Abstract
Genetic heterogeneity could reduce the power of linkage analysis to detect risk loci for complex traits such as alcohol dependence (AD). Previously, we performed a genomewide linkage analysis for AD in African-Americans (AAs) (Gelernter et al., 2009). The power of that linkage analysis could have been reduced by the presence of genetic heterogeneity owing to differences in admixture among AA families. We hypothesized that by examining a study sample whose genetic ancestry was more homogeneous we could increase the power to detect linkage. To test this hypothesis, we performed ordered subset linkage analysis (OSA) in 384 AA families using admixture proportion as a covariate to identify a more homogeneous subset of families and determine whether there is increased evidence for linkage with AD. Statistically significant increases in lod scores in subsets relative to the overall sample were identified on chromosomes 4 (P=0.0001), 12 (P=0.021), 15 (P=0.026) and 22 (P=0.0069). In a subset of 44 families with African ancestry proportions ranging from 0.858 to 0.996, we observed a genomewide significant linkage at 180 cM on chromosome 4 (lod=4.24, pointwise P<0.00001, empirical genomewide P=0.008). A promising candidate gene located there, GLRA3, which encodes a subunit of the glycine neurotransmitter receptor. Our results demonstrate that admixture proportion can be used as a covariate to reduce genetic heterogeneity and enhance the detection of linkage for AD in an admixed population such as AAs. This approach could be applied to any linkage analysis for complex traits conducted in an admixed population.
Keywords: linkage, ordered subset linkage analysis, genetic heterogeneity, alcohol dependence, admixture
Introduction
Linkage analysis can be a useful design to map disease susceptibility genes with diverse genetic effects that segregate in families, including common variants, multiple rare variants at one locus, and inherited copy number variants (CNVs). However, linkage analyses of complex traits or disorders are complicated by the presence of genetic heterogeneity. The power of linkage analysis is reduced when different families have different major risk loci (Tsuang et al. 1993).
Analysis methods that address the presence of genetic heterogeneity may increase the power of linkage analysis. Ordered subset linkage analysis (OSA) was developed as a tool for linkage analysis of complex traits characterized by genetic heterogeneity (Hauser et al. 2004). OSA uses trait-related covariate information to identify more etiologically homogeneous subsets of families that may be more similar genetically than the larger group from which they are selected, and therefore should maximize detectable evidence for linkage. In addition to providing increased power for linkage analysis, by decreasing genetic heterogeneity OSA may also reduce the linkage interval and refine gene location (Scott et al. 2003).
Alcohol dependence (AD) is extremely costly to individuals and to society throughout the world. While the etiology of AD is complex, genetic factors are known to be important to the development of AD (Gelernter and Kranzler 2009). Previously, we performed a genomewide linkage analysis for AD in African-Americans (AAs) and identified a genomewide-significant linkage on chromosome 10q23.3–24.1 at 117.2 centiMorgans (logarithm of odds score 3.32; empirical genomewide p = .033) (Gelernter et al. 2009). However, we speculated that the power of that linkage analysis in AA families could be reduced by the presence of genetic heterogeneity owing to the variation of admixture proportions across families for two reasons. First, numerous linkage studies for various complex disorders have shown evidence of different linkage signals across populations. Second, the different 12-month prevalence rates of alcohol abuse or AD between European-Americans (EAs) and AAs may indicate different genetic underpinnings of AD for the two population groups. For example, based on the data on the 12-month prevalence of alcohol abuse and dependence in the United States assessed in the National Institute on Alcohol Abuse and Alcoholism’s (NIAAA) 2001–2002 National Epidemiologic Survey on Alcohol and Related Conditions, the prevalence of alcohol abuse was significantly greater among EAs (5.10%) compared to AAs (3.29%), and the EAs (10.71%) had higher rates of AD than AAs (6.03%) for younger a group of people (ages 18–29).
In the current study, we hypothesized that a sample whose genetic ancestry was more homogeneous would increase power to detect linkage. To test this hypothesis, we used admixture proportion as a covariate and applied the OSA technique to identify a more homogeneous subset of families to determine whether it increased evidence for linkage with AD.
