Abstract
Bulge stem cells reside in the lowest permanent portion of hair follicles and are responsible for the renewal of these follicles along with the repair of the epidermis during wound healing. These cells are identified by surface expression of CD34 and the α6-integrin. When CD34 and α6 double-positive cells are isolated and implanted into murine skin, they give rise to epidermis and hair follicle structures. The current gold standard for isolation of these stem cells is fluorescence-activated cell sorting (FACS) based on cell surface markers. Here, we describe an alternative method for CD34 bulge stem cell isolation: a microfluidic platform that captures stem cells based on cell surface markers. This method is relatively fast, requiring 30 min of time from direct introduction of murine skin tissue digestate into a two-stage microfluidic device to one-pass elution of CD34+ enriched cells with a purity of 55.8%±5.1%. The recovered cells remain viable and formed colonies with characteristic morphologies. When grown in culture, enriched cells contain a larger α6+ population than un-enriched cells.
Introduction
Skin is one of the few tissue types in the body that exists in a state of constant self-renewal and repair.1 The outer layer of skin is the epidermis, a multilayered epithelium, and keratinocytes comprise 95% of all cells in this layer.2 Bulge stem cells, which reside in the lowest permanent portion of hair follicles, namely the bulge region, are responsible for the continuous regeneration of keratinocytes.1 When bulge stem cells migrate up to the deepest layer of epidermis, the basal layer, these stem cells retain their multipotency and are also called epidermal stem cells.3 Epidermal stem cells are slow-cycling cells with a high proliferative capacity.4 Several in vivo markers identify epidermal stem cells in both human and murine species, including CD34, α6-integrin, Keratin 14 (K14), Keratin 15 (K15), LGR5, LGR6, Sca-1, and Lrig1.1,2,4–8
Recent work by several groups has demonstrated the regenerative capabilities of these bulge stem cells.9–13 Murine bulge cells expressing both CD34 and α6 are capable of regenerating new hair follicles within each hair cycle.9,13 During epidermal injury, these stem cells have been observed as migrating to the wound and repair damaged tissue.3,12 When CD34+ stem cells are isolated and re-implanted into full skin defects along with neonatal dermal cells, they give rise to hair follicles, interfollicular epidermis, and sebaceous glands.9–11,13
The α6-integrin is a characteristic surface protein that is specifically expressed in all undifferentiated epidermal cells, and is, thus, a marker for basal undifferentiated keratinocytes in the epidermis as well as the resident stem cells.2 A widely used cell surface marker that is used to identify bulge and epidermal stem cells in the mouse is CD34. Interestingly, CD34 is not present in the human bulge, and the expression of the human bulge stem cell marker K15 decreases over age.14,15 The expression of CD34 by murine bulge stem cells, by contrast, is not affected by aging.16 Several negative markers are also known for human and murine bulge stem cells, including CD71, CD24, and Keratin 10 (K10).4,14,17 CD71 is a transferrin receptor and a marker of actively cycling cells. Indeed, immunostaining of epidermal keratinocytes with both α6-integrin and CD71 antibodies by Tani et al. showed that CD71+ cells comprise the majority of nonmultipotent basal keratinocytes.17
The standard methods of isolating bulge stem cells are fluorescence- and magnet-activated cell sorting (FACS and MACS, respectively).1,2,18 Both methods require preprocessing labeling of cells with antibody tags followed by centrifugation steps before cell separation. While both FACS and MACS are well established and reliable, the sample processing steps and time required present challenges when considering translational regenerative applications of resident stem cells, such as skin stem cells. Our group has demonstrated how microfluidic devices coated with antibodies can achieve positive selection capture of CD34+ cells from undiluted whole blood.19 A key element in these devices is the use of an antibody-laden hydrogel coating that is designed to selectively capture cells and then release them in a single step following the flow of sample through the device.19,20 This approach is scalable, because multiple devices can operate in parallel to process large sample amounts. Furthermore, this approach eliminates the need for sample preprocessing, thereby significantly reducing processing time and cell loss. In the context of stem and progenitor cells, an important additional requirement is the retention of phenotypic identity and functional ability of the target cells after isolation. In the present work, we describe how hydrogel-coated microfluidic devices can be utilized to enrich CD34+ bulge stem cells from digested murine skin tissue. Different device design configurations are examined along with measures of phenotypic identity and functional ability.
