Abstract
Protein–protein interaction networks mediate diverse biological processes by regulating various signaling hubs and clusters. 14-3-3 proteins, a family of phosphoserine/threonine-binding molecules, serve as major interaction hubs in eukaryotic cells and have emerged as promising therapeutic targets for various human diseases. In order to identify chemical probes for mechanistic studies and for potential therapeutic development, we have developed highly sensitive bioassays to monitor the interaction of 14-3-3 with a client protein. In this study, we describe a homogenous time-resolved fluorescence resonance energy transfer (TR-FRET) assay to detect the interaction of 14-3-3 with Bad, a proapoptotic member of the Bcl-2 family. Through a series of titration studies in which europium-labeled 14-3-3 serves as an FRET donor and a Dy647-labeled phosphorylated Bad, the peptide acts as an FRET acceptor, we have achieved a robust TR-FRET assay that is suitable for high-throughput screening (HTS) with an excellent signal-to-background ratio of >20 and Z′ values >0.7. This assay was further miniaturized to a 1,536-well format for ultra-HTS (uHTS), and exhibited a similar robust performance. The utility and performance of the assay for uHTS were validated by (i) known inhibitors, including peptide R18 and small molecule FOBISIN101, and (ii) screening of a 51,200 compound library. This simple and robust assay is generally applicable to detect the interaction of 14-3-3 with other client proteins. It provides a sensitive and easy-to-use tool to facilitate the discovery of 14-3-3 protein inhibitors as well as to study 14-3-3-mediated protein–protein interactions.
Introduction
The family of 14-3-3 phosphoserine/threonine-binding proteins consists of seven isoforms in mammalian cells.1 The isoforms are designated with Greek letters (β, ɛ, η, γ, τ, σ, and ζ) and encoded by genes that are located on different chromosomes. More than 200 proteins have been reported to interact with 14-3-3 proteins.2–5 Through interactions with client proteins, 14-3-3 binding impacts multiple signaling pathways that control diverse physiological processes, such as Bad-induced apoptosis, Raf-mediated cell proliferation, apoptosis signal-regulating kinase 1 (ASK1)-mediated stress responses, and Cdc25-regulated cell cycle progression.1,6 Given the critical role of 14-3-3 proteins in such diverse signaling pathways, it is not surprising that dysregulated 14-3-3/client protein interactions have been implicated in a wider range of human diseases, such as cancer, inflammatory diseases, and neuronal disorders.1,7–9 For instance, 14-3-3ζ has been shown to be overexpressed in patients with multiple solid tumor types, such as lung and breast cancers. Importantly, overexpression of 14-3-3 correlated with poor patient survival.10–14 Thus, studies on 14-3-3/client protein interactions and development of tools to modulate these interactions will not only provide critical insights into how intracellular signaling pathways are regulated, but also offer valuable opportunities for therapeutic intervention. The discovery of 14-3-3 inhibitors will be critical for chemical biology studies and for 14-3-3-targeting therapeutic development.
To discover 14-3-3 protein–protein interaction modulators, it is essential to develop highly sensitive methods to monitor the specific interaction of a 14-3-3 protein with its client proteins. High-throughput screening (HTS) is a widely used approach in the field of drug discovery and chemical biology to identify new chemical entities. An assay suitable for HTS requires target specificity, a robust readout, day-to-day and plate-to-plate reproducibility, technical simplicity, and suitability for automation. Assay technologies for monitoring biomolecular interactions in a homogenous format, such as fluorescence polarization (FP) and time-resolved fluorescence resonance energy transfer (TR-FRET), are extensively used in HTS campaigns for the discovery of small molecules.15 Notably, several HTS assays have already been developed for monitoring the interaction of 14-3-3 with its client proteins, including FP,16,17 AlphaScreen18 and label-free biosensor assays.19 We previously performed an HTS of the LOPAC library using an FP-based assay for the interaction of 14-3-3γ and Raf-1 protein, an interaction critical for mitogenic signal transduction,16 and identified a small molecule compound, FOBSIN101, as a 14-3-3 protein inhibitor.20 In addition to FOBOSIN101, several other small-molecule 14-3-3 inhibitors have been identified through computational-based virtual screening21 and fragment-based combinatorial small-molecule microarray.22,23 However, none of these reported compounds showed both high potency and on-target effect in any animal model systems.6,23 Since chemical modifications of existing compounds require major efforts, identifying novel chemical scaffolds that can efficiently and selectively inhibit 14-3-3 protein interactions through alternative HTS assays offers a new avenue of discovery.
The FP, AlphaScreen, and TR-FRET assay are well-established technologies for HTS.15 However, it is well-accepted that the application of different assay technologies often gives rise to different hit lists even when monitoring the same biochemical interaction.24–27 TR-FRET assay format offers several potential advantages. For example, the time-delayed measurement reduces fluorescence interference from library compounds, which presents one of the major challenges in an HTS campaign.28 TR-FRET assay also has less inter-well variation because of its ratiometric measurement.25 The primary goal of the current study is to develop an improved detection method with TR-FRET for monitoring 14-3-3/client protein interactions to facilitate the discovery of 14-3-3 protein inhibitors.
To discover new classes of small molecule 14-3-3 inhibitors, we designed and developed an HTS-suitable homogenous TR-FRET assay to monitor the interaction of 14-3-3 proteins with a pro-apoptotic protein, Bad. One of the major roles of 14-3-3 in cellular physiology is the inhibition of apoptosis through binding to critical pro-apoptotic proteins, such as Bad, a BH3-only member of the Bcl-2 family of proteins.29 In response to cell survival signals, Bad is phosphorylated by survival kinases, such as Akt, leading to the recruitment of 14-3-3. 14-3-3–bound Bad is sequestered in the cytoplasm away from Bcl-2/Bcl-XL in mitochondria, resulting in the attenuation of apoptosis.30 The effect of 14-3-3 on Bad is largely controlled by phosphorylation of Bad at S136.29 Using the interaction of 14-3-3 protein with a peptide derived from the region around the critical phosphorylated serine, pS136-Bad, as a model system, we have developed a sensitive and simple TR-FRET assay that can quantitatively measure the interaction of 14-3-3 with a client protein peptide. The homogenous and mix-and-read format allows the assay to be easily adapted to an HTS format. Results from our validation and miniaturization experiments strongly support the utility of the optimized 14-3-3/Bad interaction assay for large-scale uHTS campaigns to identify compounds that disrupt 14-3-3 protein–protein interactions. Compared with our previous assays for the interaction of 14-3-3γ with pS259-Raf1 peptide (FP assay)16 or R18 peptide (AlphaScreen assay),18 the current study based on the TR-FRET detection technology requires ∼100-fold less protein for the generation of a robust signal. In addition, the use of different 14-3-3 isoform (ζ) and binding partner (Bad) in the assay may lead to the identification of isoform- or ligand-selective 14-3-3 protein inhibitors.
Materials and Methods
Reagents
Europium (Eu) chelate-conjugated anti–HexaHis-antibody (anti–His-Eu) was purchased from PerkinElmer. All other reagents were obtained from Sigma-Aldrich, unless otherwise stated. The phosphorylated S136 Bad peptide was synthesized and labeled with Dy647 (Dyomics). The Dy647-pS136-Bad peptide consists of 17 residues: Dy647-LSPFRGRSR[pS]APPNLWA, and was dissolved in 50% dimethyl sulfoxide (DMSO)/50% H2O (v/v) as a 0.5 mM stock solution and stored at −80°C. The following peptides were synthesized and purified at the Emory Microchemistry and Proteomics Facility: pS136-Bad (LSPFRGRSR[pS]APPNLWA), S136-Bad (LSPFRGRSRSAPPNLWA), pS259-Raf (LSQRQRST[pS]TPNVHM),16 pS967-ASK1 (GSNEYLKSI[pS]LPVP),31 S967-ASK1 (GSNEYLKSISLPVP), and R18 (PHCVPKNLSWLNLEANMCLP).32
Expression and Purification of Recombinant 14-3-3 Proteins
Recombinant HexaHis-tagged 14-3-3 (His-14-3-3) isoforms (ζ, β, ɛ, η, γ, τ, and σ) were expressed and purified essentially as previously described.33 Briefly, BL21 (DE3) Escherichia coli were transformed with His-14-3-3 expression vectors and grown in LB medium (containing 100 μg/mL ampicillin) for protein auto-induction. After overnight incubation at 37°C, the cells were harvested, and the pellet was resuspended in ice-cold binding buffer (500 mM NaCl, 20 mM Tris-HCl, pH8.0, and 5 mM Imidazole, 1.0 mM phenylmethylsulfonyl fluoride, 1 μg/mL leupetin, and 1 μg/mL aprotinin). After sonication, the lysate was cleared by centrifugation (27,000 g, 20 min, 4°C) and loaded onto an Ni2+-charged histidine-binding column (His•Bind Resin; Novagen). The protein-bound column was washed with buffer (60 mM imidazole, 20 mM Tris-HCl, pH 7.9, and 500 mM NaCl). His-14-3-3 proteins were eluted (1 M imidazole, 20 mM Tris-HCl, pH 7.9, and 500 mM NaCl), and salts were removed from the pooled elution fractions using a PD-10 gel filtration column (GE Healthcare) equilibrated in HEPES buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.05% Tween-20, and 0.5 mM DTT). Protein concentration was measured using the Bradford method (BioRad). Purified proteins were stored at −80°C until use.
Assay Development and Optimization
The 14-3-3 TR-FRET assay was performed in 384-well black solid bottom plates (Corning Costar) in a total volume of 30 μL in each well. To determine the binding affinity of His-14-3-3 to the Dy647-pS-136-Bad peptide, titrations of both His-14-3-3ζ and pS136-Bad were carried out. The Dy647-pS136-Bad peptide or His-14-3-3ζ was serially diluted in FRET buffer (20 mM Tris, pH 7.0, 0.01% Nonidet P40, and 50 mM NaCl). The titrated peptide or protein was then mixed with various amounts of His-14-3-3ζ protein or the Dy647-pS-136-Bad peptide, respectively, in the presence of anti–His-Eu antibody (final, 1 nM). The mixture (final, 30 μL) was dispensed to each well of 384-well plates, and incubated at room temperature for the indicated period of time. The TR-FRET signals were recorded using an EnVision Multilabel plate reader (PerkinElmer; Eu ex 337 nm, Eu em 615 nm; Dy647 em 665/7.5 nm). A dual mirror at D400/D630 was used to simultaneously measure 665 and 615 nm emission signals. The TR-FRET signals were expressed as ratios (TR-FRET signal=F665 nm/F615 nm×104). The TR-FRET signal window was calculated by subtracting the background TR-FRET signal from the Dy647-pS136-Bad peptide and anti–His-Eu in the absence of 14-3-3 protein from that obtained from reactions in which 14-3-3 proteins were included.
