Abstract
Quorum sensing is a cell–cell communication process in bacteria that involves the production, release, and subsequent detection of chemical signal molecules called autoinducers. In Vibrio cholerae, multiple input signals activate the expression of the quorum sensing regulator HapR, which acts to repress the expression of virulence factors. We have shown that CRP, the cyclic adenosine monophosphate (cAMP) receptor protein, enhances quorum sensing by activating the biosynthesis of cholera autoinducer 1, the major signaling molecule that contributes to the activation of HapR. Thus, proquorum sensing CRP agonists could inhibit virulence and lead to new drugs to treat severe cholera. In this study, we show that expression of the quorum sensing-regulated luxCDABE operon can be used as a robust readout for CRP activity. Further, we describe and validate a highly specific cell-based luminescence high-throughput screening assay for proquorum sensing CRP ligands. A pilot screen of 9,425 compounds yielded a hit rate of 0.02%, one hit being cAMP itself. The Z′ value for this assay was 0.76 and its coefficient of variance 8% for the positive control compound. To our knowledge, this is the first cell-based assay for ligands of the highly conserved CRP protein of Gram-negative bacteria. The use of this assay to screen large chemical libraries could identify lead compounds to treat cholera, as well as small molecules to probe ligand–receptor interactions in the CRP molecule.
Introduction
CRP, the cyclic adenosine monophosphate (cAMP) receptor protein, is a member of the CRP/fumarate nitrate regulator family of bacterial transcriptional regulators which binds with cAMP to form a complex that acts at responsive promoters to activate or repress transcription.1 CRP is also a member of the cyclic nucleotide-binding (CNB) protein superfamily, which regulate signaling pathways in prokaryotic and eukaryotic cells.2 Sequence alignment of prokaryotic and eukaryotic CNB domains have identified key conserved sequence motifs that are shared by the entire superfamily.2 The CNB domain consists of a conserved eight-stranded beta barrel domain and a more divergent phosphate-binding cassette that anchors the cAMP molecule and contributes to ligand specificity.2 The CRP phosphate-binding casette shares 65% amino acid identity to eukaryotic CNB proteins, such as protein kinase A. The CRPs of Escherichia coli, Salmonella enterica (serovar Thyphimurium), Yersinia spp., and Vibrio cholerae are 209 residues long and possess >94% identity in amino acid sequences.
In Gram-negative bacteria, CRP is known for its role in carbon catabolite repression, a process by which the presence of a favorable carbon source in the medium inhibits the expression of enzymes involved in the catabolism of other carbon sources.1 This is achieved through activation of adenylate cyclase, resulting in elevated intracellular cAMP levels and formation of the cAMP-CRP complex, which binds DNA as a dimer.1 Each subunit contains one high-affinity and one low-affinity binding site for cAMP.3 The cAMP-CRP complex that is competent for activation of transcription is formed through the binding of cAMP at high-affinity sites.3 At very high cAMP concentrations, cAMP binds to low-affinity sites and prevents CRP-DNA interaction.4 The amino acid substitutions T127L/S128A lock the CRP molecule into a conformation that binds DNA and activates transcription in the absence of cAMP.5
CRP is a major regulator of virulence gene expression in numerous Gram-negative pathogens, including uropathogenic E. coli, Salmonella, Yersinia spp., and Pseudomonas aeruginosa. In a previous study, we demonstrated that CRP enhances quorum sensing in V. cholerae by activating the biosynthesis of cholera autoinducer 1 (CAI-1).6 In the V. cholerae quorum-sensing pathway, multiple input signals—such as the accumulation of two autoinducer molecules in the medium (CAI-1 and autoinducer 2); the level of the histone-like protein FIS (factor for inversion stimulation), and the global regulator CsrA contribute to the activation of the master quorum-sensing regulator HapR at high cell density.7 HapR then acts to diminish the expression of cholera toxin and the toxin co-regulated pilus.8 Hence, there is an ongoing effort to identify pro–quorum-sensing molecules to inhibit virulence in Vibrios.9 In addition to enhancing the expression of HapR, CRP negatively affects virulence by directly repressing the transcription of the transmembrane regulator TcpP/H required to activate cholera toxin gene expression.10 Thus, screening for small molecules that activate CRP could identify lead compounds with superior virulence-inhibiting potency compared to molecules that solely activate quorum sensing.