Materials and Methods
Microfluidic device design
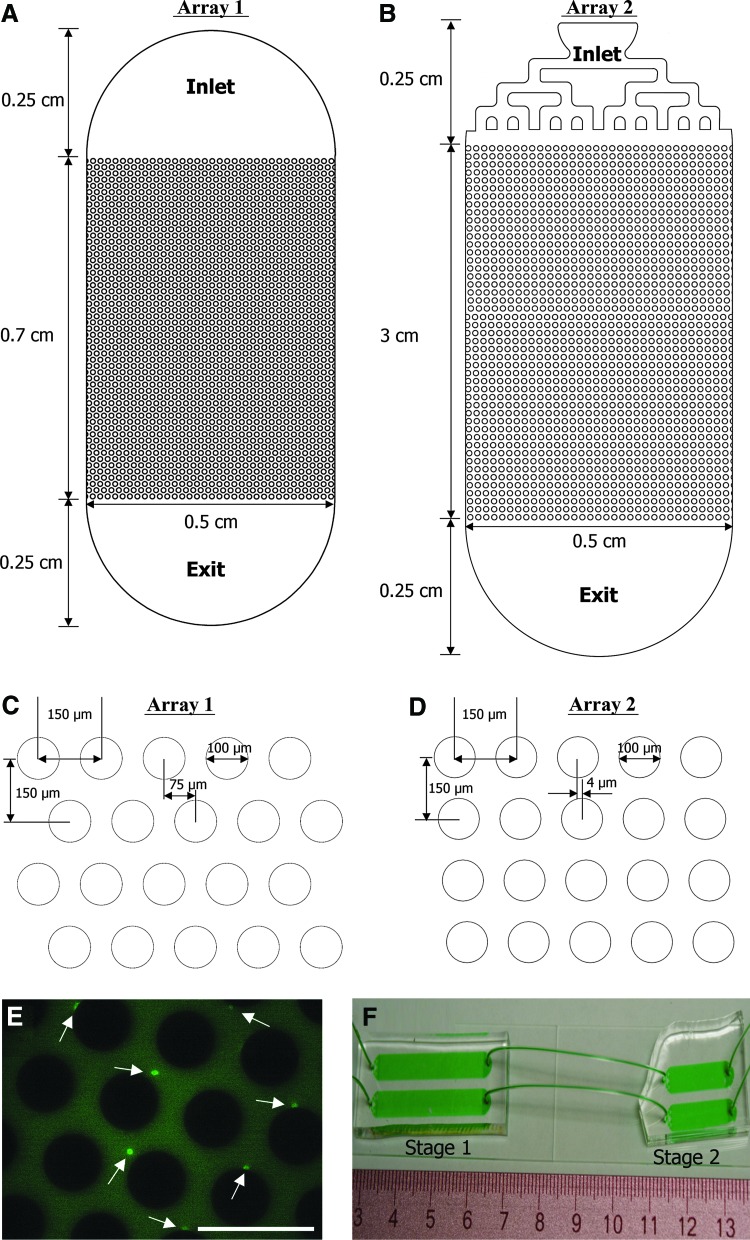
Two device designs were tested in this study, both channels with an array of pillars in the middle region (Fig. 1). The first design, referred to as “Array 1,” was adapted from a previous work by our group19 and has a hexagonal pattern with offsets modeled after those first described by Gleghorn et al.21 As shown in Figure 1A, the entrance and exit regions of this device consist of a semi-circle with a diameter of 0.5 cm, which encompasses an array of 100 μm diameter pillars with array dimensions of 0.5 cm width by 0.7 cm length (Fig. 1C). A second design, referred to as “Array 2” (Fig. 1D), was designed for improving the throughput and capture efficiency of epidermal stem cells. Array 2 has a different layout of pillars and a longer length (3 cm). A lengthened flow path was postulated to provide the epidermal stem cells with a higher likelihood of contact with the antibody-coated pillar surfaces. The maximum array length was restricted to 3 cm for ease of device fabrication and for controlling the wetted volume of devices. The layout of Array 2 was derived from calculations in which a coupled computational fluid dynamics (CFD)–particle advection code was used to track a uniform distribution of cells through the device. A range of offsets from 0 (a straight array) to 75 μm (a hexagonal array) was studied. Based on the fact that the diameter of epidermal stem cells ranged from 6 μm to 11 μm (as measured in our laboratory using a Beckman Coulter Quanta SC benchtop flow cytometer), 4 μm was identified as the optimum offset (Fig. 1D). This offset parameter resulted in cells larger than 6 μm colliding with at least 71% of the pillars. A branched channel was also used as the inlet in the Array 2 design (Fig. 1B). Each branch was sized to maintain similar hydraulic resistances across all and to, thus, achieve uniform flow rates and a uniform cell distribution across the whole inlet.22
FIG. 1.
Layouts of pillar array microfluidic devices utilized to enrich CD34+ epidermal stem cells (A, B) and array dimensions (C, D). Fluorescence micrograph showing CD34+ cells captured within a pillar array device (E). Scale bar equals to 200 μm, and arrows point to CD34+ cells. Image of two-stage configuration combining CD71+ cell depletion (Stage 1) and CD34+ positive selection capture (Stage 2) devices (F). Color images available online at www.liebertpub.com/tec
Microfluidic device fabrication–soft lithography
Microfluidic devices were fabricated via standard poly(dimethylsiloxane)-based soft lithography23 as described in an earlier work.19,20
Microfluidic device fabrication–surface functionalization
Single- and dual-stage (two devices in series) configurations of pillar array microfluidic devices were examined in this work. In two-stage configurations, the first stage was a device that was designed to deplete CD71+ cells, and the second stage was a CD34+ cell capture device (Fig. 1F). The CD71 depletion device was fabricated by the use of silane chemistry to covalently bind CD71 antibody (eBioscience; catalog number 14-0711) onto the channel surface as described in an earlier work.24 Silane chemistry was used for CD71 depletion, as the release of CD71+ cells was not needed. This method of antibody attachment to surfaces involves a permanent covalent bond but is simpler relative to the functionalized alginate method, which is needed when captured cells should be detached nondestructively.
Devices for CD34+ stem cell capture were prepared as previously described by Hatch et al.19 with the following modifications. First, the pH of the 2-(N-morpholino) ethanesulfonic acid (MES) buffer was adjusted to 6.0 using NaOH particles (Sigma) for better preservation of functional CD34 antibodies in all steps. Second, the amount of CD34-purified antibody (eBioscience; catalog number 14-0341) utilized in the reaction was increased to 40 μg. The resulting CD34 functionalized alginate solution was stored at 4°C before use. Before cell isolation experiments, solutions of PEG- and antibody- functionalized alginic acid were hand injected into each device. After incubation for 45 min, MES buffer (pH=6.0) containing 10 mM CaCl2 was rinsed through devices at a flow rate of 10 μL/min for 100 μL using a Harvard Apparatus PHD 2000 syringe pump. The devices were then immediately used in cell capture experiments.
