Abstract
Background
Crohn's disease and ulcerative colitis are the major types of chronic inflammatory bowel disease occurring in the colon and small intestine. A growing body of research has proposed that probiotics are able to attenuate the inflammatory symptoms of these diseases in vitro and in vivo. However, the mechanism of probiotic actions remains unclear.
Results
Our results suggested Lactobacillus plantarum MYL26 inhibited inflammation in Caco-2 cells through regulation of gene expressions of TOLLIP, SOCS1, SOCS3, and IκBα, rather than SHIP-1 and IRAK-3.
Conclusions
We proposed that live/ heat-killed Lactobacillus plantarum MYL26 and bacterial cell wall extract treatments impaired TLR4-NFκb signal transduction through Tollip, SOCS-1 and SOCS-3 activation, thus inducing LPS tolerance. Our findings suggest that either heat-killed probiotics or probiotic cell wall extracts are able to attenuate inflammation through pathways similar to that of live bacteria.
Keywords: Lactobacillus plantarum MYL26, Lipopolysaccharide, Caco-2 cells
Background
Inflammatory bowel disease (IBD) comprises a collection of disorders, which mainly include Crohn's disease and ulcerative colitis. These disorders cause abdominal pain, vomiting, diarrhea, and gastrointestinal (GI) inflammation [1]. To date, no effective therapy has been developed and patients may have a reduced quality of life even under proper management. It has been shown that factors related to IBD include acquired factors (e.g., smoking and diet), pathogens, genetic factors, and irregular immune system [2]. Over the past decades, the homeostatic functions of microflora on host GI tract have attracted much attention because growing numbers of clinical studies have suggested that probiotics exhibit anti-inflammatory effects on IBD patients [3,4]. Arseneau et al. [5] suggested that innate immune responses play an equally significant, even more primary character compared with adaptive immune responses in IBD initiation and progression due to the observation that probiotics elicit anti- inflammatory effects in the GI tract by means of mucosal innate immune system stimulation, instead of suppression. In this study, we put efforts on addressing the interactions between probiotics and intestinal epithelial cells, the mechanism different from the conventionally dichotomous Th1/Th2 cytokine paradigm.
Probiotics have no pharmacological actions confirmed, but numerous benefits have been proposed, such as immunomodulation [6,7], antioxidant capacities [8], hepatoprotective effects [9], maintenance of commensal microflora [10], pathogen antagonization [11], anti-allergic effects [12,13] and decreased endotoxin level in plasma [14]. Lactobacillus plantarum, one of the most commonly used probiotics, is a member of the aerotolerant group of lactobacilli found in several fermented foods [15]. It is also one of the dominant Lactobacillus species in the hosts’ intestinal tract. Recent studies have shown that some strains of Lactobacillus plantarum attenuate inflammation induced by Shigella flexneri peptidoglycan by inhibiting nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB), inactivating mitogen-activated protein kinase (MAPK), and reducing NOD2 mRNA expression as well as protein levels, the actions which in turn lead to a decrease in pro-inflammatory cytokine secretion [16]. Moreover, van Baarlen et al. [17,18] demonstrated that even dead L. plantarum can exert beneficial functions protecting the host against the enormous array of commensal bacteria in the gut via epithelial crosstalk of mucosal interface microbiota. Their research team further investigated in vivo transcriptome responses to probiotics, the work shaping that different probiotic strains induced differential gene-regulatory networks and pathways in the human mucosa [19]. This provides advanced concept that not only live probiotics can exert beneficial effects, but also dead probiotics are able to modulate GI homeostasis. Second, because of strain-dependent properties, the anti-inflammation mechanism of single strains could not be extrapolated from other specific consequences without empirical evidence.
Systemic exposure to endotoxins accompanied with elevation of interleukin (IL)-6, IL-8 and IL-12 has been recognized as representative features of IBD progression [20,21]. Endotoxins are a family of molecules that bind to many pattern recognition receptors. One of the most dominant endotoxins is lipopolysaccharide (LPS). Previous exposure to LPS leads to cells hyporesponsive to subsequent challenge with LPS. This phenomenon is regarded as LPS tolerance.
LPS tolerance is typically associated with poor signal transduction in TLR4-NFκB pathway. TLR4 recognizes LPS from Gram-negative bacteria. Myeloid differentiation primary response gene 88 (Myd88) acts as a universal adapter protein used by TLRs (except for TLR3). Interleukin-1 receptor-associated kinase 1 (IRAK1) belongs to the serine/threonine protein kinase family. This kinase mediates the TLR4 downstream signaling transduction induced by LPS, forming a kinase complex required for the activation of NFκB. TNF receptor associated factor 6 (TRAF6) kinase mediates signal transduction from IRAK1, providing a link between IRAK1 and Mitogen-activated protein kinase 7 (TAK1). TAK1 activate IκB kinase (IKK), and thus plays a role in relaying signals to NFκB. IKK is an enzyme complex involved in IκBα phosphorylation, the action that gives rise to NFκB nuclear translocation [22].
