Abstract
We previously reported the disruption of the murine gene encoding the transcription factor USF2 and its consequences on glucose-dependent gene regulation in the liver. We report here a peculiar phenotype of Usf2−/− mice that progressively develop multivisceral iron overload; plasma iron overcomes transferrin binding capacity, and nontransferrin-bound iron accumulates in various tissues including pancreas and heart. In contrast, the splenic iron content is strikingly lower in knockout animals than in controls. To identify genes that may account for the abnormalities of iron homeostasis in Usf2−/− mice, we used suppressive subtractive hybridization between livers from Usf2−/− and wild-type mice. We isolated a cDNA encoding a peptide, hepcidin (also referred to as LEAP-1, for liver-expressed antimicrobial peptide), that was very recently purified from human blood ultrafiltrate and from urine as a disulfide-bonded peptide exhibiting antimicrobial activity. Accumulation of iron in the liver has been recently reported to up-regulate hepcidin expression, whereas our data clearly show that a complete defect in hepcidin expression is responsible for progressive tissue iron overload. The striking similarity of the alterations in iron metabolism between HFE knockout mice, a murine model of hereditary hemochromatosis, and the Usf2−/− hepcidin-deficient mice suggests that hepcidin may function in the same regulatory pathway as HFE. We propose that hepcidin acts as a signaling molecule that is required in conjunction with HFE to regulate both intestinal iron absorption and iron storage in macrophages.
Iron is an essential element required for growth and survival of almost every organism. In mammals, the iron balance is primarily regulated at the level of duodenal absorption of dietary iron. Following absorption, ferric iron is loaded into apo-transferrin in the circulation and transported to the tissues, including erythroid precursors, where it is taken up by transferrin receptor-mediated endocytosis. Reticuloendothelial macrophages play a major role in the recycling of iron from the degradation of hemoglobin of senescent erythrocytes, whereas hepatocytes contain most of the iron stores of the organism in ferritin polymers. Over the past 5 years, an important body of information concerning the proteins involved in iron absorption and in the regulation of iron homeostasis has arisen from the study of inherited defects, both in humans and mice, leading to distinct iron disorders (for review see ref. 1). In the case of iron deficiency, the pathophysiological consequences of gene defects identified are well understood because they usually result in loss of function of proteins directly involved in the pathway of iron absorption. The proteins include the iron transporters DMT1 (also called Nramp2 or DCT1) (2, 3), ferroportin (also called IREG1 or MTP1) (4), and copper oxidases coupled to ferroportin, namely ceruloplasmin (5, 6) and haephastin (7). In contrast, several abnormalities associated with genetic iron overload have identified various proteins whose functional role in the control of iron homeostasis remains poorly understood. In humans, hereditary hemochromatosis (HH) is a common autosomal recessive genetic disease caused by hyperabsorption of dietary iron leading to an iron overload in plasma and multiple organs. Hemochromatosis is usually due to a mutation in the HLA-linked hemochromatosis gene (named HFE) located on chromosome 6p, and most patients are homozygous for the C282Y mutation in HFE (8). In addition, other loci have been involved in different HH families; a nonsense mutation in the transferrin receptor 2 gene (TFR2) on 7q has been reported in two HH non-HLA-linked families (9), and a locus for juvenile hemochromatosis has recently been mapped to chromosomal arm 1q (HFE2). Finally, although it has long been known that iron absorption is regulated in response to the level of body iron stores and to the amount of iron needed for erythropoiesis (10), the molecular nature of the signals that program the intestinal cells to adjust iron absorption still remains to be identified.
We previously reported the disruption of the murine gene encoding the transcription factor USF2 and its consequences on glucose-dependent gene regulation in the liver (11). We now show that Usf2−/− mice develop multivisceral iron overload that spares only the spleen and whose iron content is strikingly lower in knockout animals than in controls. Although these iron metabolic disorders resemble those observed in hereditary hemochromatosis, we demonstrate that they are not because of an alteration in genes previously identified for their implication in this pathology—e.g., HFE or TFR2. Therefore, to identify new candidate genes that may account for the abnormalities of iron homeostasis in Usf2−/− mice, we used suppressive subtractive hybridization between livers from Usf2−/− mice and wild-type mice. We isolated a cDNA encoding the peptide hepcidin. Hepcidin (also referred to as LEAP-1, for liver-expressed antimicrobial peptide) was very recently purified from human blood ultrafiltrate and from urine and was found to be a disulfide-bonded peptide exhibiting antimicrobial activity (12, 13). The protein is synthesized in the liver in the form of a propeptide that contains 83 amino acids and is converted into mature peptides of 20, 22, and 25 amino acids (13, 14). Hepcidin was recently reported to be highly synthesized in livers of experimentally or spontaneously iron-overloaded mice (14). Here, we demonstrate that hepcidin gene expression is totally silent in the iron overload Usf2−/− mice model. Taken together, these results suggest that hepcidin can act as a signaling molecule involved in the maintenance of iron homeostasis.
Materials and Methods
Generation and Genotyping of Usf2−/− Mice.
Disruption of the Usf2 gene has been described (11). The mutated allele contains the promoterless IRESβgeo cassette in exon 7 of the murine USF2 gene. All studied mice have a mixed genetic background that included contributions from C57BL/6 and 129/Sv strains. Mice were maintained on a standard laboratory mouse chow (AO3, Usine d'Alimentation Rationnelle, France) containing 280 mg of ferric carbonate per kg. Mice were killed from the ages of 2.5 months up to 19 months. Genotyping on mouse-tail DNA was performed by using a single PCR reaction to identify wild-type and USF2 knockout alleles. Genomic DNA (0.5–1 μg) was used in a 50-μl reaction that included three primers; the wild-type USF2 allele was amplified by using forward GCGAAGCCCTGGGTTCAATC (annealing in intron 6) and reverse GGGGTCCACCACTTCAAGAGG (annealing in intron 7) primers, and the knockout USF2 allele was amplified by using the forward GCGAAGCCCTGGGTTCAATC and reverse GAATTCTCTAGAGCGGCCGGAC (annealing in the Neo selection marker of the targeting construct) primers. PCR was performed as follows: 37 cycles (each cycle consisting of 30 s at 94°C, 30 s at 56°C, and 40 s at 72°C) with an initial denaturation step at 94°C for 4 min, and a final elongation step at 72°C for 5 min in 20 mM Tris⋅HCl (pH 8.4)/50 mM KCl/0.05% W-1/2 mM MgCl2/5% glycerol/0.04% bromphenol blue/0.2 mM each dNTP/0.2 μM each primer/2 units of Taq polymerase (GIBCO). The reaction was analyzed on 1.5–2% agarose gel containing ethidium bromide. This PCR method for mouse genotyping was found to give the same results as the Southern blot method reported (11).
