Abstract
We proposed to optimize the retinal differentiation protocols for human embryonic stem cells (hESCs) by improving cell handling. To improve efficiency, we first focused on the production of just one retinal precursor cell type (photoreceptor precursor cells [PPCs]) rather than the production of a range of retinal cells. Combining information from a number of previous studies, in particular the use of a feeder-free culture medium and taurine plus triiodothyronine supplements, we then assessed the values of using size-controlled embryoid bodies (EBs) and negative cell selection (to remove residual embryonic antigen-4-positive hESCs). Using size-controlled 1000 cell EBs, significant improvements were made, in that 78% CRX+ve PPCs could be produced in just 17 days. This could be increased to 93% PPCs through the added step of negative cell selection. Improved efficiency of PPC production will help in efforts to undertake shorter and larger preclinical studies as a prelude to future clinical trials.
Introduction
Age-related macular degeneration (AMD) and other retinal diseases such as retinitis pigmentosa are major causes of blindness in the developed world, with over 14 million people blind or severely visually impaired because of AMD alone.1 Currently, there are no available treatments to reverse tissue damage in these disorders. Cell transplantation, to replace lost tissue, offers the most promising approach in reversing blindness due to these conditions. With this in mind, significant advances have been reported in using cells derived from fetal tissue,2 human embryonic stem cells (hESCs),3 human adult stem cells,4,5 and reprogrammed induced pluripotent stem cells.6
In this previous work, it has been proposed that cells at least partially committed to a retinal cell fate are the best cells for retinal transplantation,2 although the optimal stage of cell fate commitment has yet to be determined. Hence, one area of particular concern in work on retinal regeneration has been to devise efficient methods to produce large quantities of partially differentiated retinal progenitor cells.7 Most work has focused on differentiating hESCs. Seminal work exposing hESCs to growth factors and modulators of signaling pathways that mimic normal retinal development has been extremely successful in directing hESCs toward either retinal pigment epithelium8,9 or the neural retinal cell fate.10,11 Using these techniques, it has been shown that most types of cells found in the neural retina, including ganglion cells, amacrine cells, horizontal cells, bipolar cells, and photoreceptor cells, can be generated using these techniques.8–11
Of the numerous challenges that remain in extrapolating early preclinical studies into clinical trials, one is the pressing need to efficiently mass produce large numbers of retinal precursor cells. For example, it has become evident that the differentiation of hESCs into cells expressing proteins, characteristic of immature and mature photoreceptors (such as CRX and NRL), is extremely time consuming, often resulting in a low cell yield.7,10,11 As a consequence, such cell production can also be extremely expensive. These practical problems have limited the amount of preclinical work that has been undertaken.
Consequently, there is an urgent need to devise more efficient methods for manufacturing retinal progenitor cells. To address this need, we have investigated new methods to improve cell handling during the differentiation period. These included the ways to synchronize differentiation through the use of size-controlled embryoid bodies (EBs), s standardized chemically defined medium to minimize the variability associated with feeder cells and conditioned media, and also cell selection so as to remove undifferentiated cells from the final product.
Materials and Methods
hESC culture
The hESC line WA09 (WiCell Research Institute) was maintained in an animal protein-free TeSR™ 2 growth medium (STEMCELL Technologies) and grown feeder-independent on six-well dishes (Nunc) coated with growth factor-reduced Matrigel™ (BD Biosciences). The medium was changed daily, and the cells were routinely passaged with 1 mg/mL dispase (STEMCELL Technologies) every 4–6 days. Spontaneously differentiated cells were manually removed, as needed. Cells from passages 34–43 were used.
EB formation and differentiation
Differentiation protocols were initially based on previously published work.10 In addition, recent studies have suggested that the size and shape of EBs used in differentiation protocols may influence the differentiation trajectory of hESCs.12,13 In previous retinal cell differentiation protocols, mixed-size EBs have been used.7,10,11 In this study, we proposed to compare progenitor cell production derived from random-sized EBs with those produced from EBs that had been sorted according to the size.
hESCs were initially incubated at 37°C with 1 mL dispase per well until the colonies began to peel off the plate (20–30 min). Colonies were gently washed with the dispase solution and collected in a 15-mL tube (colonies from up to three wells per 15-mL tube). Residual colonies were collected with 2 mL Dulbecco's modified Eagle's medium/Nutrient Mixture F-12 (DMEM/F12) (Life Technologies) per well. The colonies were allowed to settle to the bottom of the tube for ∼5 min, and the supernatant was aspirated, and the pellet was washed with 4–6 mL of DMEM/F12. After colonies had settled down and the supernatant aspirated for a second time, they were resuspended in an EB resuspension buffer containing DMEM/F12, 10% knockout serum replacement, custom B-27 and N-2 supplements (Life Technologies), 1 ng/mL mouse noggin, 1 ng/mL recombinant human DKK1, and 5 ng/mL recombinant human insulin-like growth factor (IGF-1; R&D Systems), and then placed on a nonadherent surface (Costar) in a volume of 5 mL/well for 3 days. At day 4 of incubation, EBs either were kept as a mixed-sized population or were manually separated into three size-restricted populations. From the mixed EB population, the largest EBs were isolated visually using a 200-μL tip (designated large). The remaining EB suspension was then passed over a 100-μm nylon cell strainer. The flow-through was collected and designated small, and the EBs that were trapped in the strainer were eluted and designated medium. Sizes for these sorted EBs were established by microscopic imaging with a scale bar: (1) EBs with a diameter of 400 μm or more (large), (2) EBs with a diameter of 200 μm (medium), and (3) EBs with a diameter smaller than 100 μm (small). Alternatively, to accurately manufacture EBs of specific size, EBs were produced using AggreWell plates (STEMCELL Technologies) according to the manufacturer's recommendation. Briefly, before EB formation, hESC colonies were enzymatically dissociated into single cells with Accutase (STEMCELL Technologies). The single-cell suspension was diluted in DMEM/F12 and centrifuged at 300 g for 5 min to remove the residual enzyme. The resulting cell pellet was resuspended in the EB formation medium TeSR2, supplemented with 10 μM ROCK inhibitor14 (STEMCELL Technologies), after which the viable cells were counted and added to the appropriate AggreWell. The AggreWell plate was centrifuged for 3 min at 100 g and incubated at 37°C for 24 h. EBs were then harvested from the AggreWell plate after 24 h. As with the manually formed technique, the resultant EBs were placed on a nonadherent surface for 3 days in an EB resuspension buffer as described above for the random-sized EBs.