Methods and Materials
Recruitment
Subjects were originally ascertained for genetic studies of cocaine dependence (CD) and opioid dependence (OD) (Gelernter et al. 2005; Gelernter et al. 2006) using an affected sibling pair (ASP) linkage design. The ascertained subjects include both AAs and EAs. Ordered subset linkage analysis was conducted in genetically defined AAs as described below in the Diagnosis and Study Subjects section. There were four recruitment sites: Yale University School of Medicine (APT Foundation; New Haven, Connecticut), University of Connecticut Health Center (UConn; Farmington, Connecticut), Medical University of South Carolina (MUSC; Charleston, South Carolina), and McLean Hospital (Harvard Medical School; Belmont, Massachusetts). Families were recruited based on screening information that suggested that at least two siblings would fulfill diagnostic criteria for CD or OD. There were no differences in subject recruitment protocol for the two studies except for the primary trait of the proband ascertained. Alcohol use played no role in proband selection or pedigree extension. Probands with an axis I clinical diagnosis of a major psychotic disorder such as schizophrenia or schizoaffective disorder were excluded from participation. When an ASP was recruited, additional siblings and parents were recruited whenever possible regardless of affection status. After a complete description of the study was provided to the subjects, written informed consent was obtained from all subjects. The Yale University School of Medicine institutional review board, the University of Connecticut Health Center institutional review board, Medical University of South Carolina institutional review board, and McLean Hospital institutional review board approved the study and a certificate of confidentiality for the work was obtained from the National Institute on Drug Abuse.
Diagnosis and study subjects
Subjects were accessed using the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA), a polydiagnostic instrument that has been described in detail previously (Pierucci-Lagha et al. 2005; Pierucci-Lagha et al. 2007). The diagnosis of AD was derived using a computer algorithm based on DSM-IV diagnostic criteria. Subjects were classified as AA on the basis of a Bayesian model-based clustering method implemented in STRUCTURE (Pritchard et al. 2000; Falush et al. 2003), as described previously (Gelernter et al. 2009; Panhuysen et al. 2010). Most self-reported AA subjects cluster with the AA group. A small proportion of self-reported AA subjects (1.6%) were classified as EAs and not used in the current study. Characteristics of the sample in this study are shown in Table 1. Specifically, a total of 124 families with at least 2 AD affected siblings were tested for linkage using a non-parametric, allele-sharing model (one additional family was excluded from analysis because we detected an unexpected relationship). However, other families with no or only one AD subject were also included to improve allele frequency estimation for linkage analysis.
Table 1.
Characteristics of the study sample
| No. of families | 384 |
| No. of individuals in the linkage set | 1,880 |
| No. of genotyped individuals | 1,022 |
| Mean age in yrs (SD) | |
| All genotyped individuals | 41.2 (7.3) |
| Alcohol dependence affected Males (%) | 39.9 (6.7) |
| All genotyped individuals | 43.3 |
| Alcohol dependence affected | 59.8 |
| No. of pedigrees with | |
| 0 subject with alcohol dependence | 110 |
| 1 subject with alcohol dependence | 149 |
| 2 subject with alcohol dependence | 112 |
| 3 subject with alcohol dependence | 8 |
| 4 subject with alcohol dependence | 3 |
| 5 subject with alcohol dependence | 2 |
Genotyping and Quality Control
DNA was extracted from immortalized cell lines in most cases, but for a small number of subjects DNA was extracted directly from blood or saliva. A total number of 898 subjects were genotyped at the Center for Inherited Disease Research (CIDR) for the 6,008 single nucleotide polymorphisms (SNPs) Illumina Human Linkage IVb Marker Panel. An additional 124 subjects were genotyped at the Yale Keck Center with the 6,090 SNP Illumina Infinium-12 Human Linkage Marker Panel. We limited our analyses to 4,518 autosomal SNPs available in both panels. PLINK (Purcell et al. 2007) was used to calculate allele frequencies and examine HWE based on a set of randomly selected unrelated subjects (N=384, one per family). There are 23 SNPs with genotyping rate ≤ 0.95, 318 SNPs with minor allele frequency (MAF) ≤ 0.1, and 46 SNPs with Hardy-Weinberg equilibrium p ≤ 0.01, which were excluded from analysis. Thus, 4,133 autosomal SNPs were retained for further analysis. To avoid inflated linkage signals that could be caused by linkage disequilibrium (LD) between markers, a subset of 3675 SNPs with low pairwise LD (rsq<0.1) were selected for linkage analysis.