Analysis of Assay Performance
To evaluate the quality of the assay for HTS in the absence of test compounds, we calculated the Z′ factor based on the following equation:
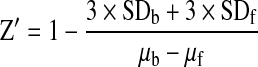 |
where SDb and SDf are the standard deviations for bound (b) and free (f) peptides, respectively, without 14-3-3 protein; whereas μb and μf are the mean TR-FRET signals for bound and free peptides, respectively. The Z′ factor reflects the quality of the assay and quantifies the suitability of a particular assay for use in HTS. A Z′ factor between 0.5 and 1.0 indicates that the assay is suitable for HTS.34 To monitor the dynamic window and sensitivity of the assay, the signal-to-background ratio (S/B) was calculated using the following equation: S/B=μb/μf.
All experimental data were analyzed using Microsoft Excel. The binding affinity was evaluated using GraphPad Prism version 5.0.
TR-FRET Competition Assay Development
For the competition assay, known 14-3-3 antagonist peptides were dissolved in HEPES buffer (10 mM, pH 7.4), NaCl (150 mM), Tween-20 (0.05%), serial diluted, and added to the reaction buffer containing His-14-3-3ζ (10 nM), Dy647-pS136-Bad peptide (20 nM), and anti–His-Eu antibody (1 nM) in a final volume of 30 μL. A small molecule 14-3-3 inhibitor, FOBISIN101,20 and the appropriate vehicle controls were included in the TR-FRET competition assay. The plates were incubated at RT for 2–18 h before the TR-FRET signal was measured. The effect of the inhibitors on the 14-3-3/Bad interaction was normalized to the vehicle control and expressed as percentage (%) of control. The % of control was calculated using the following equation:
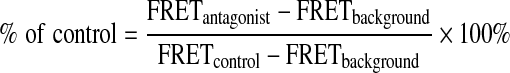 |
where FRETcontrol is the TR-FRET signal from wells containing His-14-3-3ζ (10 nM), Dy647-pS136-Bad peptide (20 nM), and anti–His-Eu antibody (1 nM), which defines the maximum signal; FRETbackground is the TR-FRET signal from wells containing Dy647-pS136-Bad and anti–His-Eu antibody without His-14-3-3ζ protein, which defines the minimal signal; and FRETantagonist is the TR-FRET signal in the presence of antagonist peptides or FOBISIB101.
Data were plotted against inhibitor concentrations and analyzed using GraphPad Prism version 5.0. IC50 values were determined by nonlinear curve fitting as the concentrations of inhibitors at which 50% of the control TR-FRET signal was inhibited.
Ultra-HTS Assay Development
The suitability of the TR-FRET assay for ultra-HTS (uHTS) was determined by performing the assay in a 1,536-well plate format. The total volume for all reactions was scaled down to 5 μL per well. His-14-3-3ζ protein was diluted in FRET buffer and mixed with Dy647-pBad peptide (20 nM) and anti–His-Eu (1 nM). The mixture (5 μL) was dispensed to 1,536-well black plates (Corning Costar; Cat# 3724). After incubation at RT for 2 h, the TR-FRET signals were recorded using an EnVision Multilabel plate reader with laser excitation at 337 nm and emissions at 615 and 665 nm. The S/B ratio and the Z′ factors were calculated as described.
uHTS
For large-scale uHTS, a subset of the Molecular Library Screening Center Network (MLSCN) library (51,200 compounds) was utilized. The assay was performed in forty (40) 1,536-well plates as described in Table 1. Appropriate controls that defined the minimal and maximal FRET signals were included in each plate. Briefly, the reaction buffer (4.5 μL/well) containing 10 nM His-14-3-3ζ, 20 nM Dy647-pS136-Bad peptide, and 1 nM anti–His-Eu was dispensed into 1,536-well assay plates using a MultiDropCombi (Thermo-Fisher Scientific). Thirty-two wells of 20 nM Dy647-pS136-Bad peptide and 1 nM anti–His-Eu and 32 wells of the control solution (10 nM His-14-3-3ζ, 20 nM Dy647-pS136-Bad peptide, and 1 nM anti–His-Eu) were included in each plate to define the minimal (background) and maximal (control) FRET signal, respectively. Using a Pin-Tool (VP Scientific) integrated with a Beckman NX liquid handler (Beckman Coulter), library compounds (0.1 μL of 1 mM stock in 100% DMSO) were then added to the reaction, yielding a final compound concentration of 21.7 μM and a final DMSO concentration of 2.2% (v/v). DMSO (0.1 μL) was added as a vehicle control into all control wells. The reactions were mixed thoroughly using a shaker integrated with a Beckman NX and incubated at RT for 2 h. The plates were delivered through integrated robotic systems to an EnVision Multimode reader for TR-FRET signal measurement.
Table 1.
Ultra-High Throughput Screening Protocol
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Dispense reaction buffer | 4.5 μL/well | A mixture containing 14-3-3ζ (10 nM), Dy647-pS136-Bad peptide (20 nM), and anti–His-Eu (1 nM), |
| 2 | Controls | 4.5 μL/well | A mixture of Dy647-pS136-Bad peptide (20 nM) and anti–His-Eu (1 nM) in one column (32 wells) |
| 3 | Library compounds | 100 nL/well | 21.7 μM final from 1 mM stock in DMSO |
| 4 | Incubation time | 2 h | Plates were incubated at room temperature for 2 h |
| 5 | Assay readout | Ex 337 nm; Em1 615 nm, Em2 665 nm; dual mirror D400/D630 | Time-resolved fluorescence; Envision Multilabel plate reader |
Step Notes
1. 1536-well black plates (Corning Costar; Cat. No. 3724) were used. The assay component was mixed together in FRET buffer (20 mM Tris, pH 7.0, 0.01% Nonidet P40, and 50 mM NaCl). The MultiDrop Combi (Thermo Scientific) was used for dispensing.
2. One column (32-wells) in a 1536-well plate was used for the background control, which defines the minimal signal.
3. Compounds were added using a Pin-Tool (VP Scientific) integrated with a Beckman NX liquid handler (Beckman Coulter).
4. Plates were stacked with the top plate lidded during incubation.
5. A 50 μs delay was used for the TR-FRET signal measurement.
DMSO, dimethyl sulfoxide; TR-FRET, time-resolved fluorescence resonance energy transfer.
All screening data were analyzed using CambridgeSoft bioassay software. The TR-FRET signals from control wells were used to calculate the S/B ratios and Z′ factors without test compounds. The percent inhibition of a compound was calculated based on each plate using the following equation:
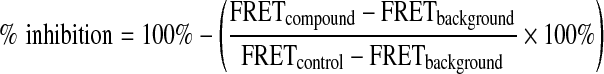 |
Results
Design of a Homogeneous TR-FRET Assay for Monitoring the 14-3-3/Bad Peptide Interaction
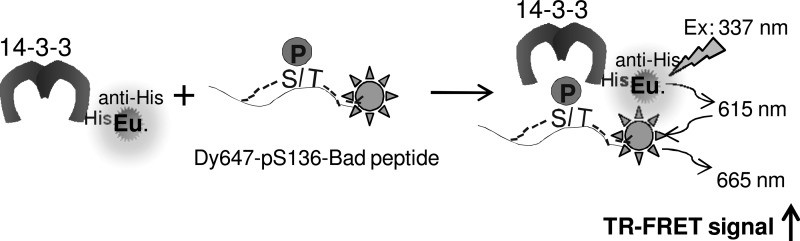
FRET can occur between two fluorophores with overlapping emission and excitation spectra. Specifically, the emission spectrum of the donor fluorophore should significantly overlap with the excitation spectrum of the acceptor fluorophore. To monitor the 14-3-3/Bad interaction, HexaHis-tagged 14-3-3 (His-14-3-3) protein was indirectly labeled with a Eu chelate through a Eu-conjugated anti-His antibody. Bad peptide phosphorylated at S136 was directly labeled with Dy647. Importantly, Eu and Dy647 comprise a fluorescence energy transfer pair. Interaction of 14-3-3 protein and the pS136-Bad peptide brings the two fluorophores into close proximity, leading to an energy transfer from Eu to Dy647 and the generation of FRET signals (Fig. 1). The use of long-lived Eu as a donor allows for a temporal delay (typically 50–150 μs) between donor excitation and detection of acceptor emission,35 resulting in the generation of a time-resolved FRET (TR-FRET) signal. The TR-FRET signal, which is expressed as the ratio of the detected Dy647 emission at 665 nm and Eu emission at 615 nm after excitation of Eu at 337 nm, is indicative of the extent of the interaction between 14-3-3 protein and the pS136-Bad peptide. The time-delayed measurement of acceptor emission makes the TR-FRET assay particularly suitable for compound screening, as it minimizes interference from short-lived background fluorescence, such as autofluorescence from screening library compounds.
Fig. 1.
Design of a time-resolved fluorescence resonance energy transfer (TR-FRET) assay for monitoring the 14-3-3/pBad peptide interaction. His-14-3-3 protein is indirectly labeled with a long-lived fluorophore, Europium (Eu), through an anti–His-Eu antibody (PerkinElmer) to serve as a FRET donor. A phosphopeptide derived from Bad (pS136-Bad) is directly labeled with Dy647 to serve as a FRET acceptor. Interaction of 14-3-3 with pS136-Bad brings two fluorophores into proximity. On excitation at 337 nm, the energy emitted from the Eu donor is transferred to the Dy647 acceptor, leading to the generation of a FRET signal.