On day 4, all EBs were then evenly distributed on growth factor-reduced Matrigel-coated dishes in a photoreceptor precursor differentiation medium containing DMEM/F12, custom B-27 and N-2 supplements, 10 ng/mL mouse noggin, 10 ng/mL recombinant human DKK1, 10 ng/mL recombinant human IGF-1, and 5 ng/mL recombinant human basic fibroblast growth factor (Life Technologies) for 13 days. On day 10, in selected experiments, the differentiation medium was modified to assess the value of supplementing with 20 mM taurine and 40 ng/mL triiodothyronine (T3; Sigma).15–17
Reverse transcription polymerase chain reaction (RT-PCR) and reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Since most preclinical and future clinical studies will principally require photoreceptor precursors we elected to focus on the production of photoreceptor precursor cells (PPCs) rather than the manufacture of retinal progenitor cells to produce a wide range of different retinal cell types.18 Although some previous work has based the definition of a PPC on the expression of a range of genes, numerous studies have suggested the expression of CRX as the critical fate-determining factor for the photoreceptor (rod and cone) lineage.19–21 We therefore proposed the expression of CRX as our endpoint in PPC production rather than, for instance, NRL expression, which is indicative of a more advanced, terminal stage of differentiation into a rod lineage.2 To identify PPCs on the 18th day of differentiation, total RNA was isolated from the cell cultures using the Aurum™ Total RNA Fatty and Fibrous Tissue kit (Bio-Rad) according to the manufacturer's instructions. For each sample, 1 μg of total RNA was reverse-transcribed with the iScript™ cDNA Synthesis kit (Bio-Rad). First-strand DNA was synthesized with 1 μg of total RNA using an iScript cDNA Synthesis kit (Bio-Rad), as per the manufacturer's protocol. Oligonucleotide primers used for CRX are forward: 5′-ATGATGGCGT ATATGAACCC, and reverse 5′-TCTTGAACCAAACCTG AACC.10 Gene expression for CRX, NANOG, SOX2, BLIMP1, NRL, RCVRN, and OPSINSW was quantified using the TaqMan primer and labeled probe system and the ViiA™ 7 Real-Time PCR system (Applied Biosystems). All reactions were performed using the TaqMan Universal Master Mix (2×), FAM-labeled TaqMan Gene Expression assays for all the genes of interest, and VIC-labeled TaqMan endogenous control GAPDH and 4.5 ng of cDNA. Thermocycling parameters were as follows: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, plus 60 s at 60°C. Real-time polymerase chain reaction (PCR) data were analyzed by the comparative CT method. Each reaction was undertaken on three occasions, and for each of these, individual samples were subdivided into three aliquots for measurement.
Magnetic removal of undifferentiated cells
It has been suggested that residual undifferentiated cells within heterogeneous precursor cell populations can lead to retinal neoplasia.22 To reduce this risk, we assessed the value of the EasySep® magnetic selection system (STEMCELL Technologies)23 to magnetically remove the cells expressing hESC surface marker stage-specific embryonic antigen-4 (SSEA-4).24 Briefly, differentiated cells were collected, counted, and resuspended in 100 μL of the recommended medium (phosphate-buffered saline [PBS], 2% fetal bovine serum [FBS], and 1 mM ethylenediaminetetraacetic acid [EDTA]) in a 5-mL tube. The SSEA-4 selection cocktail was added, and the cell suspension was incubated for 15 min at RT, followed by the addition of EasySep magnetic nanoparticles and incubation for 15 min at RT. The suspension was topped up with the recommended medium and placed into the appropriate magnet for 10 min. The magnet was picked up and inverted, and the supernatant with the desired cells was collected in a new 5-mL tube. Each sample of cells underwent two rounds of magnetic separation. To recycle the isolated cells, SSEA-4 expressing cells (attached to the magnetic beads) were resuspended in a differentiation medium and replated on Matrigel-coated plates for 2 more weeks of differentiation.
Flow cytometry
For surface labeling with SSEA-4 antibody, cells were collected and counted, and ∼200,000 cells per sample were distributed into a 1.5-mL vial and collected by centrifugation at 300 g for 5 min. Cell pellets were blocked in 100 μL PBS/10% human serum (Sigma) for 10 min on ice, followed by the addition of 1 μL SSEA-4 antibody (gift from Dr. C. Eaves) to each vial and incubation for 30 min on ice. Cells were washed once with 1 mL PBS/2% FBS, vortexed briefly, and centrifuged at 300 g for 5 min followed by incubation for 15 min on ice in 100 μL goat anti-mouse Alexa-488 secondary antibody diluted 1:400 in PBS/2% FBS. The cells were washed once as described, and each cell pellet was resuspended in 300 μL of PBS/2% FBS supplemented with propidium iodide (Life Technologies; 1:1000) and analyzed using an FACSCalibur (BD Biosciences).
For intracellular labeling, cells were dissociated with Accutase, centrifuged at 300 g for 5 min, and resuspended in PBS/2% FBS. Viable cells were counted, and 800,000 cells were distributed into 1.5-mL tubes, representing all conditions analyzed. Cells were collected by centrifugation and fixed with 250 μL of 2% paraformaldehyde for 15 min on ice. After one wash with 1 mL of PBS/2% FBS and centrifugation, cells were permeabilized for 15 min at RT in 0.5 mL of saponin permeabilization buffer (SPB; 0.2% saponin and 0.1% bovine serum albumin in PBS). Cells were centrifuged; the SPB was aspirated; and the cell pellet was resuspended in 100 μL anti-CRX polyclonal antibody developed in our laboratory,25 diluted in SPB (1:200), and incubated for 30 min at RT. Two washes, each with 1 mL of SPB followed by centrifugation, were performed to remove the primary antibody, and the cells were resuspended in 100 μL goat anti-rabbit Alexa-488 secondary antibody (diluted 1:400 in SPB) and incubated for 20 min on ice followed by two washes with SPB as described above. Cells were resuspended in 300 μL of PBS/2% FBS and analyzed using FACSCalibur (BD Biosciences). Data from all flow cytometry experiments were analyzed with CellQuest Pro Software (BD Biosciences).