Early structure activity relationship of CRP interaction with synthetic cAMP analogs have been based on in vitro binding assays or stimulation of lac operon transcription.11,12 Some cAMP analogs have been developed as biochemical tools to study eukaryotic signal transduction and prospective drugs for the treatment of a broad spectrum of human carcinomas.13 Therapeutic development of cAMP analogs has often been precluded by low membrane permeability, degradation and low target specificity.13 A major obstacle in this field is the lack of a robust cell-based assay suitable for the screening of large chemical libraries.
In this study, we show that addition of cAMP (or its analog 7-deaza cAMP) to the medium inhibits virulence gene expression in the cholera bacterium. Next, we show that expression of the V. harveyi quorum-sensing–regulated luxCDABE operon provides a robust and stringent readout of CRP activation. Finally, we describe and validate a highly specific high-throughput screening (HTS) assay to identify pro—quorum-sensing CRP ligands.
Materials and Methods
Strains, Media, and Plasmids
The strains used in this study were all derived from V. cholerae C7258 (El Tor Biotype). Construction of C7258Δcrp and C7258Δcya (adenylate cyclase deletion) have been described previously.6 Strains were grown on Luria broth (LB) medium (1% tryptone, 1% NaCl, 0.5% yeast extract, pH 6.5–7, sterilized by autoclaving 15 min at 121°C) at 30°C. The medium was supplemented with tetracycline (5 μg/mL), ampicillin (100 μg/mL) or polymyxin B (100 units/mL) as required. For measurement of cholera toxin production, strains were grown in AKI cultures as previously published.6 cAMP was obtained from Sigma-Aldrich (St. Louis, MO) and 7-deaza-cAMP from Axxora LLC (San Diego, CA).
Construction of Mutants and Reporter Strains
A derivative of C7258 containing a mutant crp allele encoding CRPT127L/S128A, hereafter denoted crp*, was constructed using the QuickChange mutagenesis kit (Agilent Technologies, Santa Clara, CA) and allelic exchange. Briefly, a DNA fragment containing the V. cholerae crp gene was amplified with primers 5′-GCC TCT AGA ATG GTT CTA GGT AAA C and 5′-GAT GCA TGC ACA CGA GAC GGG TTA T (restriction sites underlined) and ligated into XbaI and SphI digested pUC19 to generate pCRP. The mutant plasmid pCRP* containing the crp* allele was generated with the mutagenic primers 5′-CTC GTC GTC TGC AAA TGC TAG CGC AGA AAG TTG GCG (mutant codons underlined) and 5′-CGC CAA CTT TCT GCG CTA GCA TTT GCA GAC GAC GAG according to the manufacturer's instructions. The XbaI-SphI fragment containing crp* was ligated into the suicide vector pCVD44214 and the resulting plasmid was transferred to strain C7258 by conjugation. Then, the C7258 crp* mutant was obtained by sucrose selection as described previously.6 To construct a C7258ΔcrpΔcya double mutant, a cya deletion mutation was mated into C7258Δcrp by allelic exchange as described previously.6 Similarly, allelic exchange was used to transfer the crp* allele into C7258Δcya to yield C7258Δcya crp*. The pBB1 cosmid encoding the V. harveyi luxCDABE operon15 was transferred by conjugation from E. coli SM10λpir to wild type and mutant V. cholerae strains. Exconjugants were selected on LB plates containing tetracycline and polymyxin B, purified by restreaking in LB tetracycline plates, and conserved as frozen stocks in tryptic soy broth containing 25% glycerol at −80°C. All mutants were confirmed by PCR and DNA sequencing.