Preparation of single cell suspension from adult mouse epidermis
Wild type C5BL6 (male, Charles River) mice between 6 and 8 weeks of age were used in this study. All animals were housed following IACUC regulations at Northeastern University. Murine skin harvest and digestion was carried out according to the protocol described by Nowak and Fuchs.1 Next, the skin digestate was passed through a series of filters at sizes of 40 μm (twice) and 20 μm (once). The 40 μm cell strainers (Fisher Scientific) were placed onto 50 mL centrifuge tubes, and the 20 μm filters (Partec) were placed onto 15 mL centrifuge tubes. Tissue digestate was poured onto each type of filter, and the cells were collected in the tubes below the filters. After each filtration, the cell suspension was centrifuged at 500g for 8 min. The resulting cells were re-suspended in serum-free medium (Dulbecco's modified Eagle's medium [DMEM]:F12 at 1:3 ratio without calcium; Invitrogen; customized product) before cell separation experiments.
Microfluidic separation of stem cells
Tissue-derived cells at concentrations of 6.6×105 cells/mL (Array 1 design) or 13.2×105 cells/mL (Array 2 design) were loaded into 1-mL syringes (BD) filled to a level of 0.4 mL. Ten syringes of cells were clamped onto a 10-port Harvard Apparatus PHD 2000 syringe pump. Cells were flowed into either 10 one-stage devices or 10 two-stage arrangements of devices at 3 μL/min and a total injected volume of 50 μL, followed by a rinsing step using serum-free medium at 5 μL/min and a total volume of 50 μL. A total of 3.3×104 flowed through Array 1 devices, and 6.6×104 cells passed through Array 2 devices. For single-stage CD71 depletion device experiments, all cells that flowed through were collected and centrifuged at 500g for 8 min to remove supernatants. For single-stage CD34+ cell capture device and two-stage configurations, the sample effluent, that is, all cells that flowed through, was discarded. After the rinsing step, cells captured within the devices were released by flowing 50 mM EDTA (in distilled water; Sigma) through devices at 15 μL/min and a 300 μL total volume. Released cells were collected in a tube containing 3 mL culture medium supplemented with 15% chelated fetal bovine serum (FBS) to neutralize EDTA. The medium volume was set at a ratio of 10:1 relative to the EDTA volume utilized in the released step in order to minimize the effect of EDTA on cells. Collected cell suspensions were centrifuged at 500g for 8 min and resuspended in staining buffer for subsequent experiments as described next.
Flow cytometry analysis to determine cd34+ cell counts
Each cell specimen was collected from three devices, which yielded about 300 cells (100 cells per device). Cell specimens were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-mouse CD34 antibody (eBioscience; catalog number 11-0341) for 40 min in the dark at room temperature, followed by centrifugation at 500g for 8 min. The supernatant was discarded, and 300 μL of ice-cold staining buffer (phosphate-buffered saline [PBS] with 2% calcium-free chelated FBS) was added to the cells to wash out unbound fluorescent antibody. Cells were centrifuged again (500g for 8 min) and then, each specimen was re-suspended in 200 μL of ice-cold staining buffer. Flow cytometry analysis was carried out using a Beckman Coulter Quanta SC benchtop flow cytometer with a voltage and gating set as recommended by Nowak and Fuchs.1 Cell viability was assessed using propidium iodide (BD Biosciences) by adding 5 μL of dye into each cell specimen 1 min before flow cytometry.
Preparation of feeder cell layers for stem cell culture experiments
3T3-J2 cells were obtained from the laboratory of Prof. Howard Green (Harvard Medical School). These fibroblasts were grown in low glucose DMEM medium (Invitrogen) supplemented with 10% calf bovine serum (American Type Culture Collection) and 1% penicillin/streptomycin (Sigma) at 37°C with 5% CO2. Cells were passaged to just under 80% confluency to avoid contact inhibition and up to P12 to serve as feeder layers. Mitomycin treatment of cells was carried out as described by Nowak and Fuchs.1 The concentration of mitomycin C (Sigma) used in our cell treatment was 4 μg/mL, and the duration was 2 h. In tandem, collagen was coated on 24-well plates.2 Mitomycin-C-treated 3T3-J2 cells were then seeded into each plate at a density of 2.4×105 cells/well (1.26×105 cells/cm2) and cultured in DMEM supplemented with 10% calf bovine serum for up to 7 days.
Culture of mouse epidermal stem cells
Before seeding mouse epidermal cells on the feeder layers, the DMEM medium for 3T3-J2 cells was aspirated and replaced by epithelial medium, prepared as follows.1 DMEM/F-12 calcium-free medium was supplemented with 15% chelated FBS, 0.1125 nM cholera toxin, 5.625 μg/mL insulin, 5.625 μg/mL transferrin, 0.01125 nM T3, 0.45 μg/mL hydrocortisone, and 0.3 mM CaCl2 (all items from Sigma). Three study groups were tested: (1) cells released from microfluidic devices, (2) un-enriched cells directly obtained from tissue digestion, and (3) feeder cell layers only. Cell specimens collected from 10 microfluidic devices (∼100 cells from each device) were added into each well. Thus, about 1000 cells were seeded in each well of the enriched group. Epidermal cells obtained directly from tissue digestate were seeded at the same level (about 1000 cells per well) in the un-enriched group. After 3 weeks of culture at 37°C with 5% CO2, the cells formed colonies and were characterized as described next.
Rhodamine B staining and colony-forming efficiency assay
Cells in 24 well plates were washed thrice with PBS, and fixed in 2% paraformaldehyde (Pierce) for 20 min. After two additional PBS washes, the cells were incubated with 1% Rhodamine B (Sigma) in distilled water for 20 min.2 After another PBS wash, colonies were stained pink and became visible within the plates. Images of each well were captured using a Nikon D70 color camera at a resolution of 300 dpi and an image size of 500KB. All colonies were counted in each well. The colony size in each well was measured using Image J software and summed to obtain the area fraction occupied by colonies in each well (defined as colony-forming efficiency [CFE]).
