Abstract
To achieve an efficient intracellular drug and DNA delivery, attempts were made to target microparticulate drug carriers into cytoplasm bypassing the endocytotic pathway. TAT peptides derived from the HIV-1 TAT protein facilitate intracellular delivery of proteins and small colloidal particles. We demonstrated that relatively large drug carriers, such as 200-nm liposomes, can also be delivered into cells by TAT peptide attached to the liposome surface. Liposomes were fluorescently labeled with membranotropic rhodamine-phosphatidylethanolamine or by entrapping FITC-dextran. Incubation of fluorescent TAT liposomes with mouse Lewis lung carcinoma cells, human breast tumor BT20 cells, and rat cardiac myocyte H9C2 results in intracellular localization of certain liposomes. Steric hindrances for TAT peptide⋅cell interaction (attachment of TAT directly to the liposome surface without spacer or the presence of a high MW polyethylene glycol on the liposome surface) abolish liposome internalization, evidencing the importance of direct contact of TAT peptide with the cell surface. Low temperature or metabolic inhibitors, sodium azide or iodoacetamide, have little influence on the translocation of TAT liposomes into cells, confirming the energy-independent character of this process. The approach may have important implications for drug delivery directly into cell cytoplasm.
Protein transduction was demonstrated with the trans-activating transcriptional activator (TAT) protein (86-mer polypeptide) from HIV-1 shown to enter cells when added to the surrounding media (1, 2). The efficacy of TAT transduction depends on various factors, and current protocols permit the transduction into many cells (3). Some other proteins (Drosophila Antennapedia transcription factor, ANTP, and herpes simplex virus type-1 VP22 transcription factor) can also be transduced into the cytosol of many cell types (4–7). The identification of events underlying TAT protein-mediated translocation may clarify the key steps of virus penetration into cells and assist in building an effective antiviral defense. Additionally, because traversal through cellular membranes represents a major barrier for efficient delivery of macromolecules into cells, the TAT protein may serve to ferry various drugs into mammalian cells in vitro and in vivo. Schwarze et al. (8) succeeded in delivering TAT protein-attached β-galactosidase in all tissues in mice, even the brain.
Certain small regions of such proteins (10–16-mers) called protein transduction domains (PTDs), and some peptides also efficiently traverse biological membranes. This process is receptor-independent and transporter-independent, is not endocytosis-mediated, and may target the lipid layer directly (2, 9). Many of these peptides promote lipid membrane-reorganizing processes, such as fusion and pore formation, involving temporary membrane destabilization and subsequent reorganization. The minimal PTD of the TAT protein comprises residues 47–57, and that of the ANTP includes residues 43–58 (10–12). Common structural features of TAT and ANTP PTDs include the presence of many basic amino acids (arginine and lysine) as well as the ability to adopt an alpha helical conformation (6, 10–16). One can assume that the positive charge is a key property providing translocating activity to these peptides. This was confirmed by the fact that among the set of 12-mer peptide sequences synthesized by Mi et al. (17), only those with high cationic charge content facilitated transduction in different cells and provided efficient protein delivery into sinovial and tumor cells in vivo. Still, not all of the cationic peptides demonstrated this high transduction activity.
The use of peptides and protein domains with amphipathic sequences for drug and gene delivery across cellular membranes is getting increasing attention (9). Covalent hitching of proteins, drugs, or DNA onto PTDs may circumvent conventional limitations by allowing to transport these compounds into a wide variety of cells in vitro and in vivo (7, 8, 18–21). TAT peptide chemically attached to various proteins (horseradish peroxidase, β-galactosidase, and some others) was able to deliver these proteins to various cells and even in tissues in mice with high levels in heart, lung, and spleen (7). The internalization by cells was shown to proceed at 4°C, i.e., does not involve endocytic pathway (12). An energy-independent mechanism of translocation through biological membranes was also found for 60-mer homeodomain of ANTP and was not abolished by directed mutagenesis within the polypeptide C-terminal region (16). The authors were also able to derive a 16-mer polypeptide with translocating activity. Again, no classical endocytosis was assumed, as the peptide was effectively internalized at 4°C. A hypothesis was put forward that inverted micelles are formed in biological membranes in the presence of translocation peptides, facilitating their transfer through the membrane (22). Because the formation of reversed micelles in the presence of changing hydrophobic-hydrophilic balance of surroundings proceeds spontaneously, the process is energy-independent. However, amphipathicity-independent cellular uptake of cell-penetrating peptides was clearly shown in ref. 23, and this uptake was also temperature-independent. Although the actual mechanism(s) of this uptake/translocation has not yet clearly been established, it is suspected that some form of a ligand–lipid interaction plays a role, and direct contact between the translocating moiety and cell membrane is required.