Development of the TR-FRET 14-3-3 Binding Assay in a 384-well HTS Format
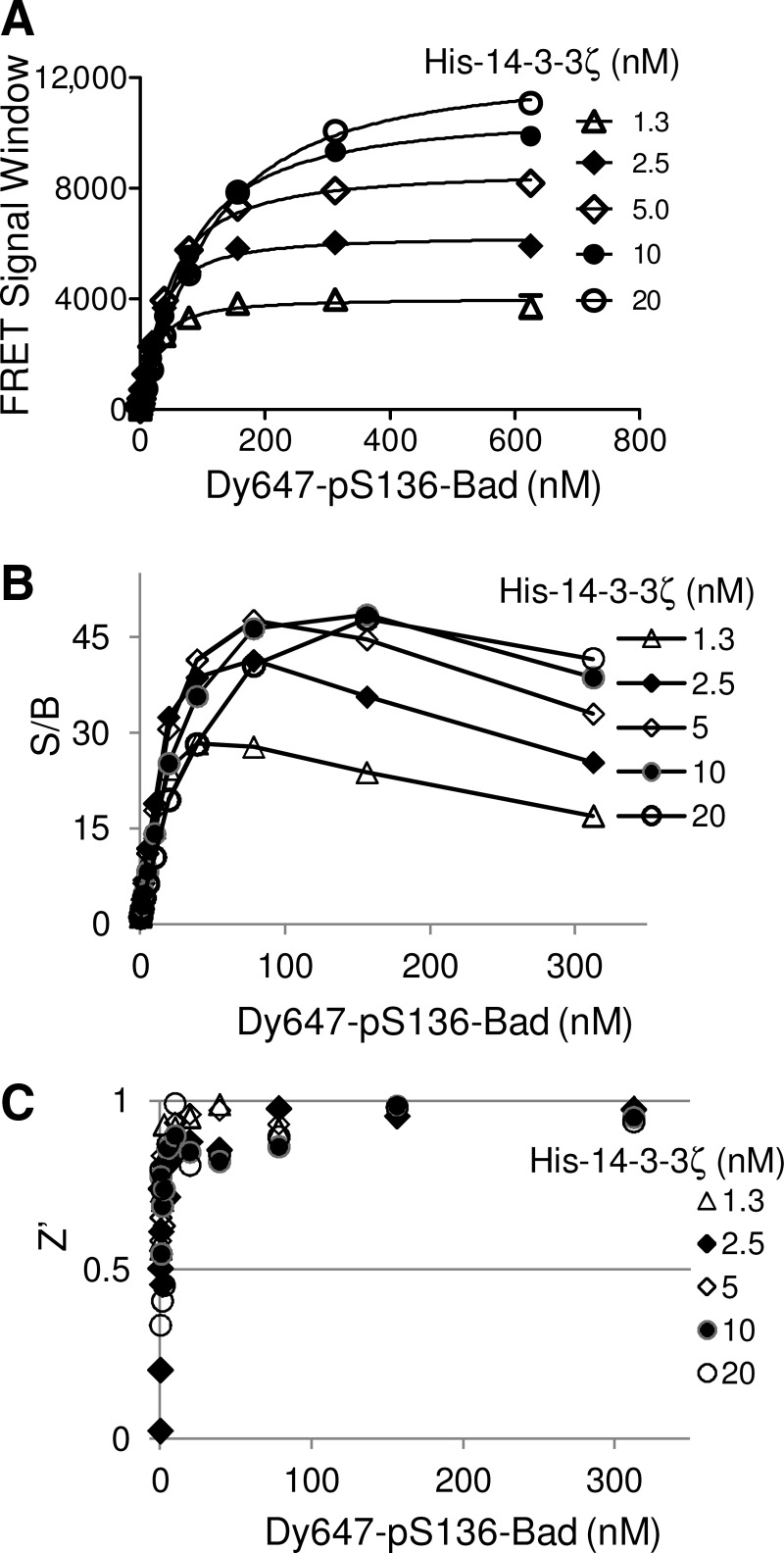
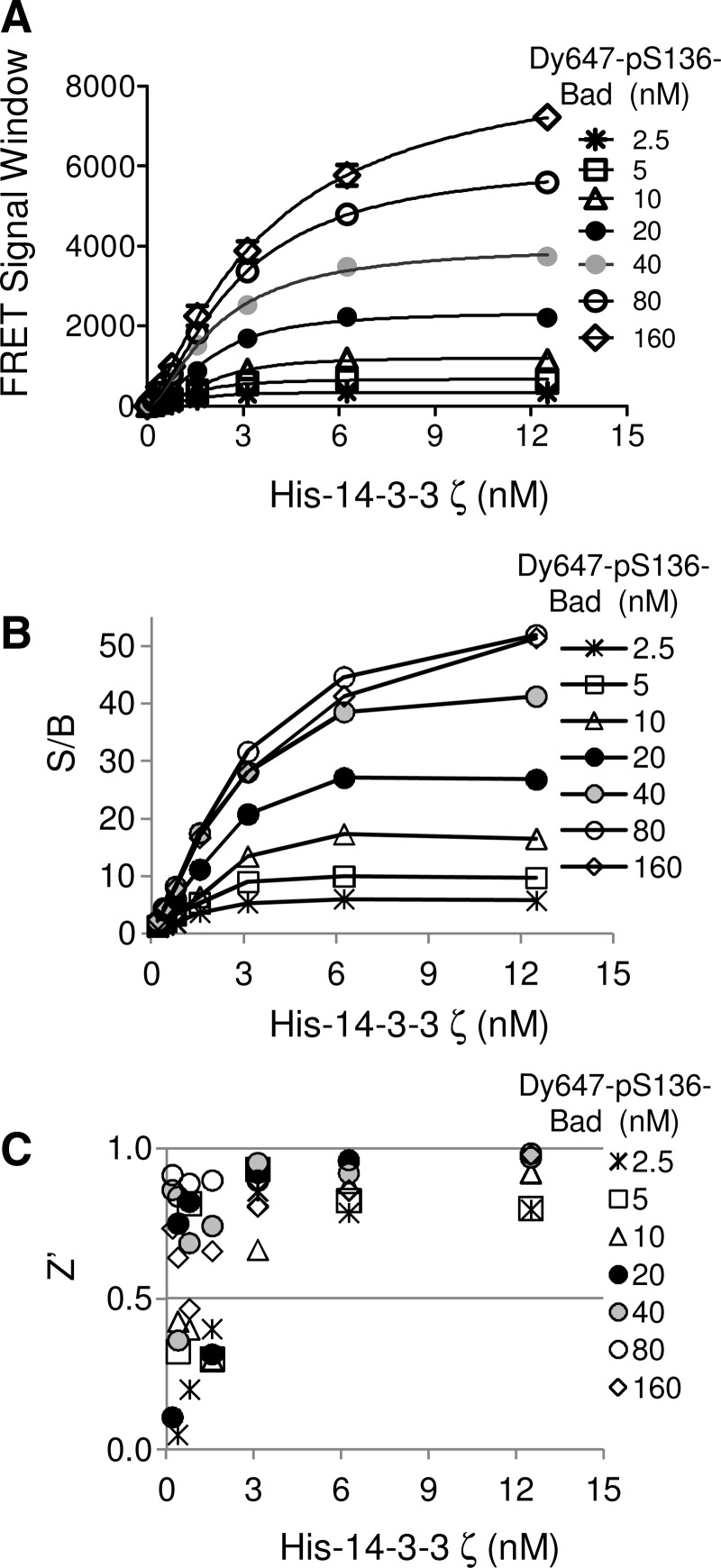
To develop a TR-FRET assay that monitors the interaction of a 14-3-3 protein and its client protein, we initially used 14-3-3ζ and a pS136-Bad peptide as assay components. In order to determine the conditions that generate an optimal TR-FRET signal, titrations of both 14-3-3ζ and pS136-Bad peptide were carried out in a 384-well format. As shown in Figure 2A, increasing concentrations of Dy647-pS136-Bad peptide resulted in a dose-dependent increase of the TR-FRET signal. The maximal TR-FRET signal was observed at approximately 300 nM of Dy647-pS136-Bad peptide for all concentrations of HexaHis-14-3-3ζ protein tested. The maximum TR-FRET signal window was ∼10,000. The quality of the assay was evaluated by calculating the S/B ratios and Z′ factors over the concentration range tested. The S/B ratio increased with increase in the concentration of Dy647-pS136-Bad peptide up to ∼100 nM Dy647-pS136-Bad, and it reached a maximum ratio of 46 at 10 nM His-14-3-3ζ and 150 nM Dy647-pS136-Bad peptide (Fig. 2B). The Z′ factors were >0.8 when the concentration of Dy647-pS136-Bad peptide was >2 nM (Fig. 2C). Similar results were obtained when His-14-3-3ζ protein was titrated against the Bad peptide. The TR-FRET signal increased dose-dependently with increasing concentrations of His-14-3-3ζ (Fig. 3A). The signal reached a plateau when the concentrations of His-14-3-3ζ and pS136-Bad peptide were 12.5 nM and 80 nM, respectively. The maximal S/B was 50 (Fig. 3B) and the Z′ factors were >0.8 when His-14-3-3ζ protein was >6 nM (Fig. 3C). The Z′ factor of an assay incorporates both the dynamic range and variability of an assay into a single measurement, and the Z′ factor should be between 0.5 and 1 for reliable and robust assays for HTS.34 Both protein and peptide titrations demonstrated consistent Z′ factors of 0.8 over the concentration ranges, indicating that the TR-FRET 14-3-3 binding assay is a robust and high-quality assay for HTS.
Fig. 2.
Evaluation of the Dy647-pS136-Bad peptide in the 14-3-3 TR-FRET assay in a 384-well high-throughput screening (HTS) format. (A) Increasing concentrations of Dy647-pS136-Bad peptide (15 μL) were mixed with the indicated concentrations of His-14-3-3ζ protein (15 μL) in the presence of anti–His-Eu antibody (1 nM, final). TR-FRET signals were recorded using an EnVision multilabel plate reader. The FRET signal window was calculated by subtracting the background signal from the Dy647-pS136-Bad peptide and anti–His-Eu only from the FRET signal obtained in the presence of 14-3-3 protein, the Dy647-pS136-Bad peptide, and anti–His-Eu (A). (B, C) The performance of the assay for HTS was evaluated by the Z′ factor and signal-to-background ratio (S/B). S/B (B) and Z′ (C) were calculated and plotted against Dy647-pS136 peptide concentrations.