It is well-studied that poor signal transduction in TLR4-NFκB pathway is mainly attributed to negative regulators [23]. Suppressors of cytokine signaling 1 (SOCS1) and suppressors of cytokine signaling 3 (SOCS3) are able to reduce JAK/STAT signal transduction, involving negative feedback to cytokine signaling [24]. Toll interacting protein (TOLLIP) interacts with many types of TLR signaling downstream pathways and potently inhibits the activity of IRAK after TLR activation. Overexpression of TOLLIP has been reported to inhibit inflammation in response to TLR4 signaling [25]. IL-1R-associated kinase 3 (IRAK3) suppresses the dissociation of IRAK1/4 from Myd88 and the connections among TRAF 6 complexes [26]. Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 1 (SHIP1) hydrolyses phosphatidylinositol 3-kinase, hence interfering with TLR4-MyD88 signaling pathway [27].
Since attenuation of pro-inflammatory cytokines secretions is IBD therapeutic targets, In this study, we co-cultured human epithelial colorectal adenocarcinoma (Caco-2) cells with probiotics and then administered LPS, which induced TNF-α, IL-6, IL-8 and IL-12 secretion, to biologically mimic the inflammatory situation of IBD. With the purpose of determining how L. plantarum weakens the downstream signal transduction of TLR4, the mRNAs that encode proteins participating in TLR4-NF-κB pathway were detected by RT-qPCR. Five negative regulator genes, SOCS1, SOCS3, TOLLIP, IRAK3 and SHIP1, which may result in inactivation of TLR4-NF-κB pathway, were also examined whether or not to be affected by probiotic treatment. Moreover, in order to explore which cellular parts contribute mostly to the anti-inflammatory properties, we tested the anti-inflammatory efficacies of live bacteria, heat-killed bacteria, cell wall extract, intracellular extract and bacterial genomic DNA in terms of negative regulator activation capacity.
Methods
Lactic acid bacterial strains
Isolation and identification of Lactobacillus plantarum from newborn infant feces and breast milk were performed in the Microbiology Laboratory of the Department of Food Science and Biotechnology of National Chung Hsing University, Taichung, Taiwan. Our preliminary data showed L. plantarum MYL26, L. plantarum MYL31, and L. plantarum MYL68 have better anti-inflammation abilities than those of other strains isolated in our laboratory.
Ethics statement
The samples from infants and adult subjects were approved employing in this study by Jeng-Yuan Hsu, Chairman of Institutional Review Board of the Taichung Veterans General Hospital. We obtained informed consent from both adult subjects and these infants’ guardians for collection of sample.
Preparation of cell wall, intracellular extracts and heat-killed lactic acid bacteria
All bacterial strains used in this study were stored at -80°C. Lactobacillus plantarum MYL26, Lactobacillus plantarum MYL31, and Lactobacillus plantarum MYL68 were cultured in MRS broth at 37°C for 16 h and collected by centrifugation at 2500 g for 8 min. For preparation of cell wall and intracellular extracts, cells were adjusted to 107 cfu/mL, washed twice with deionized water and suspended in phosphate-buffered saline (PBS). FRENCH® Pressure Cells Press (Thermo Electron, Waltham, USA) was used for cell disruption. Cell wall was removed by centrifugation at 5000 g for 10 min, and the supernatant was filtered through 0.22 μm filters as intracellular extract. The protein contents of intracellular extracts were adjusted to 1 mg/mL. The weight of cell wall extracts processed according to this protocol is about 10 ± 0.2 mg/107 cfu. For preparation of heat-killed cells, cells were suspended in PBS and adjusted to 107 cfu/mL followed by killing at 65°C for 30 min.
Preparation of bacterial genomic DNA
Lactic acid bacteria genomic DNA was extracted by tissue and cell genomic DNA purification system (GeneMark, Taichung, Taiwan). Nucleic acid concentration was measured at a wavelength of 260 nm and adjusted to 10 μg/mL.
Cell culture
Human intestinal epithelial-like cells (Caco-2) were obtained from the Bioresource Collection and Research Center (BCRC, Hsinchu, Taiwan) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 units/mL) and streptomycin (100 mg/mL) at 37°C in a humidified (95%) atmosphere with 5% CO2.
Cytokine secretions by stimulation of Caco-2 cells with L. plantarum MYL26 followed by LPS challenge
Caco-2 cells (106 cells/mL) were treated with live L. plantarum MYL26 (107 cfu/mL), heat-killed bacteria (107 cfu/mL), intracellular extracts (100 μg/mL), cell wall extracts (10 ± 0.2 mg/mL) and genomic DNA (1 μg/mL) at 37°C for 10 hours. After stimulation, cells were challenged with 1 μg/mL LPS for 18 hours. The supernatants were removed and IL-6, IL-8, IL-12p70 and TNF-α secretions were assayed by enzyme-linked immunosorbent assay (eBioscience ELISA system, California, USA).
siRNA silencing technique
Silencing of human SOCS1, SOCS3 and TOLLIP expressions was carried out in Caco-2 cells by using Dharmacon Human siGENOME® SMARTpool® siRNA Libraries for antisense oligonucleotides (AO) design. AO were transfected with DharmaFECT 2 reagent (Thermo Fisher Scientific, Massachusetts, USA) according to the manufacturer’s instructions. The siRNA experiment was conducted for 48 h and cells were collected to analyze total RNA for knockdown effect.
