Abstract
It is not known whether the mammalian mechanism of coagulation initiation is conserved in fish. Identification of factor VII is critical in providing evidence for such a mechanism. A cDNA was cloned from a zebrafish (teleost) library that predicted a protein with sequence similarity to human factor VII. Factor VII was shown to be present in zebrafish blood and liver by Western blot analysis and immunohistochemistry. Immunodepletion of factor VII from zebrafish plasma selectively inhibited thromboplastin-triggered thrombin generation. Heterologous expression of zebrafish factor VII demonstrated a secreted protein (50 kDa) that reconstituted thromboplastin-triggered thrombin generation in immunodepleted zebrafish plasma. These results suggest conservation of the extrinsic coagulation pathway between zebrafish and humans and add credence to the zebrafish as a model for mammalian hemostasis. The structure of zebrafish factor VIIa predicted by homology modeling was consistent with the overall three-dimensional structure of human factor VIIa. However, amino acid disparities were found in the epidermal growth factor-2/serine protease regions that are present in the human tissue factor–factor VIIa contact surface, suggesting a structural basis for the species specificity of this interaction. In addition, zebrafish factor VII demonstrates that the Gla-EGF-EGF-SP domain structure, which is common to coagulation factors VII, IX, X, and protein C, was present before the radiation of the teleosts from the tetrapods. Identification of zebrafish factor VII significantly narrows the evolutionary window for development of the vertebrate coagulation cascade and provides insight into the structural basis for species specificity in the tissue factor–factor VIIa interaction.
Factor VII is a vitamin K-dependent plasma protein that plays a pivotal role in the extrinsic pathway of blood coagulation in mammals (1). It circulates in both zymogen (factor VII) and active protease (factor VIIa) forms, but the protease form does not possess full catalytic activity until bound to tissue factor on a phospholipid surface (2, 3). The factor VIIa–tissue factor complex can activate additional factor VII, factor X to Xa, and factor IX to IXa in the initiation of coagulation. Factor VIIa–tissue factor activity is regulated by tissue factor pathway inhibitor (TFPI) in a factor Xa-dependent manner (4, 5). Ex vivo modeling suggests that initiation of coagulation is a threshold-mediated event related to levels of tissue factor, factor VIIa, and TFPI (6, 7). However, significant questions remain regarding the initiation of coagulation, including identification of the factor(s) responsible for generation of circulating factor VIIa and the in vivo expression levels of factor VII, tissue factor, and TFPI activity. Furthermore, compared with mammals, the mechanism for blood coagulation in other vertebrate classes is poorly understood.
Fish occupy an important evolutionary position with regard to development of vertebrate hematologic characteristics, including the hemostatic response (8). Although there is evidence for fibrinogen (9, 10) and prothrombin (11) in primitive fish species, the presence of a protease cascade similar to mammals is unproven. The zebrafish is a representative bony fish (class Osteichthyes) and a powerful model for vertebrate genetics, allowing both saturation mutagenesis of the genome with phenotypic screening and specific gene-knockdown approaches (12, 13). Morphologic, immunologic, and functional characterization of thrombocytes, biochemical analysis of plasma coagulation, and molecular characterization of prothrombin in zebrafish suggest that the major hemostatic pathways are similar to those found in mammals (14–18). Likewise, the inhibition of zebrafish coagulation by warfarin, a vitamin K antagonist, suggests a role for γ-carboxylation of glutamic acids in the γ-carboxyglutamic acid (Gla) domain of zebrafish coagulation proteins (15). However, identification of factor VII is critical in providing strong relevance of the zebrafish as a genetic model for mammalian hemostasis. This information also should yield significant insights into the evolution of the vertebrate coagulation cascade.
We report here the identification of zebrafish factor VII, which significantly narrows the evolutionary window for development of the vertebrate coagulation cascade, provides insight into the structural basis for species specificity in the tissue factor–factor VIIa interaction, and establishes the zebrafish as a relevant genetic model for mammalian hemostasis.
Experimental Procedures
Materials.
One-month-old adult zebrafish 5′ stretch plus cDNA library in λgt11 vector was purchased from CLONTECH. Oligonucleotide primers, G418, and Trizol were from Life Technologies (Gaithersburg, MD). Restriction endonucleases were obtained from New England Biolabs. ReadyMix Taq PCR Reaction, GenElute Agarose Spin Columns, vitamin K1, DMEM, FBS, penicillin-streptomycin solution, protein A, and alkaline phosphatase-conjugated mouse anti-rabbit IgG were from Sigma. Wizard Lambda Preps DNA Purification System was from Promega. pCR 2.1-TOPO vector, pCDNA3.1+ vector, and cDNA Cycle Kit were purchased from Invitrogen. Horseradish peroxidase-conjugated goat anti-rabbit IgG, biotinylated anti-mouse/anti-rabbit IgG, and horseradish peroxidase-conjugated streptavidin were from Dako. SuperSignal chemiluminescent reagent was from Pierce. Zebrafish were purchased from Ekkwill Tropical Fish Farm (Gibsonton, FL). Recombinant human thromboplastin (Dade Innovin), rabbit brain thromboplastin (Dade Thromboplastin C Plus), and rabbit brain partial thromboplastin reagent (Dade Actin) were obtained from Baxter Diagnostics (McGaw Park, IL). The purified factor X activator from Russell viper venom (RVV-Xa) was purchased from Hematologic Technologies (Essex Junction, VT). Human plasma fibrinogen was purchased from Calbiochem. All other chemicals were at least reagent-grade and purchased from major suppliers.