Measurement of α6 and CD34 in colonies
Colonies were stained with FITC-conjugated α6 antibody (Biolegend; catalog number 313605), and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The stained cells were visualized using a Nikon Eclipse TE2000-U fluorescence microscope, and images were collected using a Nikon Digital Sight (DS-2MBWc) camera. For quantitative analysis, cells in each well were washed twice with PBS, and then, both feeder layer and colonies were taken off the plates with trypsin (0.25% with 0.2 g/L EDTA) (HyClone). Cells were treated with trypsin-EDTA twice with 5 min incubation each time, and then centrifuged at 500g for 8 min. Next, the cells were resuspended in 100 μL of staining buffer and incubated with FITC-conjugated α6 antibody (Biolegend; catalog number 313605) or FITC-conjugated CD34 antibody (eBioscience; catalog number 11-0341). The number of CD34+ and α6+ cells was then determined via flow cytometry.
Data analysis and statistics
All quantitative data were taken from three experiments, and within each experimental group, three replicates were carried out. Error bars were obtained as standard deviations. A p-value from two-tailed unpaired Student's t-test <0.05 was considered statistically significant.
Results
CD34+ cell enrichment with Array 1 microfluidic devices
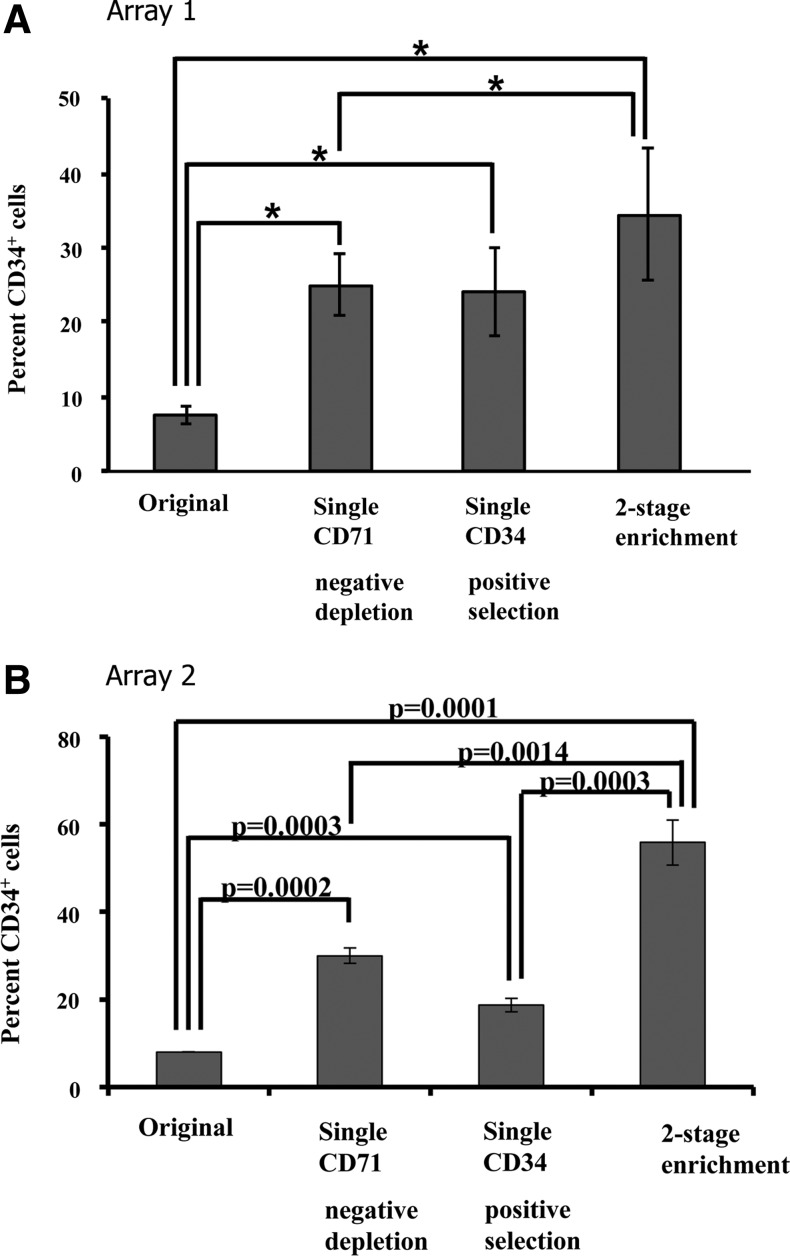
The first series of experiments employed microfluidic devices with the Array 1 design (Fig. 1A, C) for CD34+ cell enrichment. After the tissue-derived epidermal cells had been passed through the device, the captured cells were stained with FITC-conjugated anti-CD34 antibody to reveal adherent CD34+ cells (Fig. 1E). In separate experiments, the outputs from CD71 depletion devices, the CD34+ alginate-coated devices, or the two-stage devices were each collected in a tube and stained with FITC-conjugated CD34 antibody for enumeration via flow cytometry. As shown in Figure 2A, there were 7.5%±1.1% CD34+ cells in the initial cell population. The suspensions exiting the single-stage CD71 depletion devices contained 24.9%±4.1% CD34+ cells. The cells released from single-stage CD34 alginate devices contained 24.0%±5.9% CD34+ cells. When the un-enriched suspension was first passed through CD71 depletion devices and then to CD34 alginate devices connected in series, the resulting cell suspensions contained 34.4%±9.0% CD34+ cells. We conclude that by using microfluidic devices of the Array 1 design, we can enrich CD34+ cells from the original epidermal cell population by 4–5-fold via either CD71 depletion or CD34+ enrichment (p<0.05). However, the two-stage configuration provided significantly greater CD34+ enrichment than the single CD71 depletion device (p<0.05).
FIG. 2.