Fig. 3.
Optimization of the 14-3-3/pBad peptide binding assay in a 384-well HTS format. (A) The TR-FRET signal increased by increasing concentrations of His-14-3-3ζ protein. His-14-3-3ζ protein was titrated against various concentrations of the Dy647-pS136-Bad peptide in the presence of anti–His-Eu antibody (1 nM). The S/B (B) and Z′ factor (C) were calculated and plotted against His-14-3-3ζ protein concentrations.
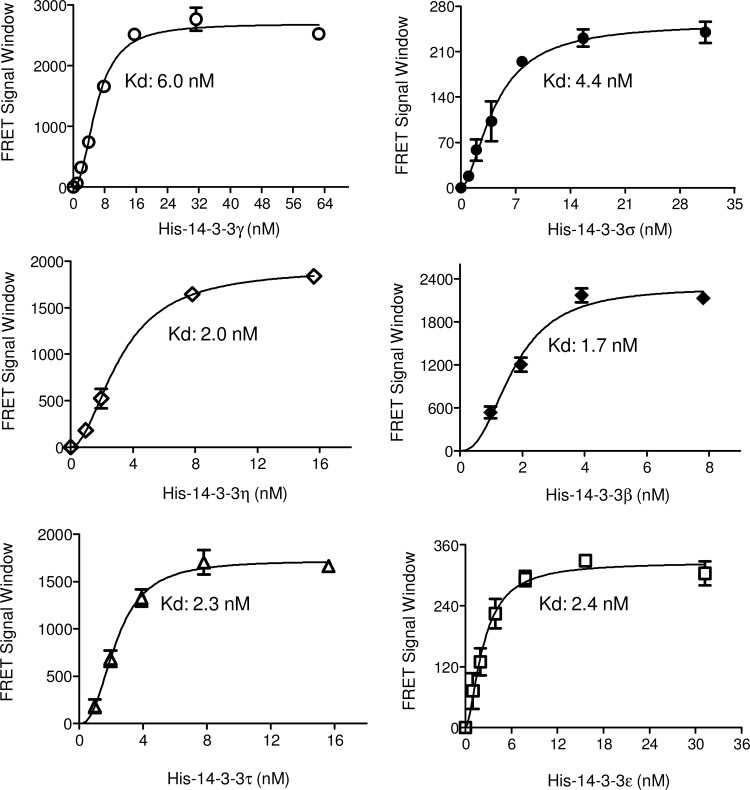
The 14-3-3 proteins are a family of homologous eukaryotic molecules with seven distinct isoforms in mammalian cells. 14-3-3 protein binding to Bad is conserved among the mammalian isoforms.36 For identification of isoform-specific 14-3-3 modulators, we tested the utility of the TR-FRET assay for monitoring the interaction of Dy647-pS136-Bad with the additional six isoforms of 14-3-3 proteins. As shown in Figure 4, increasing concentrations of all six isoforms of 14-3-3 proteins with 20 nM Dy647-pS136-Bad peptide resulted in dose-dependent increases in TR-FRET signals, which eventually reached plateaus at high concentrations. The maximal TR-FRET signals obtained varied across the six isoforms of 14-3-3 proteins. However, the Kds of the interactions of 14-3-3 proteins with Bad were similar between all isoforms. The Kd obtained for the interactions of the 14-3-3γ, σ, η, β, τ, and ɛ isoforms with the Dy647-pS136-Bad peptide were 6.0, 4.4, 2.0, 1.7, 2.3, and 2.4 nM, respectively. The Kds (low nM) obtained here are consistent with what has been previously reported for the interaction of 14-3-3 isoforms with phosphorylated Bad determined by the surface Plasmon resonance technique.37 These data indicate that the TR-FRET assay can be utilized to monitor the interaction of all 14-3-3 isoforms with the pS136-Bad peptide.
Fig. 4.
The interaction of 14-3-3 isoforms with pS136-Bad as detected by the TR-FRET assay. Dy647-pS136 Bad peptide (20 nM) was mixed with increasing concentrations of the indicated His-14-3-3 isoforms in the presence of anti–His-Eu (1 nM). The FRET signal was measured, and the FRET signal window was calculated by subtracting the background signal without 14-3-3 protein from that in the presence of 14-3-3 protein.
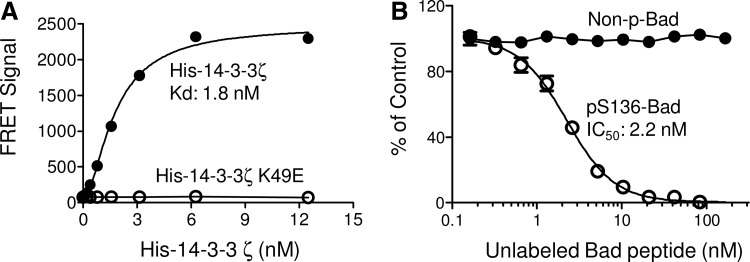
To examine the specificity of the TR-FRET assay for 14-3-3 binding, a ligand-binding defective mutant of His-14-3-3ζ, K49E, was utilized in place of wild-type 14-3-3. As shown in Figure 5A, increasing concentrations of His-14-3-3ζ K49E with Dy647-pS136-Bad failed to generate any TR-FRET signal; while incubation of His-14-3-3ζ with the Dy647-pS136-Bad peptide resulted in a dose-dependent increase of TR-FRET signal. The Kd for the 14-3-3ζ/pS136-Bad interaction was about 1.9 nM. These data indicate that the TR-FRET signal obtained in our assay is dependent on 14-3-3 binding to the Dy647-pS136-Bad peptide.
Fig. 5.
The specificity of the 14-3-3/pS136-Bad TR-FRET assay. (A) His-14-3-3ζ, but not mutant His-14-3-3ζ K49E, interacts with Dy647-pS136-Bad as measured by the TR-FRET assay. Increasing concentrations of His-14-3-3ζ, or His-14-3-3ζ K49E protein, was incubated with the Dy647-pS136-Bad peptide (20 nM) and anti–His-Eu (1 nM), and the TR-FRET signal was recorded. (B) The effect of unlabeled pS136-Bad peptide and unphosphorylated Bad peptide on the TR-FRET signal generated from the interaction of 14-3-3ζ with the Dy647-pS136-Bad peptide.
Assay Validation with Known 14-3-3 Inhibitors
To validate the TR-FRET binding assay for HTS, we developed a competition assay with several known peptide antagonists of 14-3-3 proteins. A viable HTS assay for the identification of 14-3-3 inhibitors demands the ability to detect the reversibility of the 14-3-3/Bad interaction in a competition mode, where 14-3-3 antagonist peptides or a small molecule inhibitor should be able to displace the Dy647-pS136-Bad peptide by binding to 14-3-3. Based on the protein and peptide titration results, reaction conditions (10 nM His-14-3-3ζ, 20 nM Dy647-pS136-Bad peptide, and 1 nM anti–His-Eu) that gave rise to robust TR-FRET signals (Figs. 2 and 3) were selected for the competition experiments. The effect of an unlabeled pS136-Bad peptide on the TR-FRET signal was first tested. A Bad peptide lacking phosphorylation at Ser136 (S136-Bad) was used as a negative control. As shown in Figure 5B, unlabeled pS136-Bad peptide successfully completed the binding of the Dy647-pS136-Bad peptide to His-14-3-3ζ and dose dependently decreased the TR-FRET signal (IC50 ∼2.2 nM). In contrast, the unphosphorylated S136-Bad peptide was unable to compete the binding of the Dy647-pS136-Bad peptide to 14-3-3ζ. The TR-FRET signal remained unchanged across all peptide concentrations tested. These results support the specific disruption of the 14-3-3/Bad interaction by phosphorylated Bad peptide for competitive inhibitor studies.
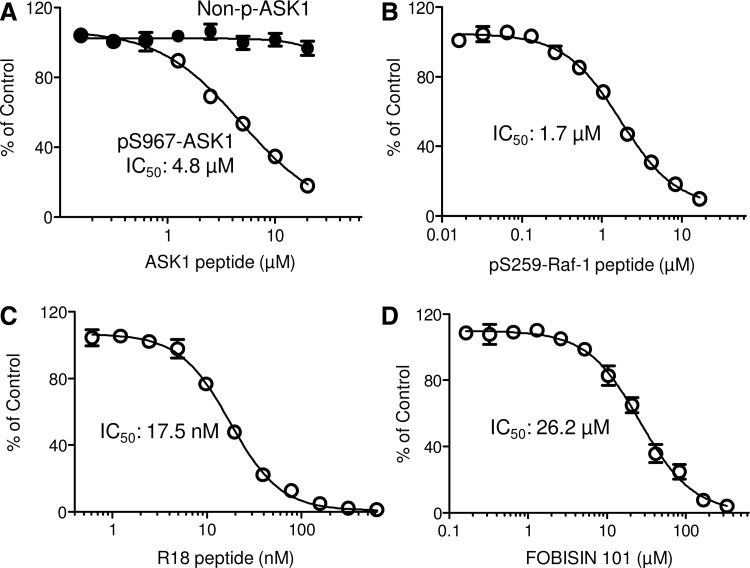
To further validate the assay for HTS, additional well-characterized 14-3-3 peptide antagonists were tested, including phosphorylated ASK1 (pS967-ASK1) and Raf-1 (pS259-Raf) peptides, and an unphosphorylated 14-3-3 peptide antagonist, R18.16,30,38 The pS259-Raf-1 peptide and pS967-ASK1 peptide are derived from the 14-3-3 binding sites in Raf-1 and ASK1, respectively. R18 is a 20-amino-acid unphosphorylated peptide obtained from phage display libraries that exhibits a high affinity for 14-3-3 proteins in a solid-phase assay (Kd of 70-90 nM).32 All of these peptides have previously been shown to inhibit 14-3-3/client protein interaction.16 As shown in Figure 6, the addition of increasing concentrations of pS967-ASK1 peptide (Fig. 6A), pS259-Raf-1 peptide (Fig. 6B), and R18 peptide (Fig. 6C) led to decreased TR-FRET signals in a dose-dependent manner, suggesting inhibited interaction of His-14-3-3ζ with Dy647-pS136-Bad. The IC50 was 1.7 μM for pS967-ASK1, 4.8 μM for pS259-Raf-1, and 17.5 nM for the R18 peptide. As expected, the negative ASK1 control peptide without phosphorylation (S967-ASK1) did not affect the TR-FRET signal (Fig. 6A).
