screen (Amsterdam et al., 2004). We obtained this mutant line, hi3394a, and determined via sequence analysis that the viral insertion resulted in the production of two transcripts, a presumptive wild-type transcript and a second aberrant transcript in which the viral insertion between the 1st and 2nd exons results in the creation of a premature stop codon in the 2nd exon (Fig. 4K). We also found that full length Sf3b1 protein levels in hi3394a mutants was reduced to 13.5+/−6.2% of wildtype levels at 24hpf (see Fig. 3C). We determined that hi3394a live phenotype is very similar but slightly less severe than that of tstb460 mutants particularly in the case of melanophores which are entirely absent in tstb46 mutants whereas a small number of melanophores usually develop in hi3394a mutants (Amsterdam et al., 2004). Critically, complementation analysis demonstrated that tstb460 and hi3394a fail to complement. Taken together with all of the results described, we conclude that the tstb460 locus corresponds to sf3b1 and that it is a severe but hypomorphic allele. Thus, we designate tstb460as sf3b1b460.
Expression of zebrafish sf3b1
Analysis of the zebrafish genome indicates that the sf3b1 gene is approximately 17 kb in size and that it is a single-copy gene. The full-length sf3b1 is 4393 bp derived from 25 exons. The sf3b1 cDNA predicts a protein comprised of 1315 amino acids. The amino acid sequence of zebrafish Sf3b1 is highly conserved when compared to the human, mouse and Xenopus Sf3b1 homologs (calculated 92% identity) and the carboxy-terminal three-fourths of Sf3b1 shows even higher conservation (97.5%; see Supplemental Fig. 1). In addition, the amino acid sequences of C. elegans and S. pombe Sf3b1 also show a high degree of conservation compared to vertebrates (Isono et al., 2001).
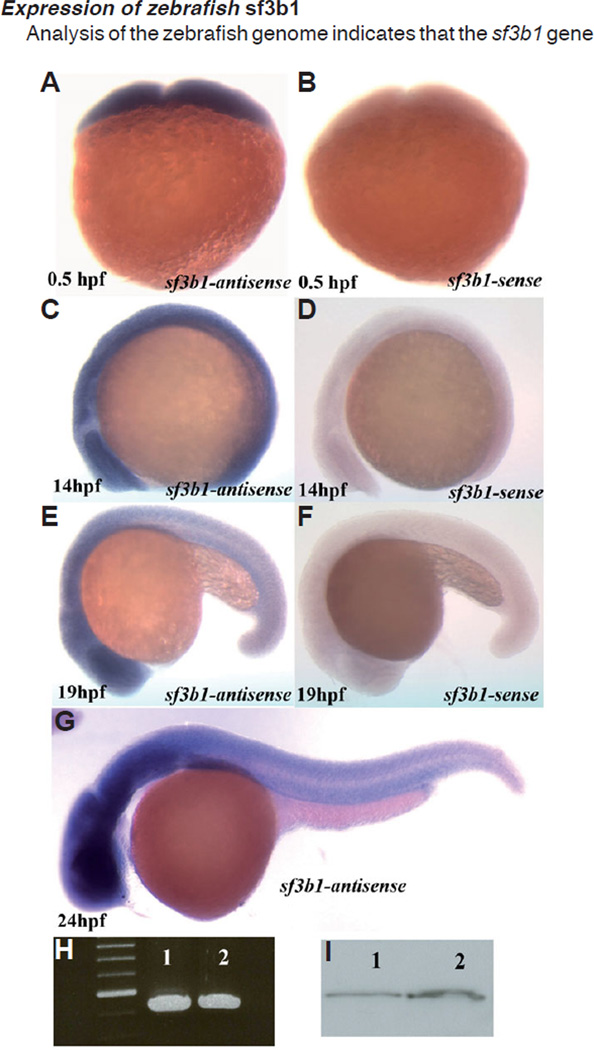
To examine the expression pattern of zebrafish sf3b1 we synthesized a 1.6kb antisense RNA probe complementary to the 2934–4393 bp 3’ region of sf3b1 and performed wholemount in situ hybridization (WISH) on embryos from 0–48 hpf (Fig. 6 A–G and not shown). We found using WISH that sf3b1 is maternally expressed (Fig. 6 A , B) and confirmed this observation using RT-PCR and Western analyses (Fig. 6 H,I). Zygotically, in general, we found that sf3b1 mRNA is expressed ubiquitously at all stages examined (Fig. 6 C–G). Expression levels appear enhanced in the brain, dorsal neural tube and ventral trunk regions of later-stage embryos (Fig. 6 E–G), although this observation may be the result of differences in the cellular composition and architecture of these regions at these stages compared to other regions of the embryo. No qualitative differences in the pattern or level of sf3b1 mRNA expression between wildtype and sf3b1b460 mutant embryos were observed using the 3’ riboprobe employed, as would be expected based on the 5’ locations of the splicing errors in sf3b1b460 mutants.
Fig. 6. Whole-mount in situ hybridization with sf3b1 antisense probe and sense probes in wildtype embryos.
(A,C) sf3b1 is ubiquitously expressed at 0.5 hpf and 14 hpf. (E,G) The expression of sf3b1 appears elevated in the brain, neural tube and ventral trunk region at 19 hpf and 24 hpf (lateral view, anterior to left). (B,D,F) sf3b1 sense controls. (H) RT-PCR products using total RNA from 0.5hpf (lane1) and 24hpf (lane2) wildtype embryos as templates and Sf3b1aa(46)F and Sf3b1aa(579)R as primers. (I) Western blot analysis shows Sf3b1 protein present maternally. Lane1: protein from 24 hpf wildtype embryos as control. Lane2: protein from 1 hpf wildtype embryos.
The development of neural crest-derived cell subpopulations fails in sf3b1 b460mutant embryos
As stated previously, a prominent visible phenotype ofs-f3b1b460mutant embryos is the complete absence of differentiated neural crest-derived melanophores (Fig. 1). Therefore, we examined the development of melanogenic precursors in sf3b1b460mu-tant embryos. To assess melanogenic sublineage development, we examined the expression of mitfa, a gene necessary for melanoblasts specification and an early marker of melanophore precursors (Opdecamp et al., 1997; Lister et al., 1999) and dct, whose expression is diagnostic of later stage melanoblasts (Kelsh et al., 2000). The initial neural crest expression of mitfa in sf3b1b460mutant embryos is delayed by several hours and then significantly reduced in expression compared to stage matched wildtype embryos (Fig. 7 A , B). In contrast, the timing and pattern of PRE expression is qualitatively normal. The expression of neural crest dct is significantly reduced with only occasional dct+ cells in the trunk of a minority of mutant embryos (Fig. 7 C , D). As is the case for mitfa, PRE dct expression in sf3b1b460 mutants was not significantly different than wildtype embryos. Thus, we conclude that the specification and early development of the melanophore sublineage is severely disrupted in sf3b1b460 mutants resulting in the absence of differentiated melanophores.
Fig. 7. Neural crest progenitor and cranial ganglia phenotypes of sf3b1b460 mutants.
There is reduced melanoblast expression of mitfa (A,B) and nearly absent melanoblast expression of dct (C,D) in sf3b1b460 mutants. (E–J) Pharyngeal arches are dramatically reduced in sf3b1b460 mutant embryos. (E–H) Lateral view of whole-mount in situ embryos with dlx2 antisense RNA probe, arch progenitors denoted by arrows. (I,J) Dorsal view of flat-mounted in situ embryos with dhand antisense RNA probe, pharyngeal arches 1 and 2 are decreased and 3, 4 and 5 are absent in sf3b1b460 mutants. (K–N) the cranial ganglia are also affected in sf3b1b460 mutants. (K,L) Whole-mount antibody staining with zn-12. Arrows in (K,L) show the trigeminal ganglia. (M,N) Whole-mount antibody staining with anti-acetylated tubulin antibody. Arrows in (M,N) show the decreased and disorganized trigeminal ganglia in sf3b1b460 mutants compared to wildtype siblings. (O,P) Crestin expressing cells are present in sf3b1b460 mutant embryos. However, these cells are reduced in numbers, particularly in the cranial region and fail to migrate normally in the sf3b1b460 mutant (P) compared to wildtype embryos (O).
We also assessed the development of neural crest-derived craniofacial cartilage progenitors by comparing the expression of dlx2 (Akimenko et al., 1994) and dhand (Yelon et al., 2000) in the developing pharyngeal arches in wildtype and sf3b1b460mutant embryos. We found that neural crest expression of both genes in the pharyngeal arches was dramatically reduced in mutant embryos, being absent in posterior arches and strongly reduced in the first and second arches (Fig. 7 E–J). These results indicate that the development of the neural crest component of the pharyngeal arches is severely compromised in sf3b1b460 mutants.
Another subpopulation of neural crest-derived cells that is specified and differentiates relatively early during embryogenesis is a subset of cranial ganglion neurons (Schilling and Kimmel, 1994). Other neurons that comprise the cranial ganglia are derived from the ectodermal placodes. In sf3b1b460mutants, the numbers of neurons and organization of trigeminal (Fig. 7 K–N) and posterior lateral line ganglia (not shown) are reduced and disorganized, respectively, compared to wildtype siblings. Although we cannot be sure that this phenotype results from a selective disruption of cranial neural crest development, as opposed to a disruption of placode development, the pattern of pan-neural crest crestin expression in the head at 22hpf (Fig. 7 O , P) is consistent with the possibility of a neural crest defect in the development of the cranial ganglia phenotype of sf3b1b460 mutant embryos.