Reverse Transcription–PCR of a Zebrafish FVII Probe.
Approximately 10 mg of adult zebrafish livers and attached internal organs were dissected and homogenized in 1 ml of Trizol reagent by using a Brinkman polytron homogenizer. Total RNA was prepared according to the manufacturer's protocol, followed by poly(A)+ mRNA isolation on oligo(dT)-cellulose columns. First-strand cDNA synthesis was performed by using an oligo(dT)10N primer and avian myeloblastoma virus reverse transcriptase (Invitrogen cDNA Cycle Kit). PCR amplification of this cDNA template with degenerate primers based on a rainbow trout “factor X” expressed sequence tag (GenBank accession no. T23100) GGNGARWGYCCNTGGCAGGCNCT (forward) and conserved regions of the sodium-binding site of prothrombin ACRTGNGTGTARAANCCRTA (reverse) generated an ≈700-bp fragment. The 700-bp fragment was gel-purified and subcloned into the pCR 2.1-TOPO vector, and the DNA sequence was determined by cycle sequencing.
Screening of the Zebrafish cDNA Library.
Approximately 2 × 106 clones from a 1-month-old adult zebrafish 5′ stretch plus cDNA library (CLONTECH) were screened with a random primer, 32P-labeled, 600-bp probe derived from the cloned 700-bp fragment. Positive plaques were purified by secondary and tertiary screenings, yielding three positive clones. DNA from these plaques was isolated, and the largest insert of these clones (1,671 bp) was subcloned into the EcoRI site of the pCR 2.1-TOPO vector. The complete nucleotide sequence was determined by the dideoxynucleotide chain-termination method.
Sequence Alignment and Homology Modeling.
Alignment of factor VII protein sequences was performed by using the clustalw program as described previously (18).
Knowledge-based, comparative protein modeling was performed with the Swiss-Model Internet-based tool (http://www.expasy.ch/swissmod/swiss-model.html), using the pro-mod ii program (19, 20). The three-dimensional structures were visualized with the SWISS PDB-VIEWER Version 3.5 computer program (21). The predicted mature zebrafish factor VII protein sequence was submitted to Swiss-Model in the First Approach Mode. The crystal structure of human factor VIIa (1DAN) was selected as the primary template for modeling the serine protease, epidermal growth factor (EGF), and Gla domains. Secondary templates for the serine protease domain [human factor Xa (1XKA, 1HCG, and 1FAX) and bovine factor Xa-TAP (1KIG)] and EGF domains [porcine factor IXa (1PFX), human factor Xa (1XKA and 1XKB), and human Gla-domainless activated protein C (1AUT)] also were selected by the program. Modeled structures for the Gla domain and EGF domains were fused into a single polypeptide chain and superimposed with the serine protease domain model on the human factor VIIa crystal structure (1DAN).
Generation of Antibody to a Zebrafish Factor VII Peptide.
The peptide QAGKAADHQVDLRSRIVGGS (based on the predicted amino acid sequence of zebrafish factor VII) was synthesized and HPLC-purified by Alpha Diagnostic (San Antonio, TX). The peptide was coupled to keyhole limpet hemocyanin and injected into rabbits i.m. at 15-day intervals. Before injections, 2 ml of preimmune serum was obtained from the same animal. Seven weeks after injection, 15–20 ml of blood was obtained and antisera was prepared. The titer of the anti-rabbit zebrafish factor VII antibody was determined by conventional solid-phase ELISA by using immobilized synthetic peptide as the target antigen.
Western Blotting and Immunohistochemistry.
Approximately 1.5 μl of zebrafish plasma was subjected to SDS/9% PAGE and transferred to nitrocellulose membrane for Western blotting. Zebrafish factor VII was detected by using the rabbit anti-zebrafish factor VII peptide antisera (1:1,000 dilution) as the primary antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10,000 dilution) as the secondary antibody. Bands were detected with SuperSignal chemiluminescent reagent according to the manufacturer's protocol.
Immunostaining was performed on 5-μm sections taken from a formalin-fixed, paraffin-embedded adult zebrafish block. Sections were deparaffinized and hydrated by passing through xylene and a graded series of ethanol. Antigen retrieval was performed for 20 min at 98°C in 0.01 M sodium citrate buffer, pH 6.4, in a microwave oven followed by incubation in 0.3% hydrogen peroxide. Sections were blocked with 10% bovine serum for 30 min and incubated with the rabbit anti-zebrafish factor VII peptide antisera (1:500 dilution) under a coverslip at 4°C overnight. The sections were washed in PBS, pH 7.3, incubated with premixed biotinylated anti-mouse/anti-rabbit IgG for 30 min, followed by peroxidase-conjugated streptavidin for an additional 30 min. Reaction sites were visualized by using diaminobenzidine as the chromogen, and nuclei were counterstained with hematoxylin.
Zebrafish Coagulation Assays.