A challenging task is to use PTDs for intracellular delivery of drug carriers, such as micelles, liposomes, and nanoparticles. So far, dextran-coated iron oxide colloidal particles with the size of about 40 nm and containing several attached molecules of TAT peptide per particle were delivered into lymphocytes much more efficiently than free particles (24–26). This raises an interesting question on what may be the maximum size of particulate drug carriers that still would be translocated into cells by TAT peptide if a sufficient number of its molecules is attached to a particle surface.
In this study, we were able to demonstrate that relatively large 200-nm plain and polyethylene glycol-coated (PEGylated) liposomes, can be delivered into various cells by multiple TAT peptide molecules attached to the liposome surface via the new spacer, p-nitrophenylcarbonyl-PEG-phosphatidylethanolamine (pNP-PEG-PE) (27). We also confirmed the noninvolvement of the cell respiratory chain and cytoskeleton in the internalization process, as liposome uptake proceeds at low temperature and in the presence of metabolic inhibitors: sodium azide is known to inhibit oxidative respiration (28), and the alkylating agent iodoacetamide (IAA) is known to modify the sulfhydryl groups in tubulin, thus affecting normal assembly of the cytoskeleton (29, 30).
The covalent coupling of TAT peptides to microparticulate drug carriers via a single terminus-reactive spacer group may provide an efficient tool for cytosolic delivery of drug-loaded plain and sterically protected nanoparticles in vitro and in vivo.
Materials and Methods
Egg phosphatidylcholine, cholesterol, PEG-PE with different sizes of PEG units (PEG2000-PE or PEG5000-PE), N-glutaryl-PE (NGPE), PE, and amphiphilic fluorescent dye rhodamine (Rh)-PE were obtained from Avanti Polar Lipids. pNP-PEG-PE was synthesized in-house. Diethylenetriaminepentaacetic acid anhydride (DTPA), PEG-nitophenylcarbonyl [(pNP)2], triethylamine, octyl glucoside, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, N-hydroxysulfosuccinimide, FITC-dextran (molecular mass, 4,400 Da), sodium azide, IAA, and TLC plates were from Sigma–Aldrich. TAT peptide (11-mer: TyrGlyArgLysLysArgArgGlnArgArgArg; molecular mass, 1,560 Da; three reactive amino groups) was prepared by Research Genetics (Huntsville, AL), and Indium111 (111-In) was from NEN. Cell culture media and FBS were from Cellgro. All solvents and buffer components were analytical grade preparations.
The cells used were BT20 (human breast tumor), Lewis lung carcinoma (LLC, mouse lung carcinoma) and H9C2 (rat embryonic cardiac myocytes). BT20 cells were maintained in Eagle's MEM (with 10 mM pyruvate, nonessential amino acids, L-glutamine, and 10% FBS). LLC cells were maintained in RPMI 1640 medium (with 10% FBS). H9C2 cells were maintained in Dulbecco's MEM (with 10% FBS). All cells were maintained under 6% CO2 at 37°C.
Preparation of Liposomes.
Liposomes prepared are listed in Table 1. For visualization, all liposomes were labeled with Rh-PE (λex568/λem590). In some experiments, Rh-PE liposomes were prepared that contained also entrapped FITC-dextran (λex494/λem518).
Table 1.
Compositions of liposomes used with corresponding abbreviations
| PC:Ch:Rh-PE | LIP |
| PC:Ch:Rh-PE:NGPE + TAT-peptide | LIP-NGPE-TAT |
| PC:Ch:Rh-PE:PEG(2000)-PE(5% mol) | LIP-PEG(2000) |
| PC:Ch:Rh-PE:PEG(5000)-PE(5% mol) | LIP-PEG(5000) |
| PC:Ch:Rh-PE:pNP-PEG(3000)-PE(0.5% mol) | LIP-pNP-PEG(3000) |
| PC:Ch:Rh-PE:pNP-PEG(3000)-PE(0.5% mol) + TAT-peptide | LIP-pNP-PEG(3000)-TAT |
| PC:Ch:Rh-PE:PEG(2000)-PE(4.5% mol):pNP-PEG(3000)-PE(0.5% mol) + TAT-peptide | LIP-PEG(2000)/pNP-PEG(3000)-TAT |
| PC:Ch:Rh-PE:PEG(5000)-PE(4.5% mol):pNP-PEG(3000)-PE(0.5% mol) + TAT-peptide | LIP-PEG(5000)/pNP-PEG(3000)-TAT |
| PC:Ch:Rh-PE:pNP-PEG(3000)-PE(0.5% mol) + TAT-peptide (+ FITC-dextran) | LIP-pNP-PEG(3000)-TAT/FITC-dextran |
For all preparations, the lipid film was first obtained from a mixture of phosphatidylcholine, cholesterol, and Rh-PE (7:3:0.1 molar ratio) in chloroform, with possible addition of other components (PEG, pNP-PEG-PE, or NGPE). Chloroform was removed on a rotary evaporator followed by freeze drying on a Freeze Dry System Freezone 4.5 (Labconco, Kansas City, MO). The film was hydrated with an appropriate buffer and vortexed at room temperature for 5 min. The lipid dispersion was extruded 40 times through polycarbonate filters (pore size 200 nm) by using a LiposoFast System (Avestin). Vesicle size was measured by using a Coulter N4 Plus submicron particle analyzer.