Sodium dodecyl sulfate/polyacrylamide gel electrophoresis and western immunoblotting
Differentiated cells were collected by centrifugation and lysed in 100 μL lysis buffer (10 mM Tris–HCl [pH=7.4], 2 mM EDTA, 150 mM sodium chloride, 0.875% Brij-96, 0.125% NP-40, 1 μg/mL Aprotinin, 1 μg/mL Leupeptin, and 174 μg/mL phenylmethylsulfonyl fluoride). The lysates were subjected to three freeze–thaw cycles after which they were centrifuged for 1 min at 10,000 g, and the supernatant was collected in a fresh tube. Samples of equal protein concentration were boiled for 5 min in 1×sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris–HCl [pH 6.8], 2% SDS, 5% glycerol, 0.005% bromophenol blue, and 1.6% β-mercaptoethanol) and separated on 12% polyacrylamide Tris–glycine gels for 2 h at 120 V. Proteins were electrophoretically transferred to a polyvinylidene fluoride membrane (4 h at 80 V), blocked for 1 h in 5% (w/v) nonfat milk powder in PBS, 0.1% Tween-20 (PBST), and incubated overnight at 4°C with our anti-CRX antibody25 (diluted 1:250 in 5% milk/PBST) or for 1 h at RT with anti-GAPDH antibody (Abcam; diluted 1:2000). The membranes were washed three times in PBST, incubated for 1 h at RT with anti-rabbit or anti-mouse secondary antibodies (Rockland Immunochemicals; diluted 1:10,000 in 5% milk powder/PBST), and washed three times in PBST before being imaged on the Odyssey® Imaging System (LI-COR Biosciences). CRX protein was identified as a band corresponding to 37 kDa.
Cytogenetic analysis
G-banded karyotyping (WiCell Research Institute) was undertaken in the WA09 cell line, and PPCs derived from these WA09 cells to determine whether chromosomal abnormalities had been acquired during expansion and differentiation.
Results
Differentiation of hESCs toward PPC fate
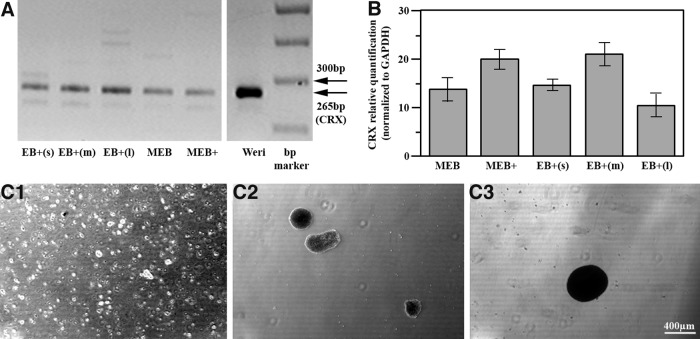
Although CRX+ve cells (PPCs) were identified after 17 days by augmenting previously published differentiation protocols10 (Fig. 1A), reverse transcription quantitative polymerase chain reaction (RT-qPCR) suggested that some improvement could be achieved by adding 20 mM taurine and 40 ng/mL T3 at day 10 (Fig. 1B). Further assessment of CRX levels using manually manufactured EBs subdivided into small (<100 μm), medium (200 μm), and large (>400 μm) EBs (Fig. 1C) suggested that within a mixed-size population, cells derived from EBs ∼200 μm in size were most efficacious, and EBs larger than this were less efficacious at CRX expression (Fig. 1B).
FIG. 1.
CRX expression in manually manufactured embryoid bodies (EBs) after 17 days in the differentiation medium. (A) Reverse transcription polymerase chain reaction (RT-PCR) analysis showing CRX expression in EBs. MEB: mixed-size EBs, nonsupplemented medium; MEB+: mixed-size EBs, medium supplemented with taurine and triiodothyronine (T3); EB+s: manually selected EBs <100 μm/200 cells; EB+m: manually selected EBs ∼200 μm/1000 cells; EB+l: manually selected EBs >400 μm/5000 cells. WERI-Rb1 cells served as a positive control. (B) RT-qPCR comparing quantitative CRX expression. Expression in MEB and MEB+cells compared with EBs supplemented with taurine and T3 and manually sorted by size. Values represent the mean±standard error of the mean (SEM) from three independent samples, each repeated in triplicate. Change in the gene expression calibrated to expression in human embryonic stem cells. (C) Representative images of size-restricted EBs, generated manually. (C1) Small EBs (<100 μm/200 cells); (C2) medium EBs (∼200 μm/1000 cells); (C3) large EBs (>400 μm/>5000 cells).
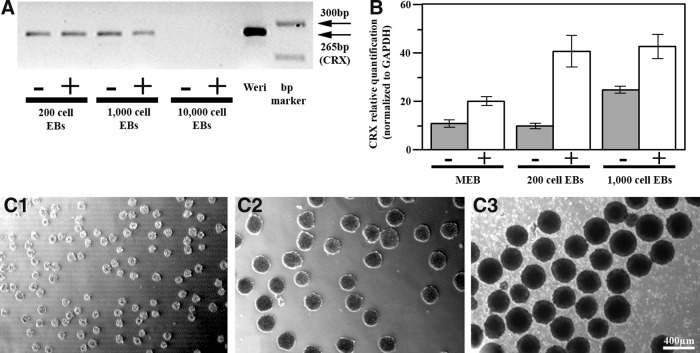
We therefore explored the value of EB size control further by looking at CRX expression in cells derived from the AggreWell plates (Fig. 2). Most strikingly, after 17 days in the differentiation medium, RT-PCR results showed no detectable CRX expression in cells derived from larger AggreWell EBs (>300 μm in size, containing 10,000 cells) (Fig. 2A). Cells derived from smaller AggreWell EBs showed significant improvement in CRX expression compared with mixed EBs, but importantly only if the medium was supplemented with taurine and T3 (Fig. 2B). It was noted that using smaller EBs (100 μm, 200 cells) also resulted in a significant improvement in CRX expression after 17 days of differentiation, but only if 10 μM ROCK inhibitor was added to the medium for the entire differentiation period (data not shown).