Measurement of lux Gene Expression
The quorum sensing regulator HapR activates the promoter of the lux operon on the pBB1 cosmid when cultures reach a high cell density (>2×108 cells/mL).15 To maximize light output, V. cholerae strains were grown in 50 mL Falcon tubes containing 5 mL of LB medium supplemented with tetracycline to stationary phase (∼16 h). Light was measured in a BioTek Synergy 2 plate reader (BioTek U.S., Winooski, VT) using Costar 96-well clear bottom black wall microtiter plates (Corning, Tewksbury, MA).
HTS Assay for Pro–Quorum-Sensing CRP Ligands
Strain C7258Δcya containing the pBB1 cosmid was grown to high cell density (OD600≈3) in LB medium containing tetracycline and 20 μL aliquots of this culture were transferred to Corning 384-well opaque wall microtiter plates containing the desired compounds dissolved in dimethyl sulfoxide (DMSO), carrier control wells containing DMSO alone or positive control wells containing 10 μM cAMP. The final concentration of DMSO in all wells was 0.5%. The plates were incubated 6 h at 30°C and luminescence measured in a Perkin Elmer (Waltham, MA) Envision plate reader with a 0.1 s integration time. The screening compound set consisted of 1,120 FDA approved or known biologically-active compounds from the Prestwick chemical collection (www.prestwickchemical.org) and 8,304 compounds from the chemically diverse Molecular Libraries Small Molecule Repository (MLSMR; https://mlsmr.evotec.com/MLSMR_HomePage). For the primary screen, compounds were tested at a concentration of 50 μM for the MLSMR set and 10 μg/mL for the Prestwick collection. Active compounds, defined as those producing a light emission three standard deviations above the average of all compounds, were tested in duplicate in 6 twofold dilutions from 100 to 3 μM. Positive compounds confirmed by dose-response were manually retested in strain C7258ΔcyaΔcrp containing pBB1 to verify that induction of light was contingent upon the presence of the CRP. A summary of the HTS protocol is provided in Table 1.
Table 1.
Summary of High-Throughput Screening Protocol
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Plate bacteria | 20 μL | Overnight culture grown to saturation |
| 2 | Carrier control | 125 nL | DMSO |
| 3 | Positive control | 5 μL | 5× concentration (2.5% DMSO, 50 μM cAMP) |
| 4 | Library compounds | 125 nL | 50 μM MLSMR/10 μg/mL Prestwick final concentration |
| 5 | Backfill | 5 μL | Carrier control and compound wells |
| 6 | Incubation | 6 h | 30°C, high humidity |
| 7 | Assay readout | 700 nm | PMT plate reader-luminescent mode |
1. Solid white tissue culture treated plates, 8-tip dispense.
2. Columns 1–2, 0.5% DMSO, acoustic dispense.
3. Columns 23–24, 10 μM cAMP, 0.5% DMSO, displacement dispense using disposable tips.
4. Columns 3–22, 10 μM, 0.5% DMSO, acoustic dispense.
5. Columns 1–22, 8-tip dispense to standardize volume.
6. Plates incubated with lids.
7. Luminescence 400–700 nm, 0.1 s integration time.
DMSO, dimethyl sulfoxide; MLSMR, Molecular Libraries Small Molecule Repository; PMT, photomultiplier tube.