Blood was harvested from anesthetized adult zebrafish, and pooled, citrated plasma was obtained as described (15). Zebrafish plasma was immunodepleted with rabbit antisera against the zebrafish factor VII peptide (see above) for use in kinetic coagulation assays. Briefly, equal volumes of antiserum and zebrafish plasma were incubated for 45 min on ice with occasional mixing. An equal volume of insoluble Protein A solution then was added, incubated for 45 min on ice, and centrifuged for 5 min in a microfuge. Five microliters of the supernatant was used in kinetic coagulation assays. For control zebrafish plasma, incubations were performed by using PBS in place of the antiserum. Human plasma similarly was immunodepleted with rabbit anti-human factor VII antibody.
Tissue thromboplastin reagent was prepared from zebrafish muscle and aliquots were stored at −25°C for use in the kinetic prothrombin time assay (15). Kinetic coagulation assays were performed in 96-well, flat-bottom (VWR Scientific) microtiter dishes as described (17). Coagulation was triggered by adding 5 μl of thromboplastin reagent (human recombinant, rabbit brain, or zebrafish muscle), partial thromboplastin reagent (Dade Actin), or RVV-Xa (0.26 mg/ml) to each well before recalcification. Thrombin generation over time (detected by fibrinogen cleavage) by the above reagents was referred to as the kinetic prothrombin time (kPT), kinetic-activated partial thromboplastin time (kPTT), and kinetic Russell viper venom activation time (kRVVT), respectively. Negative controls were performed by replacing the clotting reagent with PBS.
Expression of Zebrafish Factor VII.
The zebrafish cDNA was subcloned into the EcoRI site of the pCDNA3.1+ vector, and correct orientation was confirmed by restriction endonuclease digest. An alternative chimeric cDNA containing the 5′ end of the human factor IX cDNA (including the entire prepropeptide sequence) and the 3′ end of the zebrafish factor VII cDNA (including the predicted mature protein and 3′ noncoding sequences) was constructed by overlapping PCRs. The fidelity of PCR-amplified regions was confirmed by DNA sequencing of the final construct. Plasmid DNA from each of these expression vectors was linearized with ScaI and transfected into human embryonic kidney 293 cells by the calcium phosphate method. Clones expressing high levels of recombinant protein were selected by limiting dilution in growth medium (50% DME/50% F-12/10% FCS) containing 0.5 mg/ml G418 and screened by Western blot analysis. Cell lines expressing each of the zebrafish factor VII constructs were expanded in T-225 cm2 flasks. At confluence, cells were washed and incubated in serum-free medium containing 10 μg/ml vitamin K and insulin-transferrin-selenite and harvested every 48 h for 10 days. Benzamidine (5 mM) was added, and the conditioned medium was frozen at −25°C. Each recombinant protein was purified to homogeneity from conditioned medium by sequential barium chloride precipitation and calcium-dependent elution from an anion-exchange column, methods that rely on vitamin K-dependent γ-carboxylation of the protein. On thawing, ≈480 ml of the conditioned medium was pooled, filtered, and subjected to barium chloride precipitation (22). The precipitate was dissolved completely in 0.2 M EDTA, pH 8.0, clarified by centrifugation, and dialyzed overnight against 0.1 M NaCl/20 mM Tris⋅HCl, pH 7.4. The eluate was applied to a Mono Q HR 5/5 column equilibrated with 0.15 M NaCl/20 mM Hepes/0.1% PEG-8000 and eluted with a calcium chloride gradient (0–45 mM). Fractions from the initial peak containing fully γ-carboxylated protein were pooled and concentrated in a Centricon-30. The recombinant zebrafish factor VII demonstrated high purity by 4–20% gradient SDS/PAGE with silver staining, and identity of the expressed protein was confirmed by Western blot by using the rabbit anti-zebrafish factor VII peptide antisera (1:500 dilution) and an alkaline phosphatase-conjugated mouse anti-rabbit IgG (1:7,500 dilution).
Results
Analysis of Zebrafish Factor VII cDNA Sequence.
The cloned, 1.67-kb zebrafish cDNA sequence predicted an ORF of 1,299 bp corresponding to a predicted protein of 433 aa and a 3′ untranslated region of 372 bp [the sequence is shown in Fig. 7, which is published as supplemental data on the PNAS web site (www.pnas.org)]. The amino terminus of the protein sequence predicts a prepropeptide (residues 1–36) that includes an initiator methionine, a putative signal peptide, a conserved vitamin K-dependent carboxylase-recognition peptide (residues 22–35), and a dibasic amino acid processing site (residues 36–37). Sequentially from the amino terminus, the predicted mature protein contains Gla (residues 44–73), EGF-1 (residues 84–121), EGF-2 (residues 126–167), and serine protease (SP) domains (residues 196–433). Based on the warfarin studies and presence of the carboxylase-recognition peptide, γ-carboxylation of the glutamic acids in the Gla domain of the mature protein is expected (15).