FIG. 2.
CRX expression in EBs formed with the AggreWell plates after 17 days in the differentiation medium. (A) RT-PCR analysis showing CRX gene expression in EBs made from 200, 1000, but not 10,000 cells. +/−: with or without taurine and T3 supplements. (B) RT-qPCR comparing CRX expression in mixed-size EBs with expression in 200 and 1000 cell EBs, suggesting a significant increase in expression in size-controlled EBs if supplements are added. +/−: with or without taurine and T3 supplements. Values represent the mean±SEM from three independent samples, each repeated in triplicate. Change in gene expression calibrated to expression in human embryonic stem cells. (C) Representative images of AggreWell EBs. (C1) 100 μm/200 cells; (C2) 200 μm/1000 cells; (C3) 300 μm/10,000 cells.
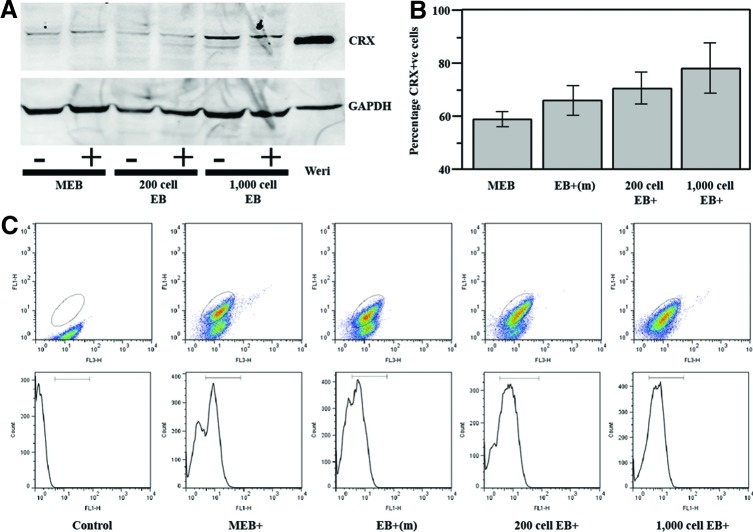
CRX protein expression in PPCs
To corroborate RT-PCR expression results, we found that western blot analysis showed CRX protein was detectable in PPCs (Fig. 3A). To determine the percentage of cells that were CRX+ve, flow cytometry was carried out on cells from mixed, manually sorted medium-sized EBs, and AggreWell-derived EBs (Fig. 3B, C). With the same 17-day differentiation protocol, we found that up to 78% CRX+ve cells could be achieved with AggreWell size-controlled EBs (200 μm, 1000 cell AggreWell EBs) compared with just 59% CRX+ve cells with mixed EBs. This was statistically significant by the Student t-test analysis (n=6; p<0.05).
FIG. 3.
CRX protein levels in photoreceptor precursor cells (PPCs). (A) Western immunoblot analysis demonstrating CRX in cell lysates. MEB: mixed-size EBs compared with 200 cell and 1000 cell AggreWell EBs.+indicates the medium supplemented with taurine and T3. (B) Comparative flow cytometry analysis showing a highest proportion of CRX+ve cells in 1000-cell AggreWell EBs. Values represent the mean±SEM (n=6). (C) Representative dot plots and histograms from one flow cytometry experiment. Elliptical gate identifies CRX+ve cells. Color images available online at www.liebertpub.com/tec
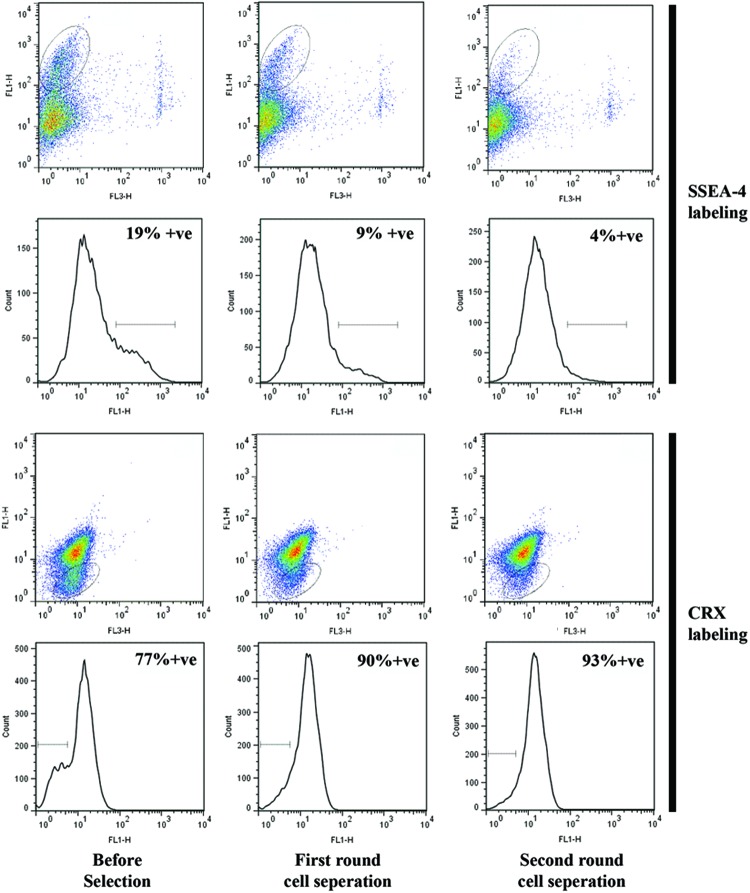
Exclusion of residual hESCs from the PPC population and subsequent recycling
To further purify the PPC population with CRX+ve cells, we used a negative selection protocol. Typically, negative cell selection using the SSEA-4 antibody depleted the total cell numbers by 50% per round. Flow cytometry established that the SSEA-4 antibody protocol decreased hESCs from 19% to ∼4% (Fig. 4, upper panels). This demonstrated that negative selection could be used to significantly reduce the number of hESCs in the cell population derived from our differentiation protocol. Residual cells, after negative selection, were then assessed for CRX expression (Fig. 4, lower panels). This indicated that after two rounds of selection, the percentage of CRX+ve cells had significantly improved from ∼77% to 93%. These data therefore suggest that after a 17-day differentiation protocol using AggreWell size-controlled EBs and negative cell selection, a cell population can be obtained containing ∼93% CRX+ve cells, 4% undifferentiated hESCs, and 3% other cells (intermediate-staged cells).