We used PedCheck (O'Connell and Weeks. 1998) and Merlin (Abecasis et al. 2002) to identify Mendelian inconsistencies. Merlin was also used to identify potentially incorrect genotypes based on estimation of the probability of double-crossover events using the “--error” option. Mendelian inconsistencies and genotyping errors were set as missing. We used the Pedigree RElationship Statistical Test (PREST) (McPeek et al. 2000) to verify family relationships. Pedigree errors were detected in two families: in one family, the relationship was corrected based on the shared IBD patterns, with the re-assigned family relationships verified by PREST and in the other family the relationship could not be resolved, so it was excluded from further analysis.
Statistical analysis
Admixture proportion estimation
We estimated the admixture proportion for each individual based on markers that were informative with respect to ancestry using the STRUCTURE program. We selected 1,574 such markers from the SNP linkage panel. Selection of markers was on the basis of the genetic information downloaded from the Hapmap CEU and YRI samples. Markers were chosen based on the following criteria: 1) absolute difference (δ) in allele frequency between the two HapMap populations > 0.2; 2) r2 between each pair of SNPs < 0.1 within each population; and 3) p-value > 0.01 for testing HWE within each population. In addition, STRUCTURE was developed to estimate ancestry proportion for unrelated individuals; we therefore divided the whole family dataset to create several subsets with each subset including only randomly selected unrelated subjects, and STRUCTURE was run separately on each subset. The log-likelihood of each analysis for a different number of population groups was estimated from the average of three independent runs (20,000 burn in and 30,000 iterations) and, as expected, the result favored a two-ancestry population model. Family-specific admixture proportion was derived by taking the mean of the African proportion from all the family members who contributed to the linkage analysis, which was used as the covariate score for the subsequent OSA.
Ordered subset linkage analysis (OSA)
The purpose of OSA is to evaluate evidence for linkage even in the presence of genetic heterogeneity by using trait-related covariates. Specifically, in the current OSA, we ranked families by their family-specific African ancestry proportion, with those having the same value given the same rank. Linkage analysis was then performed on all contiguous subsets of families with the k smallest or k largest African ancestry proportion. The subset of families that yielded the largest lod score was determined for each chromosome. The statistical significance of the increased linkage evidence for each chromosome was assessed by a permutation test under the null hypothesis that the family linkage scores were independent of the family-specific African ancestry proportion. Specifically, the families were randomly ordered 10,000 times regardless of the value of the family-specific African ancestry proportion. Each randomly ordered set of families was analyzed by OSA, and a maximum lod score was recorded for each chromosome. The empirical p-value was then derived by counting the proportion of times the permutation lod scores were equal to or larger than the observed lod score. OSA was performed using the software program FLOSS (Browning 2006), and the linear allele sharing model to generate Kong-Cox lod scores (Kong and Cox 1997). FLOSS will take the family-specific linkage score (one family by one family) as the input to identify the subset of families with the most increased evidence of linkage. The input file that includes the family-specific linkage score for each position was prepared by Merlin software using the “--npl” and “--perFamily” options.
After we identified the ordered subsets of families with increased evidence for linkage, further non-parametric linkage analysis was performed on each ordered subset using Merlin to confirm the results from FLOSS. We also derived the genomewide empirical significance for the new maximum lod score in each ordered subset by 1,000 random simulations using Merlin. Briefly, we used the gene-dropping algorithm implemented in Merlin to simulate 1,000 data sets conditional on the observed family structure, marker spacing, allele frequencies, and missing data pattern. Each simulated replicate was then analyzed by the same procedure as the observed data. The highest lod score across the whole genome was recorded for each simulated replicate. The genomewide empirical significance was estimated by counting how often the entire genome had a maximum lod score greater than or equal to the observed score across the 1,000 simulated replicates.
Results
The individual African ancestry, estimated by STRUCTURE, varied from 0.327 to 0.998 with a mean of 0.837 (standard deviation (SD) = 0.105). The family-specific African ancestry varied from 0.443 to 0.996 with a mean of 0.841 (SD = 0.0946).