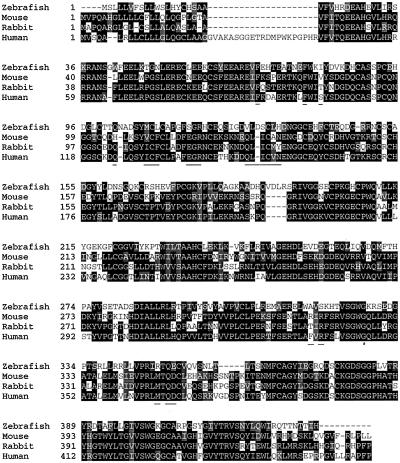
The predicted protein demonstrated a high degree of sequence similarity to mammalian factor VII sequences (Fig. 1). The amino acid sequence of the putative zebrafish factor VII showed 48% identity (I) and 63% similarity (S) overall with human factor VII. Zebrafish factor VII demonstrated a modestly lower sequence similarity with human factor X (42% I and 57% S), factor IX (42% I and 56% S), and protein C (40% I and 56% S). Similar to the mouse and rabbit, zebrafish factor VII demonstrates a shorter prepropeptide sequence than the human protein. Domain comparison between the mature zebrafish and human factor VII proteins revealed a relatively higher degree of conservation in the Gla (62% I and 83% S) and EGF-1 domains (55% I and 66% S) compared with the EGF-2 (43% I and 67% S) and serine protease domains (48% I and 62% S). Cysteine residues predicted to participate in disulfide bridges within the EGF domains (Cys-88–Cys-99, Cys-93–Cys-109, Cys-111–Cys-120, Cys-131–Cys-142, Cys-138–Cys-151, Cys-153–Cys-166), between EGF-2 and the protease domain (Cys-174–Cys-304), and within the protease domain (Cys-202–Cys-207, Cys-221–Cys-237, Cys-352–Cys-366, Cys-377–Cys-405) are conserved. A single activation site at Arg-195–Ile-196 is conserved, predicting an activated protease consisting of a 158-aa light chain (residues 38–195) and a 238-aa heavy chain (residues 196–433). No activation peptide is apparent; however, a unique 4-aa insertion (residues 190–193) is present proximal to the predicted activation cleavage site. Additional conserved features within the protease domain include: the catalytic triad of Asp-284, His-236, and Ser-381 {102, 57, 195}‖; Asp-375 {189} in the S1 substrate-binding pocket; Glu-252 {70} and Glu-262 {80} in the protease calcium-binding site; and the sodium ion-binding motif Tyr-411 {225} (23). In contrast, the amino acid residues Ser-278–Glu-279–Thr-280–Ala-281–Asp-282 {96–100}, which contribute to the S2–S3 substrate-binding pocket, are not conserved (24, 25). Likewise, the flexible insertion loop Val-353–Ser-362 {170–178} implicated in the human factor VIIa–soluble tissue factor interaction is not conserved among the factor VII proteins and demonstrates a 5-aa deletion (between Thr-359 and Leu-360) in the zebrafish (Fig. 1; ref. 24).
Figure 1.
Multiple alignment of zebrafish, mouse, rabbit, and human factor VII. The alignment was performed by using the clustalw program. Aligned sequences are shaded according to percentage of similar residues among the species. The black and shaded regions correspond to identical and similar amino acids, respectively, when 70% of the sequences share a common residue. Amino acid residues involved in the contact regions between human soluble tissue factor and human factor VIIa (ref. 24) are underlined.
Homology Modeling.
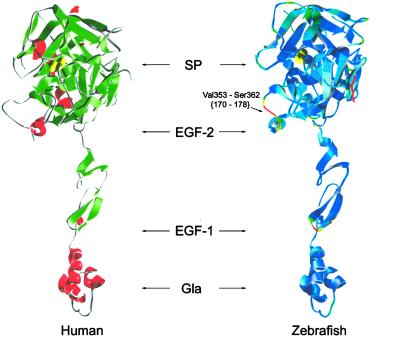
The zebrafish factor VIIa model was compared with the crystal structure of human factor VIIa in the factor VIIa–soluble tissue factor complex (Fig. 2) (24). The model was consistent with conservation of the overall three-dimensional structure between zebrafish and human factor VIIa. However, significant differences were found in the Val-353–Ser-362 {170–178} insertion loop, and the C terminal α-helix extension in the protease domain (Fig. 2). However, the tissue factor–factor VIIa interaction demonstrates a significant degree of species specificity, as demonstrated by the lack of response in zebrafish plasma to human or rabbit thromboplastin (15). To evaluate structural features that might contribute to species specificity, a detailed comparison of the putative intermolecular contact surfaces on zebrafish vs. human factor VIIa was undertaken based on the human soluble tissue factor–factor VIIa structure (24). The three major contact regions involving the Gla, EGF-1, and EGF-2/SP domains were compared (Table 1). Surface residues that participate in intermolecular contacts are largely conserved within the Gla and EGF-1 regions of zebrafish factor VIIa. However, major potential disruptions in the EGF-2/SP intermolecular contact region are evident in zebrafish vs. human factor VIIa. In particular, amino acids involved in hydrophobic interactions with EGF-2 (zebrafish residues Val-127–His-133) and the protease domain (zebrafish Arg-348) are poorly conserved.
Figure 2.
Homology model of zebrafish factor VII. The three-dimensional structure of zebrafish factor VIIa (Right) was generated by homology modeling (see Materials and Methods) compared with the crystal structure of human factor VIIa (Left) from the soluble tissue factor–factor VIIa complex. rms agreement of the alpha carbon backbone of zebrafish with human factor VIIa is color-coded (blue < 2 Å, yellow = 2–4 Å, red > 4 Å).
Table 1.