FIG. 4.
Immunomagnetic cell separation. Stage-specific embryonic antigen-4 (SSEA-4+ve) cells removed in two rounds from the PPC cohorts using EasySep magnetic cell separation. Flow cytometry results: (upper panels) decline in SSEA-4+ve cells as identified by elliptical gate, and (lower panels) proportional increase in CRX+ve cells (elliptical gate identifies CRX-ve cells). Color images available online at www.liebertpub.com/tec
We then looked at whether negatively selected cells were viable for reintroduction to our differentiation protocol to produce more CRX+ve cells rather than being discarded. This SSEA-4-enriched cell population was reapplied to Matrigel-coated dishes and subjected for a second round of differentiation. Cell colonies could still be identified after 2 weeks, suggesting that despite multiple rounds of selection and further handling, these cells were still viable (data not shown).
Gene expression profile in the PPC population
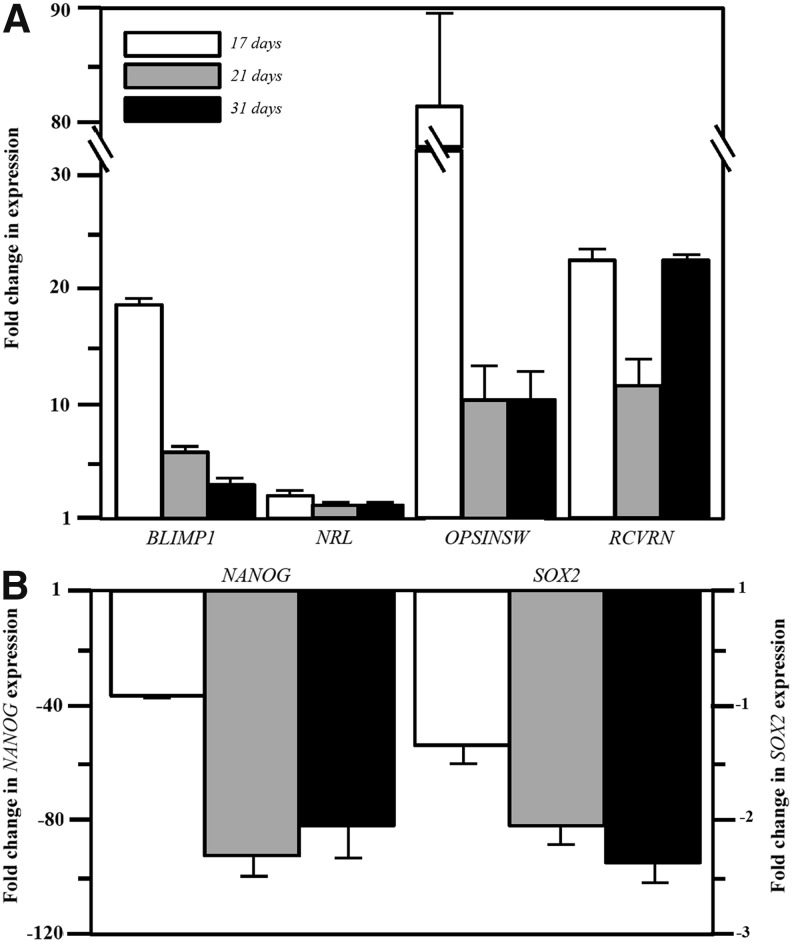
To further establish the identity of PPCs, we looked at the expression profile of a number of genes in cells kept in the differentiation medium for up to 31 days (Fig. 5). In addition to upregulation of CRX (Figs. 1 and 2), we then demonstrated upregulation of the PPC genes BLIMP1, OPSINSW (s-opsin), and RCVRN (recoverin), which was time dependent (Fig. 5A). Maintaining cells in the differentiation medium for this extended period did not, however, lead to a significant upregulation of NRL (Fig. 5A) or rhodopsin (by RT-PCR, data not shown). We did, however, also see a significant downregulation of the pluripotency genes NANOG and SOX2, indicative of cell differentiation (Fig. 5B).
FIG. 5.
RT-qPCR expression profiles of genes associated with photoreceptor differentiation (A, upper panel) and cell pluripotency (B, lower panel) in PPCs at 17, 24, and 31 days in the differentiation medium. All fold changes in gene expression normalized to GAPDH and calibrated to expression in human embryonic stem cells.