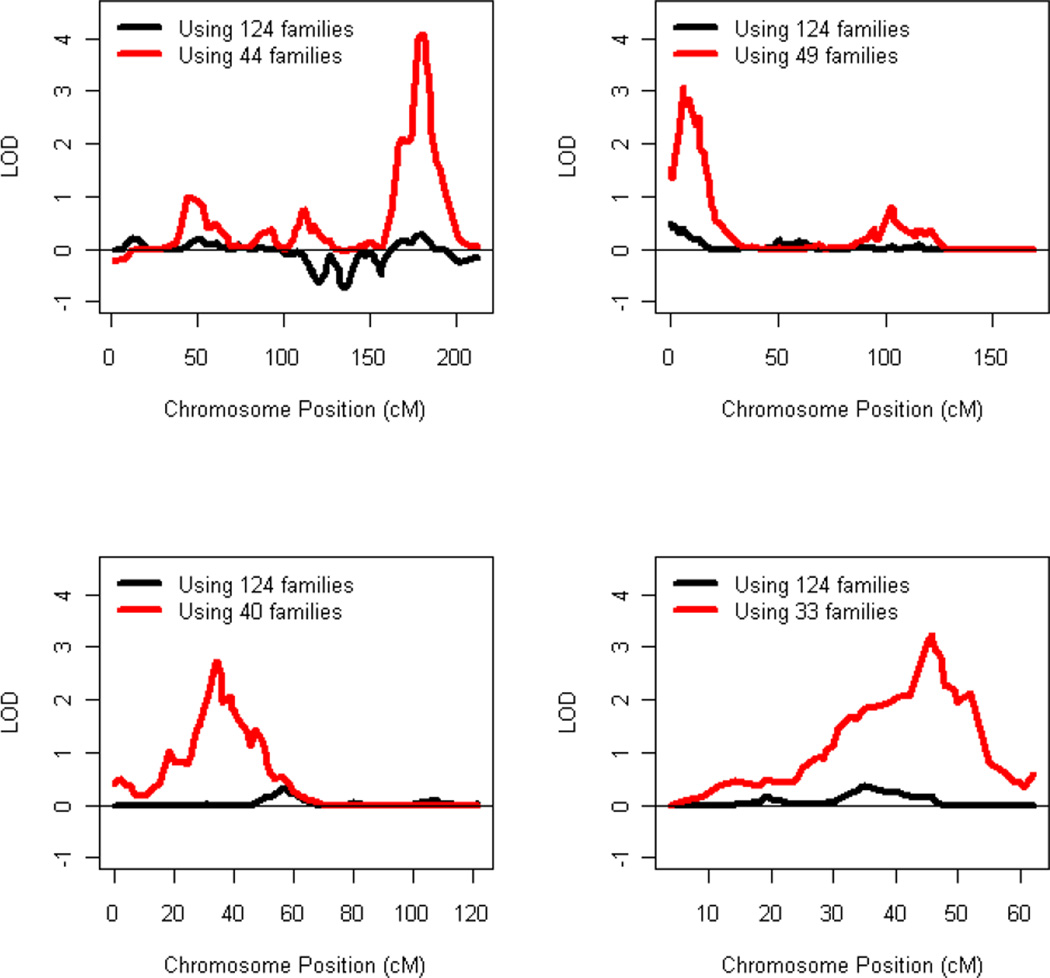
The results of OSA along with the other relevant statistics are provided in Table 2. Statistically significant increases in lod scores were identified for chromosomes 4 (P=0.0001), 12 (P=0.021), 15 (P=0.026) and 22 (P=0.0069). In a subset of 44 families (with African ancestry proportions ranging from 0.858 to 0.996), the lod score at 180 cM on chromosome 4 reached genomewide significance (lod=4.24, pointwise P<0.00001, empirical genomewide P=0.008). A second genomewide significant linkage result was observed at 46 cM on chromosome 22 in a subset of 33 families (lod=3.23, pointwise P=0.00006, empirical genomewide P=0.04). Figure 1 illustrates a comparison of the nonparametric linkage analysis results across the SNP marker positions between ordered subsets with increased linkage evidence and the overall families. The histograms of African ancestry admixture proportion for each subset of families were shown in Supplementary Figure 1.
Table 2.
Summary statistics for ordered subset linkage analysis
| Chr | Linkage peak (cM)a |
New lodb |
Old lodc |
Delta lodd |
No. of families |
Admixture Range, Mean, Variancee |
Empirical P-value for changef |
NPL pointwise P valueg |
NPL genomewide P valueh |
|---|---|---|---|---|---|---|---|---|---|
| 4 | 180.1 | 4.24 | 0.28 | 3.96 | 44 | [0.858, 0.996], 0.92, 0.0013 | 0.0001 | <0.00001 | 0.008 |
| 12 | 6.0 | 3.07 | 0.49 | 2.58 | 49 | [0.443, 0.813], 0.73, 0.0079 | 0.021 | 0.00009 | 0.052 |
| 15 | 34.8 | 2.72 | 0.33 | 2.39 | 40 | [0.443, 0.801], 0.71, 0.0080 | 0.026 | 0.0002 | 0.11 |
| 22 | 45.9 | 3.23 | 0.35 | 2.88 | 33 | [0.885, 0.996], 0.93, 0.00085 | 0.0069 | 0.00006 | 0.041 |
location of maximum lod score, maximized over all loci and ordered subsets.
maximum lod score, maximized over all loci and ordered subsets.
maximum lod score, maximized over all loci in the overall families.