Comparison of the tissue factor intermolecular contact sites for human and zebrafish factor VIIa based on the crystal structure of the human factor VIIa–tissue factor complex
| Tissue factor | Human factor VIIa | Zebrafish factor VIIa | ± | Interaction |
|---|---|---|---|---|
| GIa domain | GIa domain | |||
| V207/W158/C209 | F91 | F69 | + | Hydrophobic |
| V207 | F100 | F78 | + | Hydrophobic |
| Q110 | S103 | I81 | − | H bond |
| EGF-1 domain | EGF-1 domain | |||
| F140/L133/R131 | F131 | L110 | ± | Hydrophobic |
| L133/K20/T17 | I129 | M108 | ± | Hydrophobic |
| I22 | R139 | R118 | + | Hydrophobic |
| K20 | C130/G138 | C109/G117 | + | H bond |
| K48 | E137 | S116 | − | H bond |
| D58 | G138 | G117 | + | H bond |
| E56 | R139 | R118 | + | H bonds (two) |
| E24 | R139 | R118 | + | H bond |
| Q110 | Q124 | Q102 | + | H bond |
| EGF-2/SP domain | EGF-2/SP domain | |||
| F50 | Q148–N153 (QLICVN) | V127–H133 (VLDSCLH) | − | Hydrophobic |
| F76/Y94/P92/R74 | M366 {164} | R348 {164} | − | Hydrophobic |
| Y94 | Q368/D369 {166/167} | Q350/E351 {166/167} | ± | H bond (three) |
| W45 | R337 {134} | S319 {134} | − | Packing, H bond |
| Q37 | F335 {129F} | A317 {129F} | − | H bond |
| N96 | D369 {167} | E351 {167} | ± | H bond |
| S39 | R337 {134} | S319 {134} | − | H bond |
| G43 | R337 {134} | S319 {134} | − | H bond |
| W45 | F335 {129F} | A317 {129F} | ± | H bond |
Residues participating in the human tissue factor–factor VIIa interaction were compared with the corresponding zebrafish factor VIIa residues, and apparent conservation of the intermolecular contact is indicated by plus and minus signs (+ fully, ± partially, or − not conserved). Amino acid residues are numbered from the amino terminus of soluble tissue factor or preprofactor VII (Fig. 1). Numbers in curly brackets correspond to chymotrypsinogen-based numbering.
Expression Studies.
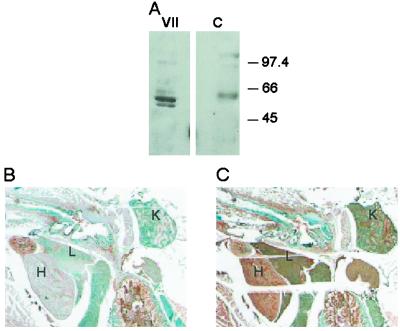
To examine expression of the predicted factor VII protein in zebrafish plasma, generation of anti-factor VII antibodies was undertaken. Rabbit polyclonal antisera was raised against a peptide (QAGKAADHQVDLRSRIVGGS) derived from the predicted sequence (Gln-181–Ser-200) of the zebrafish factor VII protein. Western blot of zebrafish plasma incubated with the antipeptide antisera demonstrated a band with an apparent molecular mass of 50 kDa (Fig. 3A). Likewise, examination of thin sections of adult zebrafish tissue with the antipeptide antibody demonstrated specific immunoreactivity in the liver, blood within the heart, and the kidney (Fig. 3 B and C). Qualitative reverse transcription–PCR to detect the factor VII transcript in tissue mRNA was positive in both the liver and kidney and negative in the heart (data not shown). The synthesis of factor VII in the liver is similar to the previously demonstrated synthesis of prothrombin in the developing zebrafish liver (26). However, the extrahepatic synthesis of factor VII in zebrafish appears to be novel.
Figure 3.
Detection of factor VII in zebrafish plasma and tissues. (A) Western blot of zebrafish plasma detected with rabbit anti-zebrafish factor VII antisera (VII) or preimmune sera (C). Molecular mass markers (in kDa) are indicated on the right. (B and C) Immunohistochemical stain of zebrafish tissue sections with preimmune (B) or rabbit anti-zebrafish factor VII antisera (C). H, heart; L, liver; K, kidney.
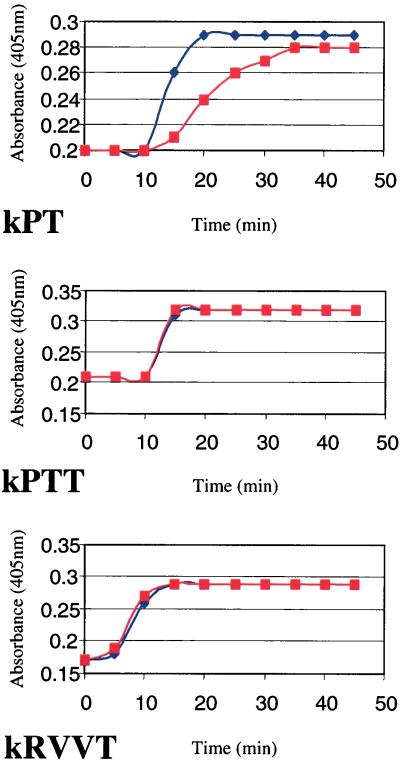
To assess the potential hemostatic function of the factor VII protein, kinetic coagulation assays were performed in zebrafish plasma immunodepleted with the antifactor VII peptide antisera. To demonstrate the specificity of the immunodepletion procedure, mock immunodepletion with protein A in the absence of specific antisera was performed as a control. Kinetic coagulation assays (kPT, kPTT, and kRVVT) then were compared in factor VII and mock-depleted zebrafish plasma. The factor VII-depleted plasma demonstrated a significant delay, relative to mock-depleted plasma, in fibrin generation initiated by zebrafish thromboplastin (Fig. 4).