G-banded karyotyping
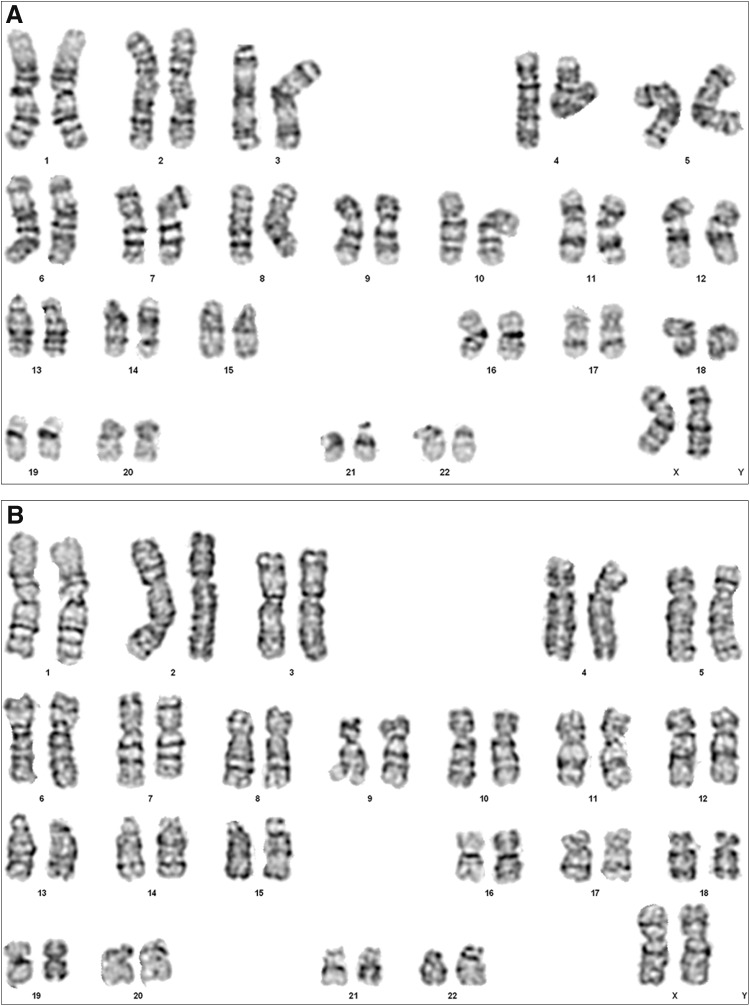
To determine whether the differentiation and negative cell selection protocol had led to chromosomal abnormalities, 20 cells, each of WA09 and CRX+ve PPCs (collected after the negative selection procedure), were analyzed for cytogenetic abnormalities. Both cell lines demonstrated a normal karyotype, and no clonal abnormalities were detected at the 400–500-kb band level (Fig. 6).
FIG. 6.
Karyotype analysis. Normal karyotype in (A) WA09 human embryonic stem cells and (B) PPCs after completing the 17-day differentiation protocol.
Discussion
Improving hESC differentiation protocols have emerged as a common theme in regenerative medicine. For example, based on previous, longer protocols to manufacture pancreatic β-cells,26–28 a 14-day procedure has recently been proposed for the differentiation of hESCs into pancreatic progenitor cells (expressing NGN3, NEUROD1, PTF1a, and PDX1). In this work, a novel combination of inhibitors of TGF-β and BMP signaling and a PKC activator was used.29 In other work, on differentiating cardiomyocytes from hESCs, a recent study optimized numerous variables, including number of EBs per well, well size and shape, medium formulation, and days of media change to propose a universal differentiation protocol generating a cell population of >90% contracting cells in just 9 days.30 This is in comparison to ∼11% contracting cells in 20 days using traditional methods.31
In parallel with this, a significant progress has also been made in improving retinal cell differentiation protocols using hESCs. Early work on hESCs using mouse feeder cell systems demonstrated only 10%–12% CRX+ve cell yield after a 21-day period.10 Others reported a 11% CRX+ve cell yield, but after a longer 120-day protocol and also using mouse feeder cells, in studies designed to terminally differentiate a range of retinal progenitor cells.11 In a novel 3-dimensional (3D) tissue construct, a range of retinal progenitor cells, including CRX+ve cells, were also reported, but after a 90-day period.17 In another 3D model (optic vesicle-like spheres), again using mouse feeder cells, an yield of ∼56% CRX+ve cells was achieved after 80-day incubation.32 A far-reaching study combining the approaches from a number of previous studies, but still using hESCs expanded on mouse feeder cells, resulted in ∼16% CRX+ve cell yield after a 45-day period.7 Most recently, a feeder-free differentiation protocol has been reported, yielding some CRX+ve cells after a 21-day period, although the relative percentage of cells was not reported.3
Although improvements in retinal cell differentiation are being reported, these are unlikely to be enough for larger-scale preclinical or ultimately clinical studies. Therefore, we tested a novel protocol combining very recent innovations from a number of different studies, such as the use of the feeder-free TeSR2 growth medium3 and taurine plus T3 supplements,17 and added these to a protocol of enhanced cell handling. This resulted in an increase of yield of CRX+ve cells from 10%–56%3,10,11,32 to 78%. This could be achieved in cycles of just 17 days representing a substantial improvement in efficiency. This improvement was achieved using 200-μm (1000 cells) EBs. Although results using 100-μm (200 cells) EBs were not statistically different from 1000 cell EBs, in practice, we concluded that 200-μm (1000 cells) EBs were optimal for use, since they proved easier to handle and did not require the addition of the ROCK inhibitor. It has been proposed that such CRX+ve cells can go on to mature into functional photoreceptors that will integrate into the diseased retina, and this has been established in a number of previous studies.3,11,33 We explored this with our cells by extending exposure in the differentiation medium up to 31 days and confirmed progressive downregulation of the pluripotency genes NANOG and SOX2. Gene expression suggestive of early photoreceptor differentiation (BLIMP1) was also seen, but as expected, declined with time. Expression suggestive of more mature photoreceptors (RCVRN, recoverin) was detectable. Interestingly, high OPSINSW (s-opsin) expression declined with time. However, s-opsin gene expression is not necessarily indicative of cone maturation, but can be a stage in the development of all photoreceptor rods and cones.34 In addition, limited expression of the NRL transcripts correlates with our observation that rhodopsin expression is not detectable up to 31 days in the differentiation medium. This is comparable with the work of others, suggesting that full rod photoreceptor maturation in vitro takes a prolonged time. This may not, however, reflect maturation in vivo when PPCs have been implanted subretinally.
An important consideration however is whether CRX expression in differentiating hESCs is sufficient to consider these cells as actual photoreceptor precursors. Strategies in the past have focused on showing expression of a range of precursor transcription factors.17,10,11 However, no study has yet suggested that hESCs or cells in normal development can progress into a photoreceptor lineage without CRX expression.19–21 Although further work needs to be done on how to best define a photoreceptor precursor, our work can be compared with that of others, since CRX expression has been a key feature in all such studies.