(A) Enrichment of CD34+ cells from mouse tissue-derived epidermal cells using microfluidic cell capture devices with the Array 1 design. Single stage CD71+ cell depletion and single-stage CD34+ positive selection result in comparable, 4–5-fold enrichment. The two-stage configuration only provides modest improvement. *indicates p<0.5. (B) Enrichment of CD34+ cells from mouse tissue-derived epidermal cells using microfluidic cell capture devices with the Array 2 design. Unlike the Array 1 design, the two-stage configuration shows an additive enrichment.
CD34+ cell enrichment with Array 2 microfluidic devices
Figure 2B shows the enrichment data from three groups of Array 2 devices. The CD71 depletion device enriched the CD34+ cell population to 30.0%±1.8% from ∼8% in the original tissue-derived suspension, a level comparable to its Array 1 counterpart. The CD34-positive selection device enriched CD34+ cell population to 18.7%±1.6%, a level lower than its Array 1 counterpart. However, with a two-stage configuration of these devices, the CD34+ cell enrichment was more than an additive, with a final enrichment of 55.8%±5.1% CD34+ cells, which was significantly higher than the 34.4%±9.0% CD34+ enrichment level obtained with the Array 1 dual stage configuration (p<0.05). It is also noteworthy that in general, the scatter in the enrichment data for Array 2 devices, as reflected by error bar size, is smaller relative to the Array 1 devices in all groups.
Viability of cells following microfluidic enrichment
Microfluidic isolation did not alter the viability of cells relative to the starting sample, as reflected by propidium iodide staining (p>0.05). Specifically, the original cell population had a viability of 51.6%±6.5%. Cells released from a single CD71 depletion device had a viability of 57.0%±12.5%, and cells enriched from a two-stage device had 57.0%±5.0% viable cells.
CFE of CD34+ enriched cells
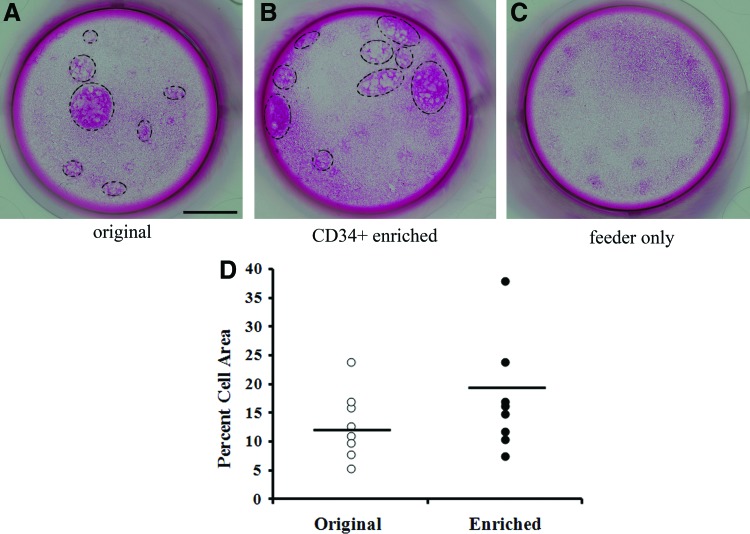
Both the un-enriched group (Fig. 3A) and enriched group (Fig. 3B) formed colonies in culture, whereas the feeder-only group did not form any colonies in wells (Fig. 3C). Colonies formed by enriched CD34+ cell populations covered 19.1%±9.6% of wells (Fig. 3D). This coverage efficiency is slightly higher than the coverage efficiency for the original un-enriched cells (12.9%±5.8%), but the difference is not statistically significant (p=0.14).
FIG. 3.
Rhodamine B staining of colonies formed on feeder layers by un-enriched epidermal cells (A), cell suspension enriched for CD34+ cells via two-stage microfluidic separation (B), and feeder cells only (C) after 3 weeks in culture. Scale bar in (A)=4 mm. Colony formation by the original and enriched groups after 3 weeks of cell culture, calculated as relative area coverage (D). Colonies are highlighted by black circles. Color images available online at www.liebertpub.com/tec
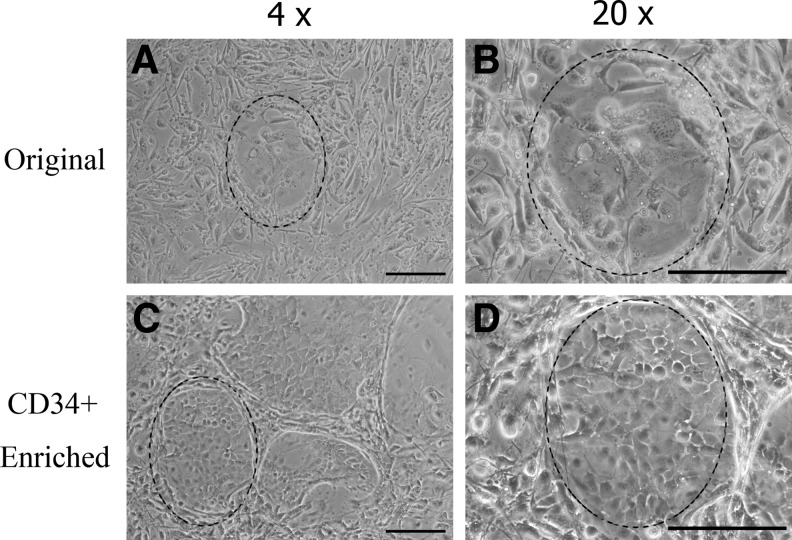
Phase-contrast micrographs of colonies grown from the un-enriched skin cell population are shown in Figure 4A and B at different magnifications. Figure 4C and D show colonies formed by CD34+ enriched obtained from the two-stage microfluidic separation. Colonies formed by the enriched epidermal cells were observed as having small and compact shapes. In comparison, colonies from the un-enriched population had an irregular shape, and the cells were loosely connected to each other.
FIG. 4.