Measurement of Cholera Toxin Production
The amount of cholera toxin secreted to the medium was determined by a ganglioside GM1 enzyme-linked immunosorbent assay (ELISA), using a peroxidase-conjugated rabbit anticholera toxin B IgG (Pierce Biotechnology, Rockford, IL) and a standard curve constructed with pure CT (Sigma-Aldrich) as described previously.6
Results and Discussion
Exogenous CRP Ligands Diminish Expression of Cholera Toxin
The role of CRP as repressor of virulence gene expression in V. cholerae has been postulated based on the effect of null cya and crp mutations on virulence gene expression.16 However, no data is available on the effect of exogenous activators of CRP on virulence gene expression. As shown in Table 2, deletion of crp or adenylate cyclase (cya) results in elevated cholera toxin production while introduction in V. cholerae of a mutant crp gene (crp*) encoding a CRP active in the absence of cAMP exhibited the opposite effect. We also show that addition of exogenous cAMP or its analog 7-deaza-cAMP to the culture medium significantly inhibited production of cholera toxin (Table 2), the main virulence and secretogenic factor produced by the cholera bacterium. These results highlight the potential application of exogenous CRP ligands as antivirulence drugs to treat severe cholera and diminish the duration of purging. In addition to their affinity for CRP, the potency of such compounds can be affected by cell permeability, succeptibility to degradation by cellular phosphodiesterases, and efflux. The availability of an HTS-ready cell-based assay, however, could facilitate identifying and optimizing, through medicinal chemistry, new and more effective pro–quorum-sensing CRP ligands to inhibit virulence gene expression.
Table 2.
Inhibition of Cholera Toxin Expression by Exogenous Cyclic Adenosine Monophosphate Receptor Protein Ligands
|
Effect of Genetic Mutations |
Effect of Exogenous CRP Ligands |
|||
|---|---|---|---|---|
| Culture | Cholera toxin (μg/mL/OD600)a | Culture | Cholera toxin (μg/mL/OD600)a | % Inhibition |
| C7258 | 2.54±0.14 | C7258+cAMP | 0.86±0.05 | 66b |
| C7258Δcrp | 6.41±0.76 | C7258+7-deaza-cAMP | 0.56±0.06 | 78b |
| C7258Δcya | 10.52±0.12 | C7258Δcya+cAMP | 2.87±0.44 | 73c |
| C7258crp* | 0.64±0.02 | C7258Δcya+7-deaza-cAMP | 2.06±0.26 | 80c |
| C7258Δcya crp* | 0.64±0.03 | |||
aEach value is the mean of at least three independent cultures.
bInhibition relative to strain C7258.
cInhibition relative to C7258Δcya. Final concentrations for cAMP and 7-deaza-cAMP were 50 μM.
cAMP, cyclic adenosine monophosphate; CRP, cAMP receptor protein.
Expression of the luxCDABE Operon Is a Robust Reporter of CRP Activity
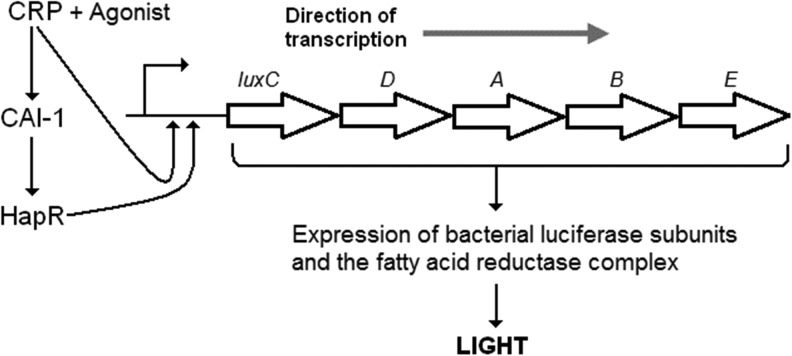
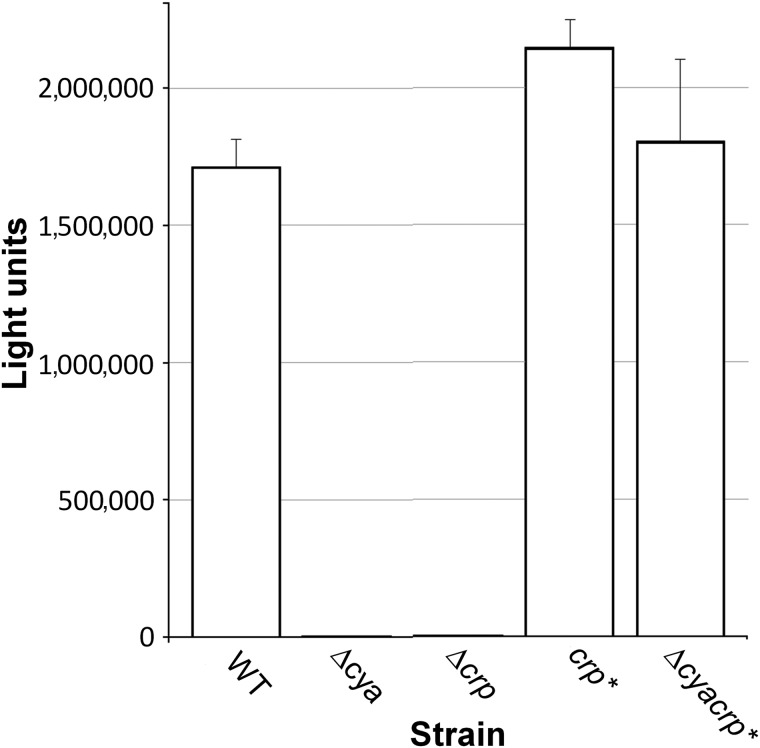