RT-qPCR
RNA isolation was conducted using EZ-RNA total RNA isolation kit (Biological Industries, Beit Haemek, Israel). Reverse transcription was carried out according to manufacturer’s instruction (Bio-Rad iScript™ cDNA synthesis kit, USA). Comparisons of gene expressions via qPCR were performed by adopting the following primer designs: SOCS3 (5′-CAA ATG TTG CTT CCC CCT TA-3′ and 5′-ATC CTG GTG ACA TGC TCC TC-3′), SHIP1 (5′-TCC AGC AGT CTT CCT CAC CT-3′ and 5′-GCT TGG ACA CCA TGT TGA TG-3′), IRAK3 (5′-GGG TGC CTG TAG CAG AGA AG-3′ and 5′-ATC TGG AGG AGC CAG GAT TT-3′), SOCS1 (5′-CTG GGA TGC CGT GTT ATT TT-3′ and 5′-TAG GAG GTG CGA GTT CAG GT-3′), TOLLIP (5′-CCA CAG TGT GAG GGA TTG TG-3′ and 5′-TCT CCT TCT CAT GCC GTT CT-3′), MyD88 (5′-GCA CAT GGG CAC ATA CAG AC-3′ and 5′-GAC ATG GTT AGG CTC CCT CA-3′), IKKβ (5′-GCT GCA ACT GAT GCT GAT GT-3′ and 5′- TGT CAC AGG GTA GGT GTG GA-3′), TAK1 (5′-TTT GCT GGT CCT TTT CAT CC-3′ and 5′-TGC CCA AAC TCC AAA GA ATC-3′), TLR4 (5′-TGA GCA GTC GTG CTG GTA TC-3′ and 5′-CAG GGC TTT TCT GAG TCG TC-3′), IκBα (5′-GCA AAA TCC TGA CCA GGT GT-3′ and 5′-GCT CGT CCT CTG TGA ACT CC-3′), GAPDH (5′-GAG TCA ACG GAT TTG GTC GT-3′ and 5′-TTG ATT TTG GAG GGA TCT CG-3′), TRAF6 (5′-CTG CAA AGC CTG CAT CAT AA-3′ and 5′-GGG GAC AAT CCA TAA GAG CA-3′), IRAK1 (5′-GGG TCC AGG TGC TTC TTG TA-3′ and 5′-TGC TAG AGA CCT TGG CTG GT-3′). Quantitative PCR was carried out according to the manufacturer’s protocol. After reverse transcription of mRNA, 5 μl of the reverse transcription product were added to a BioRad iCyclerTM PCR system containing 0.3 μM of each primer. One-fold QuantiTect SYBR Green PCR Master Mix was used as a fluorescent reporter (QuantiTect SYBR Green PCR, Qiagen). The condition was programmed as follows: (1) Denaturation at 94°C for 10 min; (2) Amplification for 40 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 20 s.
Cell viability assay
3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, which is based on the cleavage of the tetrazolium salt by mitochondrial dehydrogenases in viable cells. In order to determine toxicity concentration, approximately 105 cells were plated onto each well of 96-well plates for 24 h, followed by treatment with different probiotic agents for 6, 8, 10, 12 and 14 hours. After incubation, 200 mL of MTT solution (0.5 mg/mL) were added to each well for 4 h after washing by PBS. Finally, the supernatant was removed and 200 μL of dimethyl sulphoxide (DMSO) were added to each well to dissolve the dark blue formazan crystals. The absorbance was measured by ELISA plate reader (Jupiter, ASYS Hitech, Austria) at 570 nm. To compare the results, the relative cell viability was expressed as the mean percentage of viable cells compared with untreated cells (100%).
Statistical analysis
Each value is the mean of triplicate experiments in each group. Means comparison was carried out by Student's t-test. P < 0.05 was considered significantly different.
Results
Lactobacillus plantarum MYL26/ MYL31/ MYL68 treatment did not affect the Caco-2 cell viability within 10 hours
Due to excellent lactic acid production capacities of Lactobacillus plantarum, we perform MTT assay to assess the most appropriate incubation time. As Figure 1 showed, cell viability was not influenced within 10 hours. Incubated with 12 and 14 hours, Caco-2 cell viability showed significant decrease. As a result, we co-cultured Caco-2 cells and Lactobacillus plantarum for 10 hours in the following experiments.
Figure 1.

Approximately 1 × 105 cells were plated onto 96-well plates for 24 h, followed by treatment with live/ heat-killed L. plantarum MYL26 (L. plantarum MYL31/ MYL68 data not shown) and different cellular parts for 6, 8, 10, 12 and 14 hours. Symbol * represents P-value smaller than 0.05 analyzed by t-test in comparison with negative control group. (n = 3). Negative control: Caco-2 cells were not treated with probiotics.
Lactobacillus plantarum attenuates LPS-induced cytokine secretion
Three different strains of Lactobacillus plantarum (MYL26, MYL31 and MYL68) were tested and the most potent strain, in terms of refractoriness to subsequent LPS stimulation, was selected. As shown in Figure 2, L. plantarum MYL26 attenuated TNF-α, IL-6, IL-8, and IL-12 production more effectively than those of other strains.
Figure 2.