Another key concern in work to develop photoreceptor precursors has been the heterogeneous nature of cell populations that have so far been produced and implanted in preclinical trials. Ultimately, homogeneous cell populations would be attractive, and as a step toward this, we found that the CRX+ve cell yield could be improved further to 93% through cell sorting to reduce residual hESCs. The most efficient cell selection protocols usually involve positive selection35,36; however, we elected to undertake a negative selection strategy. The advantage of negative selection is that it avoids manufacturing PPCs that have surface antibodies attached to them, which could potentially hinder in vivo maturation or long-term survival.37 Such cell selection protocols can nonetheless be inefficient, since large numbers of cells are discarded. Although, we found that such selected cells are still viable and can therefore be reintroduced into the differentiation medium for further use. Finally, karyotyping confirmed that the differentiation and selection protocol did not appear to induce the chromosomal abnormalities that have been reported by others in hESC work.38 This might be due to the shortened time span of the protocol compared to other studies.
In summary, we have developed a method (summarized in Fig. 7) for producing PPCs in a relatively short time and in large numbers. This is in contrast to previous studies, although these were principally aimed at proof of concept or aimed at producing a range of retinal progenitor cells rather than on improving efficiency.3,10,11,17 Since blindness due to retinal disease is common in developed countries such as the United States,1 efficient production of retinal precursor cells or, more specifically, PPCs could have a major impact in preclinical and future clinical trials.
FIG. 7.
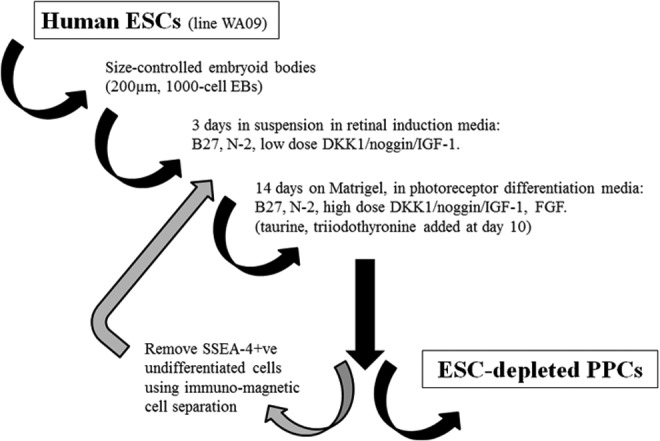
Schematic representation of the 17-day PPC differentiation protocol.
Acknowledgments
The authors wish to acknowledge technical assistance from Melanie Kardel and provision of the SSEA-4 antibody from Dr. Connie Eaves, University of British Columbia. Financial support for this work was from the Canadian Institutes of Health Research (Team Grant No. 222728).
Disclosure Statement
No competing financial interests exist.
References
- 1.Gehrs K.M. Anderson D.H. Johnson L.V. Hageman G.S. Age-related macular degeneration—emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLaren R.E. Pearson R.A. MacNeil A. Douglas R.H. Salt T.E. Akimoto M. Swaroop A. Sowden J.C. Ali R.R. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444:203. doi: 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
- 3.La Torre A. Lamba D.A. Jayabalu A. Reh T.A. Production and transplantation of retinal cells from human and mouse embryonic stem cells. Methods Mol Biol. 2012;884:229. doi: 10.1007/978-1-61779-848-1_16. [DOI] [PubMed] [Google Scholar]
- 4.Wohl S.G. Schmeer C.W. Isenmann S. Neurogenic potential of stem/progenitor-like cells in the adult mammalian eye. Prog Retin Eye Res. 2012;31:213. doi: 10.1016/j.preteyeres.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Inoue T. Coles B.L. Dorval K. Bremner R. Bessho Y. Kageyama R. Hino S. Matsuoka M. Craft C.M. McInnes R.R. Tremblay F. Prusky G.T. van der Kooy D. Maximizing functional photoreceptor differentiation from adult human retinal stem cells. Stem Cells. 2010;28:489. doi: 10.1002/stem.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips M.J. Wallace K.A. Dickerson S.J. Miller M.J. Verhoeven A.D. Martin J.M. Wright L.S. Shen W. Capowski E.E. Percin E.F. Perez E.T. Zhong X. Canto-Soler M.V. Gamm D.M. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci. 2012;53:2007. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellough C.B. Sernagor E. Moreno-Gimeno I. Steel D.H. Lako M. Efficient stage-specific differentiation of human pluripotent stem cells toward retinal photoreceptor cells. Stem Cells. 2012;30:673. doi: 10.1002/stem.1037. [DOI] [PubMed] [Google Scholar]
- 8.Idelson M. Alper R. Obolensky A. Ben-Shushan E. Hemo I. Yachimovich-Cohen N. Khaner H. Smith Y. Wiser O. Gropp M. Cohen M.A. Even-Ram S. Berman-Zaken Y. Matzrafi L. Rechavi G. Banin E. Reubinoff B. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Lu B. Malcuit C. Wang S. Girman S. Francis P. Lemieux L. Lanza R. Lund R. Long-term safety and function of RPE from human embryonic stem cells in preclinical models of macular degeneration. Stem Cells. 2009;27:2126. doi: 10.1002/stem.149. [DOI] [PubMed] [Google Scholar]
- 10.Lamba D.A. Karl M.O. Ware C.B. Reh T.A. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:12769. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osakada F. Ikeda H. Mandai M. Wataya T. Watanabe K. Yoshimura N. Akaike A. Sasai Y. Takahashi M. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26:215. doi: 10.1038/nbt1384. [DOI] [PubMed] [Google Scholar]
- 12.Bauwens C.L. Peerani R. Niebruegge S. Woodhouse K.A. Kumacheva E. Husain M. Zandstra P.W. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 13.Bratt-Leal A.M. Carpenedo R.L. McDevitt T.C. Engineering the embryoid body microenvironment to direct embryonic stem cell differentiation. Biotechnol Prog. 2009;25:43. doi: 10.1002/btpr.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe K. Ueno M. Kamiya D. Nishiyama A. Matsumura M. Wataya T. Takahashi J.B. Nishikawa S. Nishikawa S. Muguruma K. Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 15.Levine E.M. Fuhrmann S. Reh T.A. Soluble factors and the development of rod photoreceptors. Cell Mol Life Sci. 2000;57:224. doi: 10.1007/PL00000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley M.W. Turner J.K. Reh T.A. Regulation of proliferation and photoreceptor differentiation in fetal human retinal cell cultures. Invest Ophthalmol Vis Sci. 1995;36:1280. [PubMed] [Google Scholar]