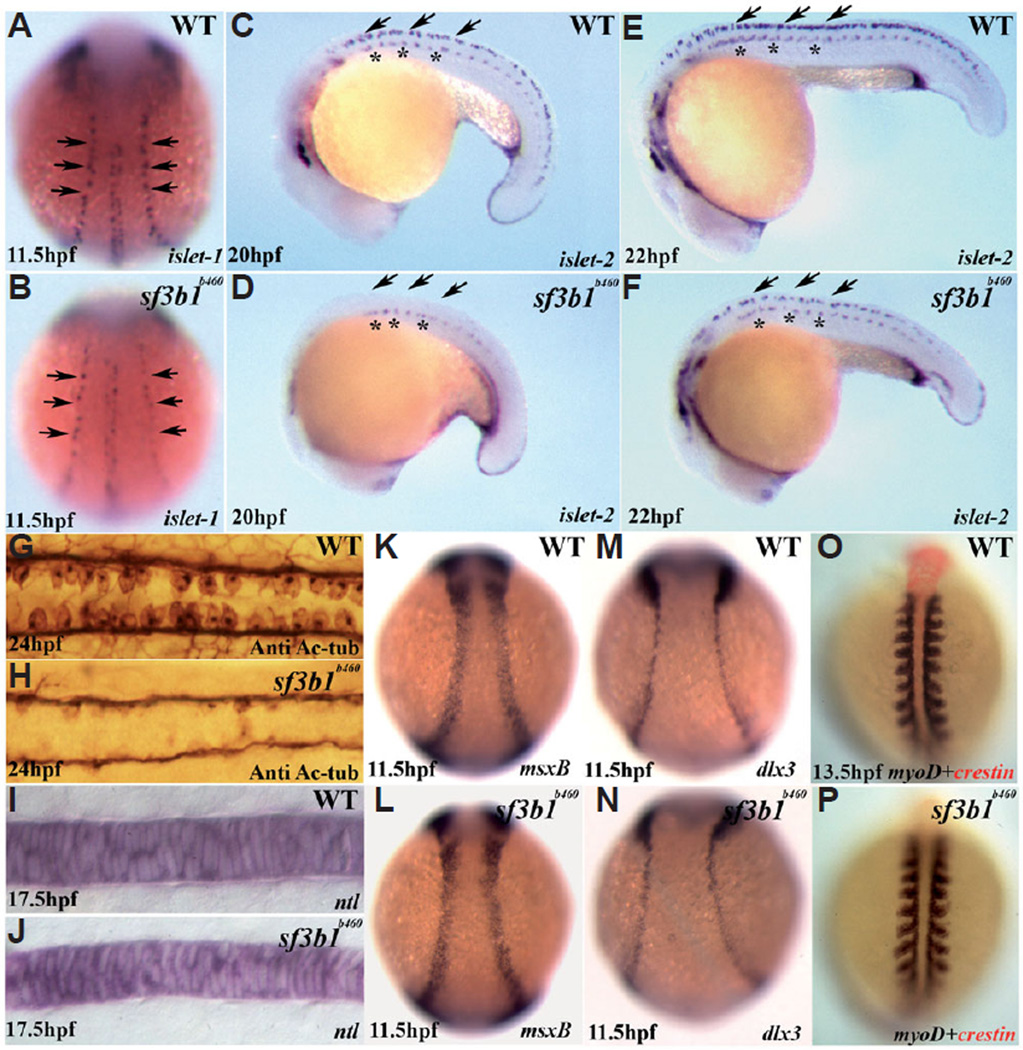
Within the developing neural plate border in zebrafish embryos, the neural crest and Rohon-Beard sensory neurons are thought to comprise an equivalence group (Artinger et al., 1999; Cornell and Eisen, 2000). At early somitogenesis stages, differentiated Rohon-Beard sensory neurons in the neural plate border can be identified based on the expression of huC as well as islet1 (Artinger et al., 1999; Cornell and Eisen, 2000). Examination of Rohon-Beard sensory neuron development at 11.5, 20 and 22hpf in sf3b1b460mutant and wildtype embryos revealed that the number of neurons was significantly reduced in mutant embryos compared to wildtype siblings (Fig. 8 A–F). Rohon-Beard sensory neuron numbers in sf3b1b460mutant embryos do not recover as revealed by immunostaining with acetylated tubulin and zn-12 antibodies at later developmental stages (Fig. 8 G , H). Thus, along with severe defects in the development of neural crest-derived cell subpopulations, sf3b1b460mutant embryos display abnormal development of another neural plate border cell subpopulation, Rohon-Beard sensory neurons. However, even given the early defects in neural crest and Rohon-Beard neuron development, the timing and pattern of the establishment of the neural plate border generally is unaffected in sf3b1b460 mutant embryos as assessed by the expression of msxb and dlx3 (Fig. 8 K–N). Further, the early development of somites (Fig. 8 O.P) and notochord (Fig. 8 I , J) were also normal in mutants. Lastly, development of primary motorneurons was also comparatively normal in mutant embryos (Fig. 8 C–F and not shown). Thus, early ectodermal developmental defects in sf3b1b460 mutant embryos are by and large confined to the neural crest/Rohon Beard neurons of the neural plate border.
Fig. 8. Early development of ectodermal and mesodermal derivatives in sf3b1b460 mutants.
(A,B) Whole-mount in situ hybridization with islet-1 antisense RNA probes shows that the number of Rohon-Beard precursor cells (arrows) is decreased in sf3b1b460 mutants (dorsal view, anterior to top). (C–F) Whole-mount in situ hybridization with islet-2 antisense RNA probe shows deficiency in the development of Rohon-Beard sensory neurons (arrows), whereas the development of primary motor neurons is comparatively less affected in mutant embryos (asterisks). (G,H) Dorsal view of spinal cords labeled with anti-acetylated tubulin (Anti Ac-tub), anterior to left, showing that the Rohon-Beard sensory neuron phenotype persists at later stages in sf3b1b460 mutants. K–N: Induction of the neural plate border occurs normally in sf3b1b460 mutants. Whole-mount in situ hybridization with msxB (K,L) and dlx3 (M,N). (I,J;O,P) The derivatives from paraxial mesoderm and chordamesoderm are not affected in sf3b1b460 mutants. (I,J) Lateral view of whole-mount in situ with ntl, anterior to left. (O,P) Dorsal view of double in situ with crestin (red) and myoD (blue), anterior to top.
A number of transcription factors, including foxd3, tfap2a, snai1b, sox10 and sox9b, have been shown to play important roles in the fate-specification, survival, migration and/or differentiation of distinct and overlapping neural crest sublineages in multiple species (Gammill and Bronner-Fraser, 2003). Therefore, because of the miscues exhibited in NC development in sf3b1b460mutant embryos, we examined the neural crest expression of these genes in sf3b1b460mutants. We also examined the expression of the neural crest marker gene crestin (Luo et al., 2001).
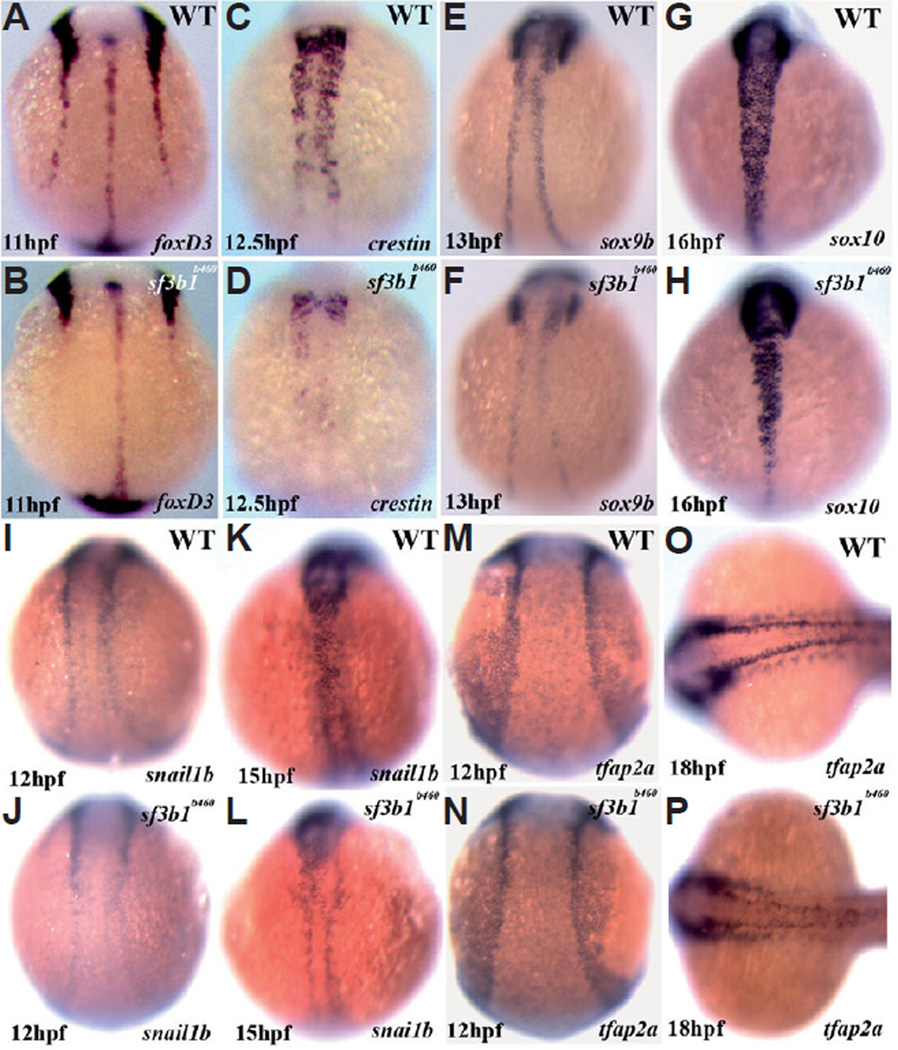
At 12.5hpf, the expression of crestin is nearly absent in the trunk of sf3b1b460 mutant embryos, whereas expression is present but reduced in cranial neural crest (Fig. 9 C , D). Later in development, at 22hpf, crestin is expressed by trunk neural crest in mutant embryos although by noticeably fewer cells and, other than in the most rostral somitic regions, is confined to the dorsal neural tube suggesting defective trunk neural crest migration. At the same stage, and qualitatively in contrast to earlier embryonic stages, cranial neural crest expression of crestin is nearly absent in sf3b1b460 mutant embryos (Fig. 7 O , P).