Generation of a Subtracted Library by Suppression Subtractive Hybridization.
Total RNA was prepared as described (15). Polyadenylated RNA was isolated by using oligo(dT) cellulose (Boehringer Mannheim). Suppression subtractive hybridization was performed between three pooled liver RNA from 5-month-old homozygous USF2-deficient mice (“driver”) and liver RNA from a 5-month-old wild-type mouse (“tester”) by using the PCR-select cDNA subtraction kit (CLONTECH) according to the manufacturer's recommendations for all steps. Briefly, 14 ng of the ligated tester and 420 ng of nonligated driver cDNAs were mixed, denatured, and allowed to re-anneal. After subtractive hybridization, 1 μl of cDNA was amplified by two rounds of PCR. The subtracted cDNA library was cloned into the pT-Adv vector by using the AdvanTAge PCR cloning kit (CLONTECH). After the secondary PCR (15 cycles) with the Advantage cDNA polymerase mix (CLONTECH), the subtracted PCR cDNA mix was incubated for a further 10 min at 72°C with 1 unit of Taq DNA polymerase (GIBCO/BRL) to maximize the cloning efficiency and purified with the QIAquick PCR purification kit (Qiagen, Chatsworth, CA). The ligation mixture was introduced into the Electromax bacterial strain DH10B (GIBCO/BRL) by electroporation (1.8 kV) using a Cell-Porator (GIBCO/BRL). The library was plated onto 22 × 22-cm agar plates containing ampicillin (100 μg/ml) and spread with 80 μl of 5-bromo-4-chloro-3-indolyl β-D-galactoside (40 mg/ml) and 80 μl of isopropyl β-D-thiogalactoside (0.1 M). Bacteria were grown at 37°C until colonies were visible and kept at 4°C until blue/white staining could be clearly distinguished.
Reverse Northern High Density Blots and Screening.
A total of 400 individual clones were collected, resuspended into 30 μl of water, heated at 100°C for 10 min, and then placed in ice for 5 min and centrifuged for 5 min. PCR was performed by using 3 μl of clear supernatant with 5′-CAGGAAACAGCTATGACCATGATTAC-3′ (forward) and 5′-TAATACGACTCACTATAGGGCGA-3′ (reverse). The PCR products were blotted onto Hybond-N+ filters (Amersham Pharmacia). Blots were hybridized overnight at 72°C with [32P]dCTP-labeled double-stranded cDNA (RTS RadPrime DNA labeling system, GIBCO) synthesized with 2 μg of polyadenylated RNA from wild-type or Usf2−/− mouse liver, as described below. Blots were washed four times in 2 × SSC/0.1% SDS at 68°C for 20 min and two times in 0.2 × SSC/0.1% SDS at 68°C for 20 min.
Reverse Transcription (RT) and RT-PCR.
Double-stranded cDNA was synthesized in 20 μl, with 2 μg total RNA (or polyA RNA for the subtracted library), in the presence of 0.25 mM of each dNTP, 200 ng of random hexanucleotide primers, 20 units of RNasin (Promega), 10 mM DTT, and 200 units Moloney murine leukemia virus reverse transcriptase (GIBCO). After denaturation of RNA at 70°C for 10 min in a therman cycler (Perkin–Elmer), the reaction was performed for 1 h at 42°C before reverse transcriptase was inactivated for 6 min at 96°C. At the end of the reaction, 80 μl of 10 mM Tris⋅HCl (pH 8.0) and 0.1 mM EDTA (pH 8.0) were added. PCR amplification was performed with 5 μl of reverse transcriptase reaction mixture in 50 μl of 20 mM Tris⋅HCl (pH 8.4)/50 mM KCl/2 mM MgCl2/0.05% (vol/vol) W-1/0.2 mM of each dNTP/1 pmol of forward and reverse specific primers (listed below)/1 pmol of forward and reverse control β-actin primers/2 units of Taq polymerase (GIBCO). PCR conditions were 25 cycles of denaturation at 94°C for 20 s, annealing at 50°C for 20 s, and primer extension at 72°C for 20 s. Following PCR, the amplified products (171 bp for HEPC1 or HEPC2 and 250 bp for β-actin) were separated by electrophoresis on 1.5% agarose gel. Sequences of the primers were as follows: HEPC1, 5′-CCTATCTCCATCAACAGATG-3′ (forward) and 5′-AACAGATACCACACTGGGAA-3′ (reverse); HEPC2, 5′-CCTATCTCCAGCAACAGATG-3′ (forward) and 5′-AACAGATACCACAGGAGGGT-3′ (reverse); β-actin, 5′-AGCCATGTACGTAGCCATCC-3′ forward) and 5′-TTTGATGTCACGCACGATTT-3′ (reverse).
The primers used for amplification of DMT1 were as follows: DMT1 isoform without IRE, 5′-TCCTGGACTGTGGACGCT-3′ (forward) and 5′-GGTGTTCAGAAGATAGAGTTCAGG-3′ (reverse); DMT1 with IRE, 5′-TGTTTGATTGCATTGGGTCTG-3′ (forward) and 5′-CGCTCAGCAGGACTTTCGAG-3′ (reverse); normalization with 14S, 5′-CAGGACCAAGACCCCTGGA-3′ (forward) and 5′-ATCTTCATCCCAGAGCGA-3′ (reverse).