Figure 4.
Effect of immunodepletion of factor VII from zebrafish plasma on kinetic coagulation assays. Fibrin formation over time was compared for mock-depleted (♦) and immunodepleted (■) zebrafish plasma in the presence zebrafish thromboplastin (kPT), Actin (kPTT), and RVV-X activator (kRVVT). The time in minutes is plotted against absorbance at 405 nm.
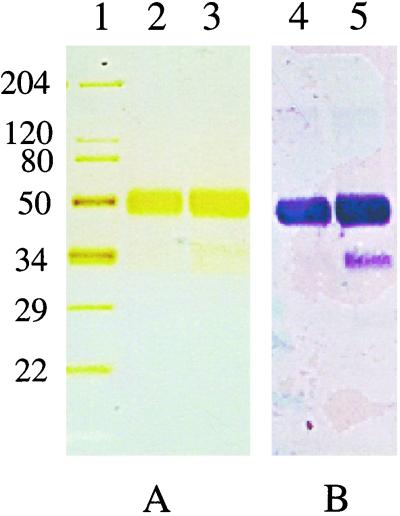
To establish whether the isolated cDNA clone could direct expression of the predicted zebrafish factor VII protein, heterologous expression in 293 cells was undertaken. Because of concerns about whether the zebrafish prepropeptide could direct the synthesis of the zebrafish factor VII in human 293 cells, a modified cDNA encoding the prepropeptide of human factor IX and the predicted mature zebrafish factor VII protein also was constructed. Recombinant protein was purified to homogeneity from the conditioned medium. Both constructs expressed soluble proteins with an apparent molecular mass of 50 kDa (Fig. 5). Western blot analysis with the anti-factor VII peptide antisera confirmed the identity of the recombinant proteins.
Figure 5.
SDS/PAGE (4–20% gradient) of purified recombinant factor VII detected by silver staining (A) or Western blot with rabbit anti-zebrafish factor VII antisera (B). Lanes: 1, molecular mass markers; 2 and 4, zebrafish factor VII; 3 and 5, human factor IX/zebrafish factor VII. Sizes of the molecular mass markers (in kDa) are indicated on the left.
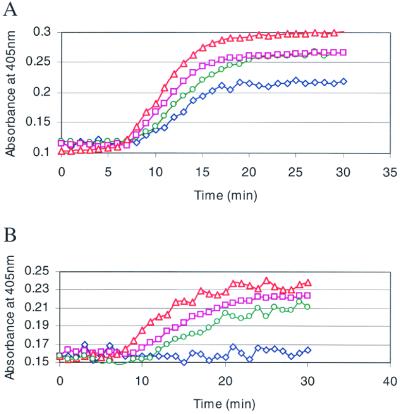
Hemostatic function of the purified recombinant proteins was assessed in the kinetic coagulation assays. Addition of increasing amounts of the purified factor VII to zebrafish plasma shortened the delay time and enhanced the rate for fibrin generation initiated with zebrafish thromboplastin in a dose-dependent fashion (Fig. 6A). No significant effect of zebrafish factor VII was seen on fibrin generation initiated with the kPTT or kRVTT reagents (data not shown). In zebrafish plasma immunodepleted with the antifactor VII peptide antibody, purified factor VII demonstrated a dose-dependent increase in fibrin generation initiated with zebrafish thromboplastin (Fig. 6B). The purified human factor IX/zebrafish VII protein demonstrated activity similar to the zebrafish factor VII protein in the kinetic coagulation assays (data not shown).
Figure 6.
The effect of recombinant zebrafish factor VII depletion on fibrin formation in the kinetic prothrombin time (kPT). Fibrin formation over time in normal (A) or immunodepleted (B) zebrafish plasma was determined without (⋄) or with addition of 1 (○), 4 (□), or 8 μl (▵) of 176.4 μg/ml recombinant factor VII. The time in minutes is plotted against absorbance at 405 nm.
Discussion
This investigation demonstrates the presence of coagulation factor VII in the zebrafish, providing molecular evidence for an extrinsic coagulation pathway in a nonmammalian species. Similarities between zebrafish and human primary protein sequence, domain organization (Gla-EGF-EGF-SP), and postulated three-dimensional structure suggest that this cDNA is orthologous to human factor VII. These sequence similarities include conservation of a single activation cleavage site, the cysteines participating in disulfide bonds, and residues that contribute to the formation of specific binding sites for sodium and calcium ions. The presence of the predicted protein was demonstrated in zebrafish plasma and liver with specific rabbit antisera vs. a unique zebrafish factor VII peptide. Immunodepletion of zebrafish plasma with the antipeptide antisera selectively inhibited thromboplastin-triggered thrombin generation. Finally, the zebrafish cDNA directed the expression of a secreted mature protein that was similar in size to human factor VII, which reconstituted thromboplastin-triggered thrombin generation in immunodepleted plasma. These structural and functional correlates reflect the conservation of factor VII gene function between zebrafish and man.