Fig. 9. Significant abnormalities in trunk neural crest expression of critical transcriptional regulators as well as crestin at early developmental stages in sf3b1b460 mutants, but grossly normal expression in the cranial neural crest.
Whole-mount in situ hybridization with foxd3 (A,B), crestin (C,D), sox9b (E,F), sox10 (G,H), snai1b (I–L) and tfap2a (M–P) antisense RNA probes. All embryos are dorsal view, anterior to top with the exception of (O,P) which is anterior to left.
At 11hpf, the expression of foxd3 is essentially absent in trunk neural crest in sf3b1b460 mutant embryos, whereas expression by cranial neural crest is very similar to that observed in wildtype embryos (Fig. 9 A , B). The expression of sox9b at 13hpf is also very strongly reduced in trunk neural crest and is reduced to a lesser extent in cranial neural crest in sf3b1 b460mutant embryos (Fig. 9 E , F), an abnormal neural crest expression pattern similar to that of foxd3. Strong trunk neural crest as well as cranial neural crest expression of snai1b at 12 hpf and 15 hpf and sox10 at 16hpf is evident in wildtype embryos (Fig. 9 G,I,K). The expression of both genes in sf3b1 b460mutant embryos was markedly reduced in trunk neural crest and was similar but slightly reduced in cranial neural crest (Fig. 9 H,J,L). At 12hpf, neural crest expression of tfap2a was indistinguishable between sf3b1b460mutant and wildtype embryos (Fig. 9 M,N). In contrast, a noticeable reduction in neural crest expression of tfap2a, particularly in the trunk, is evident in sf3b1b460mutant embryos at 18hpf (Fig. 9 O.P). The abnormal neural crest expression patterns of foxd3, sox9b, snai1b and sox10 observed at earlier developmental stages in sf3b1b460mutant embryos are maintained at 18hpf (not shown). The fact that neural crest tfap2a expression is initially normal indicates that neural crest induction occurs normally in sf3b1b460 mutants. Thus, the neural crest expression of multiple other transcriptional regulators of neural crest development are disrupted in sf3b1b460mutant embryos in an asynchronous manner and share a comparatively more severe trunk neural crest as opposed to cranial neural crest phenotype. As these transcription factors have been implicated as regulators of neural crest sublineage specification, survival and/or migration, these results suggest that the severe deficiencies in later neural crest development in sf3b1b460 mutants is, at least in part, a consequence of the disregulated expression of early transcriptional regulators of neural crest development.
sf3b1 is required cell autonomously for neural crest development
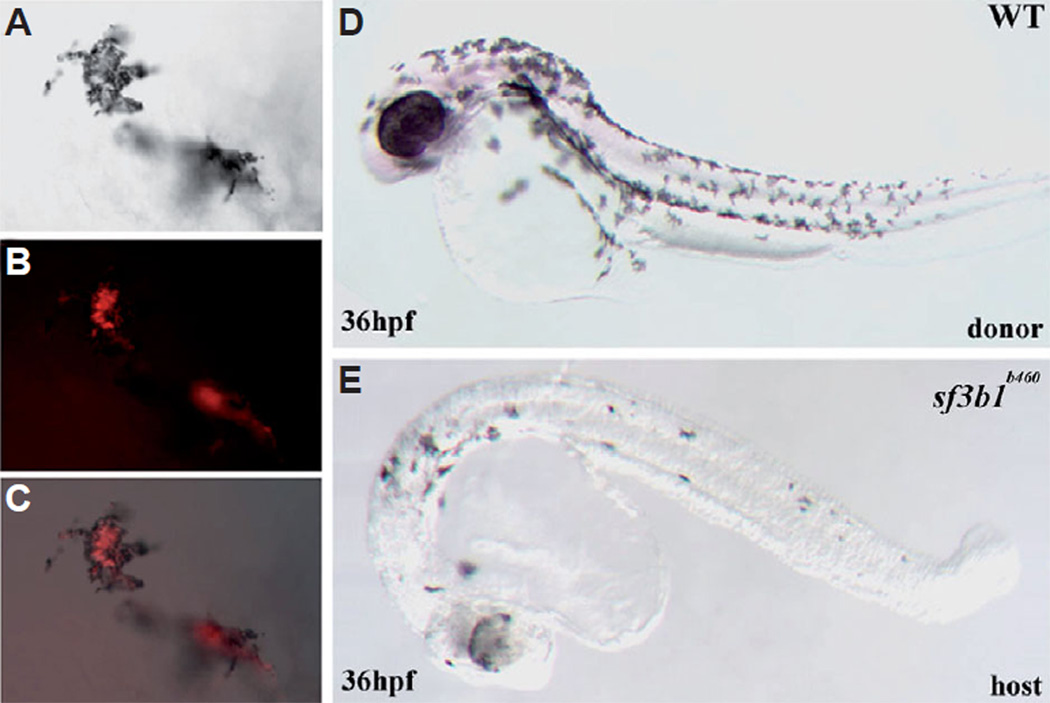
To determine whether or not sf3b1 function is required cell autonomously for neural crest development, we analyzed genetically mosaic embryos generated by cell transplantation between mutant and wildtype embryos. Because sf3b1b460mutant embryos completely lack neural crest-derived me-lanophores, we scored mosaic embryos for the presence or absence of donor-derived melanophores. Embryos generated from crosses of sf3b1b460 heterozygous adults were separated into two groups, half of which were injected at the 1–4 cell stage with lyseinated rhodamine dextran (LRD) to label all cells. Cells from LRD-labeled embryos were transplanted to unlabeled siblings at the late blastula stage (Ho and Kane, 1990) and according to the zebrafish embryo fate map (Kimmel et al., 1990). Host and donor embryos were grown to 24–36hpf when the visible phenotype of absent melanophores in sf3b1b460 mutant embryos is readily apparent. All embryos were also genotyped with linked markers as described previously. We found that wildtype donor neural crest cells readily generate melanophores in sf3b1b460 mutant hosts (Fig. 10; n=23) while we never observed mosaic embryos in which sf3b1b460mutant neural crest cells generated melanophores in wildtype hosts (not shown; n=29). We conclude that sf3b1 is required cell autonomously for neural crest development.
Fig. 10. Genetic mosaics of sf3b1b406 mutants and wildtype embryos.
(A) Melanophores derived from wildtype donor cells on the yolk of a sf3b1b460 mutant host (DIC image). (B) Fluorescence image of the same (donor-derived) cells. (C) (A,B) merged. (D) A wildtype donor. (E) An sf3b1b460 mutant host with wildtype donor-derived melanophores.
Increased neural crest apoptosis and migratory defects in sf3b1b460 embryos
Because the transcription factors foxd3, tfap2a, snai1b, sox9b and sox10 have been shown to be required to differing degrees for crucial aspects of neural crest development including the specification of neural crest sublineage fates, neural crest cell survival and neural crest migration and the expression of all of these genes is disrupted in sf3b1b460 mutants, we further investigated general aspects of neural crest development in mutant embryos. As described earlier, the specification of neural crest sublineage fates is disrupted in sf3b1b460 mutants, at least in the case of the melanophore and craniofacial cartilage sublineages.
Whereas the neural crest expression of crestin is nearly absent in the trunk and reduced in the head at 12.5hpf (Fig. 9 C,D) in sf3b1b460mutants, we found that crestin is expressed by trunk neural crest, albeit in fewer cells than wildtype, at 22hpf (Fig. 7 O,P). Cranial neural crest expression of crestin, in contrast to earlier stages, is nearly absent in mutants at this stage. In addition to a reduction in the number of crestin expressing trunk neural crest cells in mutants, those that are present are located predominantly along the dorsal neural tube indicating a severe disruption in trunk neural crest migration in sf3b1b460 mutants compared to wildtype embryos.