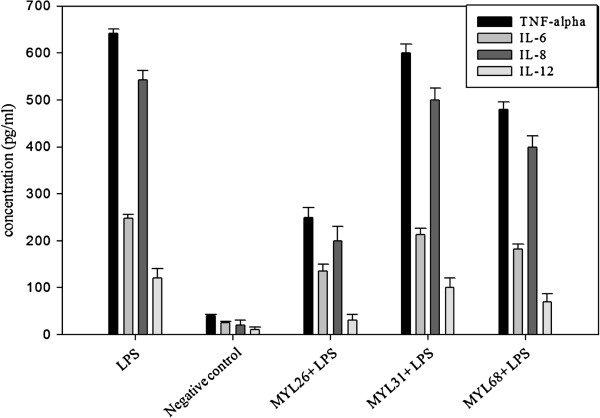
Caco-2 cells (106 cells/mL) were treated with live L. plantarum MYL26/ MYL31/ MYL68 (107 cfu/mL) at 37°C for 10 hours, followed by 1 μg/mL LPS challenge. Negative control: Caco-2 cells were not treated with LPS and probiotics. (Cytokine secretion baseline).
Lactobacillus plantarum MYL26 attenuates downstream signal transduction of TLR4-NFκB pathway
The results of RT-qPCR (Figure 3) indicated that there are no significant differences in the expressions of TLR4, MyD88 and IRAK1 in comparison with those of LPS treatment group. The expressions of TRAF6, TAK1 and IKKβ decreased more significantly under L. plantarum MYL26 treatment than those under LPS treatment alone.
Figure 3.
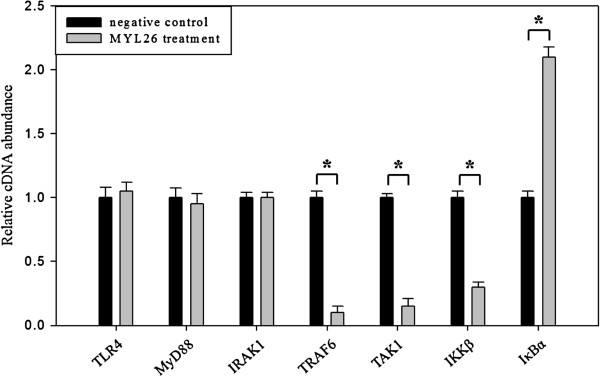
Caco-2 cells (106 cells/mL) were treated with live L. plantarum MYL26 (107 cfu/mL) at 37°C for 10 hours followed by 1 μg/mL LPS challenge. Gene expressions were assayed by RT-qPCT normalized by GAPDH. Symbol * represents P-value smaller than 0.05 analyzed by t-test in comparison with negative control group. (n = 3). Negative control: Caco-2 cells were challenged by LPS without pretreatment with probiotics.
Lactobacillus plantarum MYL26 pretreatment elicits anti-inflammatory properties by enhancing the expressions of TOLLIP, SOCS1 and SOCS
Since TRAF6, TAK1 and IKKβ were down-regulated, five potential negative regulator gene expressions were examined. As shown in Figure 4, there were no considerable differences in the expressions of IRAK3 and SHIP1 while the expressions of TOLLIP, SOCS1 and SOCS3 were higher than those in the control groups.
Figure 4.

Caco-2 cells (106 cells/mL) were treated with live L. plantarum MYL26 (107 cfu/mL) at 37°C for 10 hours. Gene expressions were assayed by RT-qPCR normalized by GAPDH. Symbol * represents P-value smaller than 0.05 analyzed by t-test in comparison with negative control group. (n = 3). Negative control: Caco-2 cells were not treated with probiotics.
TOLLIP, SOCS1 and SOCS3 knockdown gave rise to impaired anti-inflammation abilities
We then used gene knockdown technique to silence TOLLIP, SOCS1 and SOCS3. Prior tests have shown that silencing of target genes does not decrease the expression of non-target genes (Figure 5). TOLLIP, SOCS1 and SOCS3 were silenced separately and subsequently challenged by LPS. The silencing of these three genes resulted in the partial loss of anti-inflammatory function of L. plantarum MYL26 (Figure 6).
Figure 5.
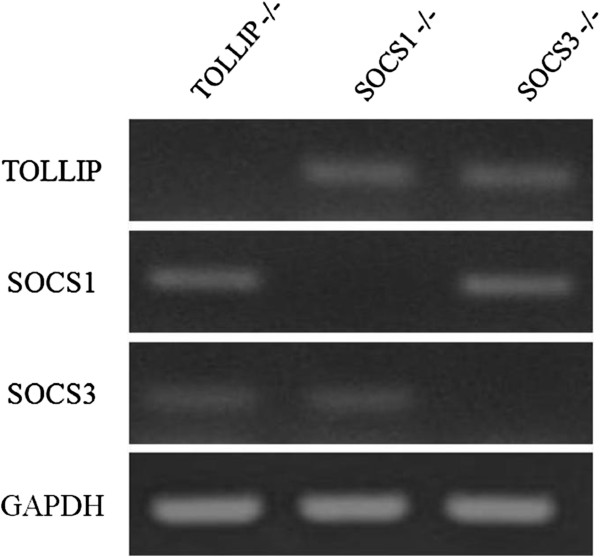
Human SOCS1, SOCS3 and TOLLIP gene expressions were not off-targeted. The siRNA experiment was conducted for 48 h.
Figure 6.