Northern Blot.
The primers used for amplification of probes used to detect specific mRNAs were as follows: for mouse hemochromatosis (HFE) cDNA amplification (1080 bp), 5′-ATGAGCCTATCAGCTGGGCT-3′ (forward) and 5′-TCACTCACAGTCTGTTAAGA-3′ (reverse); for mouse transferrin receptor (TfR) cDNA amplification (285 bp), 5′-GAAATCCCTGTCTGTTATAC-3′ (forward) and 5′-GGCAAAGCTGAAAGCATTTC-3′ (reverse); for mouse transferrin receptor 2 (TFR2) cDNA amplification (333 bp), 5′-TACAGCTCGGAGCGGAACG-3′ (forward) and 5′-TTACAATCTCAGGCACCTCC-3′ (reverse); for mouse ceruloplasmin cDNA amplification (350 bp), 5′-ACTTATTTCAGTTGACACGG-3′ (forward) and 5′-GCAGCACATACACATACTGT-3′ (reverse); and for mouse heme oxygenase 1 (Hmox1) cDNA amplification (258 bp), 5′-ATGGAGCGTCCACAGCCCG-3′ (forward) and 5′-CCTTCGGTGCAGCTCCTCAG-3′ (reverse). Each fragment was amplified by using Taq polymerase and hepatic total cDNA, purified from agarose gel (QIAquick PCR purification kit, Qiagen) and subcloned into TA vector (AdvanTAge cloning kit, CLONTECH). Recombinant plasmid was selected according to the protocol and amplified into LB medium containing 100 μg/ml ampicillin and purified (QIAprep Spin Miniprep, Qiagen). Each cDNA was purified from the vector after EcoRI digestion and migration on agarose gel. The probe used to detect HEPC1 mRNA was prepared from the EcoRI digestion of the pT-Adv/HEPC1 isolated by suppressive subtractive hybridization. Twenty micrograms of RNA from each source was denatured in formaldehyde-containing buffer and electrophoresed in 1% agarose, 2.2 M formaldehyde gels. Northern blot was performed as described (11). Each blot was stripped and reprobed with ribosomal 18S cDNA to check for the integrity and the amount of loaded RNAs.
Southern Blot.
Southern blots were performed as described (11). The HEPC1 probe was prepared from a 1437-bp mouse genomic DNA fragment amplified with forward 5′-GAGCAGCACCACCTATCTCCA-3′ and reverse 5′-AACAGATACCACAGGAGGGT-3′ primers. After digestion with PvuII, a 545-bp fragment was purified from agarose gel and used as probe for Southern blot. Note that this HEPC1 probe showed 95% identity with the homologous HEPC2 region.
Hematological Analysis of Mice.
Blood was obtained by retroorbital phlebotomy before sacrifice of mice and collected in heparinized tubes (Capiject T-MLH, Terumo) (Elkton, MD). Blood cell counts and erythrocyte parameters were determined by using a MaxM Coulter automatic analyzer.
Iron Measurements and Histology.
Quantification of iron level was performed (5) on fragments or total organs by using IL test (Instrumentation Laboratory, Lexington, MA). For histology, tissues were fixed in 4% formaldehyde, embedded in paraffin, mounted onto slides, and stained with Prussian blue and nuclear red counterstain by using standard procedures.
Results
Massive Iron Overload in Liver and Pancreas of Usf2−/− Mice.
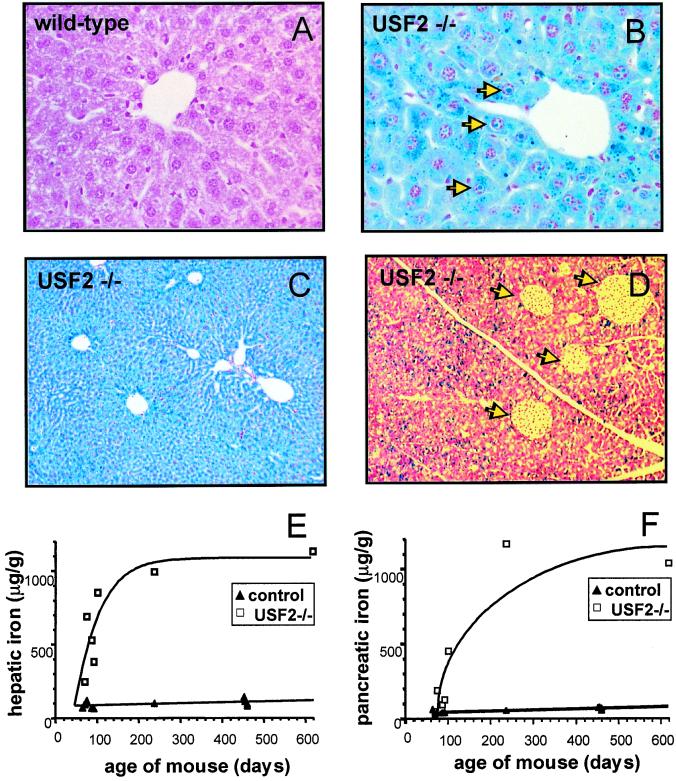
All Usf2−/− mice exhibit after the third month of life a dense brown pigmentation of the liver and a more or less pronounced bronze pigmentation of the pancreas. As this phenotypic trait is characteristic of hemochromatosis, the inherited disorder of iron absorption, we decided to analyze the iron status of the Usf2−/− mice. First, to assess the level of iron accumulation, Perls' Prussian blue staining was performed on liver and pancreas of wild-type and Usf2−/− mice maintained on a standard diet. Whereas control mice showed very little or no positive iron staining in the liver (Fig. 1A), Usf2−/− mice displayed iron accumulation in hepatocytes (Fig. 1 B and C). This iron deposition was primarily confined to periportal hepatocytes; then, with age, the number of stained hepatocytes increased. By 19 months of age, as shown in Fig. 1C, iron accumulation was considerable, and the staining was homogeneous throughout the liver parenchyma. Furthermore, a strong nuclear iron accumulation was detected in some hepatocytes (Fig. 1B). For the pancreas, similar results were obtained, i.e., no