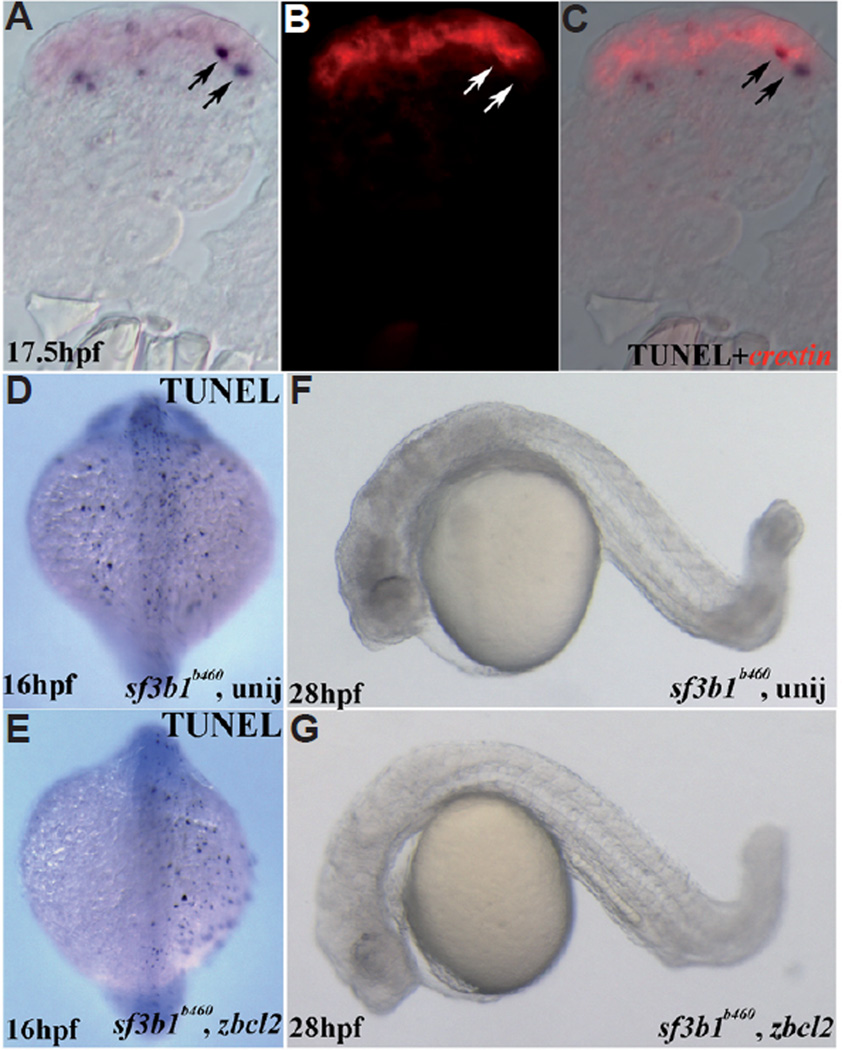
We then examined cell death during development in sf3b1b460mutant embryos using TU NEL. At early stages of trunk neural crest development (12–14hpf) the numbers of dying cells in mutants is moderately increased compared to wildtype embryos, particularly in the anterior CNS (not shown). Subsequently, numerous TUNEL-positive cells along the dorsal neural tube of mutant embryos, but not wildtype embryos, are observed. We employed WISH with the neural crest marker crestin of 17.5hpf embryos previously processed by TUNEL and examined cryosectioned samples of the trunk region. We found that many of the TUNEL-positive cells co-labeled with crestin indicating increased neural crest cell death in sf3b1b460 mutants compared to wildtype siblings (Fig. 11 A–C). At subsequent stages of development, in whole mount, TUNEL processed embryos, we observed a progressive wave, from rostral to caudal, of substantial dorsal CNS cell death (not shown) that presumably leads to the demise of sf3b1b460 mutant embryos by 40 hpf.
Fig. 11. Analysis of cell death in sf3b1b460 mutants.
(A–C) Double labeling with TUNEL (NBT/BCIP) and crestin whole-mount in situ hybridization (fast red). Transverse section is from the trunk region of a sf3b1b460 mutant, dorsal is up. (A) DIC image/TUNEL; (B) fluorescence image/crestin of the same section; (C) (A,B) merged. (D) Dorsal view of uninjected sf3b1b460 mutant at 16 hpf with TUNEL staining. (E) Dorsal view of zbcl2 mRNA injected sf3b1b460 mutant at 16 hpf with TUNEL staining. Reduced TUNEL staining is evident on the left side. (F) Lateral view of live uninjected sf3b1b460 mutant at 28 hpf. (G) Lateral view of live zbcl2 injected sf3b1b460 mutant at 28 hpf, showing less CNS cell death as compared with uninjected sf3b1b460 mutants but no melanophore rescue.
The elevated levels of neural crest cell death in sf3b1b460 mutant embryos raised the possibility that the abnormal development of the neural crest in sf3b1b460 mutants results from neural crest cell apoptosis as a consequence, directly or indirectly, of abrogated Sf3b1 function. To test this possibility, we first investigated whether misexpression of bcl2 mRNA in sf3b1b460 mutant embryos significantly attenuates cell death as detected by TUNEL. We found that injection of 1–4 cell stage embryos resulted in a dramatic reduction in TUNEL-positive cells in sf3b1b460 mutant embryos (Fig. 11 D,E) including cells along and in the immediate vicinity of the neural tube previously shown to be neural crest cells (see above). While the extent of CNS cell death observed in sf3b1b460 mutants, confirmed by genotyping, was dramatically reduced (Fig. 11 D,E), we did not observe any rescue of neural crest-derived tissues such as craniofacial cartilage anlagen or in neural crest-derived cells such as melanophores when embryos were observed between 28 (Fig. 11 F,G) and 40hpf (not shown). Thus, although we cannot conclude that apoptosis plays no role in the development of the neural crest phenotype of sf3b1b460 mutant embryos, these results strongly suggest that much of the abnormal neural crest development observed in mutant embryos results from miscues in the function of other key regulatory components of neural crest development that are dependent upon normal Sf3b1 function.
Aberrant pre-mRNA processing of a subset of transcriptional regulators of neural crest development
Because Sf3b1 is essential for pre-mRNA processing, and wild-type Sf3b1 protein levels in sf3b1b460mutant embryos are reduced by 90% due to aberrant pre-mRNA processing of sf3b1 itself (Fig. 3C; see above), we examined the pre-mRNA processing of the essential transcriptional regulators of neural crest development whose neural crest expression is abnormal in sf3b1b460mutant embryos (Fig. 9; see above). To do so, we isolated RNA from 18–24hpf sf3b1b460 mutant embryos and performed RT-PCR using gene-specific primers. Samples were then sequenced and compared to published sequence data. We found that only normal (identical to wildtype in size and sequence) transcripts for foxd3, tfap2a and sox10 were present in sf3b1b460 mutants suggesting that the relatively low levels of predicted functional Sf3b1 protein present in mutant embryos is sufficient for the correct pre-mRNA processing of these gene products. In contrast, we found that the pre-mRNA processing of snai1b and sox9b transcripts was severely disrupted in sf3b1b460 mutants (Fig. 12).
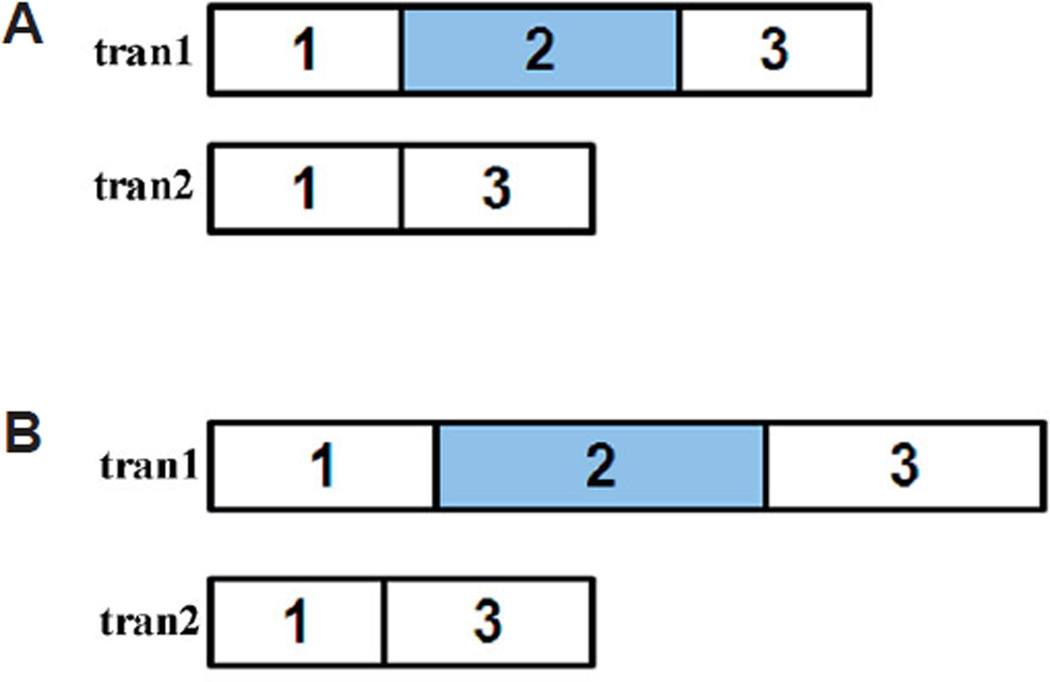
Fig. 12. Abnormal pre-mRNA processing in sf3b1b460 mutant embryos.