(New lod) – (Old lod).
range, mean and variance of African ancestry proportion.
empirical P value of increased evidence for linkage obtained by 10000 times permutation test.
NPL (non-parametric linkage analysis) P value obtained by performing NPL analysis using the subsets of families identified from OSA.
genomewide empirical P value obtained by 1000 times simulations using the subsets of families identified from OSA.
Figure 1.
A comparison of the nonparametric linkage analysis results across the SNP marker positions between ordered subset families and the overall families on four chromosomes
Discussion
We conducted a genomewide ordered subset linkage analysis for AD using admixture proportion as a covariate in AAs. Our results showed that admixture proportion can be a useful covariate to identify the subset of families which are more homogeneous and thereby increase power for linkage detection in AAs. We identified four ordered subsets with statistically significant increases in lod scores on chromosomes 4, 12, 15 and 22.
Our most interesting finding was on chromosome 4, where we observed the strongest linkage signal (lod = 4.2) at 180 cM in a subset of 44 families with an African ancestry proportion ranging from 0.858 to 0.996. Linkage evidence for alcohol-related phenotypes on chromosome 4 has been reported in many previous studies. However, the locus identified here does not appear to overlap with any chromosome 4 linkage signals reported previously, with the exception of the study by Wilhelmsen (Wilhelmsen et al. 2003), which yielded modest evidence for linkage to the trait of “level of response to alcohol” (peak lod = 1.4, 170–190 cM). The 1-lod support interval (177–185 cM) for the chromosome 4 peak region contains 50 genes and harbors a particularly promising AD risk candidate gene, GLRA3. This gene encodes the alpha-3 subunit of the neuronal glycine receptor, a primary mediator of neuronal inhibition in the brain regions known to be sensitive to ethanol (Perkins et al. 2010).
The second strongest linkage signal (lod = 3.2) obtained by the current OSA is located at 46 cM on chromosome 22 in a subset of 33 families with a high proportion of African ancestry ranging from 0.885 to 0.996. Wilhelmsen et al. (Wilhelmsen et al. 2003) found a suggestive linkage (lod = 2.9) peak to the trait of “level of response to alcohol” in the region of 20–30 cM on chromosome 22; however, this linkage region does not overlap the 1-lod support interval (45–49 cM) for our finding. However, considering the small number of families contributing to our observed linkage signal, and the low mapping location precision possible with ASPs, it is possible that these findings are in fact reflective of the same risk gene. Three candidate genes located in the 1-lod support interval of the linkage peak on chromosome 22 are particularly interesting: PACSIN2, which encodes protein kinase C and casein kinase substrate in neurons; NPTXR, which encodes the neuronal pentraxin receptor; and SYNGR1, which encodes an integral membrane protein associated with presynaptic vesicles in neurons.
We also identified two regions with suggestive evidence of linkage on chromosomes 12 and 15 in two ordered subsets with lower proportions of African ancestry (i.e., ranging from 0.443 to 0.813 and from 0.443 to 0.801 for chromosomes 12 and 15, respectively). We did not identify compelling candidate genes from the 1-lod support interval for these two regions.
We also examined the characteristics of the 124 families and the families in the subsets to evaluate whether the increased linkage signals in the subset families could be due to the basic demographic difference between the overall families and the subsets. Supplementary Table 1 illustrates sex and age distribution among the total set of families and the subset families. We did not find any significant difference in age or sex distribution between the subsets and the overall families, suggesting the increased linkage signals in the subsets may not be accounted for by these basic family characteristics.