FITC-dextran loaded liposomes were prepared by adding 45 mg/ml FITC-dextran (molecular mass, 4,400 Da) in HBS, pH 7.4, to the lipid film during hydration. The nonincorporated FITC-dextran was removed on a BioGel 1.5 M column (0.7 × 24 cm).
Synthesis of pNP-PEG-PE.
The synthesis of pNP-PEG-PE was performed as described in ref. 27, by interaction of PE with a 10-fold molar excess of PEG-(pNP)2 in chloroform in the presence of triethylamine. Organic solvents were removed, and pNP-PEG-PE micelles were formed in 0.01 M HCl/0.15 M NaCl by bath sonication, separated from free PEG and pNP on a CL-4B column, and freeze-dried. pNP-PEG-PE was extracted with chloroform and stored at −80°C.
Attachment of TAT Peptide to Liposomes.
To attach TAT peptide to liposomes, two protocols were used. TAT peptide was coupled to preformed liposomes containing 1% mol NGPE via carbodiimide (31). Briefly, 0.2 ml of liposomes (2-μmol lipids) in MES, pH 5.5, 40 μl of 0.25 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, and 40 μl of 0.1 M N-hydroxysulfosuccinimide were incubated for 10 min. Then, 400 μl of TAT peptide solution (“cold” or 111-In labeled) in 50 mM Na tetraborate with 0.1 M NaCl, pH 7.4 (1 mg/ml), was added to activated liposomes, and the mixture was incubated for 3 h at 20°C. Free TAT peptide was separated on a BioGel 1.5A column.
Alternatively, nonmodified TAT peptide was attached via pNP groups to the distal ends of liposome-grafted pNP-PEG-PE chains (27). One milligram of TAT peptide was added to 10 mg of pNP-PEG-PE-containing liposomes stored at pH 5.1 (Na citrate), and pH was raised to 8.5. A 2-h incubation period resulted in TAT peptide binding and simultaneous hydrolysis of nonreacted pNP groups (27). Liposomes were purified by gel filtration on a Bio-Gel A15 M column.
To check for possible formation of TAT peptide aggregates, TLC of various solubilized TAT peptide-containing liposomes was performed on Silica Gel 60 F plates in chloroform/methanol/water (10:6:1, vol/vol/vol). Free TAT peptide and plain liposomes were used as controls. TLC runs have been done in duplicate and developed with 0.2% ninhydrin in ethanol or with iodine vapors.
Radiolabeling of TAT Peptide with 111-In and Determination of the Quantity of Liposome-Bound TAT Peptide.
To estimate the quantity of TAT peptide bound to a single liposome, TAT peptide was labeled with 111-In. First, TAT peptide was modified with DTPA residues as in ref. 32 by incubating it overnight with 50–100-fold molar excess of DTPA anhydride in 0.1 M NaHCO3, pH 8.0. The product was purified by dialysis. The quantity of DTPA covalently linked to the peptide was assessed by assay of free amino groups (33). In average, around one DTPA group was attached to a single TAT peptide molecule, which made us believe that the majority of TAT peptide molecules contained at least one chelating group, leaving two amino groups available for the interaction with the reactive groups on the liposome surface. Then, DTPA-TAT peptide (100 μg) was supplemented with 1 mCi of 111-InCl3 in 0.1 M Na citrate (transchelator), pH 5.5, and incubated for 30 min. Free 111-In was removed on Sephadex-G10 or by dialysis. Because the quantities of TAT peptide and lipid were known, as well as the number of phospholipid molecules forming a liposome of a given size (34), we could estimate an approximate quantity of TAT peptide associated with a single liposome by measuring the liposome-associated 111-In radioactivity.
To assess a possible nonspecific association of TAT peptide with plain and PEG liposomes, the 111-In labeled TAT peptide was incubated with reactive group-free liposome (LIP) and LIP-PEG; after the same purification procedures as described above, the liposome-associated radioactivity was measured.
Cell Treatment Protocols.
We have tested TAT peptide-mediated uptake of liposomes in vitro by using three different cell lines (see Materials and Methods). To determine whether the TAT peptide-mediated uptake into the cells was energy-dependent or, more generally, cell function-dependent, we incubated liposomes with cells under varying metabolic conditions. The translocation experiments were performed at 37°C and 4°C, in the presence of 0.1% sodium azide or 1 mM IAA.