staining in the control tissue and a strong iron accumulation in the exocrine pancreas of Usf2−/− mice (Fig. 1D). To quantify more accurately the iron overload during the life of animals, iron levels were measured in liver and pancreas of mice from 2.5 to19 months of age. As shown in Fig. 1E, iron accumulated in the liver of mice between 60 and 100 days after birth and reached a plateau corresponding approximately to a 10-fold greater iron content than in control mice. In the pancreas (Fig. 1F), iron accumulation was more progressive, with levels in Usf2−/− mice a maximum of 20-fold higher than in control mice. Iron accumulation was also measured in kidney and heart showing a 2-fold and 4-fold accumulation, respectively. Finally, we found a 1.7-fold higher iron level in serum of Usf2−/− compared with control mice (3.550 ± 259 μg of iron/liter in controls, n = 15, vs. 6.274 ± 514 μg of iron/liter in Usf2−/− mice, n = 13, P < 0.0001), but this increase did not appear to be age-dependent. This increase in serum iron level in Usf2−/− mice was correlated with a 1.6-fold increase in transferrin saturation (61 ± 9% saturation in controls, n = 6, vs. 95 ± 9% saturation in Usf2−/− mice, n = 6, P < 0.0004). Finally, in the oldest female analyzed so far (19 months), the iron overload became widespread with increased iron level in all tissues tested including muscle, uterus, lung, and pituitary gland (data not shown).
Figure 1.
Iron accumulation in liver and pancreas of Usf2−/− mice. Liver and pancreas were fixed in formaldehyde and stained with the Perls' stain for iron. Nonheme iron stains blue. Liver sections are from an 8-month-old wild-type mice (×50) (A), an 8-month-old Usf2−/− littermate (B), and a 19-month-old Usf2−/− mouse (×10) (C). Pancreas section in D is from an 8-month-old Usf2−/− mouse (×12.5). Arrowheads in C indicate iron in the nucleus of the hepatocyte. Arrowheads in D point to islets of Langerhans scattered throughout the exocrine tissue. (E and F) Age-dependent hepatic and pancreatic nonheme iron concentration (micrograms of iron per gram dry tissue) as measured in control (wild-type and heterozygote mice, ▴) and Usf2−/− mice (□).
The Spleen of Usf2−/− Mice Is Resistant to Natural Iron Deposition.
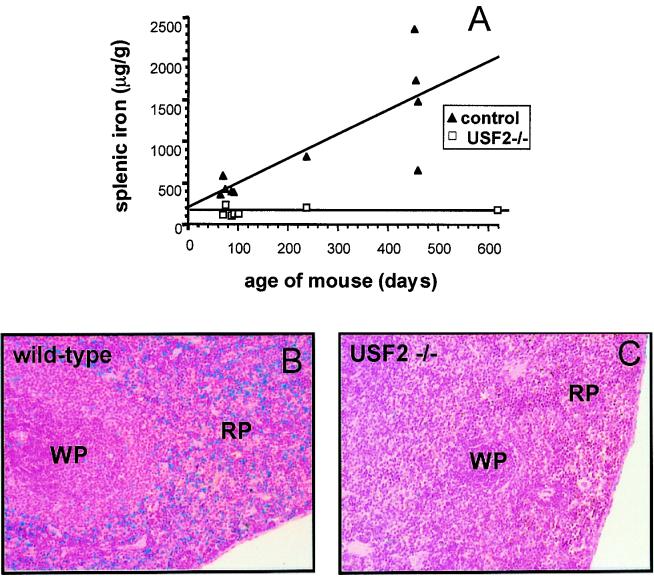
In contrast to all other tissues tested, we observed an age-dependent iron accumulation in the spleen of wild-type mice (Fig. 2A). Granules that gave a positive reaction with Perls' Prussian blue staining were observed, primarily scattered between cells of the red pulp (Fig. 2B). We found this accumulation to fluctuate greatly between mice, suggesting that it may depend on the 129/Sv × C57BL/6 hybrid strain background of each animal. This natural iron storage has been previously reported in C57BL/6 mice and was described to occur mainly in splenic macrophages (16, 17). Surprisingly, in spleen of Usf2−/− mice, iron levels remained very low (Fig. 2A), with a complete absence of Perls' Prussian blue staining (Fig. 2C).
Figure 2.
Iron content in spleen of Usf2−/− mice. (A) Age-dependent splenic nonheme iron concentration (micrograms of iron per gram dry tissue) as measured in control (wild-type and heterozygote mice, ▴) and Usf2−/− mice (□). Spleen section from a representative 8-month-old wild-type mouse (×20) (B) and an 8-month-old Usf2−/− littermate (×20) (C) stained with the Perls' stain for iron. RP, red pulp; WP, white pulp.
Erythroid Parameters Are Not Affected in Usf2−/− Mice.
To rule out the possibility that the increased iron accumulation in Usf2−/− mice might result from dyserythropoietic anemia, we measured erythroid parameters in control and Usf2−/− mice at different ages. Values of red blood cell count (106/ml), hemoglobin concentration (g/dl), and mean corpuscular volume (femtoliter) were normal: RBC, Hb, and mean corpuscular volume of 10.3 ± 0.3, 16.73 ± 0.49, and 48.27 ± 0.67 for wild-type mice (n = 3) and 10.0 ± 0.3, 15.67 ± 0.06, and 48.63 ± 1.36 for Usf2−/− mice (n = 3), respectively.