Fig. 6.
Validation of the 14-3-3/pS136-Bad interaction assay with known 14-3-3 inhibitors. An increasing concentration of 14-3-3 peptide antagonists or a small-molecule inhibitor was added to a reaction containing the Dy647-pS136-Bad peptide (20 nM), His-14-3-3ζ (10 nM), and anti–His-Eu (1 nM). After incubation at room temperature for 2 h, the TR-FRET signals were recorded. The effect of the 14-3-3 inhibitors on the TR-FRET signal is expressed as % of control as described in the Materials and Methods section. The effect of the pS976-ASK1 peptide, or a nonphosphorylated ASK1 peptide (A), the pS259-Raf-1 peptide (B), R18 peptide (C), or FOBISIN101 (D), on the TR-FRET signals from the 14-3-3/pS136-Bad interaction is shown.
We also tested the effect of a small molecule 14-3-3 protein inhibitor, FOBISIN 101, in the TR-FRET competition assay. FOBISIN 101 was discovered by an FP-based HTS campaign for 14-3-3 protein modulators.20 As shown in Figure 6D, FOBISIN 101 dose dependently decreased the TR-FRET signal with an IC50 of ∼26.2 μM. Together, these data validate the TR-FRET 14-3-3 binding assay as specific and sensitive for HTS. Furthermore, the established conditions were shown to be suitable for HTS application to identify 14-3-3 protein inhibitors.
Examination of Assay Stability and DMSO Tolerance
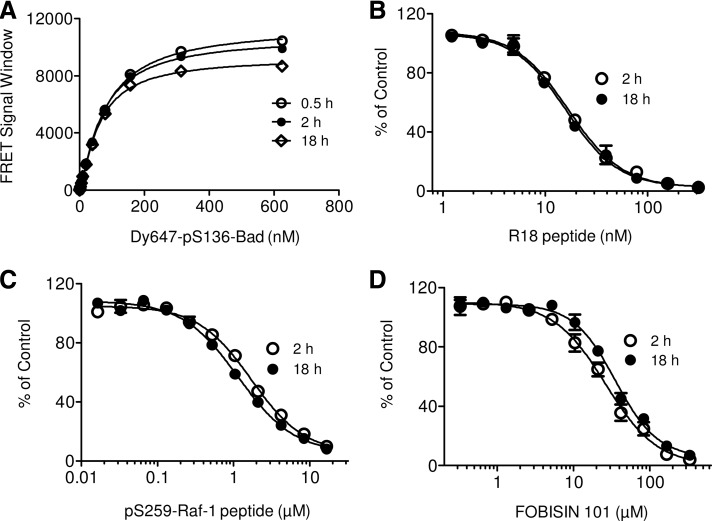
An assay suitable for HTS should show good stability over a reasonable period of time. To evaluate the stability of the TR-FRET 14-3-3 binding assay, we monitored the binding of His-14-3-3ζ to increasing concentrations of the Dy647-pS136-peptide for various incubation times. As shown in Figure 7A, The TR-FRET signal did not change significantly for ∼18 h. This stability eliminates concerns associated with dispensing large numbers of assay plates for scheduled readings during the high-throughput screen. In addition to the binding assay, we also examined the stability of the competition assay with the 14-3-3 peptide antagonists and small-molecule inhibitor over time. The inhibitory effect of the R18 peptide (Fig. 7B), pS259-Raf peptide (Fig. 7C), and FOBISIN101 (Fig. 7D) on the TR-FRET signal remained similar between 2 and 18 h of incubation. The stability of the competition assay renders great flexibility for large-scale compound screening.
Fig. 7.
Evaluation of assay stability. The FRET reactions were performed as in Figure 5 and incubated without peptide antagonist (A) or with increasing amounts of peptide antagonists R18 (B), pS259-Raf-1 (C), or FOBOSIN101 (D). The TR-FRET signals were measured after incubation for the indicated time at room temperature (RT). The data were normalized to vehicle control and expressed as % of control.
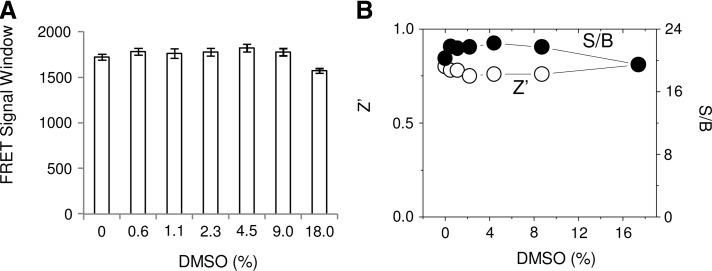
Compounds for HTS are generally dissolved in DMSO; therefore, the HTS assay components should tolerate the presence of a certain amount of DMSO. In the presence of increasing amounts of DMSO, the TR-FRET signals did not appreciably decrease. The assay performance remains robust in 9% DMSO with very little decrease observed in 18% DMSO (Fig. 8A). The Z′ values remained above 0.7 throughout the DMSO titration, and the S/B ratios decreased only slightly with 18% DMSO (Fig. 8B). Thus, the TR-FRET 14-3-3 binding assay is stable in the presence of at least 18% DMSO.
Fig. 8.
The effect of dimethyl sulfoxide (DMSO) on 14-3-3/Bad TR-FRET assay. (A) Increasing amounts of DMSO (0%–18%) were added to the TR-FRET reaction and incubated at RT for 2 h. The TR-FRET signals were measured, and the FRET signal windows were calculated and plotted against the amount of DMSO. (B) The quality of assay performance for HTS was evaluated by Z′ and S/B in the presence of increasing amounts of DMSO. Z′ and S/B were calculated as described in the Materials and Methods section.
Development and Validation of an uHTS Assay in a 1,536-Well Format
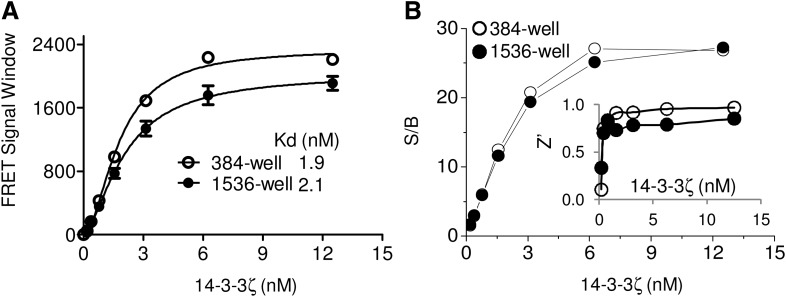
The use of high-density microplates allows increased throughput with reduced reagent cost in a miniaturized format. We, therefore, examined the performance of the TR-FRET assay in a 1,536-well uHTS format. Using the conditions established for 384-well plates, we did not observe significant changes in the TR-FRET signals from the His-14-3-3ζ/Dy647-pS136-Bad peptide interaction in a 1,536-well plate format (Fig. 9A). The Kds obtained were 1.9 and 2.7 nM from titration experiments in 384-well and 1,536-well plates, respectively. The Z′ values were >0.7 and the S/B was >20 when the His-14-3-3ζ concentration was >6 nM in both 384- and 1,536-well format (Fig. 9B). This indicates that the optimized TR-FRET assay conditions for monitoring 14-3-3/pBad peptide binding are suitable for uHTS in a 1,536-well plate format. The assay protocol is described in Table 1.
Fig. 9.
Miniaturization of the 14-3-3/Bad TR-FRET assay to a 1,536-well format for ultra-HTS (uHTS). Increasing concentrations of His-14-3-3ζ were incubated with the Dy647-pS136-Bad peptide (20 nM) in the presence of His-Eu (1 nM) in a 384-well (30 μL/well) or a 1,536-well (5 μL/well) plate. The TR-FRET signal was measured after incubation at RT for 2 h. The FRET signal window was plotted against 14-3-3ζ concentration (A). The S/B and Z′ factors were calculated and plotted against 14-3-3ζ concentration (B).
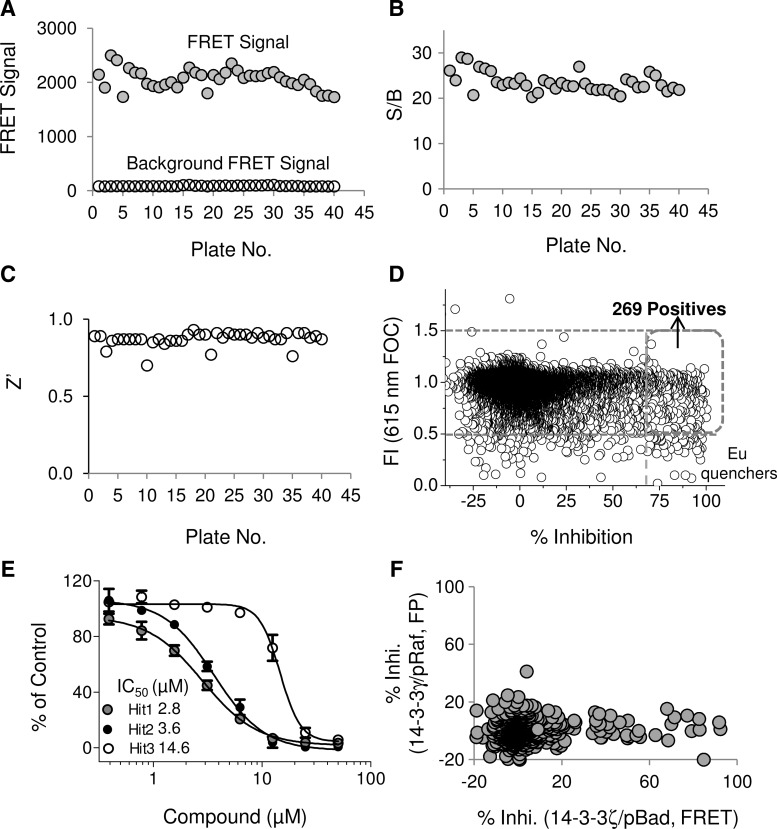
To demonstrate the utility of the TR-FRET 14-3-3 binding assay in a 1,536-well plate format, we carried out a large-scale screening campaign. Assay performance results are summarized from an example set of uHTS data for 51,200 compounds from a total of 40 1,536-well plates. The day-to-day, plate-to-plate, and well-to-well variations were determined by performing the screen on different days. Various controls were included in each plate, including 32 wells of blank control containing the Dy647-pS136-Bad peptide (20 nM) and anti–His-Eu (1 nM), and 32 wells of negative control with a mixture of the Dy647-pS136-Bad peptide (20 nM), His-14-3-3ζ (10 nM), and anti–His-Eu (1 nM). The blank control defines the minimal TR-FRET signal, and the negative control defines the maximal TR-FRET signal. The average TR-FRET signal of each plate from the blank and negative control wells was consistent across the 40 plates and was consistent from day-to-day (Fig. 10A). The average TR-FRET signal from the 40 plates was 2060.4 with a standard deviation (SD) of 181.6. The S/B ratios, as determined from each plate, were consistently higher than 20, and the average S/B from the 40 plates was 23.5±2.1 (Fig. 10B). The Z′ factor ranged from 0.7 to 0.9, with an average Z′ of 0.87 (Fig. 10C). Together, these data demonstrate a robust and consistent assay for uHTS.