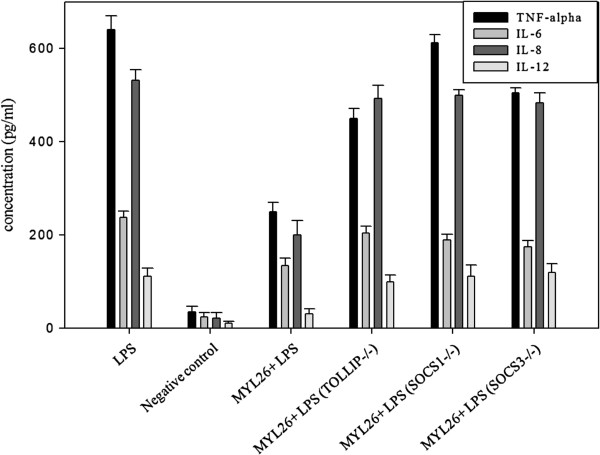
TOLLIP, SOCS1 and SOCS3-silenced Caco-2 cells (106 cells/mL) were treated with live L. plantarum MYL26 (107 cfu/mL) at 37 ±°C for 10 hours, followed by 1 μg/mL LPS challenge. Negative control: Caco-2 cells were not treated with LPS and probiotics. (Cytokine secretion baseline).
The physiologically active components that affect SOCS1/3, TOLLIP and IκBα expression might be located in the cell walls
To investigate the involvement of different cellular parts in reducing LPS-induced inflammation, live bacteria, heat-killed bacteria, cell wall extract, intracellular extract and bacterial genomic DNA were tested to assess which cellular parts activate TOLLIP, SOCS1, SOCS3 and IκBα. The results showed that dead L. plantarum MYL26 activate gene expressions as well as live bacteria. Cell wall extract, intracellular extract and genomic DNA also stimulated gene expression, but not as well as the whole cell (Figure 7).
Figure 7.
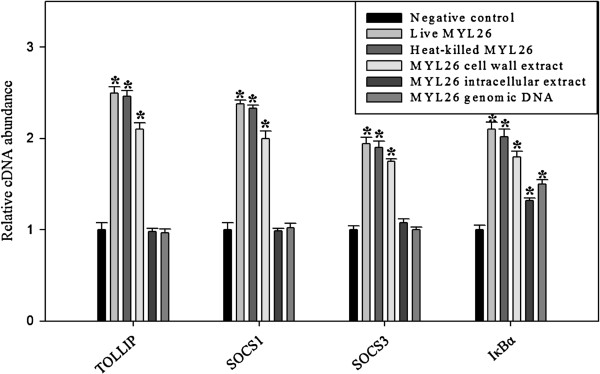
The candidate anti-inflammation gene expressions were induced in different degrees by diverse cellular components. Caco-2 cells (106 cells/mL) were treated with live L. plantarum MYL26 (107 cfu/mL), heat-killed bacteria (107 cfu/mL), intracellular extracts (100 μg/mL), cell wall extracts (10 ± 0.2 mg/mL) and genomic DNA (1 μg/mL) at 37°C for 10 hours. Symbol * represents P-value smaller than 0.05 analyzed by t-test in comparison with negative control group. (n = 3). Negative control: Caco-2 cells were not treated with probiotics.
Discussion
Almost all of the IBD medicines are associated with decrease of inflammation signal pathways. On the other hand, pro-inflammatory cytokines play imperative character in mediating the progression of IBD. Numerous clinical trials have shown that better control of pro-inflammatory cytokine production is an essential method for improving symptoms [28-30].
Due to sustained contact with pathogen-associated molecular patterns (PAMPs), the epithelial cells act as the first barrier of defense against invading microbes. Intestinal epithelial cells take part in mediating balanced immune actions, as well as stimulating immune cells that dwell in the lamina propria. In this respect, Baumgart et al. [31] suggested that IBD results from a collapse of tolerance towards the commensal microbiota. An aberrant LPS response results in an inflammatory phenotype. As a consequence, elevated attention to probiotics for the treatment of GI tract disorders has shed light on new therapeutic regimens.
LPS tolerance may occur as the host’s defense system that confines an inflammatory break upon successive stimulation [32]. In our study, it is expected to reveal the mechanism by which prolonged contact of lactic acid bacteria with intestinal epithelial cells leads to hyporesponsive to the following inflammatory stimuli. It helps establish a probiotic screen criteria for selection of the best LPS tolerance induction bacterial strains, rather than traditional criteria focused on bile-acid resistant ability.
Until now, many possible anti-inflammatory mechanisms of probiotic actions have been proposed and it is observed that probiotic effect is both strain dependent and dose dependent [33]. Although different strains of lactic acid bacteria possess different properties, there have been the most publications reported on L. plantarum when searching by key words “dead probiotics” or ”killed probiotics”. As a result, we examined three different strains of L. plantarum and used the most potent strain MYL26, as a study object researching the underlying molecular mechanisms.
In this research, upon L. plantarum MYL26 treatment, the expression of genes that encode proteins participating in LPS-induced inflammation was compared with that of untreated group and found that TRAF6, TAK1 and IKKβ expressions were suppressed. We also observed that expression of IκBα was increased. It was perhaps attributed to prior probiotic stimulation on Caco-2 cells, the action that caused mild inflammation (data not shown) as well as slightly NFκB nuclear translocation which encoded not only cytokines but also IκBα. This observation was similar to the results Wahlstrom et al. reported [34]. They suggested that low-dose LPS pretreatment changed subsequent LPS-activated signal transduction pathways by means of up-regulation of IκBα that acted as a feedback control inhibitor.