Thus, interestingly, the iron abnormalities observed in Usf2−/− mice, including the resistance of spleen to iron accumulation and normal hematological parameters, strikingly resemble the phenotype of HFE−/− mice (18, 19), the murine model of hereditary hemochromatosis.
Expression of HFE and TFR2 Genes Is Not Modified in the Liver of Usf2−/− Mice.
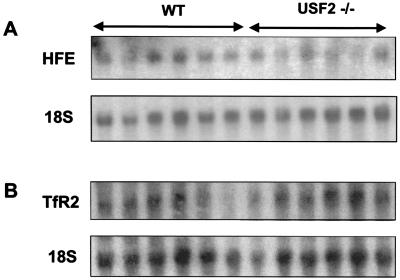
Because USF2 is a transcription factor, we sought to determine whether USF2 could be involved in the regulation of genes encoding proteins related to iron metabolism. Because of the similarity between HFE−/− mice and our model, we first checked for the expression of the HFE gene. As shown in the Northern blot of Fig. 3A, abundance of HFE mRNA in liver of Usf2−/− mice is comparable with that of wild-type mice. We also looked at the gene encoding transferrin receptor-2, a mutation that was recently reported in HH (9). Northern blot analysis demonstrated that the hepatic expression of this gene was not modified in Usf2−/− mice compared with wild-type mice (Fig. 3B). The level of ceruloplasmin, heme oxygenase 1, and transferrin receptor mRNAs was also monitored in Usf2−/− mice, as the abundance of these mRNAs has been reported to be modified in disorders that disturb iron balance (for review, see ref. 20). Again, we found that the level of these mRNAs was comparable in Usf2−/− and control mice (not shown). Finally, we analyzed the expression of the DMT1 gene (also referred to as Nramp2), the major transmembrane iron uptake protein that actively transports reduced dietary iron into intestinal enterocytes. Duodenal expression of DMT1 was analyzed by relative quantification using RT-PCR (seven Usf2−/− vs. six control mice). No statistically significant differences were found between the two groups of mice (data not shown).
Figure 3.
HFE and TFR2 mRNA content in liver of wild-type and Usf2−/− animals as determined by Northern blot analysis. Twenty micrograms of total liver RNAs from wild-type mice and Usf2−/− mice (from 3 to 11 months old) were electrophoresed and blotted. Blots were hybridized with a 32P-labeled probe (made by PCR, as described in Materials and Methods) for HFE (A) and RTf2 (B).
Analysis of Subtraction cDNA Libraries: Identification of Hepcidin as a Putative Candidate for Hemochromatosis.
To identify genes whose level of expression is modified in Usf2−/− mice, we performed a subtracted cDNA library between liver from Usf2−/− (driver) and wild-type (tester) mice (21). Among 400 clones analyzed, we isolated several clones that were down-regulated in the liver from Usf2−/− mice as analyzed by reverse Northern blot (data not shown). One of these clones contained a full-length cDNA encoding the recently characterized peptide hepcidin (12–14). This cDNA is of particular interest with regard to iron homeostasis because it was recently reported that the liver-specific hepcidin gene was overexpressed during iron overload (14).
Murine Organization of Usf2 and Hepcidin Genes on Chromosome 7.
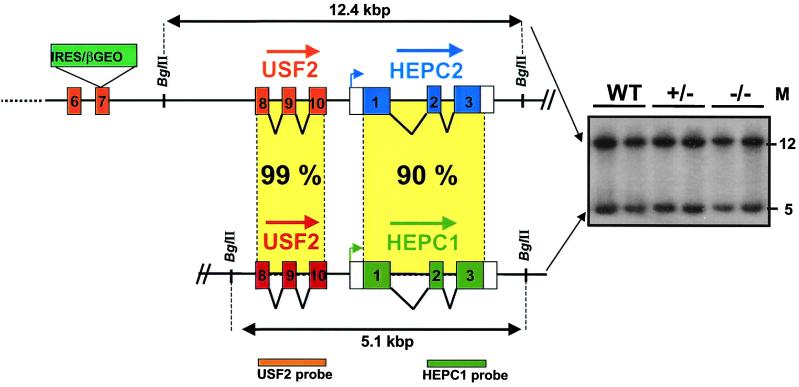
The murine genome contains two closely related hepcidin genes that colocalize on the same mouse genomic clone (GenBank clone CTZ8N15, accession no. AC020841). These genes were designated HEPC1 and HEPC2 by Pigeon et al. (14). Interestingly, the genomic CT7–8N15 clone also revealed that HEPC1 is situated in close proximity to the Usf2 gene on murine chromosome 7. Pigeon et al. reported that HEPC1 was located directly downstream of the Usf2 gene (14). By analyzing another genomic clone, RP23–22G9 (GenBank, accession number AC087143), we found that part of the Usf2 gene (encompassing exons 8, 9, and 10) was also duplicated and that, in fact, HEPC1 lies downstream of the truncated Usf2 gene. The genomic organization of Usf2 and hepcidin genes is shown in Fig. 4. The HEPC2 gene is located downstream of the functional complete Usf2 gene, and the HEPC1 gene is located downstream of the partial Usf2 gene. At present, no information is available concerning the relative orientation 5′–3′ of the HEPC1 and HEPC2 genes and the distance between them.
Figure 4.
Genomic organization of Usf2 and hepcidin genes. Schematic representation (not to scale) of the locus region encompassing the Usf2 and hepcidin genes. The targeted allele is represented with the betageo cassette insertion in exon 7 (11). Data are the result of genomic RP23–22G9 clone (GenBank). So far, no data are available concerning the orientation and the distance between the two hepcidin genes. The Southern blot in the right of the figure is from tail DNA of wild-type, heterozygote, and homozygote mice digested by BglII and hybridized with the HEPC1 probe. Two bands of the expected size, 12.4 and 5.1 kbp, were detected, whatever the genotype. The same bands were revealed by using the Usf2 probe.