After initial passage in tissue culture flasks, all cells were grown on coverslips in 6-well tissue culture plates. After the cells had reached semiconfluence, the plates were washed twice with PBS, pH 7.4, and then treated with liposome samples in serum-free medium (2 ml/well, 10 mg total lipid/ml) with or without sodium azide or IAA. For 4°C experiments, cells were cooled right before sample addition. The control groups for all sets were plain LIP-treated cells. After a 1-h incubation period, the media were removed and the plates washed with PBS three times. Cells were fixed with 4% paraformaldehyde in PBS for 1 h at 20°C and washed with PBS. Individual coverslips were mounted cell-side down onto fresh glass slides with fluorescence-free glycerol-based mounting medium (Fluoromount-G; Southern Biotechnology Associates). Cells were viewed with a Nikon Eclipse E400 microscope under bright light, or under epifluorescence with rhodamine/TRITC and fluorescein/FITC filters.
For an additional control of intracellular localization of internalized liposomes, the representative samples (slides) were viewed with an Axioplan 2 microscope (Zeiss) using differential interference contrast (confocal) microscopy. After capturing the images at 100× (oil) using the relevant fluorescence filters, pseudocoloring and layering were performed by using a digital imaging system, openlab (Improvision, Boston).
Results and Discussion
Liposome/TAT Liposome Properties.
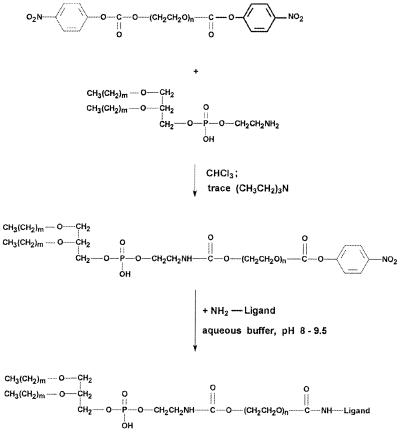
Recently, we introduced a new amphiphilic single-terminus reactive PEG derivative, pNP-PEG-PE (27), which is stable at slightly acidic pH, incorporates into liposomes via PE residue, and binds primary amino group-containing ligands via water-exposed pNP groups, forming stable urethane (carbamate) bonds. The reaction proceeds at pH 7.5–8.0, and nonreacted pNP groups are eliminated by spontaneous hydrolysis (see the scheme of pNP-PEG-PE synthesis and ligand attachment in Fig. 1). In our experiments, we have used 0.5% mol of pNP-PEG-PE because it was shown that as little as 0.5 to 1% mol (of total lipid) of pNP-PEG-PE provides a sufficient number of reactive groups on the liposome surface to bind several hundred protein molecules to a single 200-nm liposome (27).
Figure 1.
Synthesis of pNP-PEG-PE and its interaction with amino-containing ligand.
The average number of TAT peptides attached per liposome was calculated from radioactivity data, assuming that practically all TAT peptides are labeled with DTPA residues, and about 3 × 105 molecules of lipid form a liposome with an average diameter of 200 nm (34). Approximately 500 TAT peptide molecules were coupled to a single liposome. This corresponds well to the number of reactive pNP groups available on the liposome surface, assuming an even distribution of pNP-PEG-PE between the inner and outer lipid monolayers. This also corresponds well to the range of protein molecules bound to liposomes via pNP-PEG-PE (27) and provides a hope that even such large particles as liposomes can be translocated through the plasmic membrane as a result of the multipoint TAT peptide-to-membrane interaction.
Because of low concentration and random distribution of reactive groups on the liposome surface, one can hardly expect TAT peptide cross-linking on the surface. TLC of various solubilized TAT LIP confirms this yielding a single spot different from the spot of free TAT peptide and corresponding to a monomeric TAT peptide⋅lipid (or PEG-lipid) conjugate. The attachment of TAT peptide to the surface of LIP-PEG(5000)/pNP-PEG(3000) under similar conditions results in slightly decreased binding, probably because of steric hindrances created by the liposome-grafted PEG(5000). Still, the binding was on the level of 100+ molecules of TAT peptide per single liposome.
In the case of TAT peptide attachment to NGPE-containing liposomes, the binding efficacy was also determined by measuring the liposome-associated radioactivity after the interaction of 111-In labeled TAT peptide with NGPE liposomes and the removal of free TAT peptide. The used peptide-to-NGPE ratio provided the same number of liposome-bound TAT peptide molecules (400+) as in pNP-PEG-PE-mediated coupling protocol.
Control experiments on the incubation of the labeled TAT peptide with reactive group-free LIP and LIP-PEG demonstrated only background levels of radioactivity associated with liposomes. The result seems to be quite natural, assuming a random penetration rate of TAT peptide through the liposome membrane in both directions at equilibrium conditions and the fact that the intraliposomal volume constitutes only about 1% of the total volume of the liposome suspension.