(A) diagram of the exons of snai1b. tran1: shows the normal snai1b transcript. tran2: shows the abnormally spliced transcript in sf3b1b460 mutant embryos, in which exon2 is deleted. Both transcripts were detected in mutant embryos. (B) diagram of the exons of sox9b. There are two types of transcripts, wildtype transcript (tran1) and the abnormally spliced transcript (tran2) whose partial exon1, whole exon2 and partial exon3 were deleted in sf3b1b460 mutant embryos. Only the aberrant transcript was detected in mutant embryos.
In sf3b1b460 mutant embryos, we were unable to detect normal sox9b transcripts (GenBank Accession No. NM131644 and AF277097). Instead the only sox9b transcript detected was devoid of the 138–1379 bp coding region and thus not expected to encode any protein product (Fig. 12). While we cannot rule out the presence of small amounts of normal sox9b transcripts in sf3b1b460mutants present at levels below the limit of detection of methods employed, these results suggest that sf3b1b460 mutants lack sox9b functional activity. The ability to detect sox9b mRNA by WISH in sf3b1b460 mutants is likely due to the fact that the antisense probe used consists of 541 bp of the 3’-UTR present in the abnormal transcripts in addition to 404 bp corresponding to missing coding sequence. Thus, some specific hybridization may be expected in sf3b1b460 mutants albeit at reduced efficiency.
We detected two species of snai1b transcripts in sf3b1b460 mutants (Fig. 12). One species corresponded to the wildtype transcript (GenBank Accession. No. NM130989) whereas the other species contained a 513 bp deletion (106–618 bp) of coding sequence corresponding to a region encoding three of the five zinc finger motifs found in snai1b (Thisse et al., 1995) and as such predicting that if a protein product is produced from the aberrant transcript that its function would be severely compromised. The antisense riboprobe used for WISH detection of snai1b is approximately 400 bp (Thisse et al., 1995) corresponding to 5’-UTR sequence and coding sequence up to the first zinc finger. Thus, the riboprobe used would be predicted to recognize both normal and abnormal transcripts in sf3b1b460 mutants. Therefore, the reduced levels of neural crest snai1b expression observed in sf3b1b460 mutants using WISH is likely to underestimate actual functional Snailb protein levels. In any case, the aberrant pre-mRNA processing of snai1b in sf3b1b460 mutant embryos strongly suggests that Snai1b function during neural crest development in sf3b1b460 mutants is severely disrupted.
Differential sf3b1b460 mutant neural crest phenotype rescue activity of transcriptional regulators of neural crest development
Because the neural crest expression of the transcription factors foxd3, tfap2a, sox10, sox9b and snai1b are severely disrupted in sf3b1b460 mutants and because each of these transcription factors have been shown to be differentially required for aspects of early neural crest development in multiple species including zebrafish, we asked whether partial restoration of the function of each of these genes could rescue the neural crest phenotype of sf3b1b460 mutants when misexpressed. To do so, we individually misexpressed each transcription factor via mRNA or inducible cDNA injected into 1–4 cell stage sf3b1b460 mutant embryos. We allowed injected embryos to develop until 36hpf and examined each for the presence or absence of differentiated melanophores, which are entirely absent in uninjected mutant embryos, followed by genotyping using linked markers.
We found that misexpression of hs>sox10 resulted in efficient (41% of injected sf3b1b460 mutant embryos) melanophore rescue in sf3b1b460 mutants (Fig. 13). In the vast majority of individual injected mutant embryos, multiple melanophores were observed in the head and trunk as well as over the yolk. Similarly, we ob-served a similar melanophore rescue when tfap2a mRNA was misexpressed in mutant embryos (Fig. 13), although qualitatively fewer melanophores were present in each rescued mutant embryos compared to hs>sox10 injected mutants and rescue was slightly less efficient (34%). As was the case with hs>sox10 injected mutants, melanophores in tfap2a injected mutants were observed in the head, trunk and over the yolk. In contrast to the melanophore phenotype rescue activities of sox10 and tfap2a in sf3b1b460 mutant embryos, we did not observe any differentiated melanophores in mutant embryos in which foxd3, snai1b or sox9b mRNAs were misexpressed.
Fig. 13. Misexpression of genes that regulate neural crest development results in melanophore rescue in sf3b1b460 mutant embryos.
(A) Lateral view of live uninjected sf3b1b460 mutant at 28 hpf (anterior to left). (B) Dorsal view of live uninjected sf3b1b460 mutant at 28hpf (anterior to left). (C) hs>sox10 injected sf3b1b460 mutant embryo at 28 hpf (dorsal view), showing partial melanophore rescue (arrows). (D) tfap2a mRNA injected sf3b1b460 mutant embryo at 28hpf (dorsal view), showing partial melanophore rescue (arrows)
Discussion
In a screen for ENU-induced mutations that result in defects in neural crest development in zebrafish (Henion et al., 1996), we isolated the tstb460 mutant based on the absence of neural crest-derived melanophores. tstb460 was found to be a recessive, embryonic lethal mutation with embryos dying at approximately 40hpf presumably due to prominent CNS cell death and/or the lack of circulation.
Multiple lines of evidence indicate that the pre-mRNA splicing factor sf3b1 is the locus mutant in tstb460. We found that; tstb460maps to a critical region of chromosome 9 containing sf3b1; a BAC containing sf3b1 displayed phenotype rescue activity when misexpressed in mutant embryos; tstb460 sf3b1 contains a T>G point mutation in the 5’ splice site of the 4th intron and that tstb460 embryos produce 3 aberrant, likely non-functional transcripts resulting from defective pre-mRNA processing that are consistent with this mutation; Sf3b1 protein levels are reduced by approximately 90% in mutant embryos; knockdown of Sf3b1 in wildtype embryos using two different morpholinos phenocopies tstb460 mutants; morpholinos can increase phenotype severity in mutant embryos suggesting that tstb460 is hypomorphic; misexpression of sf3b1 in tstb460 mutant embryos results in melanophore phenotype rescue; tstb460 failed to complement a previously identified (Amsterdam et al., 2004) viral insertion mutant in sf3b1. Taken together, we conclude that tstb460is a strong hypomorphic sf3b1 mutant thus designated sf3b1b460.
A prominent phenotype of sf3b1b460 mutants is the absence of neural crest-derived melanophores that is preceded by the abnormal neural crest expression of genes required for the development of and/or diagnostic of melanoblasts. This phenotype is in turn preceded by abnormal neural crest expression of transcription factors known to be required for early aspects of neural crest development (Gammill and Bronner-Fraser, 2003). These include foxd3, tfap2a, snai1b, sox9b and sox10. A subset of these, sox9b and snai1b, undergo defective pre-mRNA processing while a different subset, sox10 and tfap2a, display neural crest rescue activity when misexpressed in sf3b1b460 mutants.
In zebrafish, mutants for foxd3, tfap2a, sox9b and sox10 have been isolated and characterized (Dutton et al., 2001; Knight et al., 2003; Barrallo-Gimeno et al., 2004; Yan et al., 2005; Carney et al., 2006; Montero-Balaguer et al., 2006; Stewart et al., 2006; Arduini et al,,2009). Mutants for each of these transcription factors display both discreet and overlapping neural crest development phenotypes. The neural crest phenotypes of sf3b1b460mutant embryos with respect to the expression patterns of these transcription factors, errors in pre-mRNA processing of a subset of these and the melanogenesis rescue activity of a different subset of these transcription factors provides multiple insights into the functional interactions among these genes in regulating early neural crest development and the role of sf3b1 in this context.
Specifically, neural crest expression of sox10 is reduced, particularly in the trunk, of sf3b1b460 mutants and misexpression of sox10 rescues melanogenesis in mutant embryos. Further, while sox10 pre-mRNA appears to be processed normally in mutant embryos, melanoblast specification is severely disrupted ultimately resulting in the complete absence of differentiated melanophores in sf3b1b460 mutants. Because sox10 is known to be globally necessary for melanogenesis, including in zebrafish (Kelsh and Eisen, 2000; Dutton et al., 2001), it appears that the reduction in neural crest sox10 expression levels in sf3b1b460 mutants is sufficient to prevent melanogenesis. But, since sox10 pre-mRNA processing is normal in mutants, the role of sf3b1 in the regulation of sox10 expression is indirect. Importantly, the neural crest expression of sox9b, foxd3 and tfap2a are also reduced in sf3b1b460 mutants. Neural crest expression of sox10 has been shown to be reduced in both sox9b (Yan et al., 2005) and foxd3 (Montero-Balaguer et al., 2006; Stewart et al., 2006) mutants and entirely abolished in foxd3;tfap2a double mutants (Arduini et al., 2009). These results indicate that sox9b and foxd3 regulate neural crest sox10 expression while combined foxd3 and tfap2a function also synergistically regulates sox10. However, foxd3 and tfap2a transcripts are normally generated in sf3b1b460 mutants, albeit at reduced levels or fail to be maintained, respectively. Importantly though, sox9b pre-mRNA processing is severely disrupted in sf3b1b460 mutants leading to the absence of functional transcripts and sox9b has been shown to regulate the neural crest expression of both foxd3 and sox10 (and snai1b, see below; Yan et al., 2005). In turn, maintenance of neural crest tfap2a expression is dependent upon foxd3 (Stewart et al., 2006). Taken together, these results suggest the possibility that failure in the sf3b1-dependent pre-mRNA processing of sox9b in sf3b1b460 mutants results in downregulation of neural crest foxd3 and sox10 expression, and that the resulting reduction in foxd3 expression levels results in decreased neural crest expression of tfap2a. Thus, sf3b1-dependent pre-mRNA processing of sox9b is necessary for the establishment and/or maintenance of the transcription factor network that ultimately promotes sox10-dependent melanogenesis.