Since the results showed that anti-inflammatory effects of L. plantarum MYL26 on Caco-2 might be through interfering with TLR4 downstream pathway, it is reasonable to infer that the activation of the negative regulators of TLR4-NFκb pathway contributes to the anti-inflammatory effect. We investigated TLRs-associated negative regulators, including TOLLIP, SOCS1, SOCS3, IRAK3 and SHIP1, and found TOLLIP and SOCS1/3 expressions were enhanced by L. plantarum MYL26 treatment. However, the consequence that TOLLIP and SOCS1/3 knockdown gave rise to impaired anti-inflammatory ability further supported the hypothesis that activation of the negative regulators of TLR4-NFκb pathway is a primary exploit for the anti-inflammatory effect L. plantarum MYL26 exerts. As numerous literatures have revealed that different components of the bacterial cells can result in different activities on the human GI tract, such as cell wall components [35], genomic DNA [36], and intracellular extract [37], we further researched the TOLLIP and SOCS1/3 activation ability of live/ heat-killed whole bacterial cells, cell wall extract, intracellular extract and genomic DNA from L. plantarum MYL26 to see which cellular parts contributed mostly to LPS tolerance induction.
In contrast with our expectations, although intracellular extract and genomic DNA induced IκBα expression more significantly than that of control group, they failed to activate TOLLIP, SOCS1, and SOCS3. There are five TLRs (TLR2/ 4/ 5/ 7/ 9) sharing similar downstream signal pathway (MyD88, IRAK, TRAF, IKK, NFκb) [38]. Except for IκBα which directly binds to NFκb, the negative regulators TOLLIP, SOCS1, and SOCS3 are well-established having abilities in interference with recruitment of MyD88 and IRAK. It has been reported that TOLLIP, SOCS1, and SOCS3 not only attenuate TLR4 signaling, but also have impact on TLR2/5/7/9 signaling [39,40]. Briefly, L. plantarum MYL26 intracellular extract and genomic DNA activate TLRs-NFκb pathways other than TLR4 (TLRs cross-tolerance), but they did not attenuate inflammation through induction of TOLLIP, SOCS1, and SOCS3. Taken together, we proposed that L. plantarum MYL26 intracellular extract and genomic DNA induced LPS tolerance through pathways different from induction of Tollip, SOCS-1 and SOCS-3, which were key negative regulators activated by live/dead L. plantarum MYL26 and cell wall components.
One of the limitations of this study is that the causes of IBD, other than breakdown of LPS tolerance, are multifaceted. Several lines of evidence has pointed out that in addition to inherited factors, pollution, drugs, diets, breastfeeding, even emotional stress, could be responsible for genetically failing to interpret molecular microbial patterns appropriately, thus leading to irregular innate and adaptive immune responses [41,42]. The second limitation is that PAMPs other than LPS induce GI inflammation through different pathways. Criteria for probiotic selection of LPS tolerance induction strains might be not suitable with respect to inflammation symptoms caused by other PAMPs.
Conclusions
The administration of lactic acid bacteria in patients suffering from GI disorders regularly depends on try-error methods, and numerous probiotics treatment applied to clinical trials showed frustrated results, which perhaps might be related to the fact that the probiotic screening criteria is generally based on susceptibility to artificial GI environments (acid and bile resistance) or adhesive properties instead of on immunomodulatory capacities, for instance, induction of LPS tolerance. Our research provided a new insight to describe the L. plantarum strain-dependent characterization in terms of anti-inflammatory effects, and suggested an essential role for Lactobacillus plantarum and Lactobacillus plantarum-derived constituents in the induction of LPS tolerance.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
Chiu YH and Lin MY conceived and designed the experiments. Tsai CC and Huang CT performed the experiments. Lu YC, Ou CC and Lin SL analyzed the data and performed the computational analysis, producing the figures and tables. Chiu YH drafted the manuscript and Lin MY revised it. All authors read and approved the final manuscript.
Contributor Information
Yi-Heng Chiu, Email: yihengchiu@gmail.com.
Ying-Chen Lu, Email: biolyc@mail.ncyu.edu.tw.
Chu-Chyn Ou, Email: occ@csmu.edu.tw.
Shiao-Lin Lin, Email: lilquiet1231@hotmail.com.
Chin-Chi Tsai, Email: b942010009@student.nsysu.edu.tw.
Chien-Tsai Huang, Email: g9843021@nchu.edu.tw.
Meei-Yn Lin, Email: mylin@dragon.nchu.edu.tw.
Acknowledgements
We thank Chung CD for excellent technical support and helpful discussions of the data. This work was funded by grant from National Science Council of Taiwan.