All of the liposomal preparations used in our experiments had approximately the same average diameter of 150–200 nm (see the data on liposome size and size distribution in Fig. 2).
Figure 2.
Size distribution patterns for various liposomal preparations used in the study: LIP (A), LIP-NGPE-TAT (B), LIP-PEG(5000) (C), LIP-pNP-PEG(3000) (D).
Interaction of Different TAT Peptide Liposomes with Cells.
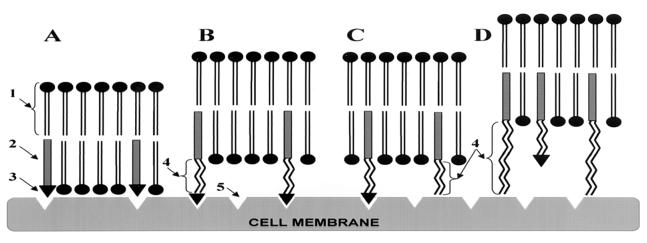
To investigate the TAT peptide-mediated liposome internalization and significance of a direct nonhindered interaction of TAT peptide moiety with the cell membrane for this process, we prepared several types of TAT liposomes shown in Fig. 3. In A, TAT peptide was attached directly to the surface of the liposome, via the liposomal NGPE (LIP-NGPE-TAT). In B, TAT peptide was attached to the liposome surface via the PEG spacer by coupling it via pNP-PEG-PE [LIP-pNP-PEG(3000)-TAT]. In C, in addition to the TAT peptide coupled to the liposome surface via PEG(3000) spacer, the liposome was also modified with a “short” PEG(2000)-PE [LIP-PEG(2000)/pNP-PEG(3000)-TAT]. And in D, in addition to the TAT peptide coupled to the liposome surface via PEG(3000), the liposome was modified with a “long” PEG(5000)-PE [LIP-PEG(5000)/pNP-PEG(3000)-TAT]. From Fig. 3, one can assume that, in A and D, steric hindrances created by the liposome surface or by long PEG chains attached to the liposome should interfere with TAT peptide-to-cell surface interaction, whereas in B and C there are no obstacles for such interaction.
Figure 3.
Schematic pattern of the interaction of TAT peptide-containing liposomes with cell surface: LIP-NGPE-TAT (TAT peptide is located directly on the surface of liposome) (A), LIP-pNP-PEG(3000)-TAT (TAT peptide is coupled via PEG spacer) (B), LIP-PEG(2000)/pNP-PEG(3000)-TAT (TAT peptide is attached to PEGylated liposomes; the length of protecting PEG is smaller that the length of PEG spacer) (C), and LIP-PEG(5000)/pNP-PEG(3000)-TAT (TAT peptide is attached to PEGylated liposomes; the length of protecting PEG is bigger that the length of PEG spacer) (D). 1, phospholipid constituting liposomes; 2, PE anchor for TAT peptide or TAT peptide-bearing PEG spacer; 3, TAT peptide; 4, PEG chains of variable length; and 5, hypothetical sites of TAT peptide interaction with the cell surface.
The interaction of different liposomal preparations with all used cell lines (LLC, BT20, and H9C2) at 37°C with no metabolic inhibitors added yielded practically identical results, partially presented in Figs. 4 and 5 and reflecting the typical data for various cells. None of the cells treated with Rh-PE-labeled LIP, LIP-PEG(5000), or LIP-pNP-PEG(3000)-PE (these three samples represent negative controls) and LIP-NGPE-TAT or LIP-PEG(5000)/pNP-PEG(3000)-TAT shows intracellular fluorescence after incubation with liposomes and subsequent washing (Fig. 4 A–C and Fig. 5 A–C). At the same time, all cells treated in similar conditions with Rh-PE-labeled LIP-pNP-PEG(3000)-TAT or with LIP-PEG(2000)/pNP-PEG(3000)-TAT demonstrate high intracellular fluorescence, i.e., liposome internalization (Fig. 4 D and E and Fig. 5 D and E). The results of confocal microscopy (Fig. 4 F and G) clearly confirm the intracellular localization of TAT liposomes, as the evident majority of the liposome-associated fluorescence can be seen inside the cell (when a single cell is viewed at the midpoint of the cell thickness) and not on the cell surface. Thus, steric hindrances created by the liposome surface or by PEG molecules of higher MW than that of pNP-PEG-PE to which TAT peptide is attached do not permit TAT peptide-to-cell interaction (Fig. 3 A and D) and prevent liposome internalization. Contrary to that, liposomal preparations shown in Fig. 3 B and C permit the multipoint interaction between the liposome-attached TAT peptide and the cell surface and subsequent liposome internalization. Thus, even such relatively large particles as 150–200-nm plain and PEGylated (long-circulating) liposomes can be effectively translocated inside various cells if a sufficient number of TAT peptide molecules is attached to the liposome surface in a fashion permitting free interaction between TAT peptide moieties and the cell surface.