Phase-contrast microscopy of colonies in wells. Colonies from the un-enriched epidermal cells (A, B) have no distinctive form, whereas colonies formed by suspensions enriched for CD34+ cells via two-stage microfluidic separation show characteristic cobblestone morphology (highlighted region; C, D). Scale bar=200 μm for all images.
α6 integrin expression in colonies
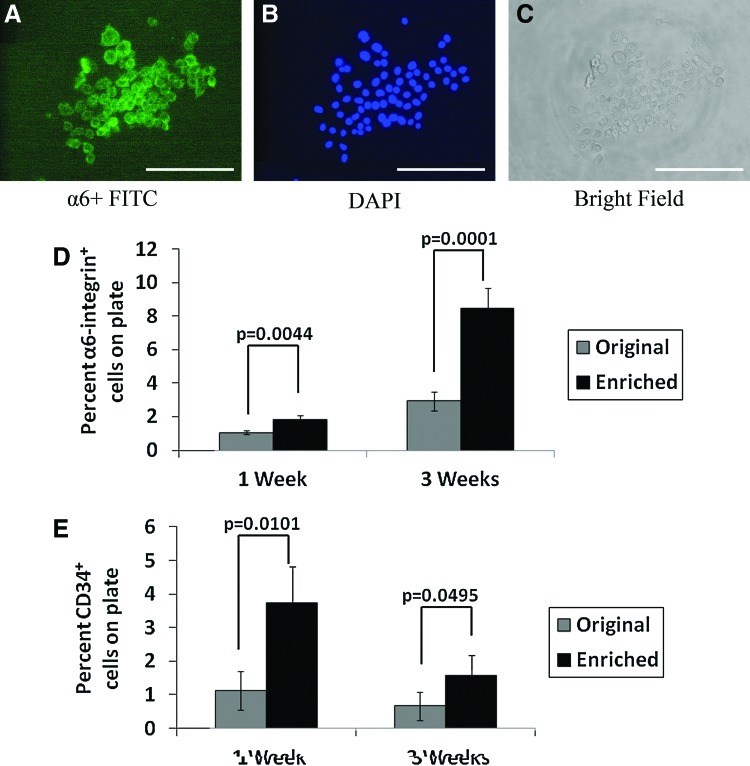
The α6 integrin is expressed in keratinocytes and stem cells in the basal layer of skin but not in 3T3-J2 fibroblasts. To show that these colonies were formed by basal keratinocytes, we stained colonies with FITC-conjugated anti α6-integrin antibody. Cells inside any given colony were considered α6-positive if the outline of the cells stained green (Fig. 5A). Figure 5B shows the DAPI counter-stain of the nuclei, and Figure 5C shows cells in bright field. We then released colonies along with the 3T3-J2 feeder layers from each well and determined the number of α6+ cells via flow cytometry. In this mixed suspension of CD34+ enriched cells and feeder cells, the proportion of α6+ cells was 8.5%±1.2%, which was significantly higher than the percentage of α6+ cells in cultures of the un-enriched cell population (2.9%±0.6%, p<0.01) after 3 weeks in culture (Fig. 5D).
FIG. 5.
α6 integrin immunostaining in colonies formed by suspensions enriched for CD34+ cells using FITC-conjugated anti-α6 (A). DAPI counterstaining and brightfield images are shown in (B) and (C), respectively scale bar equals to 200 μm. Flow cytometry data from a total of nine colonies (three colonies from each of three independent experiments) are quantified in (D) to illustrate α6 expression and in (E) to show CD34 expression after 1 and 3 weeks of culture. The percentages of α6 and CD34 cells plotted in (D) and (E) were obtained by subtracting the contribution of feeder cells whose readings have a few α6+ and CD34+ cell counts. FITC, fluorescein isothiocyanate; DAPI, 4′,6-diamidino-2-phenylindole. Color images available online at www.liebertpub.com/tec
CD34 expression in colonies
Lastly, we examined whether colonies formed by enriched keratinocytes retained more CD34+ cells in culture. For this purpose, we released all cells in wells (both colony and feeder layers) by trypsinization and stained with FITC-conjugated anti-CD34 to allow quantification of CD34+ population by flow cytometry. As shown in Figure 5E, 3.7%±1.0% cells in the enriched group were CD34 positive after 1 week in culture. By comparison, there were only 1.1%±0.6% CD34+ cells in the original un-enriched group at the same time point. Therefore, more CD34+ cells were preserved in colonies of the enriched cell population in comparison to the control group (p<0.05) after a week of in vitro culture.
Discussion
The gold standard for the isolation of epidermal stem cells from tissue is FACS.1,2 This method provides purities ≥90% but requires expensive instrumentation typically found only in core facilities and a minimum run time of over 2 h, from incubation of cells with fluorescent tags to running through the FACS instrument. MACS, which is another well-developed cell isolation technique utilized in epidermal stem cell isolation,18 requires similar run times. By contrast, our microfluidic technique is capable of providing an enriched suspension of epidermal stem cells in less than 30 min and does not require preprocessing labeling of the sample with fluorescent or magnetic tags or the associated incubation and centrifugation steps that may contribute to cell loss. Furthermore, this microfluidic technique is readily scalable; in the present study, 500 μL of epidermal cells were processed by 10 microfluidic systems (in one- or two-stage configurations) in parallel. There is no intrinsic limit to the number of such systems that can be run in parallel to process larger volumes of digested tissue. The collective advantages of versatility, simplicity, and shorter run times of the microfluidic approach become significant when considering translational or clinical applications of tissue-derived stem cells. In the present work, we demonstrate how microfluidic systems can enrich CD34+ cells in epidermal tissue-derived cell suspensions with retention of viability, phenotypic identity, and function.