Our previous studies demonstrating that CRP activates the biosynthesis of CAI-16 suggested that quorum sensing could be used as readout for CRP activity. The principle of our assay for proquorum sensing CRP agonists is shown in Figure 1. The V. harveyi lux operon consists of genes encoding luciferase α (luxA) and β (luxB) subunits and the fatty acid reductase complex composed of a reductase (luxC), transferase (luxD), and synthase (luxE). The fatty acid reductase complex generates the luciferase aldehyde substrate to generate light in the presence of oxygen and reduced flavin mononucleotide. Activation of the luxCDABE operon to produce light in the C7258Δcya genetic background is predicted to be strongly dependent on the presence of cAMP or a CRP agonist. Formation of the CRP-agonist complex is required for the biosynthesis of CAI-1 and expression HapR, which subsequently activates the luxC promoter. In addition, Lux activity is enhanced by direct binding of the CRP-agonist complex to the luxC promoter.17 To determine the maximal signal to background window and ratio of this assay, strains C7258, C7258Δcrp, C7258Δcya, C7258crp*, and C7258Δcya crp* were grown to saturation in LB medium and light production was measured. As shown in Figure 2, no light production could be detected in the crp and cya deletion mutants. Further, the dark phenotype produced by the cya deletion mutant could be fully reversed by introducing the crp* gene encoding a CRP that does not require cAMP for DNA binding.5 These results indicate that expression of the lux operon is stringently dependent on cAMP activation of CRP and that this requirement can be bypassed in a strain that makes a mutant CRP that could bind the lux promoter and activate transcription in the absence of cAMP. In Figure 3, we show that light production could be restored in the cya deletion mutant by exogenous cAMP or its analog 7-deaza-cAMP. Under these experimental conditions, light production could be detected at 62 μM cAMP (∼4,492 light units) and 4 μM 7-deaza-cAMP (∼502 light units). Lower concentrations of both compounds could be detected by increasing the incubation time. Interestingly, 7-deaza-cAMP induced light production at a significantly lower concentration than cAMP but exhibited substrate inhibition at higher concentration. We suggest that the observed substrate inhibition could result from the binding of 7-deaza-cAMP to CRP low affinity cAMP binding sites to inhibit CRP-DNA interaction.4 Light production by wild type strain C7258 could be inhibited by cGMP, which binds to CRP but does not induce the allosteric change required to activate transcription (Fig. 3). As expected, little cGMP inhibition was observed in strain crp* encoding a CRP protein locked in a transcriptionally active conformation (data not shown). It was not surprising that inhibition of light production by exogenous cGMP in the wild type strain, which produces endogenous cAMP, required high concentrations of inhibitor. In this case, cell permeability to cGMP might limit its capacity to compete with endogenous cAMP for binding to CRP. Taken together, we conclude that expression of the lux operon provides a highly specific and robust indicator of CRP activity. It is noteworthy that, when screening for CRP agonists in a Δcya reporter, light production is a gain of function strictly dependent on the presence of exogenous cAMP or an agonist regardless of other environmental or cellular factors that could modulate quorum sensing activation of the lux operon.