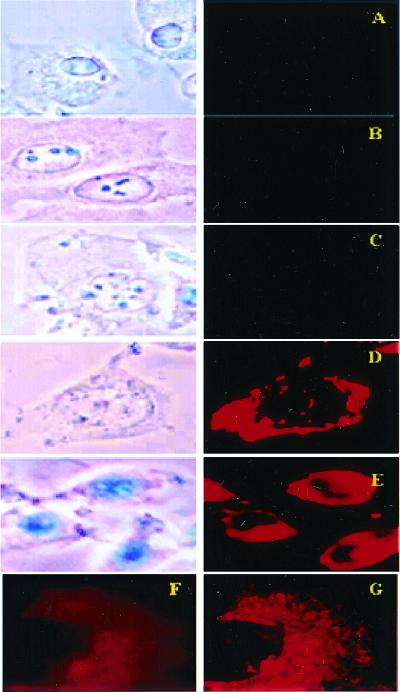
Figure 4.
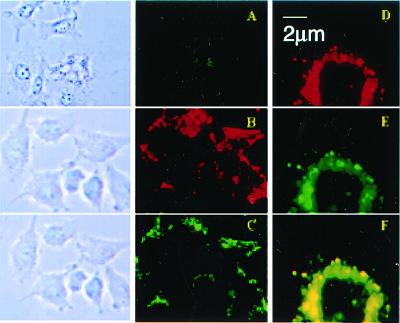
Bright field (Left) and fluorescent (Right) microscopy of cell cultures treated with different preparations of Rh-PE labeled liposomes (37°C; ×600): H9C2 and LIP (A), BT20 and LIP-pNP-PEG(3000) (B), BT20 and LIP-NGPE-TAT (C), BT20 and LIP-pNP-PEG(3000)-TAT (D), and LLC and LIP-pNP-PEG(3000)-TAT (E). (Blue spots seen in pictures under light microscopy are because of variations in light intensity, camera capture settings, and processing.) (F and G) Images represent the results of confocal microscopy of BT20 treated with LIP-pNP-PEG(3000)-TAT at 37°C: single cell viewed from the top cell surface (F); single cell viewed at the mid-point of the cell thickness (≈0.8-μm distance) (G).
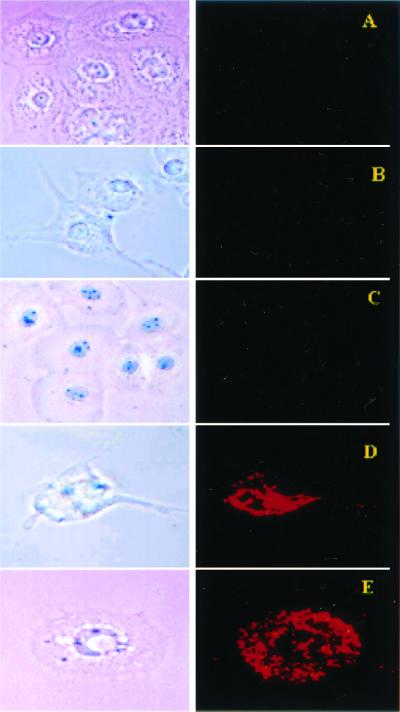
Figure 5.
Bright field (Left) and fluorescent (Right) microscopy of cell cultures treated with Rh-PE-liposomes (37°C; ×400): BT20 and LIP-PEG(2000) (A), H9C2 and LIP-PEG(5000)/pNP-PEG(3000)-TAT (B), BT20 and LIP-PEG(5000)/pNP-PEG(3000)-TAT (C), H9C2 and LIP-PEG(2000)/pNP-PEG(3000)-TAT (D), and BT20 and LIP-PEG(2000)/pNP-PEG(3000)-TAT (E).
It is interesting to note that after 1 h of observation (time interval used in our experiments) LIP-TAT are still seen in the cytoplasm and show only minimal association with cell nuclei, whereas free TAT peptide frequently demonstrates nuclear localization. We can explain it by slower intracellular migration of larger liposomes or by their size-hindered interaction with nuclei. Still, the time dependence of LIP-TAT intracellular trafficking is an important problem to be addressed in the future.
Metabolic Inhibitors Do Not Influence Internalization of TAT Peptide Liposomes by Cells.