Samples included in the current study were ascertained mostly for DSM-IV cocaine or opioid dependence. Therefore, we had to consider the possibility that the linkage signals detected for AD in AAs could be attributable to other substance dependence (SD), such as cocaine dependence (CD) and opioid dependence (OD). We examined this alternate explanation for the findings by directly testing linkage for CD or OD in the same subset of families identified for AD on chromosomes 4, 12, 15 and 22.We found a significant linkage on chromosome 4 at 180 cM for CD (LOD = 3.49). Another suggestive linkage signal for CD was also identified on chromosome 22 at 46 cM (LOD = 2.93).The linkage results from other SD might indicate that the linkage signal is not specific for AD and could reflect a shared liability between different substances
Although genome-wide association studies (GWAS) have successfully identified common risk variants associated with complex disorders, the heritability of most complex disorders remains largely unexplained. It is now widely believed that the “missing heritability” could be attributable to rare variants of high penetrance or structural variation that is not well tagged by the current GWAS approach (Manolio et al. 2009). Compared with GWAS design, linkage analysis has the advantage of enriching signals for the same gene/functional unit that may harbor multiple rare variants of large effect or inherited CNVs segregating in families. With the growing evidence for the role of rare variants and CNVs in psychiatric disorders (Stefansson et al. 2008; Walsh et al. 2008; Williams et al. 2010), the regions discovered by linkage may complement the GWAS approach in pinpointing rare variants of high penetrance that cannot be captured using the current GWAS design. Thus, further investigation through targeted deep sequencing studies of the linkage regions identified by the current OSA may identify novel genes containing multiple rare or uncommon AD risk variants with large effects.
In summary, our results demonstrate that the genetic ancestry proportion can be used as a covariate to reduce genetic heterogeneity and enhance the detection of linkage for AD in AA samples. The current OSA approach based on admixture proportion could be applied to any linkage study for complex traits conducted in AA or other admixed populations.
Supplementary Material
Supplementary Figure 1. The histograms of admixture proportion (African Ancestry) for each subset of families that show increased evidence for linakge
Acknowledgements
The authors are grateful to the volunteer families and individuals who participated in this research study. This work was supported by the U.S. National Institutes of Health (R01 AA11330, R01 AA017535, K24AA013736, R01 DA12849, R01 DA12690, R01 DA018432, R01DA030976, K01 DA024758, and M01 RR06192) and by the US Department of Veterans Affairs (VA CT REAP and New England MIRECC Center; VA CT Alcohol Research Center).
Dr. Kranzler reports consulting arrangements with Lundbeck, GlaxoSmithKline, Gilead, and Alkermes and research support from Merck ACTIVE). Dr. Kranzler also reports associations with Eli Lilly, Janssen, Schering Plough, Lundbeck, Alkermes, GlaxoSmithKline, Abbott, and Johnson & Johnson, as these companies provide support to the ACNP Alcohol Clinical Trials Initiative (ACTIVE) and Dr. Kranzler receives support from ACTIVE.
Footnotes
Financial Disclosures
Drs. Yang, Han, and Gelernter report no competing interests.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Browning BL. FLOSS: flexible ordered subset analysis for linkage mapping of complex traits. Bioinformatics. 2006;22:512–513. doi: 10.1093/bioinformatics/btk012. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, Farrer L. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry. 2009;65:111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, Kranzler HR. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M. Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol. 2004;27:53–63. doi: 10.1002/gepi.20000. [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ. Allele-sharing models: lod scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuysen CI, Kranzler HR, Yu Y, Weiss RD, Brady K, Poling J, Farrer LA, Gelernter J. Confirmation and generalization of an alcohol-dependence locus on chromosome 10q. Neuropsychopharmacology. 2010;35:1325–1332. doi: 10.1038/npp.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Crawford DK, Alkana RL, Davies DL. Molecular targets and mechanisms for ethanol action in glycine receptors. Pharmacol Ther. 2010;127:53–65. doi: 10.1016/j.pharmthera.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, Kranzler HR. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Hauser ER, Schmechel DE, Welsh-Bohmer KA, Small GW, Roses AD, Saunders AM, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am J Hum Genet. 2003;73:1041–1051. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Möller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Mühleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nöthen MM, Peltonen L, Collier DA, St Clair D, Stefansson K GROUP. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Faraone SV, Lyons MJ. Identification of the phenotype in psychiatric genetics. Eur Arch Psychiatry Clin Neurosci. 1993;243:131–142. doi: 10.1007/BF02190719. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, Kalmijn J. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, Stefansson H, Stefansson K, Magnusson P, Gudmundsson OO, Gustafsson O, Holmans P, Owen MJ, O'Donovan M, Thapar A. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The histograms of admixture proportion (African Ancestry) for each subset of families that show increased evidence for linakge