Fig. 10.
Validation of the 14-3-3/Bad TR-FRET assay in a uHTS format. The uHTS assay was validated by screening a library of 51,200 compounds. Average FRET signal (A), S/B (B), and Z′ factor (C) from the uHTS were evaluated across the forty (40) 1,536-well plates. Background FRET signals from wells containing the Dy647-pS136-Bad peptide and anti–His-Eu in the absence His-14-3-3ζ is shown in (A). (D) Summary of uHTS results. Percent inhibition of the 14-3-3ζ/pBad peptide interaction was plotted against compound fluorescence, expressed as fluorescence intensity (FI; Eu 615 nM; fold over control (FOC) for 51,200 compounds. Positive hits from the uHTS were defined as those with % inhibition >70 and 0.5<FOC<1.5. (E) Dose-response curves of three representative positive hits. (F). Comparison of the screening results for the overlapping set of 843 compounds between the current TR-FRET (14-3-3ζ/pBad peptide) assay with those from the FP (14-3-3γ/pRaf peptide) assay.
We next evaluated the potential hit rate by excluding signals resulting from fluorescent compounds. Compound interference is one of the major causes for false positives in a primary screening campaign for the discovery of small-molecule modulators. Due to the ratiometric measurement of the TR-FRET signal (F665 nm/F615 nm×104), compounds that increase or decrease donor (Eu) emission signals at the 615 nm channel may interfere with the assay readout, leading to false positives. To address this issue, in addition to analyzing the screening data with FRET signal ratios, we also analyzed the fluorescence intensity (FI) of the 615 nm channel for Eu emission across the screen. The FI is expressed as fold over control (FOC) and is calculated by the following equation:
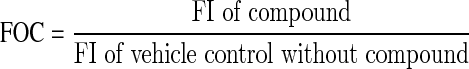 |
After excluding compounds with possible fluorescence interference, for example, fluorescence (FOC>1.5) and fluorescence quencher (FOC<0.5), compounds that inhibited the binding of His-14-3-3ζ to the Dy647-pS136-Bad peptide by more than 70% were considered potential positives. The representative results for the screening of 51,200 compounds are shown in Figure 10D, and plotted as FI (615 nm FOC) against % inhibition. A total of 317 compounds showed % inhibition >70 as primary positive hits. However, among those positives, 108 compounds decreased Eu emission signal at 615 nm with FOC<0.5. After excluding fluorescence interference compounds (most Eu quenchers), a total of 269 compounds have been identified as positives (shown as boxed in Fig. 10D). The resulting primary hit rate is 0.53%. It is, thus, desirable to exclude false positives early in the HTS process to maximize productivity.
Dose-response experiments were carried out to test the effect of 269 primary positives on the interaction of 14-3-3ζ with Dy647-pBad peptide. Out of these 269 TR-FRET primary hits, 134 compounds were confirmed active in the dose-response with IC50<30 μM. As an example shown in Figure 10E, three hit compounds exhibited dose-dependent inhibition of the interaction of 14-3-3 with Dy647-pBad peptide with varying potency (IC50s of 2.8, 3.6, and 14.6 μM, respectively). This result further supports the application of the optimized TR-FRET assay for screening of 14-3-3 protein–protein interaction inhibitors. The detailed results of a large-scale uHTS with our optimized TR-FRET assay and the follow-up studies will be reported in separate publications.
Among the 51,200 compounds screened as summarized in Figure 10D, 843 compounds were tested in the previous FP assay for the interaction of 14-3-3γ and pS259-Raf-peptide. This overlapping set of 843 compounds allows us to compare the results of the current TR-FRET (14-3-3ζ/pBad peptide) assay with that from the FP (14-3-3γ/pRaf peptide) assay. As shown in Figure 10F, the TR-FRET assay revealed more hits (16 with % inhibition >70) than the FP assay did (one hit with % inhibition >30). There were no overlapping hits from this small set of compounds in these two assays.
The TR-FRET Assay Exhibits Favorable Performance for the Discovery of 14-3-3 Protein Inhibitors
To examine whether the observed differences in hit sets between the TR-FRET and the FP assays are due to the assay technology employed, we directly compared the sensitivity of these two assay formats with the same pair of interacting proteins, 14-3-3ζ and pS136-Bad (Fig. 11). First, we optimized the assay conditions for both TR-FRET and FP assay by titrating each assay component for comparison purpose. Through these experiments, robust TR-FRET signals with Z′>0.5 for all assays were achieved for the interaction of 14-3-3ζ and 14-3-3γ with Dy647-conjugated pS136-Bad and pS259-Raf peptides, respectively.
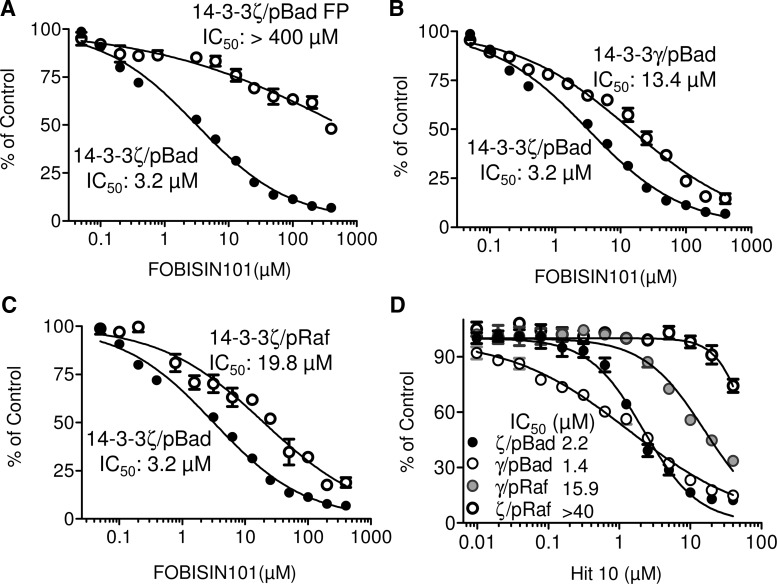
Fig. 11.
Performance evaluation of the TR-FRET and FP assays for 14-3-3 inhibitor discovery. The dose-response effect of FOBISIN101 was examined on the interaction of 14-3-3ζ with pBad-Dy647 peptide in TR-FRET and FP assay (A), the interaction of 14-3-3ζ or 14-3-3γ with pBad-Dy647 in the TR-FRET assay (B), and the interaction of 14-3-3ζ with pBad-Dy647 or pRaf-Dy647 peptide (C). (D) The dose-response effect of Hit 10 on the interaction of 14-3-3ζ or γ with pBad-Dy647 or pRaf-Dy647 in the TR-FRET assay. Increasing concentrations of FOBOSIN101 or Hit 10 compound were added to the reaction buffer containing 14-3-3ζ and pBad-Dy647 for the FP assay. Four optimized TR-FRET reactions contain the combinations of anti–His-Eu with (●) 14-3-3ζ and pBad-Dy647, (◯) 14-3-3γ and pBad-Dy647, ( ) 14-3-3ζ and pRaf-Dy647, and (
) 14-3-3ζ and pRaf-Dy647, and ( ) 14-3-3γ and pRaf-Dy647. The inhibitory effect of the 14-3-3 inhibitors on the FP or TR-FRET signal was expressed as% of control as described in the Materials and Methods section. IC50 values were obtained using GraphPad Prism version 5.0.
) 14-3-3γ and pRaf-Dy647. The inhibitory effect of the 14-3-3 inhibitors on the FP or TR-FRET signal was expressed as% of control as described in the Materials and Methods section. IC50 values were obtained using GraphPad Prism version 5.0.
In the FP assay for the same interaction pair, ∼10-fold of 14-3-3 proteins were required to obtain the mP window of ∼80 mP with S/N>8 and Z′>0.5 for screening.16 It is clear that the TR-FRET assay consumes fewer protein reagents than the FP assay in order to achieve a similar robust signal window.
To directly compare the sensitivity between the TR-FRET and FP assay for 14-3-3 inhibitor discovery, we tested the effect of the known 14-3-3 small-molecule inhibitor, FOBISIN101, on the interaction of 14-3-3ζ with pBad-Dy647 peptide in both TR-FRET and FP assays (Fig. 11A). FOBISIN101 led to a dose-dependent decrease in both TR-FRE and FP signal. The IC50 in TR-FRET assay was 3.2 μM, while the IC50 in FP assay was >400 μM. These data suggest that the TR-FRET assay is more sensitive than the FP assay for detecting the inhibition of the 14-3-3ζ/pBad interaction.