In an agreement with the previous data (12, 13, 16, 23), the internalization of LIP-pNP-PEG(3000)-TAT and LIP-PEG(2000)/pNP-PEG(3000)-TAT still proceeds quite effectively at 4°C (Fig. 6A), i.e., the transduction process does not seem to be energy-dependent and influenced by the temperature (although the fluorescence-based method does not permit us to exactly determine whether any decrease in the efficacy of internalization takes place). This process is also not inhibited by sodium azide or IAA (Fig. 6 B and C), demonstrating a noninvolvement of oxidative respiration (30) or cytoskeleton (31, 32) in the TAT peptide-mediated liposome internalization by cells. Slightly different patterns of the intracellular liposomal fluorescence distribution in different cells may reflect certain cell type-specific phenomena.
Figure 6.
Bright field (Left) and fluorescent (Right) microscopy of viable cells treated with Rh-PE-liposomes in the presence of metabolic inhibitors (×400): BT20 and LIP-pNP-PEG(3000)-TAT, 4°C (A); BT20 and LIP-pNP-PEG(3000)-TAT, 0.1% sodium azide, 37°C (B); and LLC and LIP-pNP-PEG(3000)-TAT, 1% IAA, 37°C (C).
Liposome Integrity on Intracellular Delivery.
To investigate whether TAT peptide-mediated internalization results in the intracellular delivery of intact liposomes, Rh-PE labeled liposomes with entrapped FITC-dextran were used. Upon the interaction of such liposomes with cells, cells were studied for both Rh and FITC fluorescence (Fig. 7). As can be clearly seen from the typical data presented here, whereas free FITC-dextran shows only minor intracellular accumulation, the incubation of cells with Rh-PE-labeled LIP-pNP-PEG(3000)-TAT/FITC-dextran results in the intracellular appearance of a colocalized Rh and FITC fluorescence, i.e., both the membrane and internal space markers are delivered into the cell cytoplasm and the majority of liposomes remains intact during a 1-h observation time. The data of confocal microscopy (Fig. 7 D–F) support this conclusion, as the superposition of Rh- and FITC-based images also yielded identical patterns. The fate of intracellular liposomes at later times is a subject of our current studies.
Figure 7.
Bright field (Left) and fluorescent (Center) microscopy of H9C2 treated with free FITC-dextran and FITC-dextran-loaded Rh-PE labeled liposomes (37°C; ×400): H9C2 and free FITC-dextran (A); H9C2 and LIP-pNP-PEG(3000)-TAT/FITC-dextran, TRITC filter (B); and H9C2 and LIP-pNP-PEG(3000)-TAT/FITC-dextran, FITC filter (C). (Right) Images represent the results of confocal microscopy of BT20 treated with FITC-dextran-loaded Rh-PE liposomes (37°C; pseudocolor enhancement): single cell under FITC filter (D); single cell under Rh filter (E); and composite of superimposed layers from D and E (F).
Our experiments provide proof of principle for efficient TAT peptide-mediated transduction of plain and sterically protected liposomes and their contents into cells. TAT peptide attached to the liposome surface needs to be nonshielded and accessible for cell membrane interaction. Although the actual mechanism of this process still remains to be established, our results seem to favor the hypothesis on the spontaneous formation of membrane structures (reverse micelles) sharply, although temporarily, increasing the cell membrane permeability under the action of TAT peptide (22). The inability of metabolic inhibitors to block the translocation process confirms the energy-independent character of this TAT peptide-mediated process as well as noninvolvement of oxidative respiration or cytoskeleton. The approach described may have important implications for drug and DNA delivery directly into the cell cytoplasm in different protocols of local drug administration or ex vivo cell treatment.