Fig. 1.
Cell-based assay for proquorum sensing CRP ligands. Expression of the lux genes in Vibrio cholerae requires transcriptional activation of lux promoter by the quorum sensing regulator HapR and the cAMP-CRP complex. The cAMP-CRP complex is also required for Vibrio to make HapR. cAMP, cyclic adenosine monophosphate; CRP, cAMP receptor protein.
Fig. 2.
Expression of the luxCDABE operon in V. cholerae is a robust reporter of CRP activity. Strains were grown with agitation to stationary phase in Luria broth medium at 30°C and light production measured in a Synergy BioTek plate reader. Each value is the mean of three independent experiments. Error bars indicate the standard deviation. WT, wild-type.
Fig. 3.
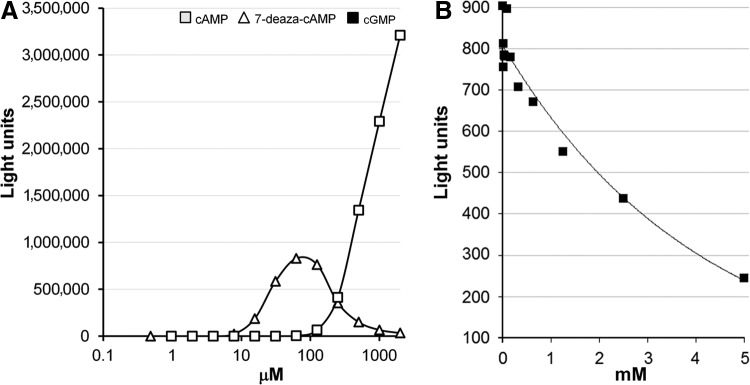
Effect of exogenous cAMP and cAMP analogs on light production by V. cholerae quorum sensing reporter strains. (A) Dose response for cAMP and 7-deaza-cAMP. Strain C7258Δcya was grown to stationary phase in LB medium; 100 μL of culture were added to triplicate wells of 96-well microtiter plates containing increasing concentration of cAMP or 7-deaza-cAMP, and the plates incubated 6 h at 30°C. (B) inhibition of light production by cGMP. Overnight cultures of C7258 containing the pBB1 cosmid were diluted 1:1,000 in fresh LB containing and 100 μL of culture transferred to triplicate wells containing different concentrations of cGMP. Plates were incubated 6 h at 30°C and light production read.