To evaluate whether 14-3-3 isoforms or binding partners utilized in the assay contributed to the observed sensitivity difference of the TR-FRET and FP screening to reveal positive hits (Fig. 10F), we tested the effect of FOBSIN101 on the interaction of different 14-3-3 isoforms (ζ and γ) with the same peptide (pBad or pRaf) in the same TR-FRET assay format. FOBISIN101 exhibited a different effect on the interaction of pBad with either 14-3-3ζ (IC50 of 3.2 μM) or 14-3-3γ (IC50 of 13.4 μM; Fig. 11B), showing a 14-3-3 isoform effect. Similarly, FOBISIN101 showed a differential effect on the interaction of 14-3-3ζ with either pBad (IC50 of 3.2 μM) or pRaf (IC50 of 19.8 μM; Fig. 11C), showing a 14-3-3 binding ligand effect. These data suggest that different 14-3-3 isoforms and ligands used in the assay are expected to impact assay sensitivity for the identification of 14-3-3 inhibitors. To confirm this observation, we tested the effect of one of the 16 hit compounds, Hit 10, in the TR-FRET assay with different 14-3-3 isoforms and different ligands. As shown in Figure 11D, Hit 10 exhibited different potencies when different combinations of 14-3-3 isoforms and ligands were utilized. Thus, it is likely that our observed different set of hits in Figure 10F could be due to a combination of both factors. TR-FRET assay appears to be more sensitive than the FP assay for the 14-3-3 inhibitor discovery. Different 14-3-3 isoforms along with different ligands also affect the sensitivity of the detection.
It remains possible that the TR-FRET assay is prone to compound artifacts and, therefore, more likely to generate promiscuous hit compounds. To address this issue, we compared the performance of the top 16 hit compounds from the TR-FRET assay against that of known promiscuous compounds by mining the PubChem database. As shown in Table 2, the average number of bioassays tested for each of these 16 compounds is 619. The average number of bioassays with active flags was 14.4, which represents an average of 2.3% of active in all bioassays tested for each hit compound. Remarkably, two compounds showed activity only in our 14-3-3 inhibitor screening among more than 600 bioassays profiled (Table 2). For comparison, we also analyzed the data for 37 well-defined promiscuous compounds, which are known redox cycling compounds (RCCs) that are capable of generating H2O2 in buffers containing dithiothreitol39 and summarized the data with updated information from the PubChem database (Table 3). For these 37 compounds, the average number of total bioassays tested for each compound is 563. The average activeflag of bioassays was 93, and the resulting average percentage of active bioassays for each compound was 16.3%. It is obvious that the active rate (2.3%) of the 16 14-3-3 hits from the TR-FRET assay is significantly lower than that of the 37 RCCs compounds from published PubChem bioassays (16.3%), supporting further experimental validation of these 16 hit compounds. To confirm their true nature as 14-3-3 inhibitors, these hit compounds have to be evaluated in a panel of bioassays, including the test of their direct binding to 14-3-3 or client proteins with technologies such as isothermal calorimetry or surface plasma resonance, and an examination of the antagonistic effect on 14-3-3/ligand interaction-mediated functions such as Bad-mediated apoptosis or Raf-mediated ERK activation and cell proliferation. Taken together, these results demonstrate that this TR-FRET assay provides a simple, sensitive, reliable, and robust readout for monitoring the 14-3-3/p-Bad peptide interaction to discover 14-3-3 protein modulators.
Table 2.
Target Query of the PubChem Database: Promiscuity Profile of 16 Hits from 14-3-3ζ/pBad TR-FRET Assay
| Compound no. | No. of total bioassays | No. of active flags | % activea |
|---|---|---|---|
| 1 | 548 | 9 | 1.6 |
| 2 | 632 | 1 | 0.2 |
| 3 | 537 | 42 | 7.8 |
| 4 | 686 | 65 | 9.5 |
| 5 | 475 | 12 | 2.5 |
| 6 | 766 | 7 | 0.9 |
| 7 | 738 | 3 | 0.4 |
| 8 | 472 | 8 | 1.7 |
| 9 | 684 | 19 | 2.8 |
| 10 | 695 | 21 | 3.0 |
| 11 | 523 | 17 | 3.3 |
| 12 | 770 | 12 | 1.6 |
| 13 | 451 | 6 | 1.3 |
| 14 | 736 | 1 | 0.1 |
| 15 | 480 | 5 | 1.0 |
| 16 | 726 | 2 | 0.3 |
The % active was calculated from (# of active flags/# of total bioassays)×100.
Table 3.
Updated Cross-Target Query of the PubChem Database: Promiscuity Profile of Confirmed Redox Cycling Compounds from Soares et al.39
| PubChem-SID | No. of total bioassays | No. of active flags | % activea | PubChem-SID | No. of total bioassays | No. of active flags | % activea |
|---|---|---|---|---|---|---|---|
| 14727570 | 245 | 34 | 13.9 | 17510616 | 343 | 63 | 18.4 |
| 14727734 | 480 | 68 | 14.2 | 14727791 | 680 | 141 | 20.7 |
| 22404392 | 549 | 93 | 16.9 | 17510946 | 568 | 129 | 22.7 |
| 16952991 | 579 | 130 | 22.5 | 24813200 | 540 | 103 | 19.1 |
| 14719117 | 706 | 119 | 16.9 | 22400632 | 464 | 32 | 6.9 |
| 14730721 | 777 | 215 | 27.7 | 17433753 | 529 | 68 | 12.9 |
| 24809506 | 498 | 55 | 11.0 | 17513322 | 570 | 46 | 8.1 |
| 14742005 | 552 | 66 | 12.0 | 17505825 | 415 | 58 | 14.0 |
| 24812833 | 540 | 50 | 9.3 | 14742003 | 523 | 76 | 14.5 |
| 14723875 | 570 | 159 | 27.9 | 17516177 | 605 | 26 | 4.3 |
| 845167 | 815 | 219 | 26.9 | 24825938 | 594 | 33 | 5.6 |
| 24809205 | 554 | 149 | 26.9 | 14742458 | 671 | 57 | 8.5 |
| 14726526 | 756 | 251 | 33.2 | 14742370 | 679 | 48 | 7.1 |
| 24814503 | 538 | 92 | 17.1 | 11532976 | 440 | 86 | 19.5 |
| 17507336 | 539 | 106 | 19.7 | 22412622 | 609 | 105 | 17.2 |
| 17508465 | 571 | 146 | 25.6 | 847359 | 457 | 94 | 20.6 |
| 17511877 | 505 | 43 | 8.5 | 857446 | 771 | 91 | 11.8 |
| 17510947 | 341 | 69 | 20.2 | 17504835 | 618 | 61 | 9.9 |
| 22405508 | 645 | 73 | 11.3 |
The % active was calculated from (# of active flags/# of total bioassays)×100.
Discussion
Here, we present details of the design, development, optimization, and validation of a homogeneous TR-FRET assay to monitor the interaction of 14-3-3 proteins with a client protein for the discovery of 14-3-3 protein inhibitors. Using the interaction of His-14-3-3ζ and a Dy647-pS136-Bad peptide as a model system, we established assay conditions for an optimized TR-FRET assay in a 384-well format. The 14-3-3 TR-FRET assay exhibited a robust performance, good stability, and DMSO tolerance. Importantly, wild-type 14-3-3ζ, but not mutant 14-3-3ζ K49E, generated robust TR-FRET signals when paired with the Dy647-pS136-Bad peptide, which demonstrates the specificity of the assay. This is consistent with previous findings that a singlecharge-reversal mutation, K49 to E, in the amphipathic groove of 14-3-3 is sufficient to disrupt 14-3-3/client protein interactions.40 In addition to 14-3-3ζ, similar TR-FRET assays can be developed for the other six isoforms of 14-3-3 proteins with a phosphorylated Bad peptide as a binding partner. The similar binding affinity of all isoforms of 14-3-3 proteins with the phosphorylated Bad peptide reflects the highly conserved nature of the peptide binding site on the amphipathic groove of 14-3-3 proteins.40
The suitability and sensitivity of the HTS assay for the identification of small molecule 14-3-3 inhibitors have been validated with a competition assay with several known 14-3-3 antagonist peptides and a small molecule 14-3-3 inhibitor. Unlabeled phosphor-peptides derived from well-defined 14-3-3 client proteins, such as pS136-Bad, pS967-ASK1, and pS259-Raf-1, were able to compete with the Dy647-pS136-Bad peptide for 14-3-3 binding. Conversely, unlabeled unphosphorylated peptides, such as S136-Bad and S967-ASK1, failed to reduce TR-FRET signals from the 14-3-3/pS136-Bad peptide interaction. These date further support the specificity of the FRET signal from the interaction of 14-3-3 with the phosphorylated Bad peptide. In addition, R18, a well-characterized 14-3-3 peptide antagonist, and FOBOSIN101, a small molecule 14-3-3 inhibitor, were able to effectively inhibit the FRET signal. Taken together, these data validate the specificity of the TR-FRET assay and its utility for identifying competitive 14-3-3/Bad binding inhibitors.
To enhance screening efficiency and further reduce costs, the HTS assay has been miniaturized for uHTS in a 1,536-well format. Notably, the uHTS assay maintains the same vigorous assay performance as that of the 384-well format. The utility of the miniaturized TR-FRET assay for uHTS has been further validated with a large-scale screen. The TR-FRET signals were consistent across the 40 plates tested during the screen with minimal signal variation. The Z′ factors were all greater than 0.5 and S/B ratios were above 20, indicating a robust assay in a uHTS format.
To obtain some insight into different detection technologies for the identification of 14-3-3 protein–protein interaction inhibitors, we compared the performance of the developed TR-FRET assay with that of a previously reported FP assay16,20 through the use of a set of 843 overlapping compounds screened in both assays (Fig. 10F). It appears that more positives (16 compounds) were identified in the TR-FRET assay than in the FP assay (1 compound), raising the possibility of a superior performance of the TR-FRET assay. The higher number of positives from the TR-FRET screening could be due to its increased assay sensitivity compared with the FP assay. Indeed, the direct comparison studies show that the TR-FRET assay is more sensitive than the FP assay in showing the inhibitory effect of a known small molecule 14-3-3 inhibitor, FOBISIN101 (Fig. 11A). One additional advantage of the TR-FRET assay compared with the FP assay is clear that the TR-FRET assay consumes ∼100-fold less 14-3-3 protein than the FP assay does (10 nM vs. 1 μM).