Abbreviations
- PE
phosphatidylethanolamine
- PEG
polyethylene glycol
- PTD
protein transduction domain
- IAA
iodoacetamide
- NGPE
N-glutaryl-PE
- Rh
rhodamine
- DTPA
diethylenetriaminepentaacetic acid anhydride
- LLC
Lewis lung carcinoma
- LIP
liposome
- 111-In
Indium111
References
- 1.Green M, Loewenstein P M. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 2.Frankel A D, Pabo C O. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 3.Vocero-Akbani A, Lissy N A, Dowdy S F. Methods Enzymol. 2000;322:508–521. doi: 10.1016/s0076-6879(00)22046-6. [DOI] [PubMed] [Google Scholar]
- 4.Verhoef K, Koken S E C, Berkhout B. Anal Biochem. 1993;210:210–214. doi: 10.1006/abio.1993.1176. [DOI] [PubMed] [Google Scholar]
- 5.Joliot A, Pernelle C, Deagostini-Bazin H, Prochiantz A. Proc Natl Acad Sci USA. 1991;88:1864–1868. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott G, O'Hare P. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 7.Fawell S, Seery J, Daikh Y, Moore C, Chen L L, Pepinsky B, Barsoum J. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarze S R, Ho A, Vocero-Akbani A, Dowdy S F. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 9.Plank C, Zauner W, Wagner E. Adv Drug Delivery Rev. 1998;34:21–35. doi: 10.1016/s0169-409x(98)00005-2. [DOI] [PubMed] [Google Scholar]
- 10.Mann D A, Frankel A D. EMBO J. 1991;10:1733–1739. doi: 10.1002/j.1460-2075.1991.tb07697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezhevsky S A, Nagahara H, Vocero-Akbani A M, Gius D R, Wei M C, Dowdy S F. Proc Natl Acad Sci USA. 1997;94:10699–10704. doi: 10.1073/pnas.94.20.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vives E, Brodin P, Lebleu B. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 13.Derossi D, Joliot A H, Chassaing G, Prochiantz A. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 14.Calnan B J, Biancalana S, Hudson D, Frankel A D. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 15.Loret E P, Vives E, Ho P S, Rochat H, Van Rietschoten J, Johnson W C. Biochemistry. 1991;30:6013–6023. doi: 10.1021/bi00238a027. [DOI] [PubMed] [Google Scholar]
- 16.Derossi D, Joliot A H, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 17.Mi Z, Mai J, Lu X, Robbins P D. Mol Ther. 2000;2:339–347. doi: 10.1006/mthe.2000.0137. [DOI] [PubMed] [Google Scholar]
- 18.Bonifaci N, Sitia R, Rubartelli A. AIDS. 1995;9:995–1000. doi: 10.1097/00002030-199509000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Huang S Y, Pooyan S, Wang J, Choudhury I, Leibowitz M J, Stein S. Bioconjugate Chem. 1998;9:612–617. doi: 10.1021/bc980038p. [DOI] [PubMed] [Google Scholar]
- 20.Gius D R, Ezhevsky S A, Becker-Hapak M, Nagahara H, Wei M C, Dowdy S F. Cancer Res. 1999;59:2577–2580. [PubMed] [Google Scholar]
- 21.Schwarze S R, Dowdy S F. Trends Pharmacol Sci. 2000;21:45–48. doi: 10.1016/s0165-6147(99)01429-7. [DOI] [PubMed] [Google Scholar]
- 22.Prochiantz A. Ann NY Acad Sci. 1999;886:172–179. doi: 10.1111/j.1749-6632.1999.tb09410.x. [DOI] [PubMed] [Google Scholar]
- 23.Scheller A, Weisner B, Melzig M, Bienert M, Oehlke J. Eur J Biochem. 2000;267:6043–6050. doi: 10.1046/j.1432-1327.2000.01681.x. [DOI] [PubMed] [Google Scholar]
- 24.Josephson L, Tung C-H, Moore A, Weissleder R. Bioconjugate Chem. 1999;10:186–191. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 25.Lewin M, Carlesso N, Tung C-H, Tang X-W, Cory D, Scadden D T, Weissleder R. Nat Biotech. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- 26.Bhorade R, Weissleder R, Nakakoshi T, Moore A, Tung C H. Bioconjugate Chem. 2000;11:301–305. doi: 10.1021/bc990168d. [DOI] [PubMed] [Google Scholar]
- 27.Torchilin V P, Levchenko T S, Lukyanov A N, Khaw B A, Klibanov A L, Rammohan R, Samokhin G P, Whiteman K R. Biochim Biophys Acta. 2001;1511:397–411. doi: 10.1016/s0005-2728(01)00165-7. [DOI] [PubMed] [Google Scholar]
- 28.Speert D P, Wong S Y, Macdonald M, Sargeant R. Behring Inst Mitt. 1997;98:274–282. [PubMed] [Google Scholar]
- 29.Luduena R F, Roach M C. Biochemistry. 1981;20:4437–4444. doi: 10.1021/bi00518a031. [DOI] [PubMed] [Google Scholar]
- 30.Putnam A J, Schultz K, Mooney D J. Am J Physiol. 2001;280:C556–C564. doi: 10.1152/ajpcell.2001.280.3.C556. [DOI] [PubMed] [Google Scholar]
- 31.Bogdanov A A, Jr, Klibanov A L, Torchilin V P. FEBS Lett. 1988;231:381–384. doi: 10.1016/0014-5793(88)80854-8. [DOI] [PubMed] [Google Scholar]
- 32.Hnatowich D J, Layne W W, Childs R L, Lanteigne D, Davis M A, Griffin T W, Doherty P W. Science. 1983;220:613–615. doi: 10.1126/science.6836304. [DOI] [PubMed] [Google Scholar]
- 33.Habeeb A F. Anal Biochem. 1966;14:328–336. doi: 10.1016/0003-2697(66)90275-2. [DOI] [PubMed] [Google Scholar]
- 34.Enoch H G, Strittmatter P. Proc Natl Acad Sci USA. 1979;76:145–149. doi: 10.1073/pnas.76.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]