We also found that misexpression of tfap2a rescued melanogenesis in sf3b1b460 mutants. While neural crest tfap2a expression is initially normal, expression is not maintained at wildtype levels. Analysis of transcripts indicates that pre-mRNA processing of tfap2a is normal in sf3b1b460 mutants. In zebrafish tfap2a mutants (Knight et al., 2003; Barrallo-Gimeno et al., 2004), melanogenesis is significantly delayed, although appears to recover by 48–72hpf. Lethality in sf3b1b460 mutants (40hpf) corresponds to the same timeframe in which melanogenesis is delayed in tfap2a mutants. Further, in addition to the delay in overt melanophore differentiation (pigmentation), the expression of both mitfa, and to a greater extent the levels and pattern of the expression of c-kit are also delayed in tfap2a mutants. In the latter case, c-kit is known to be required for melanoblast/melanophore migration and survival in zebrafish (Parichy et al., 1999), suggesting that the reduction in tfap2a expression in sf3b1b460 mutants could contribute to a delay in mitfa-dependent melanoblast specification as well as c-kit-dependent melanophore migration/survival thus contributing to the sf3b1b460 mutant melanophore phenotype that can be partially rescued upon tfap2a misexpression.
Although misexpression of snai1b in sf3b1b460 mutants in and of itself failed to rescue the melanogenesis phenotype of mutant embryos, the fact that snai1b pre-mRNA processing is severely disrupted in mutants resulting in a significant reduction in the levels of neural crest snai1b expression may contribute to the overall neural crest phenotype of sf3b1b460 mutants. Studies in multiple species indicate a role for snai1b in epithelial-mesenchymal transitions (EMT), including that which generates the premigratory neural crest population (Gammill and Bronner-Fraser, 2003; Nelms and Labosky, 2010). It is plausible that reduced neural crest snai1b expression in sf3b1b460 mutants, due at least in part to aberrant pre-mRNA processing, could disrupt neural crest EMT and contribute to the increased levels of neural crest apoptosis observed in mutant embryos. A consequence could be a reduction in the overall neural crest population that could contribute to the derivative phenotypes observed in sf3b1b460 mutants.
It should also be noted that while our discussion concerning neural crest derivatives has focused on melanphores with respect to the sf3b1b460 mutant phenotype, the disruptions in the neural crest expression of the transcription factors analyzed as a consequence, directly or indirectly of disruption of Sf3b1 function, may also be involved in the development of the other neural crest derivative phenotypes including craniofacial progenitors and cranial ganglion neurons. In addition, there is a strong possibility that additional genes that regulate neural crest development, whose expression was not analyzed, may also be disrupted directly or indirectly in sf3b1b460 mutants and contribute to the expression defects in the transcription factors analyzed and/or neural crest development.
Our results provide new insights into sf3b1 function during development. For example, we have shown that the 90% reduction in Sf3b1 levels in sf3b1b460 mutants results in selective defects in development rather than global early developmental arrest observed in mouse null mutants and when sf3b1 is further knocked down in sf3b1b460 mutants. These results are similar to previous reports (Isono et al., 2005; Lim et al., 2006; Massiello et al., 2006) and to recent reports indicating that inhibition of Sf3b1 function can lead to selective effects, in this case anti-proliferative effects of a Sf3b1 inhibitor due to selective aberrant pre-mRNA processing of cell cycle genes as opposed to a global disruption of pre-mRNA processing (Corrionero et al., 2011; Folco et al., 2011). Our results are also similar to recent findings indicating selective defects in early neural crest development when the functions of genes thought to be generalized regulators of transcriptional elongation are disrupted (Nguyen et al., 2010; White et al., 2011).
Lastly, we found that the development of neural crest is differentially affected in sf3b1b460 mutants, potentially revealing new functional insights. We previously elucidated a framework gene regulatory regulating neural crest cell diversification in which foxd3 and tfap2a are globally and synergistically required for neural crest cell diversification and this function is due in large part to their regulation of the SoxE family genes sox9a, sox9b and sox10 (Arduini et al., 2009). Our results here have revealed new levels of interactions within this network and with additional genes (e.g. snai1b). Taken together, our results and those of others suggest that alterations in the function of the ubiquitous and essential pre-mRNA processing gene sf3b1 may have implications for specific aspects of development as well as clinically relevant conditions in humans.
Materials and Methods
Zebrafish strains and maintenance
Zebrafish embryos and adults were raised and maintained in the Ohio State University zebrafish facility as described (Westerfield, 1993), and staged according to Kimmel et al., 1995. The toastb460 (tst) mutation was induced in the wild-type *AB genetic background by ENU (ethyl-N-nitrosourea) mutagenesis (Henion et al., 1996). tst WIK lines were obtained by outcrossing with wild-type WIK background fish for genetic mapping and genotyping. Heterozygous mating pairs (*AB background or WIK background) were used to produce embryos (wild-type and mutant) for all experiments. The hi3394a (sf3b1) mutant line was kindly provided by the Hopkins’s lab from MIT (Amsterdam et al., 2004).
Whole-mount in situ hybridization and immunohistochemistry
Whole-mount single and double in situ hybridization were performed as described (Jowett and Lettice, 1994; Jowett, 2001) with minor modifications. Embryos were fixed overnight in 4% PFA (paraformaldehyde) in 1X PBS (phosphate-buffered saline solution) at 4°C. Embryos older than 24 hpf were treated with PTU (1-phenyl-2-thiourea) to prevent melanin synthesis. Prehybridization and hybridization were carried out at 65°C. The following digoxigenin (DIG) labeled antisense RNA probes were used in single in situ hybridization: crestin (Luoet al., 2001), foxd3 (Odenthal and Nusslein-Volhard, 1998), snai1b (snail-2, Thisse et al., 1995), sox10 (Dutton et al., 2001), tfap2a (Knight et al., 2003), sox9b (Chiang et al, 2001), dlx2, dlx3 (Akimenko et al, 1994), islet-1 (Inoue et al., 1994), islet-2 (Appel et al., 1995), huC (Park et al, 2000), dct and mitfa (Lister et al., 1999), nkx2.5 (Lee et al., 1996), and myoD (Weinberg et al, 1996). Whole-mount double in situ hybridization was performed by using DIG labeled myoD antisense RNA probe and fluorescein labeled crestin antisense RNA probe as described (Jowett, 2001) with minor modifications.
sf3b1 antisense and sense RNA probes were synthesized corresponding to 3’-sf3b1 around 1.6 kb (2934–4393 bp) from a PCRIIsf3b1c construct linearized with NotI or BamHI separately and transcribed with SP6 or T7 separately. Other sf3b1 antisense and sense RNA probes were made corresponding to 5’-sf3b1 around 1.6 kb (1–1659 bp) from a PCRIIsf3b1a construct linearized with NotI and HindIII and transcribed with SP6 and T7 separately.
Immunohistochemistry was performed as described (Henion et al., 1996). Embryos were fixed in 4% PFAfor 2 h at room temperature or overnight at 4°C. Antibodies were used at the following dilutions: zn-12 (Trevarrow et al., 1990), 1:4000; acetylated tubulin (Sigma), 1:200; goat anti-mouse, 1:200; PAP, 1:200.
TUNEL assays
Whole-mount TUNEL staining was performed on zebrafish embryos fixed overnight with 4% PFAin 1X PBS at 4°C, permeabilized with methanol, and rehydrated. TdT/digoxigenin or fluorescein-dUTP reactions (Roche) were done for 1 h on ice, followed by 1 h at 37°C. After labeling with digoxigenin or fluorescein-dUTP, anti-digoxigenin or anti-fluorescein-AP (Roche) was used to bind digoxigenin or fluorescein. NBT/BCIP was used for color development. For whole-mount in situ hybridization and TUNEL double labeling, the TUNEL assay was performed first by using fluorescein-dUTP and NBT/BCIP for color development. After TUNEL staining, the embryos were incubated in 2mg/ml glycine (pH 2.9) for 10 min at room temperature to inactivate anti-fluorescein-AP, washed with 1X PBS to recover pH and then, WISH was performed as described. Fast red was used for detection.