The sensitivity of the TR-FRET assay to small-molecule inhibitors is also affected by the specific 14-3-3 isoform used along with a particular binding partner, as shown in Figure 11. Therefore, HTS campaigns with different 14-3-3 isoforms and different client proteins may lead to the discovery of different sets of 14-3-3 inhibitors. Although the TR-FRET assay shows high sensitivity for hit discovery, the identified positive hits are unlikely to be those frequently revealed promiscuous compounds (Tables 2 and 3). However, the follow-up secondary binding and functional assays are essential to confirm the direct binding of the hit compounds to 14-3-3 or its ligand and to validate their antagonistic activity for future development.
In summary, we have developed and validated a simple “mix-and-read” TR-FRET assay for measuring the interaction of 14-3-3 and a client protein peptide. We have also demonstrated that the assay can be applied to the interaction of all isoforms of 14-3-3. The ability of multiple peptide antagonists and a small molecule 14-3-3 inhibitor to disrupt the interaction of 14-3-3 with a pS136-Bad peptide demonstrates the specificity of the TR-FRET assay for 14-3-3 and exhibits the feasibility of revealing 14-3-3/Bad interaction inhibitors. Furthermore, the assay has been successfully miniaturized to a 1,536-well uHTS format without sacrificing signal window or assay performance quality. Due to the high cost of large-scale compound screening, the ability to perform the assay in a 1,536-well format renders our TR-FRET assay particularly valuable for screening. The concept and development of the TR-FRET assay for 14-3-3 protein/ligand interaction will not only greatly facilitate the discovery of 14-3-3 protein inhibitors, but also provide a sensitive and easy-to-use tool to study 14-3-3-mediated protein–protein interactions.
Abbreviations
- ASK1
apoptosis signal-regulating kinase 1
- DMSO
dimethyl sulfoxide
- Eu
Europium
- FI
fluorescence intensity
- FOC
fold-over-control
- FP
fluorescence polarization
- HTS
high-throughput screening
- RCCs
redox cycling compounds
- TR-FRET
time-resolved fluorescence resonance energy transfer
- uHTS
ultra-HTS
Acknowledgments
The authors thank their colleagues at Emory University for contributions to this report: Ray Dingledine for helpful scientific discussions; Brian Revennaugh, Lian Li, and Iestyn Lewis for technical support; and Cheryl Meyerkord for valuable comments and editing. This work was supported in part by the National Institutes of Health Grants P01 CA116676 (to F.R.K. and H.F.) and U54 HG003918 (to R.D. and H.F.), the Georgia Cancer Coalition (to F.R.K. and H.F.), the Emory URC award (to Y.D.), Emory University's SPORE in Head and Neck Cancer Career Development award (P50 CA128613, to Y.D.), and an Emory Winship Cancer Institute Kennedy seed grant (to Y.D). H.F. is a Georgia Research Alliance Distinguished Investigator.
Disclosure Statement
No competing financial interests exist.
References
- 1.Fu H. Subramanian RR. Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Tinti M. Johnson C. Toth R. Ferrier DE. Mackintosh C. Evolution of signal multiplexing by 14-3-3-binding 2R-ohnologue protein families in the vertebrates. Open Biol. 2012;2:120103. doi: 10.1098/rsob.120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pozdeyev N. Taylor C. Haque R, et al. Photic regulation of arylalkylamine N-acetyltransferase binding to 14-3-3 proteins in retinal photoreceptor cells. J Neurosci. 2006;26:9153–9161. doi: 10.1523/JNEUROSCI.1384-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin J. Smith FD. Stark C, et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 5.Meek SE. Lane WS. Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J. Meyerkord CL. Du Y. Khuri FR. Fu H. 14-3-3 proteins as potential therapeutic targets. Semin Cell Dev Biol. 2011;22:705–712. doi: 10.1016/j.semcdb.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Dougherty MK. Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117(Pt 10):1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 9.Wilker E. Yaffe MB. 14-3-3 proteins—a focus on cancer and human disease. J Mol Cell Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Qi W. Martinez JD. Reduction of 14-3-3 proteins correlates with increased sensitivity to killing of human lung cancer cells by ionizing radiation. Radiat Res. 2003;160:217–223. doi: 10.1667/rr3038. [DOI] [PubMed] [Google Scholar]
- 11.Li Z. Zhao J. Du Y, et al. Down-regulation of 14-3-3zeta suppresses anchorage-independent growth of lung cancer cells through anoikis activation. Proc Natl Acad Sci USA. 2008;105:162–167. doi: 10.1073/pnas.0710905105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan T. Li R. Todd NW, et al. Up-regulation of 14-3-3{zeta} in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–7906. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 13.Neal CL. Yao J. Yang W, et al. 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 2009;69:3425–3432. doi: 10.1158/0008-5472.CAN-08-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin M. Morrison CD. Jones S. Mohamed N. Bacher J. Plass C. Copy number gain and oncogenic activity of YWHAZ/14-3-3zeta in head and neck squamous cell carcinoma. Int J Cancer. 2009;125:603–611. doi: 10.1002/ijc.24346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inglese J. Johnson RL. Simeonov A, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 16.Du Y. Masters SC. Khuri FR. Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay. J Biomol Screen. 2006;11:269–276. doi: 10.1177/1087057105284862. [DOI] [PubMed] [Google Scholar]
- 17.Wu M. Coblitz B. Shikano S, et al. SWTY—a general peptide probe for homogeneous solution binding assay of 14-3-3 proteins. Anal Biochem. 2006;349:186–196. doi: 10.1016/j.ab.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Du Y. Khuri FR. Fu H. A homogenous luminescent proximity assay for 14-3-3 interactions with both phosphorylated and nonphosphorylated client peptides. Curr Chem Genomics. 2008;2:40–47. doi: 10.2174/1875397300802010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu M. Coblitz B. Shikano S, et al. Phospho-specific recognition by 14-3-3 proteins and antibodies monitored by a high throughput label-free optical biosensor. FEBS Lett. 2006;580:5681–5689. doi: 10.1016/j.febslet.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J. Du Y. Horton JR, et al. Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor. Proc Natl Acad Sci USA. 2011;108:16212–16216. doi: 10.1073/pnas.1100012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corradi V. Mancini M. Manetti F. Petta S. Santucci MA. Botta M. Identification of the first non-peptidic small molecule inhibitor of the c-Abl/14-3-3 protein-protein interactions able to drive sensitive and Imatinib-resistant leukemia cells to apoptosis. Bioorg Med Chem Lett. 2010;20:6133–6137. doi: 10.1016/j.bmcl.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Wu H. Ge J. Yao SQ. Microarray-assisted high-throughput identification of a cell-permeable small-molecule binder of 14-3-3 proteins. Angew Chem Int Ed Engl. 2010;49:6528–6532. doi: 10.1002/anie.201003257. [DOI] [PubMed] [Google Scholar]
- 23.Ottmann C. Small-molecule modulators of 14-3-3 protein-protein interactions. Bioorg Med Chem. 2012 doi: 10.1016/j.bmc.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 24.Sills MA. Weiss D. Pham Q. Schweitzer R. Wu X. Wu JJ. Comparison of assay technologies for a tyrosine kinase assay generates different results in high throughput screening. J Biomol Screen. 2002;7:191–214. doi: 10.1177/108705710200700304. [DOI] [PubMed] [Google Scholar]
- 25.Glickman JF. Wu X. Mercuri R, et al. A comparison of ALPHAScreen, TR-FRET, and TRF as assay methods for FXR nuclear receptors. J Biomol Screen. 2002;7:3–10. doi: 10.1177/108705710200700102. [DOI] [PubMed] [Google Scholar]
- 26.Wu X. Glickman JF. Bowen BR. Sills MA. Comparison of assay technologies for a nuclear receptor assay screen reveals differences in the sets of identified functional antagonists. J Biomol Screen. 2003;8:381–392. doi: 10.1177/1087057103256466. [DOI] [PubMed] [Google Scholar]
- 27.Du Y. Nikolovska-Coleska Z. Qui M, et al. A dual-readout F2 assay that combines fluorescence resonance energy transfer and fluorescence polarization for monitoring bimolecular interactions. Assay Drug Dev Technol. 2011;9:382–393. doi: 10.1089/adt.2010.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorne N. Auld DS. Inglese J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masters SC. Yang H. Datta SR. Greenberg ME. Fu H. 14-3-3 inhibits Bad-induced cell death through interaction with serine-136. Mol Pharmacol. 2001;60:1325–1331. doi: 10.1124/mol.60.6.1325. [DOI] [PubMed] [Google Scholar]
- 30.Masters SC. Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal. J Biol Chem. 2001;276:45193–45200. doi: 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L. Chen J. Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc Natl Acad Sci USA. 1999;96:8511–8515. doi: 10.1073/pnas.96.15.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B. Yang H. Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry. 1999;38:12499–12504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- 33.Fu H. Coburn J. Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 35.Degorce F. Card A. Soh S. Trinquet E. Knapik GP. Xie B. HTRF: a technology tailored for drug discovery—a review of theoretical aspects and recent applications. Curr Chem Genomics. 2009;3:22–32. doi: 10.2174/1875397300903010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian RR. Masters SC. Zhang H. Fu H. Functional conservation of 14-3-3 isoforms in inhibiting bad-induced apoptosis. Exp Cell Res. 2001;271:142–151. doi: 10.1006/excr.2001.5376. [DOI] [PubMed] [Google Scholar]
- 37.Hekman M. Albert S. Galmiche A, et al. Reversible membrane interaction of BAD requires two C-terminal lipid binding domains in conjunction with 14-3-3 protein binding. J Biol Chem. 2006;281:17321–17336. doi: 10.1074/jbc.M600292200. [DOI] [PubMed] [Google Scholar]
- 38.Petosa C. Masters SC. Bankston LA, et al. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- 39.Soares KM. Blackmon N. Shun TY, et al. Profiling the NIH Small Molecule Repository for compounds that generate H2O2 by redox cycling in reducing environments. Assay Drug Dev Technol. 2010;8:152–174. doi: 10.1089/adt.2009.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L. Wang H. Liu D. Liddington R. Fu H. Raf-1 kinase and exoenzyme S interact with 14-3-3zeta through a common site involving lysine 49. J Biol Chem. 1997;272:13717–13724. doi: 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]