In microfluidic channels, while diffusion has little effect on cell movement, cells follow un-disturbed fluid streamlines. Bluff-body obstacle arrays, such as cylindrical pillar arrays, can be used to distort these streamlines, increasing cell-wall interactions in those areas in which streamlines are compressed. These cell-wall interactions lead to capture of cells of a particular kind using antibodies coated on the pillar surfaces.21 Cell–pillar collision rates are dependent on the size and position of the pillars, as well as on the cell size. In a pillar array, an optimized offset between each row can increase cell–pillar collision rates and result in enhanced rare cell capture.21,25,26 This study tested two different pillar array designs (Fig. 1) in the functional enrichment of epidermal stem cells. The Array 1 design has been employed by our group to isolate low-abundance endothelial progenitor cells from human blood.19 The Array 2 design was tailored to the need for higher throughput and capture efficiency of epidermal cells. Furthermore, in contrast to the semi-circular inlets of the Array 1 design (Fig. 1A), the Array 2 design included branched inlets that provide greater uniformity in velocity across the width of the inlets.22 Unidirectional inlet flow evenly distributed cells, reducing the potential for cell clogging both within the inlet regions and within the pillar arrays.
Although one of the objectives of the Array 2 design was higher purity, higher throughput was an additional, equally important criterion. The maximum achievable throughput through a single Array 1 device (with 0.7 cm pillar array length) was ∼33,000 cells. Here, throughput is defined as the number of cells that can be flowed through a device before clogging, resulting from cell-cell adhesion within the pillar arrays that is high enough to block access of incoming cells to a significant portion of the pillar array, thereby reducing the maximum attainable target cell purity in the output. By contrast, the design of Array 2 (with 3 cm length) was set to ensure a 70% probability of each flowing cell making contact with at least one pillar (via CFD; results not shown). This design allowed for considerably higher throughput, ∼66,000 cells. Another benefit of the Array 2 design, also a result of reduced clogging and uniform flow, was greater reproducibility, as reflected by smaller error bars in Figure 3B relative to those in Figure 3C.
Although the Array 2 design achieved reproducible, high-throughput enrichment of CD34+ cells up to a level of ∼56%, higher levels of purity may be needed for translational and clinical applications. Several approaches can be taken to enhance purity, although such measures may result in lower yield, a typical attribute of most affinity-based continuous flow separation processes.27–32 These steps could include placing additional CD34 capture devices in series and increasing the surface density of CD34 antibody within these devices. Other epidermal stem cell surface markers, such as LGR5, LGR6, Sca-1, or Lrig11,2,4–8, could also be targeted for antibody-mediated capture.
Epidermal cells enriched by use of microfluidic devices effectively formed colonies in vitro. After 3 weeks of culture, CD34+ enriched cells formed larger areas of colonies in wells than the original cell population did, although not statistically different (Fig. 4; p=0.14). This observation is consistent with the findings of Trempus et al. who observed little difference in colony size between CD34+ FACS-enriched and un-enriched groups after 2 weeks of culture.4 The CD34+-enriched cells, however, were tightly packed in colonies with regular beveled shapes (Fig. 5), which is consistent with the morphology of FACS-sorted CD34+ skin stem cell cultures described in the literature.1,2 By contrast, colonies of un-enriched skin cells were composed of loosely packed cells with a more irregular morphology. In Figure 5, the colonies of the un-enriched group occupy ∼12% of the well surface. Based on the CD34+ composition of 0.6%, the percentage of colonies in the un-enriched group that might be expected to be CD34+ would be at most 0.7% (0.6% of 12%) of the well surface, and this would explain the absence of any cobblestone morphology in the colonies of the un-enriched cells.
The similarity in size of colonies formed by the CD34+-enriched cells and un-enriched cells (Fig. 3) suggests that both groups have similar cell numbers in culture after 3 weeks. However, staining for α6 (Fig. 5D) showed that CD34+-enriched cell populations had a significantly higher (p=0.0002) proliferative capacity in terms of producing a larger population of α6+ basal cell population than un-enriched epidermal cells.
The overall expression of CD34 was higher in colonies formed by enriched CD34+ epidermal cells than in those formed by the un-enriched cells (Fig. 5E). However, this trend was only observed after 1 week of culture period, and not later on. This phenomenon is consistent with the fact that CD34 is an in vivo bulge and epidermal stem cell marker.1,2 When cultured in epidermal growth media, epidermal stem cells tend to differentiate and thereby lose surface-expressed CD34. Furthermore, since most of the literature in this area is focused on the differentiation of CD34+ epidermal stem cells in vitro as opposed to CD34-based preservation of stemness, it is hard to find a relevant comparison to our data in Figure 5E. At a minimum, however, our data indicate that the higher CD34+ proportion in the microfluidic-enriched cell populations is accompanied by higher α6+ cell content, relative to un-enriched epidermal cells, and, in turn, a higher proliferative capacity. This higher level of CD34 expression may enable greater in vitro propagation ability of the CD34+ fraction in the presence of appropriate culture medium and supplements.
Conclusions
This article describes a microfluidic approach to functionally enrich epidermal stem cells from tissue digestate. Compared with traditional separation methods of FACS and MACS, the microfluidic approach does not require preprocessing of the digestate to bind fluorescent or magnetic tags, thereby reducing overall processing time, cell loss, and viability loss. Epidermal cells enriched for CD34+ stem cells via microfluidic separation were shown to retain colony-forming and regenerative potential.
Acknowledgment
Funding was provided by National Institutes of Health under grant R01-EB009327.
Disclosure Statement
No competing financial interests exist.