- 17.Nistor G. Seiler M.J. Yan F. Ferguson D. Keirstead H.S. Three-dimensional early retinal progenitor 3D tissue constructs derived from human embryonic stem cells. J Neurosci Methods. 2010;190:63. doi: 10.1016/j.jneumeth.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Tibbetts M.D. Samuel M.A. Chang T.S. Ho A.C. Stem cell therapy for retinal disease. Curr Opin Ophthalmol. 2012;23:226. doi: 10.1097/ICU.0b013e328352407d. [DOI] [PubMed] [Google Scholar]
- 19.Freund C.L. Gregory-Evans C.Y. Furukawa T. Papaioannou M. Looser J. Ploder L. Bellingham J. Ng D. Herbrick J.A. Duncan A. Scherer S.W. Tsui L.C. Loutradis-Anagnostou A. Jacobson S.G. Cepko C.L. Bhattacharya S.S. McInnes R.R. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa T. Morrow E.M. Cepko C.L. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen S. Wang Q.L. Nie Z. Sun H. Lennon G. Copeland N.G. Gilbert D.J. Jenkins N.A. Zack D.J. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 22.West E.L. Gonzalez-Cordero A. Hippert C. Osakada F. Martinez-Barbera J.P. Pearson R.A. Sowden J.C. Takahashi M. Ali R.R. Defining the integration capacity of embryonic stem cell-derived photoreceptor precursors. Stem Cells. 2012;30:1424. doi: 10.1002/stem.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters C.E. Woodside S.M. Eaves A.C. Isolation of subsets of immune cells. Methods Mol Biol. 2005;302:95. doi: 10.1385/1-59259-903-6:095. [DOI] [PubMed] [Google Scholar]
- 24.Kannagi R. Cochran N.A. Ishigami F. Hakomori S. Andrews P.W. Knowles B.B. Solter D. Stage-specific embryonic antigens (SSEA-3 and −4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J. 1983;2:2355. doi: 10.1002/j.1460-2075.1983.tb01746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bibb L.C. Holt J.K. Tarttelin E.E. Hodges M.D. Gregory-Evans K. Rutherford A. Lucas R.J. Sowden J.C. Gregory-Evans C.Y. Temporal and spatial expression patterns of the CRX transcription factor and its downstream targets. Critical differences during human and mouse eye development. Hum Mol Genet. 2001;10:1571. doi: 10.1093/hmg/10.15.1571. [DOI] [PubMed] [Google Scholar]
- 26.D'Amour K.A. Bang A.G. Eliazer S. Kelly O.K. Agulnick A.D. Smart N.G. Moorman M.A. Kroon E. Carpenter M.K. Baetge E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 27.Kroon E. Martinson L.A. Kadoya K. Bang A.G. Kelly O.G. Eliazer S. Young H. Richardson M. Smart N.G. Cunningham J. Agulnick A.D. D'Amour K.A. Carpenter M.K. Baetge E.E. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 28.Scott C.T. McCormick J.B. Owen-Smith J. And then there were two: use of hESC lines. Nat Biotechnol. 2009;27:696. doi: 10.1038/nbt0809-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rezania A. Bruin J.E. Riedel M.J. Mojibian M. Asadi A. Xu J. Gauvin R. Narayan K. Karanu F. O'Neil J.J. Ao Z. Warnock G.L. Kieffer T.J. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61:2016. doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burridge P.W. Thompson S. Millrod M.A. Weinberg S. Yuan X. Peters A. Mahairaki V. Koliatsos V.E. Tung L. Zambidis E.T. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kehat I. Kenyagin-Karsenti D. Snir M. Segev H. Amit M. Gepstein A. Livne E. Binah O. Itskovitz-Eldor J. Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer J.S. Howden S.E. Wallace K.A. Verhoeven A.D. Wright L.S. Capowski E.E. Pinilla I. Martin J.M. Tian S. Stewart R. Pattnaik B. Thomson J.A. Gamm D.M. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamba D.A. Gust J. Reh T.A. Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell. 2009;4:73. doi: 10.1016/j.stem.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaroop A. Kim D. Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in mammalian retina. Nat Rev Neurosci. 2010;11:563. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mucci I. Legitimo A. Compagnino M. Consolini R. Migliaccio P. Metelli M.R. Scatena F. The methodological approach for the generation of human dendritic cells from monocytes affects the maturation state of the resultant dendritic cells. Biologicals. 2009;37:288. doi: 10.1016/j.biologicals.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Shin S.Y. Jang S. Park C.J. Chi H.S. Lee J.H. Lee J.H. Lee K.H. Suh C. Lim S.E. Seo E.J. Application of an immune-magnetic cell sorting method for CD138-positive plasma cells in FISH analysis of multiple myeloma. Int J Lab Hematol. 2012;34:541. doi: 10.1111/j.1751-553X.2012.01433.x. [DOI] [PubMed] [Google Scholar]
- 37.Alon R. Bayer E.A. Wilchek M. Affinity cleavage of cell surface antibodies using the avidin-biotin system. J Immunol Methods. 1993;165:127. doi: 10.1016/0022-1759(93)90114-m. [DOI] [PubMed] [Google Scholar]
- 38.Zucchelli M. Ström S. Holm F. Malmgren H. Sahlén S. Religa P. Hovatta O. Kere J. Inzunza J. In vivo differentiated human embryonic stem cells can acquire chromosomal aberrations more frequently than in vitro during the same period. Stem Cells Dev. 2012;21:3363. doi: 10.1089/scd.2012.0066. [DOI] [PubMed] [Google Scholar]