Assay Miniaturization, Automation, and Validation
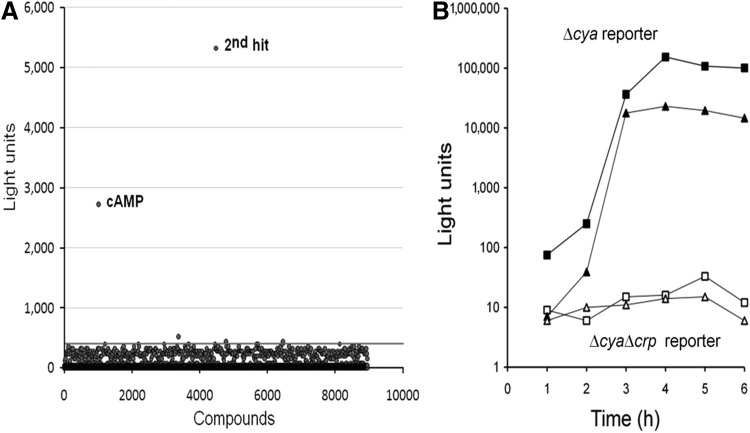
A major advantage of cell-based assays is that they select for compounds that effectively penetrate the cell. Thus, cell based assays circumvent a major hurdle in the path to identify biologically active compounds with good membrane permeability. To our knowledge, a cell-based assay for ligands of the Gram-negative bacteria CRP has not been developed. Hence, we miniaturized the assay to a 384-well format and conducted a pilot screen as described in materials and methods. For the pilot screen, compounds were screened for their capacity to induce light production in an adenylate cyclase (cya) mutant grown to high cell density. The data from this screen are shown in Figure 4A. Overall our pilot screen of 9,425 compounds yielded a hit rate of 0.02%, a Z′ value of 0.76 (0.6–0.9), a signal to background ratio >105, and a coefficient of variance of 8%±2.5% for the positive control (10 μM cAMP). The mean readouts were 404,140 light units for the positive control and 10 light units for the negative control. Interplate variability of the HTS assay, relative signal strengths and concentration-response curves are provided as supplementary material. Identification of cAMP (30 μM) within the Prestwick compound collection as one of the hits together with the low hit rate demonstrates the high specificity of this assay for molecules that bind to CRP to induce a transcriptionally active CRP molecule. The other hit, N-(2, 4-difluorophenyl)-3-{[3-trifluoromethyl)benzyl]sulfanyl}-2-thiophenecarboxamide (50 μM), was from the chemically diverse MLSMR collection and structurally unrelated to cAMP. This compound failed to induce light in a ΔcyaΔcrp reporter indicating that it was a true positive hit. This result suggests that closely fitting into the cAMP binding pocket is not a prerequisite for an agonist to trigger CRP into adopting a transcriptionally active conformation. Such molecules could be extremely valuable for investigating essential receptor-ligand interactions. Unfortunately, due to its low solubility in water, we could not test the effect of this compound in virulence gene expression. Since the optical density of the inoculum used in the HTS assay is high (∼3) and there are limitations in handling a large number of compounds using automation equipment, a fraction of this compound could have precipitated when tested for light production.
Fig. 4.
Pilot HTS for proquorum sensing CRP-ligands. (A) Representation of pilot HTS for light induction in strain C7258Δcya containing pBB1. The line indicates the 3×standard deviations cutoff value used as “hit” criteria. (B) Secondary assay. Cultures of C7258Δcya (■, ▲) and C7258ΔcyaΔcrp (□, △) containing pBB1 were grown to stationary phase in LB and 100 μL of each culture added to triplicate wells of 96-well microtiter plates containing 100 μM of cAMP (■, □) and the 2nd hit compound (▲, △). Plates were incubated at 30°C and light production measured at different times. HTS, high throughput screening.
To our knowledge, this is the first cell-based assay for ligands of the highly conserved CRP protein of Gram-negative bacteria. The use of this assay to screen large chemical libraries could identify lead compounds to treat cholera and new cAMP analogs to probe ligand-receptor interactions in the CRP molecule. The present assay could also facilitate the discovery of new cAMP analogs acting as agonists or antagonists of eukaryotic CNB proteins. We note that the members of the CNB superfamily exhibit a modular design with a conserved CNB domain linked to a functional domain, such as the CRPs DNA binding motif. Given the conservation of the CNB across the superfamily, it should be technically feasible to construct V. cholerae reporter strains expressing a CRP with a eukaryotic CNB to identify ligands of eukaryotic CNB proteins. Finally, the high specificity of this assay, reflected in the low hit rate obtained in our pilot screen suggests that the composition of the chemical library is critical for success. In our case, 88% of the compounds used in our pilot screen were designed to maximize chemical diversity. It is likely, however, that a more targeted library, enriched for structures akin to the natural ligand, could identify new cAMP analogs and yield a higher hit rate.