Because of the proximity of the Usf2 gene and hepcidin locus, we sought to determine whether the recombination event in intron 7 of the targeted Usf2 allele might have eliminated or truncated the HEPC1 and HEPC2 genes. To check this hypothesis, we performed Southern blot on genomic tail DNA from wild-type, Usf2+/−, or Usf2−/− mice with an HEPC1 probe (Fig. 4). Genomic DNA was digested by BglII. Based on the analysis of the AC087143 locus, this digestion was predicted to generate two fragments of 5.1 and 12.4 kbp, containing the HEPC1 and HEPC2 genes, respectively. Because of the close similarity (more than 95%) between the hybridizing region of HEPC1 and HEPC2, we expected both bands to be revealed by the HEPC1 probe. This is what we found, as shown on the Southern blot in Fig. 4. The same pattern was observed with DNA from Usf2−/− mice, indicating that the hepcidin genes are present in Usf2−/− mice and that they have not undergone major rearrangement. Finally, the two bands also hybridized with a USF2 probe extending from exon 8 to exon 10, demonstrating that exons 8 to 10 of USF2 are indeed duplicated.
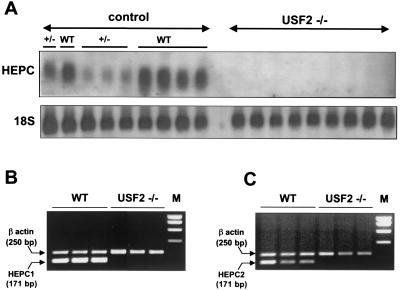
The Hepcidin Genes Are Totally Silent in the Liver of Usf2−/− Mice.
The level of expression of the hepcidin genes was measured by Northern blot analysis. In fact, hepcidin mRNA was totally undetectable in the liver of Usf2−/− mice (Fig. 5A). It is worth noting that the liver of Usf2+/− mice contained a reduced amount of hepcidin mRNA compared with wild-type mice. To further assess the specific level of HEPC1 and HEPC2 messengers, we designed specific primers for the HEPC1 and HEPC2 transcripts. By RT-PCR, we demonstrated that both genes were actively transcribed in the liver of wild-type mice (Fig. 5 B and C), whereas both HEPC1 and HEPC2 transcripts were totally absent from the liver of Usf2−/− mice (Fig. 5 B and C).
Figure 5.
Hepcidin mRNA content in liver of wild-type, Usf2+/−, and Usf2−/− animals as determined by Northern blot analysis and RT-PCR. (A) Twenty micrograms of total liver RNAs from wild-type, Usf2+/−, and Usf2−/− animals (between 3 and 11 months old) were electrophoresed and blotted. The blot was hybridized with a 32P-labeled HEPC probe (as described in Materials and Methods), which most likely recognized both HEPC1 and HEPC2 transcripts. (B) Specific HEPC1 and HEPC2 levels were measured by RT-PCR as described in Materials and Methods. Following PCR, the amplified products (171 bp for HEPC1 or HEPC2 and 250 bp for β-actin) were separated by electrophoresis on 1.5% agarose gel. Neither HEPC1 nor HEPC2 specific primers were able to reamplify HEPC2 and HEPC1 PCR products, respectively, demonstrating the high specificity of each pair of primers (not shown).
Discussion
We have found profound abnormalities of iron metabolism in Usf2-deficient mice. Although full penetrance of this phenotype was observed in homozygote Usf2−/− mice, it was never observed in heterozygote Usf2+/− mice. Iron accumulation was predominantly observed in liver and pancreas and to a lesser extent in heart. We also found a serum iron level in Usf2−/− mice that was 1.7-fold greater than in wild-type animals. This excessive iron deposition in the liver was found to be associated with an elevated level of the L-ferritin subunit in Usf2−/− mice (not shown). Finally, the Usf2−/− mice showed reduced iron stores in the spleen compared with wild-type mice. Interestingly, this phenomenon has also been observed in human HH (22, 23). In the spleen of HH patients, despite the elevation of serum iron, concentration of the metal in macrophages is minimal until late in the disease (24). Spleens from HFE knockout mice are also resistant to iron loading. Thus, the Usf2 gene knockout model shows biochemical abnormalities and histopathology similar to both human HH and HFE−/− mice. We did not find any significant increase in the DMT1 mRNA level in the Usf2−/− mice. Although some authors have reported that the DMT1 mRNA level is increased in patients with HH (25) and in HFE-deficient mice (26), DMT1 induction in HFE−/− mice remains controversial (27, 28).
To gain insights into the molecular mechanism underlying iron accumulation in Usf2−/− mice, we analyzed several genes known to control iron homeostasis and that are involved in the development of iron overload diseases when mutated. Among these genes, we found that expression of HFE and TFR2, both mutated in some human hemochromatosis (8, 9), was normal. Thus, tissue iron overload can occur in the context of an apparently functional HFE gene and can produce very similar phenotypes. Recently, three additional knockout mice models have been reported to develop iron overload phenotype: haem oxygenase 1, ceruloplasmin, and β2-microglobulin. However, we showed that none of these genes is affected in Usf2−/− mice.
By analysis of a liver-subtracted library made between wild-type and Usf2−/− mice, we identified several clones for transcripts down-regulated in Usf2−/− mice. Among these, we found an HEPC1 cDNA and confirmed by Northern blot that the HEPC1 mRNA is absent in liver of Usf2−/− mice, whereas it is highly expressed in liver of wild-type mice. Pigeon and colleagues reported in fact the existence of two hepcidin genes (14). The presence of these two genes is most likely the result of a very recent duplication as indicated by the high conservation between HEPC1 and HEPC2 nucleic sequences (90% identity) and the very similar exon–intron organization. In contrast, analysis of human nucleic databases using BLAST search analysis revealed the presence of one hepcidin gene or cDNA that corresponds to HEPC1. We found that HEPC1 and HEPC2 genes are highly expressed in the liver of wild-type mice but totally down-regulated in Usf2−/− mice. The presence of only one hepcidin gene in the human genome (75% identity on the mature peptide sequence) suggests the probable redundancy of HEPC1 and HEPC2 gene functions in the mouse. We can therefore hypothesize that inactivation of only one of the HEPC1 or HEPC2 genes would not have been sufficient to lead to the development of the iron overload phenotype.