Genetic mosaic assays
Genetic mosaic assays were performed by transplanting cells between wild-type and mutant embryos at the late blastula stage as described (Ho and Kane, 1990). The embryos obtained from sf3b1b460 heterozygous carriers were divided into two groups, a donor group and a host group. Donor embryos were injected with rhodamine dextran (103 MW; Molecular Probes) at the 1- to 4-cell stage. Rhodamine dextran-labeled cells from donor embryos were transplanted into different regions of host embryos at the same developmental stage (dome stage 3.7–4 hpf) according to the zebrafish embryo fate map (Kimmel et al., 1990). The host and donor embryos were grown under the same conditions (28.5°C). After 24 hpf, wild-type and sf3b1b460 mutants embryos were identified based on phenotype. Since sf3b1b460 mutants completely lack melanophores, it allowed us to easily assess the terminal cell fate of transplanted donor cells. Transplanted cells were visualized using fluorescence microscopy on a Zeiss Axioplan/ DIC microscope.
Genetic mapping and positional cloning
tstb460heterozygous carriers (*AB background) were crossed to WIK wild-type to produce a mapping line. Female heterozygous F1 generation fish were used for the in vitro production of parthogenetic diploid F2 embryos. Homozygocity at most loci was achieved by suppressing the second meiotic division by application of hydrostatic pressure (Westerfield, 1993). Based on live phenotype, F2 diploid embryos were divided into two groups, homozygous mutants and wild-type embryos including homozygous and heterozygous wild-type embryos. The tstb460 locus was mapped to its corresponding chromosome using linkage analysis with simple sequence length polymorphisms (SS LPs). Through recombination frequency analysis using haploid F2 mutants and WT embryos, several closely linked SSLPs and simple-strand conformation polymorphisms (SSCPs) were identified and a critical region was defined. Bacterial artificial chromosomes (BAC, commercially available from RZPD) and expressed sequence tags (ESTs, commercially available from RZPD) were searched for within this critical region through the zebrafish genome database at Welcome Trust Sanger Institute (http://www.sanger.ac.uk/Projects/D_rerio/). SSCP and SSCP markers that were used for fine genetic mapping:
- ctg9339 566337
- 5’-CTGGAAAACCCCATGTCAAA-3’
- 5’-AGGTGCAAGGCAGAGTTAGC-3’
- zk83M22SP6
- 5’-GGTTCATAGTAGCTGAAGACACCTG-3’
- 5’-GTCAGTACATTCAGCAGAACATTCG-3’
- zk83M22T7
- 5’-GGCATGCAAGCTTCCTCGTTTGAT-3’
- 5’-TCCACAACAGCTTCTAAATTCCGC-3’
- zV4.5
- 5’-CCACGTCGATCTCCGAGTA-3’
- 5’-GTGCCTCTTCACGCCTACAT-3’.
Zebrafish Microsatellite markers used were Z60982, Z21824, Z54324, z35323.
BAC microinjection and phenotype rescue analysis
BAC DNA clones overlapping with the defined critical region used for injection were isolated using a Qiagen protocol for large plasmid isolation withQiagen midiprepkit. For phenotype rescue assays, the BAC DNA was microinjected into a single blastomere of 1- to 2-cell stage embryos obtained from heterozygote intercrosses. The BAC DNA was diluted to 100–120 ng/µl as a working stock in sterile 1% phenol red. Injected embryos were scored according to live phenotype. Since tst mutant embryos completely lack melanophores and differentiated melanophores are easily observed in live embryos after 24 hpf, this allowed us to easily and accurately assess phenotypic rescue. Potential rescued mutants were genotyped with highly linked SSLP markers (z21824, Z54324) for definitive confirmation. The embryos for injection of BACs were obtained by intercrossing heterozygous parents that are generated by crossing tstb460 heterozygotes on *AB background to WIK. Therefore, tstb460 homozygous mutants could be identified by genotyping with polymorphic SSLP markers.
cDNA sequencing analysis in tstb460 mutant and wild-type embryos
Total RNA from 18–24 hpf AB wild-type embryos and tstb460 mutants was isolated using TRIzol reagent (Sigma). Qiagen one-step RT PCR kit was used for RT-PCR with *AB wild-type and tstb460 mutant total RNA as templates. Primers used for sf3b1 cDNA fragments are:
- sf3b1 (1–2703 bp)
- 5’-AAATGGCGCAGATCGCCAAA-3’
- 5’- CGGCACCAAGGTTACCCATGATTT-3’
- sf3b1a(1–1659 bp)
- 5’-AAATGGCGCAGATCGCCAAA-3’
- 5’-ACCAAACTCCCTGGCCTTATCTGT-3’
- sf3b1 b(1486–3159 bp)
- 5’-TGCTGGTCGAAGTTGATGAGTCCA-3’
- 5’-TGCAGTTCTCCTGCACCTTCTCAT-3’
- sf3b1c(2934–4393 bp)
- 5’-AAGCTTATGGGCCATTTGGGTGTG-3’
- 5’-AAGAGCGAACTTGACAGACCAGGA-3’
- sf3b1aa (46)F
- 5’-ATGACATTGAGGCTCAGATCCTGG-3’
- sf3b1aa (579)R
- 5’-TCACTGCTTTCAGCTCTCCAGCTT-3’.
The RT-PCR programs used were as described in the Qiagen protocol (OneStep RT-PCR kit). The fresh RT-PCR products were cloned into PC Rl I vector by TA cloning (Invitrogen). The cDNAs obtained from RT-PCR were sequenced by the Ohio State University Plant-Microbe Genomics Facility (PMGF). The sequencing analysis was performed using Clustal X1.81 and sequence analysis program from http://www.ncbi.nlm.nih.gov.
sf3b1 cDNA cloning and misexpression analysis
The wild-type full length cDNA of sf3b1 was assembled with wild-type cDNA fragment 1–2703 bp and EST clone fb99f09. The EST clone fb99f09 (RZPD) was sequenced by PMGF at Ohio State University and it contains a 3’- sf3b1 cDNA fragment which overlaps with the sf3b1 cDNA fragment 1–2703 bp. The 2.5 kb fragment from sf3b1 cDNA fragment 1–2703 bp in PCRII vector (digested with XhoI/ClaI) and the 1.9 kbfragmentfrom fb99f09 EST clone (digested with ClaI/XbaI) were cloned into the XhoI/XbaI site of pCS2+ vector as pCS2+sf3b1. The sf3b1 full length cDNA from construct pCS2+sf3b1 with XhoI (blunt)/XbaI site was cloned into ClaI (blunt)/XbaI site of the heatshock vector pCSHSP (Halloran et al., 2000; Dutton et al., 2001) as hsp>sf3b1. DNA of hsp>sf3b1 was diluted to a final concentration of 25–30 ng/µl in 0.5% phenol red. tstb460 mutant embryos and their wild-type siblings from intercrosses of the tst WIK line were injected with hsp>sf3b1 at the 1 - to 2- cell stage and raised at 28.5°C. Embryos were heatshocked 2 or 3 times at 6–7 hpf, 11–12 hpf, and 23–24 hpf time points for 1 hour at 37°C. Embryos were genotyped as described previously.
hs>sox10 (Dutton et al., 2001) was injected into a single blastomereof tst mutant embryos at 1- to 2- cell stages and their wild-type siblings from intercrosses of tst WIK line. The injected embryos were heatshocked as described (Dutton et al., 2001). The injected embryos were scored as in the BAC DNA injection experiments, and genotyped by the highly linked SSLP markers (Z21824 and Z54324).
Synthetic sense-polyA-capped mRNAs were transcribed in vitro from linearized templates using the mMESSEGE mMACHINE kit (Ambion). After transcription, the mRNAs were recovered as described (mMESSEGE mMACHINE kit manual). pCS2+foxd3 (Stewart et al., 2006), pCS2+tfap2a (Knight et al., 2003), pCS2+sox9a, pCS2+sox9b (Yan et al., 2005) and pCS2+bcl2 (Langenau et al., 2005), were digested with NotI. The linearized DNA was used as template for in vitro transcription by SP6 polymerase. The RNA for injection was diluted to a final concentration range from 50–100 ng/µl and injected into a single blastomere of tstb460 mutant and wild-type embryos from intercrosses of tst WIK line at the 1- to 4- cell stage. The injected embryos were scored and genotyped as described above. The embryos injected with bcl2 were fixed at 17 hpf and used for the TUNEL assay to detect programmed cell death as well as subsequently (see above) for phenotype rescue assessment.
Morpholino phenocopy analysis
Two Morpholino antisense oligonucleotides (MO) were designed and acquired from Gene Tools: sf3b1MO3rd that targets pre-mRNA splicing at the 3rd exon (ATGAATCCTCGTCATCATCCTAAA), and sf3b1ATGmo that targets the start site of sf3b1 (GGCGATCTGCGCCATTTTCGTGCTG). Each morpholino was injected separately into one blastomere of wild-type embryos at the 1- to 4- cell stage to interfere with either pre-mRNA splicing of sf3b1 or to block translation, respectively. Wild-type embryos for morpholino injections were divided into two groups: morphants (embryos injected with morpholinos) and uninjected wild-type embryos. Efficiency of sf3b1MO3rd against the splice site was evaluated by RT-PCR with primers sf3b1aa (46)F and sf3b1aa (579)R. Western blot with mouse Sf3b1 monoclonal antibody (Horie et al., 2003) was used to determined the efficiency of sf3b1 ATGmo.