Glossary
Abbreviations
- CAI-1
cholera autoinducer 1
- cAMP
cyclic adenosine monophosphate
- CNB
cyclic nucleotide binding
- CRP
cAMP receptor protein
- DMSO
dimethyl sulfoxide
- HTS
high-throughput screening
- MLSMR
Molecular Libraries Small Molecule Repository
Acknowledgments
This work was supported by PHS Research Grant AI081039 to A.J.S., the NIH Molecular Libraries Probe Production Center Network (U54 HG005034), and a Southern Research Endowment fund to J.A.B. The full dataset for this assay is available at PubChem (http://pubchem.ncbi.nlm.nih.gov, AID 686977).
Disclosure Statement
The authors declare no conflict of interests in the publication of this study.
References
- 1.Brückner R. Titgemeyer F. Carbon catabolite repression in bacteria. FEMS Microbiol Lett. 2002;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- 2.Kannan N. Wu J. Anand GS, et al. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007;8:R264. doi: 10.1186/gb-2007-8-12-r264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harman JG. Allosteric regulation of the cAMP receptor protein. Biochim Biophys Acta. 2001;1547:1–17. doi: 10.1016/s0167-4838(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay R. Parrack P. Cyclic AMP-dependent functional forms of cyclic AMP receptor protein from Vibrio cholerae. Arch Biochem Biophys. 2006;447:80–86. doi: 10.1016/j.abb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Krueger S. Gorshkova I. Brown J. Hoskins J. McKenney KH. Schwarz FP. Determination of the conformations of cAMP receptor protein and its T127L, S128A mutant with and without cAMP from small angle neutron scattering measurements. J Biol Chem. 1998;273:20001–20006. doi: 10.1074/jbc.273.32.20001. [DOI] [PubMed] [Google Scholar]
- 6.Liang W. Pascual-Montano A. Silva AJ. Benitez JA. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology. 2007;153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 7.Ng WL. Bassler BL. Bacterial quorum-sensing network architecture. Ann Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J. Miller MB. Vance RE. Dziejman M. Bassler BL. Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng WL. Cong LJ. Semmelhack MF. Bassler BL. Broad spectrum pro-quorum sensing molecules as inhibitors of virulence in vibrios. PloS Pathog. 2012;8:e1002767. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacikova G. Skorupski K. Overlapping binding sites for the virulence gene AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol. 2001;41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 11.Ebright RH. Analogs of cyclic AMP that elicit the biochemically defined conformational change in catabolite gene activator protein (CAP) but do not stimulate binding to DNA. J Mol Biol. 1985;182:91–107. doi: 10.1016/0022-2836(85)90030-0. [DOI] [PubMed] [Google Scholar]
- 12.Scholübbers HG. Van Knippenberg PH. Baraniak J. Stec WJ. Morr M. Jastorff N. Investigations on stimulation of lac transcription in vivo in Escherichia coli by cAMP analogs. Eur J Biochem. 1984;138:101–109. doi: 10.1111/j.1432-1033.1984.tb07887.x. [DOI] [PubMed] [Google Scholar]
- 13.Schwede F. Maronde E. Genieser HG. Jastorff B. Cyclic nucleotide analogs as biochemical tools and prospective drugs. Pharmacol Ther. 2000;87:199–226. doi: 10.1016/s0163-7258(00)00051-6. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg MS. Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MB. Skorupski K. Lenz DH. Taylor RK. Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 16.Skorupski K. Taylor RK. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin co-regulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee J. Miyamoto CM. Zouzoulas A. Skouris BF. Meighen EA. MetR and CRP bind to the Vibrio harveyi lux promoters and regulate luminescence. Mol Microbiol. 2002;46:101–111. doi: 10.1046/j.1365-2958.2002.03128.x. [DOI] [PubMed] [Google Scholar]