References
- Sorensen GV, Erichsen R, Svaerke C, Farkas DK, Sorensen HT. Risk of cancer in patients with inflammatory bowel disease and venous thromboembolism: a nationwide cohort study. Inflammatory bowel diseases. 2012;18(10):1859–1863. doi: 10.1002/ibd.22870. [DOI] [PubMed] [Google Scholar]
- Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369(9573):1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- Parkes GC, Sanderson JD, Whelan K. Treating irritable bowel syndrome with probiotics: the evidence. Proc Nutr Soc. 2010;69(2):187–194. doi: 10.1017/S002966511000011X. [DOI] [PubMed] [Google Scholar]
- McFarland LV, Dublin S. Meta-analysis of probiotics for the treatment of irritable bowel syndrome. World J Gastroenterol. 2008;14(17):2650–2661. doi: 10.3748/wjg.14.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arseneau KO, Tamagawa H, Pizarro TT, Cominelli F. Innate and adaptive immune responses related to IBD pathogenesis. Curr Gastroenterol Rep. 2007;9(6):508–512. doi: 10.1007/s11894-007-0067-3. [DOI] [PubMed] [Google Scholar]
- Peng S, Lin JY, Lin MY. Antiallergic effect of milk fermented with lactic acid bacteria in a murine animal model. J Agric Food Chem. 2007;55(13):5092–5096. doi: 10.1021/jf062869s. [DOI] [PubMed] [Google Scholar]
- Li CY, Lin HC, Lai CH, Lu JJ, Wu SF, Fang SH. Immunomodulatory effects of Lactobacillus and Bifidobacterium on both murine and human mitogen-activated T cells. Int Arch Allergy Immunol. 2011;156(2):128–136. doi: 10.1159/000322350. [DOI] [PubMed] [Google Scholar]
- Ou CC, Lu TM, Tsai JJ, Yen JH, Chen HW, Lin MY. Antioxidative effect of lactic acid bacteria: intact cells vs. intracellular extracts. J Food Drug Anal. 2009;17(3):209–216. [Google Scholar]
- Loguercio C, Federico A, Tuccillo C, Terracciano F, D'Auria MV, De Simone C, Del Vecchio BC. Beneficial effects of a probiotic VSL#3 on parameters of liver dysfunction in chronic liver diseases. J Clin Gastroenterol. 2005;39(6):540–543. doi: 10.1097/01.mcg.0000165671.25272.0f. [DOI] [PubMed] [Google Scholar]
- Trebichavsky I, Rada V, Splichalova A, Splichal I. Cross-talk of human gut with bifidobacteria. Nutr Rev. 2009;67(2):77–82. doi: 10.1111/j.1753-4887.2008.00141.x. [DOI] [PubMed] [Google Scholar]
- Golowczyc MA, Mobili P, Garrote GL, Abraham AG, De Antoni GL. Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar Enteritidis. Int J Food Microbiol. 2007;118(3):264–273. doi: 10.1016/j.ijfoodmicro.2007.07.042. [DOI] [PubMed] [Google Scholar]
- Ou CC, Lin SL, Tsai JJ, Lin MY. Heat-killed lactic acid bacteria enhance immunomodulatory potential by skewing the immune response toward Th1 polarization. J Food Sci. 2011;76(5):M260–M267. doi: 10.1111/j.1750-3841.2011.02161.x. [DOI] [PubMed] [Google Scholar]
- Chuang L, Wu KG, Pai C, Hsieh PS, Tsai JJ, Yen JH, Lin MY. Heat-killed cells of lactobacilli skew the immune response toward T helper 1 polarization in mouse splenocytes and dendritic cell-treated T cells. J Agric Food Chem. 2007;55(26):11080–11086. doi: 10.1021/jf071786o. [DOI] [PubMed] [Google Scholar]
- Tandon P, Moncrief K, Madsen K, Arrieta MC, Owen RJ, Bain VG, Wong WW, Ma MM. Effects of probiotic therapy on portal pressure in patients with cirrhosis: a pilot study. Liver Int. 2009;29(7):1110–1115. doi: 10.1111/j.1478-3231.2009.02020.x. [DOI] [PubMed] [Google Scholar]
- Sanni AI, Morlon-Guyot J, Guyot JP. New efficient amylase-producing strains of Lactobacillus plantarum and L. fermentum isolated from different Nigerian traditional fermented foods. Int J Food Microbiol. 2002;72(1-2):53–62. doi: 10.1016/S0168-1605(01)00607-9. [DOI] [PubMed] [Google Scholar]
- Kim HG, Lee SY, Kim NR, Lee HY, Ko MY, Jung BJ, Kim CM, Lee JM, Park JH, Han SH, Chung DK. Lactobacillus plantarum lipoteichoic acid down-regulated Shigella flexneri peptidoglycan-induced inflammation. Mol Immunol. 2011;48(4):382–391. doi: 10.1016/j.molimm.2010.07.011. [DOI] [PubMed] [Google Scholar]
- van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, Hooiveld GJ, Brummer RJ, Kleerebezem M. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci USA. 2009;106(7):2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, Brummer RJ, Kleerebezem M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4562–4569. doi: 10.1073/pnas.1000079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desreumaux P. Specific targeting of IL-6 signalling pathway: a new way to treat IBD? Gut. 2000;47(4):465–466. doi: 10.1136/gut.47.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarek D, Cibor D, Szczepanek M, Mach T. Biological therapy of inflammatory bowel disease. Pol Arch Med Wewn. 2009;119(1–2):84–88. [PubMed] [Google Scholar]
- West MA, Heagy W. Endotoxin tolerance: a review. Crit Care Med. 2002;30(1 Suppl):S64–S73. [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5(6):446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Tamiya T, Kashiwagi I, Takahashi R, Yasukawa H, Yoshimura A. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T cell inflammation by SOCS1 and SOCS3. Arterioscler Thromb Vasc Biol. 2011;31(5):980–985. doi: 10.1161/ATVBAHA.110.207464. [DOI] [PubMed] [Google Scholar]