USF2 is a member of the basic helix-loop-helix family consisting of multiple transcription factors, including USF1 and Myc. It is often considered that USF acts in living cells as USF1/USF2 heterodimers. However, several studies demonstrate that USF1 or USF2 possesses specific functions (29, 30). The iron overload phenotype described herein is specific to Usf2 knockout mice and does not occur in the USF1−/− mice model that we previously reported (31). Even if we cannot rule out the possibility that USF2 directly controls expression of the HEPC1 and HEPC2 genes, it is more likely that HEPC1 and HEPC2 deficiency is because of alteration of the Usf2 alleles by insertion of the neomycin resistance (NeoR) marker in exon 7. The reduction of HEPC gene expression observed by Northern blot in Usf2+/− mice (Fig. 5) argues for a cis effect of the mutation on hepcidin gene expression rather than a trans effect depending on the USF2 transcription factor. Such a disturbance of neighboring genes around the NeoR selection marker has already been described (32–34).
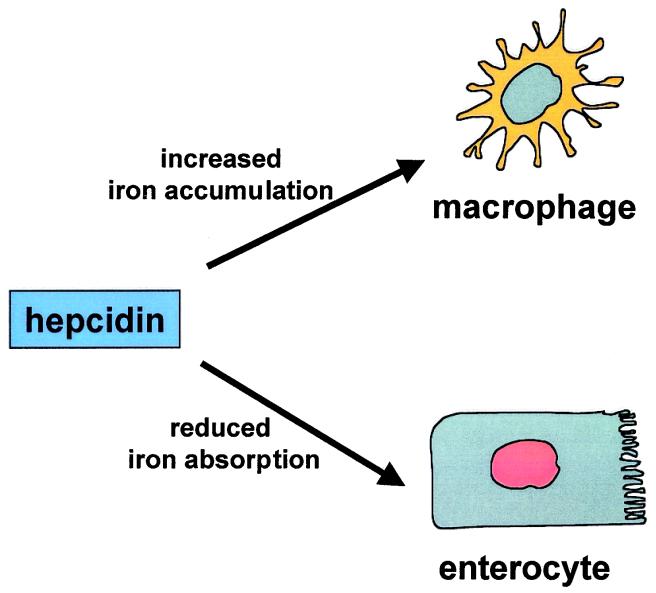
The existence of a sensor for iron homeostasis has been suspected for a long time and is thought to be a soluble component of the plasma that would signal between different organs and tissues such as liver, intestine, erythropoietic precursors, and spleen macrophages (for review, see ref. 35). Hepcidin peptide that is secreted in plasma after synthesis and maturation in the liver could fulfill this important role. Indeed, Pigeon et al. (14) demonstrated that accumulation of iron in the liver up-regulates hepcidin expression, whereas our data clearly show that a complete defect in hepcidin expression is responsible for progressive tissue iron overload. Taken together, these results allow us to propose that hepcidin could be the iron signal involved in the pathway regulating iron absorption. The similarity of the alterations in iron metabolism between HFE knockout mice and the Usf2−/− hepcidin deficient mice also suggests that hepcidin may function in the same regulatory pathway as HFE. It has been shown that HFE physically interacts with the transferrin receptor in crypt cells of the duodenal mucosa (36). It is postulated that this interaction modulates the iron status of these cells, which, in turn, controls the expression of the apical and basolateral transporters in mature cells at the tips of the villi. It is tempting to speculate that hepcidin may be required for HFE activity perhaps through direct interaction with the HFE/beta2 microglobulin/transferrin receptor complex. Similarly, hepcidin may be required for the regulation of iron storage in macrophages. The presence of a mutated HFE protein or a complete defect in hepcidin expression may be responsible for increased intestinal iron absorption and reduced macrophage iron stores (Fig. 6). Under both conditions, plasma iron overcomes transferrin binding capacity, and nontransferrin-bound iron accumulates in various tissues including heart and pancreas.
Figure 6.
Hypothetical model for hepcidin as a key regulator of iron homeostasis. In this model, hepcidin prevents iron overload by reducing iron transport in the enterocyte and by programming macrophages to retain iron. In Usf2−/− mice, the hepcidin defect would be responsible for increased intestinal iron transport and reduced macrophage iron stores.
According to our proposed role of hepcidin in iron homeostasis, hepcidin production may depend on the uptake of transferrin-bound iron mediated by TFR2 in hepatocytes. This might explain why the TFR2 defect is responsible for a form of human genetic hemochromatosis if this defect leads to a decrease in hepcidin secretion that, in turn, results in increased iron absorption. This hypothesis will be testable by measuring plasma hepcidin in patients with TFR2 deficiency or in TFR2 knockout mice.
In conclusion, our results highlight the role of hepcidin as a key regulator of iron homeostasis. We propose hepcidin as a novel candidate gene that, when mutated, could be involved in abnormal regulation of iron metabolism and development of HH. Further investigations in patients with hereditary hemochromatosis not related to HFE or TFR2 and in hepcidin-rescued Usf2−/− mice will provide definitive clues as to the implication of this peptide in iron metabolism. Finally, this new murine model of iron overload disease appears to be a suitable animal model for testing new therapeutic approaches for prevention and correction of the iron storage in HH as well as for the understanding of iron homeostasis.
Acknowledgments
We thank Marie-Elise Martin and Fathi Driss for quantification of liver ferritin and serum transferrin, respectively. We thank Dimitri Tchernitchko for help with Nramp2 amplification. We are grateful to Steven Brown and Jürgen Ripperger (University of Geneva) for fruitful discussions. We thank Alan Srickland for the careful revision of the text. This work was supported by Institut National de la Santé et de la Recherche Médicale.