Western blotting
Western blot analysis was performed as described (Monani et al., 2003) with minor modifications. Dechorionated zebrafish embryos (12 hpf −24 hpf) were dissolved in blending buffer (10% SDS, 50mMTris HCI, pH 6.8,10mM EDTA, µl/each embryo) and incubated in boiling water for 5–10 min. BAC protein assay kit (PIERCE) was used for measuring protein concentration. 80µg of protein from different embryos (Wild-type embryos, tstb460 homozygous mutants, hi3394a homozygous mutants, sf3b1ATGmo morphants, hsp>sf3b1 injected tstb460 mutant embryos with phenotypic rescue) mixed with an equal volume of sample buffer (62.5mM Tris HCI, pH 6.8, 10% glycerol, 0.1% Bromophenol blue, 10% |3-mercaptoethanol) was loaded and electrophoresed on 8% polyacrylamide gels. Then, samples were transferred to nitrocellulose membranes (Whatman GmbH). TBS Blotto A (Santa cruz biothecnology, Inc.) was used as a blocking reagent. Primary antibodies for western blotting are mouse monoclonal antibodies, anti-Sf3b1 (kindly provided by Dr. K. Isono; Horie et al., 2003) and anti-actin as protein loading control (Abcam Inc.). HRP-conjugated secondary antibody was used to bind primary antibody and detected by chemiluminescence system (ECL western blotting analysis system, Amersham Bioscience).
Supplementary Material
Acknowledgements
We thank numerous colleagues for reagents and A. Amsterdam and the Hopkins lab for the hi3394a mutant. This work was supported by grant GM074505 from the NIH to P. D. H.
Abbreviations used in this paper
- hpF
hours postfertilization
- BAC
bacterial artificial chromosome
- sf3b1
splicing factor 3b, subunit 1
- snRNPs
small nuclear ribonu-cleoprotein particles
References
- Akimenko MA, Ekker M, Wegner J, LIN W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head . J. Neuosci. 1994;14(6):3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, SUN Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned lim ho-meobox gene expression in embryonic zebrafish. Development. 1995;121(12):4117–4125. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- Arduini BL, Bosse KM, Henion PD. Genetic ablation of neural crest cell diversification. Development. 2009;136:1987–1994. doi: 10.1242/dev.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinger KB, Chitnis AB, Mercola M, Driever W. Zebraf-ish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons . Development. 1999;126:3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function . Development. 2004;131:1463–1477. doi: 10.1242/dev.01033. [DOI] [PubMed] [Google Scholar]
- Brosi R, Groning K, Behrens S, Luhrmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993a;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Brosi R, Hauri H, Kramer A. Separation of splicing factor SF3 into two componentsand purification of SF3aactivity. J. Biol. Chem. 1993b;268:17640–17646. [PubMed] [Google Scholar]
- Burge C, Tuschl T, Sharp P. The RNA World. Cold Spring Harbor, NY.: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Carney TJ, Dutton KA, Greenhill E, Delfino-Machin M, Dufourcq P, Blader P, Kelsh RN. A direct role for Sox10 for specification of neural crest-derived sensory neurons. Development. 2006;133:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Chiang EF, Pai CI, Wyatt M, Yan YL, Postlethwait J, Chung B. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev. Biol. 2001;231:149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–2882. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho P, Parsons MJ, Thomas KA, Hirst EM, Saude L, Cam-Pos I, Williams PH, Stemple DL. Differential requirements for COPI transport during vertebrate early development. Dev. Cell. 2004;7:547–558. doi: 10.1016/j.devcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and.specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Neural crest specification: migrating into genomics. Nat. Rev. Neurosci. 2003;4:795–805. doi: 10.1038/nrn1219. [DOI] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Henion PD, Raible DW, Beattie CE, Stoesser KL, Weston JA, Eisen JS. Screen for mutations affecting development of zebrafish neural crest. Dev. Genetics. 1996;18:11–17. doi: 10.1002/(SICI)1520-6408(1996)18:1<11::AID-DVG2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Horie A, Isono K, Koseki H. Generation of a monoclonal antibody against the mouse sf3b1 (sap155) gene product for u2 snrnp component of spliceosome. Hybrid Hybridomics. 2003;22:117–119. doi: 10.1089/153685903321948049. [DOI] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev. Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Isono K, Abe K, Tomaru Y, Okazaki Y, Hayashizaki Y, Koseki H. Molecular cloning, genetic mapping, and expression of the mouse Sf3b1 (SAP155) gene for the U2 snRNP component of the spliceosome. Mamm. Genome. 2001;12:192–198. doi: 10.1007/s003350010258. [DOI] [PubMed] [Google Scholar]
- Isono K, Mizutani-Koseki Y, Komori T, Schmidt-Zachmann MS, Koseki H. Mammalian polycomb-mediated repression of Hox genes requires the essential spliceosomal protein Sf3b1. Genes Dev. 2005;19:536–541. doi: 10.1101/gad.1284605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T. Double in situ hybridization techniques in zebrafish. Methods. 2001;23:345–358. doi: 10.1006/meth.2000.1147. [DOI] [PubMed] [Google Scholar]
- Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labeled probes. Trends Genet. 1994;10:73–74. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Kambach C, Walke S, Nagai K. Structure and assembly of the splieosomal small nuclear ribonucleoprotein paricles. Curr. Opin. Struct. Biol. 1999;9:222–230. doi: 10.1016/s0959-440x(99)80032-3. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Eisen JS. The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development. 2000;127:515–525. doi: 10.1242/dev.127.3.515. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Schmid B, Eisen JS. Genetic analysis of melanophore development in zebrafish embryos. Dev. Biol. 2000;225:277–293. doi: 10.1006/dbio.2000.9840. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–594. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel, Ullmann SRB, Schilling TF. Stages of embryonicdevelopment of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch GJ, Schilling TF. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Jette C, Berghmans S, Palomero T, Kanki JP, Kutok JL, Look AT. Suppression of apoptosis by bcl-2 over-expression in lymphoid cells of transgenic zebrafish. Blood. 2005;105:3278–3285. doi: 10.1182/blood-2004-08-3073. [DOI] [PubMed] [Google Scholar]
- Lee KH, Xu Q, Breitbart RE. A new tinman-related gene, nkx2.7, anticipates the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev. Biol. 1996;180:722–731. doi: 10.1006/dbio.1996.0341. [DOI] [PubMed] [Google Scholar]
- Lim DA, Suarez-Farinas M, Naef F, Hacker CR, Menn B, Take-Bayashi H, Magnasco M, Patil N, Alvarez-Buylla A. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol. Cell. Neuroscience. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish crestin throughout embryonic development. Dev. Dyn. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Massiello A, Roesser JR, Chalfant CE. SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5’ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- Monani UR, Pastore MT, Gavrilina TO, Jablonka S, Le TT, An-Dreassi C, Dicocco JM, Lorson C, Androphy EJ, Sendtner M, Podell M, Burghes AH. A transgene carrying an a2g missense mutation in the smn gene modulates phenotypic severity in mice with severe (type i) spinal muscular atrophy. J. Cell Biol. 2003;160:41–52. doi: 10.1083/jcb.200208079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Balaguer M, Lang MR, Sachdev SW, Knappmeyer C, Stewart RA, De La Guardia A, Hatzopoulus AK, Knapik EW. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev. Dyn. 2006;235:3199–32112. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- Nambiar RM, Henion PD. Sequential antagonism of early and late Wnt-signaling by zebrafish colgate promotes dorsal and anterior fates. Dev. Biol. 2004;267:165–180. doi: 10.1016/j.ydbio.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Nambiar RM, Ignatius MS, Henion PD. Zebrafish Colgate/ hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wntindependent branchiomotor neuron migration. Mech. Dev. 2007:682–698. doi: 10.1016/j.mod.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms BL, Labosky PA. Developmental Biology. San Rafael, CA: Morgan and Claypool, Publishers; 2010. Transcriptional control of neural crest development. [PubMed] [Google Scholar]
- Nilsen TW. A parallel spliceosome. Science. 1997;273:1813. doi: 10.1126/science.273.5283.1813. [DOI] [PubMed] [Google Scholar]
- Nyguyen CT, Langenbacher A, Hsieh M, Chen JN. The PAF-1 complex component Leo1 is essential for cardiac neural crest development in zebrafish. Dev. Biol. 2010;341:167–175. doi: 10.1016/j.ydbio.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J, Nusslein-Volhard C. Fork head genes in zebrafish. Dev. Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;12:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SJ. Zebrafish sparse corresponds to an or thologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- Park HC, Hong SK, Kim HS, Kim SH, Yoon EJ, Kim CH, Miki N, Huh TL. Structural comparison of zebrafish elav/hu and their differential expressions during neurogenesis. Neurosci Lett. 2000;279:81–84. doi: 10.1016/s0304-3940(99)00940-4. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Segment and cell type lineage restric-tions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol. Cell. Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev. Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Postlethwait JH. Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev. Biol. 1995;172:86–89. doi: 10.1006/dbio.1995.0007. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- Yan Y-L, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor Hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish myod in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene, OR: University of Oregon Press; 1993. [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, Chen F, Long HK, Kramer M, Datta S, Neuberg D, Granter S, Young RA, Morrison S, Wheeler GN, Zon LI. DHODH modulates transcription elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Schneider C, Reed R, Luhrmann R. Identification of both shared and distinct proteins in the major and minor splieosomes. Science. 1999;284:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.