- Bulut Y, Faure E, Thomas L, Equils O, Arditi M. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J Immunol. 2001;167(2):987–994. doi: 10.4049/jimmunol.167.2.987. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Hernandez LD, Galán JE, Janeway CA Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110(2):191–202. doi: 10.1016/S0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- Arndt PG, Suzuki N, Avdi NJ, Malcolm KC, Worthen GS. Lipopolysaccharide- induced c-Jun NH2-terminal kinase activation in human neutrophils: role of phosphatidylinositol 3-Kinase and Syk-mediated pathways. J Biol Chem. 2004;279(12):10883–10891. doi: 10.1074/jbc.M309901200. [DOI] [PubMed] [Google Scholar]
- Moustafa T, Fickert P, Magnes C, Guelly C, Thueringer A, Frank S, Kratky D, Sattler W, Reicher H, Sinner F, Gumhold J, Silbert D, Fauler G, Hofler G, Lass A, Zechner R, Trauner M. Alterations in lipid metabolism mediate inflammation, fibrosis, and proliferation in a mouse model of chronic cholestatic liver injury. Gastroenterology. 2012;142(1):140–151. doi: 10.1053/j.gastro.2011.09.051. e12. [DOI] [PubMed] [Google Scholar]
- Bondia-Pons I, Ryan L, Martinez JA. Oxidative stress and inflammation interactions in human obesity. Journal of physiology and biochemistry. 2012;68(4):701–711. doi: 10.1007/s13105-012-0154-2. [DOI] [PubMed] [Google Scholar]
- Leonard B, Maes M. Mechanistic explanations how cell-mediated immune activation, inflammation and oxidative and nitrosative stress pathways and their sequels and concomitants play a role in the pathophysiology of unipolar depression. Neurosci Biobehav Rev. 2012;36(2):764–785. doi: 10.1016/j.neubiorev.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Baumgart DC, Thomas S, Przesdzing I, Metzke D, Bielecki C, Lehmann SM, Lehnardt S, Dorffel Y, Sturm A, Scheffold A. et al. Exaggerated inflammatory response of primary human myeloid dendritic cells to lipopolysaccharide in patients with inflammatory bowel disease. Clin Exp Immunol. 2009;157(3):423–436. doi: 10.1111/j.1365-2249.2009.03981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinderola G, Matar C, Perdigon G. Role of intestinal epithelial cells in immune effects mediated by gram-positive probiotic bacteria: involvement of toll-like receptors. Clin Diagn Lab Immunol. 2005;12(9):1075–1084. doi: 10.1128/CDLI.12.9.1075-1084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maassen CB, Claassen E. Strain-dependent effects of probiotic lactobacilli on EAE autoimmunity. Vaccine. 2008;26(17):2056–2057. doi: 10.1016/j.vaccine.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Wahlstrom K, Bellingham J, Rodriguez JL, West MA. Inhibitory kappaBalpha control of nuclear factor-kappaB is dysregulated in endotoxin tolerant macrophages. Shock. 1999;11(4):242–247. doi: 10.1097/00024382-199904000-00003. [DOI] [PubMed] [Google Scholar]
- Kang SS, Ryu YH, Baik JE, Yun CH, Lee K, Chung DK, Han SH. Lipoteichoic acid from Lactobacillus plantarum induces nitric oxide production in the presence of interferon-gamma in murine macrophages. Mol Immunol. 2011;48(15–16):2170–2177. doi: 10.1016/j.molimm.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Shimosato T, Kimura T, Tohno M, Iliev ID, Katoh S, Ito Y, Kawai Y, Sasaki T, Saito T, Kitazawa H. Strong immunostimulatory activity of AT- oligo- deoxynucleotide requires a six-base loop with a self-stabilized 5′-C…G-3′ stem structure. Cell Microbiol. 2006;8(3):485–495. doi: 10.1111/j.1462-5822.2005.00640.x. [DOI] [PubMed] [Google Scholar]
- Pathmakanthan S, Li CK, Cowie J, Hawkey CJ. Lactobacillus plantarum 299: beneficial in vitro immunomodulation in cells extracted from inflamed human colon. J Gastroenterol Hepatol. 2004;19(2):166–173. doi: 10.1111/j.1440-1746.2004.03181.x. [DOI] [PubMed] [Google Scholar]
- Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16(1):3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33(9):449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Naka T, Fujimoto M, Tsutsui H, Yoshimura A. Negative regulation of cytokine and TLR signalings by SOCS and others. Adv Immunol. 2005;87:61–122. doi: 10.1016/S0065-2776(05)87003-8. [DOI] [PubMed] [Google Scholar]
- Talaei F, Atyabi F, Azhdarzadeh M, Dinarvand R, Saadatzadeh A. Overcoming therapeutic obstacles in inflammatory bowel diseases: a comprehensive review on novel drug delivery strategies. European journal of pharmaceutical sciences: official journal of the European Federation for Pharmaceutical Sciences. 2013;49(4):712–722. doi: 10.1016/j.ejps.2013.04.031. [DOI] [PubMed] [Google Scholar]
- Frolkis A, Dieleman LA, Barkema H, Panaccione R, Ghosh S, Fedorak RN, Madsen K, Kaplan GG. Environment and the inflammatory bowel diseases. Can J Gastroenterol = J Can Gastroenterologie. 2013;27(3):e18–e24. doi: 10.1155/2013/102859. [DOI] [PMC free article] [PubMed] [Google Scholar]