Abbreviations
- HH
hereditary hemochromatosis
- HFE
the protein defective in hereditary hemochromatosis
- HEPC
hepcidin
- USF
upstream stimulatory factor
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 8160.
References
- 1.Andrews N C. Nat Rev Genet. 2000;1:208–217. doi: 10.1038/35042073. [DOI] [PubMed] [Google Scholar]
- 2.Fleming M D, Trenor C C, Su M A, Foernzler D, Beier D R, Dietrich W F, Andrews N C. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 3.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Nature (London) 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 4.Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt S J, Moynihan J, Paw B H, Drejer A, Barut B, Zapata A, et al. Nature (London) 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- 5.Harris Z L, Durley A P, Man T K, Gitlin J D. Proc Natl Acad Sci USA. 1999;96:10812–10817. doi: 10.1073/pnas.96.19.10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida K, Furihata K, Takeda S, Nakamura A, Yamamoto K, Morita H, Hiyamuta S, Ikeda S, Shimizu N, Yanagisawa N. Nat Genet. 1995;9:267–272. doi: 10.1038/ng0395-267. [DOI] [PubMed] [Google Scholar]
- 7.Vulpe C D, Kuo Y M, Murphy T L, Cowley L, Askwith C, Libina N, Gitschier J, Anderson G J. Nat Genet. 1999;21:195–199. doi: 10.1038/5979. [DOI] [PubMed] [Google Scholar]
- 8.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 9.Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. Nat Genet. 2000;25:14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 10.Roy C N, Carlson E J, Anderson E L, Basava A, Starnes S M, Feder J N, Enns C A. FEBS Lett. 2000;484:271–274. doi: 10.1016/s0014-5793(00)02173-6. [DOI] [PubMed] [Google Scholar]
- 11.Vallet V S, Henrion A A, Bucchini D, Casado M, Raymondjean M, Kahn A, Vaulont S. J Biol Chem. 1997;272:21944–21949. doi: 10.1074/jbc.272.35.21944. [DOI] [PubMed] [Google Scholar]
- 12.Krause A, Neitz S, Magert H J, Schulz A, Forssmann W G, Schulz-Knappe P, Adermann K. FEBS Lett. 2000;480:147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 13.Park C H, Valore E V, Waring A J, Ganz T. J Biol Chem. 2001;276:7806–7610. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 14.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira, C., Santambrogio, P., Martin, M. E., Andrieu, V., Feldmann, G., Henin, D. & Beaumont, C. (2001) Blood, in press. [DOI] [PubMed]
- 17.Veninga T S, Wieringa R A, Morse H. Lab Anim. 1989;23:16–20. doi: 10.1258/002367789780886984. [DOI] [PubMed] [Google Scholar]
- 18.Levy J E, Montross L K, Cohen D E, Fleming M D, Andrews N C. Blood. 1999;94:9–11. [PubMed] [Google Scholar]
- 19.Zhou X Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews N C, Fleming M D, Gunshin H. Nutr Rev. 1999;57:114–123. doi: 10.1111/j.1753-4887.1999.tb06934.x. [DOI] [PubMed] [Google Scholar]
- 21.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson G J, Halliday J W, Powell L W, Fleming R E, Migas M C, Zhou X, Jiang J, Britton R S, Brunt E M, Tomatsu S, et al. Hepatology. 1987;7:967–969. [Google Scholar]
- 23.Cox T. Nat Genet. 1996;13:386–388. doi: 10.1038/ng0896-386. [DOI] [PubMed] [Google Scholar]
- 24.Yam L T, Finkel H E, Weintraub L R, Crosby W H. N Engl J Med. 1968;279:512–514. doi: 10.1056/NEJM196809052791003. [DOI] [PubMed] [Google Scholar]
- 25.Zoller H, Pietrangelo A, Vogel W, Weiss G. Lancet. 1999;353:2120–2123. doi: 10.1016/S0140-6736(98)11179-0. [DOI] [PubMed] [Google Scholar]
- 26.Fleming R E, Migas M C, Zhou X, Jiang J, Britton R S, Brunt E M, Tomatsu S, Waheed A, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1999;96:3143–3148. doi: 10.1073/pnas.96.6.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canonne-Hergaux F, Levy J E, Fleming M D, Montross L K, Andrews N C, Gros P. Blood. 2001;97:1138–1140. doi: 10.1182/blood.v97.4.1138. [DOI] [PubMed] [Google Scholar]
- 28.Levy J E, Montross L K, Andrews N C. J Clin Invest. 2000;105:1209–1216. doi: 10.1172/JCI9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews G K, Lee D K, Ravindra R, Lichtlen P, Sirito M, Sawadogo M, Schaffner W. EMBO J. 2001;20:1114–1122. doi: 10.1093/emboj/20.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong G, Fan T, Liu L. J Exp Med. 1999;189:1931–1938. doi: 10.1084/jem.189.12.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallet V S, Casado M, Henrion A A, Bucchini D, Raymondjean M, Kahn A, Vaulont S. J Biol Chem. 1998;273:20175–20179. doi: 10.1074/jbc.273.32.20175. [DOI] [PubMed] [Google Scholar]
- 32.Kaul A, Koster M, Neuhaus H, Braun T. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- 33.Moran J L, Levorse J M, Vogt T F. Nature (London) 1999;399:742–743. doi: 10.1038/21560. [DOI] [PubMed] [Google Scholar]
- 34.Tallquist M D, Weismann K E, Hellstrom M, Soriano P. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 35.Roy C N, Enns C A. Blood. 2000;96:4020–4027. [PubMed] [Google Scholar]
- 36.Waheed A, Parkkila S, Saarnio J, Fleming R E, Zhou X Y, Tomatsu S, Britton R S, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1999;96:1579–1584. doi: 10.1073/pnas.96.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]