References
- 1.Nowak J.A. Fuchs E. Isolation and culture of epithelial stem cells. Methods Mol Biol. 2009;482:215. doi: 10.1007/978-1-59745-060-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen K.B. Driskell R.R. Watt F.M. Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat Protoc. 2010;5:898. doi: 10.1038/nprot.2010.39. [DOI] [PubMed] [Google Scholar]
- 3.Alonso L. Fuchs E. Stem cells of the skin epithelium. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11830. doi: 10.1073/pnas.1734203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trempus C.S. Morris R.J. Bortner C.D. Cotsarelis G. Faircloth R.S. Reece J.M. Tennant R.W. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol. 2003;120:501. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 5.Tumbar T. Guasch G. Greco V. Blanpain C. Lowry W.E. Rendl M. Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snippert H.J. Haegebarth A. Kasper M. Jaks V. van Es J.H. Barker N. van de Wetering M. van den Born M. Begthel H. Vries R.G., et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 7.Roh C. Roche M. Guo Z. Photopoulos C. Tao Q. Lyle S. Multi-potentiality of a new immortalized epithelial stem cell line derived from human hair follicles. In Vitro Cell Dev Biol Anim. 2008;44:236. doi: 10.1007/s11626-008-9084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin K.K. Andersen B. Have hair follicle stem cells shed their tranquil image? Cell Stem Cell. 2008;3:581. doi: 10.1016/j.stem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Zouboulis C.C. Adjaye J. Akamatsu H. Moe-Behrens G. Niemann C. Human skin stem cells and the ageing process. Exp Gerontol. 2008;43:986. doi: 10.1016/j.exger.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Amici A.W. Yamato M. Okano T. Kobayashi K. The multipotency of adult vibrissa follicle stem cells. Differ Res Biol Divers. 2009;77:317. doi: 10.1016/j.diff.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Cotsarelis G. Epithelial stem cells: a folliculocentric view. J Invest Dermatol. 2006;126:1459. doi: 10.1038/sj.jid.5700376. [DOI] [PubMed] [Google Scholar]
- 12.Ito M. Liu Y. Yang Z. Nguyen J. Liang F. Morris R.J. Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 13.Blanpain C. Lowry W.E. Geoghegan A. Polak L. Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Schreder A. Pierard G.E. Paquet P. Reginster M.A. Pierard-Franchimont C. Quatresooz P. Facing towards epidermal stem cells (Review) Int J Mol Med. 2010;26:171. doi: 10.3892/ijmm_00000449. [DOI] [PubMed] [Google Scholar]
- 15.Tiede S. Kloepper J.E. Bodo E. Tiwari S. Kruse C. Paus R. Hair follicle stem cells: walking the maze. Eur J Cell Biol. 2007;86:355. doi: 10.1016/j.ejcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Triel C. Vestergaard M.E. Bolund L. Jensen T.G. Jensen U.B. Side population cells in human and mouse epidermis lack stem cell characteristics. Exp Cell Res. 2004;295:79. doi: 10.1016/j.yexcr.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 17.Tani H. Morris R.J. Kaur P. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proc Natl Acad Sci U S A. 2000;97:10960. doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang E. Lian X. Chen W. Yang T. Yang L. Characterization of rat hair follicle stem cells selected by vario magnetic activated cell sorting system. Acta Histochem Cytochem. 2009;42:129. doi: 10.1267/ahc.09016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatch A. Hansmann G. Murthy S.K. Engineered alginate hydrogels for effective microfluidic capture and release of endothelial progenitor cells from whole blood. Langmuir. 2011;27:4257. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatch A. Pesko D.M. Murthy S.K. Tag-free microfluidic separation of cells against multiple markers. Anal Chem. 2012;84:4618. doi: 10.1021/ac300496q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gleghorn J.P. Pratt E.D. Denning D. Liu H. Bander N.H. Tagawa S.T. Nanus D.M. Giannakakou P.A. Kirby B.J. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip. 2010;10:27. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirby B.J. Cambridge, United Kingdom: Cambridge University Press; 2010. Micro- and Nanoscale Fluid Mechanics: Transport in Microfluidic Devices. [Google Scholar]
- 23.Xia Y. Whitesides G.M. Soft lithography. Angew Chem Int Ed. 1998;37:550. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 24.Green J.V. Murthy S.K. Microfluidic enrichment of a target cell type from a heterogenous suspension by adhesion-based negative selection. Lab Chip. 2009;9:2245. doi: 10.1039/b906768j. [DOI] [PubMed] [Google Scholar]
- 25.Smith J.P. Barbati A.C. Santana S.M. Gleghorn J.P. Kirby B.J. Microfluidic transport in microdevices for rare cell capture. Electrophoresis. 2012;33:3133. doi: 10.1002/elps.201200263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagrath S. Sequist L.V. Maheswaran S. Bell D.W. Irimia D. Ulkus L. Smith M.R. Kwak E.L. Digumarthy S. Muzikansky A., et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Autebert J. Coudert B. Bidard F.C. Pierga J.Y. Descroix S. Malaquin L. Viovy J.L. Microfluidic: an innovative tool for efficient cell sorting. Methods. 2012;57:297. doi: 10.1016/j.ymeth.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Bhagat A.A.S. Bow H. Hou H.W. Tan S.J. Han J. Lim C.T. Microfluidics for cell separation. Med Biol Eng Comput. 2010;48:999. doi: 10.1007/s11517-010-0611-4. [DOI] [PubMed] [Google Scholar]
- 29.Didar T.F. Tabrizian M. Adhesion based detection, sorting and enrichment of cells in microfluidic lab-on-chip devices. Lab Chip. 2010;10:3043. doi: 10.1039/c0lc00130a. [DOI] [PubMed] [Google Scholar]
- 30.Gossett D.R. Weaver W.M. Mach A.J. Hur S.C. Tse H.T.K. Lee W. Amini H. Di Carlo D. Label-free cell separation and sorting in microfluidic systems. Anal Bioanal Chem. 2010;397:3249. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt E.D. Huang C. Hawkins B.G. Gleghorn J.P. Kirby B.J. Rare cell capture in microfluidic devices. Chem Eng Sci. 2011;66:1508. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radisic M. Iyer R.K. Murthy S.K. Micro- and nanotechnology in cell separation. Int J Nanomed. 2006;1:3. doi: 10.2147/nano.2006.1.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]