The coagulation serine proteases arose from the family of trypsin-like genes, characterized by protease domains that use the AGY codon for the catalytic serine {195}, and contain a specific sodium ion-binding motif, Tyr/Phe {225} (23, 27). The organization of the N terminus propeptide is unique to these proteases, composed of a γ-carboxylation (Gla) domain followed by two EGF or kringle domains. Although Gla, EGF, and SP modules exist in invertebrates such as Drosophila melanogaster, there is no evidence for this unique domain organization (28). Likewise, serine proteases responsible for hemolymph coagulation in Tachypleus tridentatus (Japanese horseshoe crab) and other invertebrates species differ significantly in N terminus domain content and organization from mammalian coagulation proteases (29–31). Evidence for prothrombin (which contains kringle domains) exists in the primitive jawless hagfish (Myxinidae) (11). However, zebrafish factor VII now provides the earliest known evolutionary appearance of the Gla-EGF-EGF-SP domain structure common to factors VII, IX, and X and protein C. Demonstration of both zebrafish prothrombin (18) and factor VII indicates that the relevant domain assembly for the coagulation proteases had occurred (presumably by exon shuffling) (32) at or before the time of the last common ancestor of humans and zebrafish, ≈450 million years ago (33).
Significant species specificity exists in the tissue factor–factor VIIa interaction between mammalian species and appears relatively complete between zebrafish and man (15). The human soluble tissue factor–factor VIIa structure demonstrates three major intermolecular contacts involving the Gla, EGF-1, and EGF-2/SP domains, respectively (24). Inspection of homologous surfaces on the zebrafish factor VIIa model suggests that intermolecular interactions are conserved largely for the Gla and EGF-1 contact regions. In contrast, surface residues in the EGF-2/SP contact region are poorly conserved, suggesting that the intermolecular interactions with human tissue factor are largely disrupted. This disruption includes the protease insertion loop Leu-371–Glu-385 {170–178}, which appears to undergo a conformational change between the bound and unbound structures of human factor VIIa (25). In the zebrafish protein, this loop demonstrates a 5-aa deletion and replacement of a neighboring conserved Met with Arg-348 {164}. These differences in the EGF-2/SP intermolecular contact region between zebrafish and man suggest a structural basis for the observed species specificity in the tissue factor–factor VIIa interaction. Hybrid recombinant human/zebrafish factor VII proteins in which the EGF-2/SP intermolecular contact regions are exchanged could directly test their contribution to the species specificity of this interaction.
Identification of zebrafish factor VII has important implications for the relevance of this powerful genetic model to the study of hemostasis and thrombosis. The ability to trigger (and selectively inhibit) tissue thromboplastin-dependent coagulation in a species-specific manner provides indirect evidence for tissue factor in the zebrafish. Functional data also suggest the presence of a contact-activated coagulation pathway that is independent of factor VII and factor X-like activity in the zebrafish (15). Factor X-like activity (based on RVV-X activator) is not affected by immunodepletion of factor VII, demonstrating that these activities are distinct. The degree of similarity between zebrafish and mammalian coagulation suggests that the zebrafish is a relevant animal model for the study of genes that affect hemostasis. Phenotypic screens of mutagenized zebrafish may identify novel genes that regulate the initiation of coagulation.
In conclusion, the structural features of factor VII in the zebrafish suggest that domain assembly for the coagulation proteases occurred before radiation of the ancestral Actinopterygii (ray-finned fishes) and Sarcopterygii (lungfish and tetrapods). The gene structure (intron/exon boundaries) of coagulation factors VII, IX, and X and protein C indicates that these genes are paralagous, suggesting a common origin via gene duplication (34). Thus, it appears likely that the vertebrate coagulation cascade arose rapidly during proposed genome duplications between ancestral chordates and the development of jawed vertebrates (35–37). Given the functional and structural similarities to mammalian coagulation demonstrated thus far, the zebrafish should be a powerful model to identify novel genes involved in vertebrate coagulation.
Supplementary Material
Acknowledgments
This research was supported by a Merit Award from the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.S.), a University of Texas Interdepartmental/Nathan Shock Aging Center grant (to J.S. and P.J.), and National Institutes of Health Grants GM 53373 and HL63792 (to P.J.).
Abbreviations
- Gla
γ-carboxyglutamic acid
- EGF
epidermal growth factor
- SP
serine protease
- RVV-X
Russell viper venom factor X activator
- kPT
kinetic prothrombin time
- kPTT
kinetic activated partial thromboplastin time
- kRVVT
kinetic Russell viper venom time
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY040345).
Numbers in curly brackets represent amino acid numbering of the protease domain based on three-dimensional topological identity with chymotrypsinogen.
References
- 1.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 2.Eichinger S, Mannucci P M, Tradati F, Arbini A A, Rosenberg R D, Bauer K A. Blood. 1995;86:3021–3025. [PubMed] [Google Scholar]
- 3.Nemerson Y. Blood. 1988;71:1–8. [PubMed] [Google Scholar]
- 4.Broze G J, Jr, Warren L A, Novotny W F, Higuchi D A, Girard J J, Miletich J P. Blood. 1988;71:335–343. [PubMed] [Google Scholar]
- 5.Rapaport S I. Blood. 1989;73:359–365. [PubMed] [Google Scholar]
- 6.Lawson J H, Kalafatis M, Stram S, Mann K G. J Biol Chem. 1994;269:23357–23366. [PubMed] [Google Scholar]
- 7.van't Veer C, Mann K G. J Biol Chem. 1997;272:4367–4377. doi: 10.1074/jbc.272.7.4367. [DOI] [PubMed] [Google Scholar]
- 8.Rowley A F, Hill D J, Ray C E, Munro R. Thromb Haemostasis. 1997;77:227–233. [PubMed] [Google Scholar]
- 9.Bohonus V L, Doolittle R F, Pontes M, Strong D D. Biochemistry. 1986;25:6512–6516. doi: 10.1021/bi00369a026. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle R F. Adv Exp Med Biol. 1990;281:25–37. doi: 10.1007/978-1-4615-3806-6_2. [DOI] [PubMed] [Google Scholar]
- 11.Banfield D K, Irwin D M, Walz D A, MacGillivray R T. J Mol Evol. 1994;38:177–187. doi: 10.1007/BF00166164. [DOI] [PubMed] [Google Scholar]
- 12.Solnica-Krezel L, Schier A F, Driever W. Genetics. 1994;136:1401–1420. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasevicius A, Ekker S C. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 14.Jagadeeswaran P, Sheehan J P, Craig F E, Troyer D. Br J Haematol. 1999;107:731–738. doi: 10.1046/j.1365-2141.1999.01763.x. [DOI] [PubMed] [Google Scholar]
- 15.Jagadeeswaran P, Sheehan J P. Blood Cells Mol Dis. 1999;25:239–249. doi: 10.1006/bcmd.1999.0249. [DOI] [PubMed] [Google Scholar]
- 16.Jagadeeswaran P, Liu Y C, Sheehan J P. Methods Cell Biol. 1999;59:337–357. doi: 10.1016/s0091-679x(08)61833-6. [DOI] [PubMed] [Google Scholar]
- 17.Jagadeeswaran P, Gregory M, Johnson S, Thankavel B. Br J Haematol. 2000;110:946–956. doi: 10.1046/j.1365-2141.2000.02284.x. [DOI] [PubMed] [Google Scholar]
- 18.Jagadeeswaran P, Gregory M, Zhou Y, Zon L, Padmanabhan K, Hanumanthaiah R. Blood Cells Mol Dis. 2000;26:479–489. doi: 10.1006/bcmd.2000.0330. [DOI] [PubMed] [Google Scholar]
- 19.Blundell T L, Sibanda B L, Sternberg M J, Thornton J M. Nature (London) 1987;326:347–352. doi: 10.1038/326347a0. [DOI] [PubMed] [Google Scholar]
- 20.Peitsch M C. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 21.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 22.Cote H C, Stevens W K, Bajzar L, Banfield D K, Nesheim M E, MacGillivray R T. J Biol Chem. 1994;269:11374–11380. [PubMed] [Google Scholar]
- 23.Dang Q D, Di Cera E. Proc Natl Acad Sci USA. 1996;93:10653–10656. doi: 10.1073/pnas.93.20.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banner D W, D'Arcy A, Chene C, Winkler F K, Guha A, Konigsberg W H, Nemerson Y, Kirchhofer D. Nature (London) 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 25.Pike A C, Brzozowski A M, Roberts S M, Olsen O H, Persson E. Proc Natl Acad Sci USA. 1999;96:8925–8930. doi: 10.1073/pnas.96.16.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jagadeeswaran P, Liu Y. Blood Cells Mol Dis. 1997;23:147–156. doi: 10.1006/bcmd.1997.0132. [DOI] [PubMed] [Google Scholar]
- 27.Brenner S. Nature (London) 1988;334:528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Yang C T, Jin D, Stafford D W. J Biol Chem. 2000;275:18291–18296. doi: 10.1074/jbc.M001790200. [DOI] [PubMed] [Google Scholar]
- 29.Iwanaga S, Kawabata S, Muta T. J Biochem (Tokyo) 1998;123:1–15. doi: 10.1093/oxfordjournals.jbchem.a021894. [DOI] [PubMed] [Google Scholar]
- 30.Jiang H, Wang Y, Kanost M R. Insect Mol Biol. 1999;8:39–53. doi: 10.1046/j.1365-2583.1999.810039.x. [DOI] [PubMed] [Google Scholar]
- 31.Shishikura F, Abe T, Ohtake S, Tanaka K. Comp Biochem Physiol B Biochem Mol Biol. 1997;118:131–141. doi: 10.1016/s0305-0491(97)00217-4. [DOI] [PubMed] [Google Scholar]
- 32.Patthy L. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Hedges S B. Nature (London) 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 34.Ichinose A, Davie E. In: Hemostasis and Thrombosis: Basic Principles and Clinical Practice. 3rd Ed. Coleman R, Hirsch J, Marder V J, Salzman E, editors. Philadelphia: Lippincott; 1994. [Google Scholar]
- 35.Holland P W, Garcia-Fernandez J, Williams N A, Sidow A. Dev. Suppl. 1994. 125–133. [PubMed] [Google Scholar]
- 36.Pebusque M J, Coulier F, Birnbaum D, Pontarotti P. Mol Biol Evol. 1998;15:1145–1159. doi: 10.1093/oxfordjournals.molbev.a026022. [DOI] [PubMed] [Google Scholar]
- 37.Bailey W J, Kim J, Wagner G P, Ruddle F H. Mol Biol Evol. 1997;14:843–853. doi: 10.1093/oxfordjournals.molbev.a025825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.