Abstract
Nutrient absorption is the basic function that drives mammalian intestinal biology. To facilitate nutrient uptake, the host’s epithelial barrier is composed of a single layer of cells. This constraint is problematic, as a design of this type can be easily disrupted. The solution during the course of evolution was to add numerous host defense mechanisms that can help prevent local and systemic infection. These mechanisms include specialized epithelial cells that produce a physiochemical barrier overlying the cellular barrier, robust and organized adaptive and innate immune cells, and the ability to mount an inflammatory response that is commensurate with a specific threat level. The autophagy pathway is a critical cellular process that strongly influences all these functions. Therefore, a fundamental understanding of the components of this pathway and their influence on inflammation, immunity, and barrier function will facilitate our understanding of homeostasis in the gastrointestinal tract.
Keywords: inflammation, immunity, intestinal epithelium, Paneth cell, xenophagy, inflammatory bowel disease, Crohn’s disease
INTRODUCTION
The process of autophagy was first described in 1963 by Christian de Duve. Since this time, more than 30 autophagy (Atg) genes, most of which are required for the process of autophagy, have been identified in yeast genetic studies (predominantly in Saccharomyces cerevisiae) (1, 2). The proteins that compose the core machinery of autophagy are highly conserved in all eukaryotes (3). Autophagy functions as a bulk degradation system for cellular components that is classically induced by starvation or nutrient deprivation. In multicellular organisms, autophagy genes are also intimately connected to many fundamental cellular processes such as regulation of cell death, cell proliferation, inflammation, and numerous innate and adaptive immune functions. Because of these wide-ranging effects, perturbations in autophagy have been associated with numerous diseases and disease processes, including neurodegeneration, cancer, aging, cardiovascular infection/inflammation, and autoimmune conditions such as inflammatory bowel disease (IBD) (4, 5). The relationship between autophagy and disease pathogenesis is an area of intense investigation complicated by the fact that autophagy can be either protective or harmful, depending on the biological context.
The primary functions of the intestine are nutrient digestion and absorption. The process of nutrient digestion involves a large and diverse population of indigenous microbes. Thus, the intestine must also serve as a barrier to these microbes as well as to other environmental microbes and antigens that pass through the intestinal lumen. Multiple cell types, including specific epithelial and immune cell lineages, participate in the production and maintenance of this physiochemical barrier (6, 7). Recently, several genome-wide association studies (GWAS) have shown that mutations in multiple autophagy genes are highly linked to Crohn’s disease (CD), a spontaneous and chronic inflammatory condition that affects the gastrointestinal tract (8). Given the many potential roles of autophagy, it is still unclear which downstream effects of autophagy are required for intestinal homeostasis and in which cell types these functions occur. As there are still only a few studies of autophagy proteins in the intestine, this review also explores the diverse biological functions of the autophagy pathway in other systems. An important future direction will be the application of the general principles outlined here to mechanisms of intestinal homeostasis.
CORE MACHINERY OF THE AUTOPHAGY PATHWAY AND ITS REGULATION
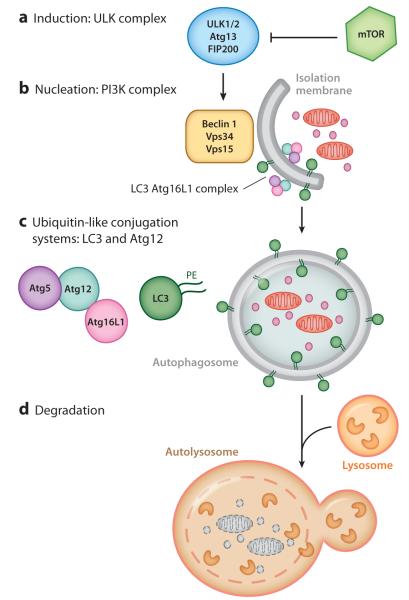
During autophagy (the focus here is on macroautophagy), subcellular components are engulfed by a double-membraned vacuole termed the autophagosome. The contents are then delivered to the lysosome for degradation. The core machinery of the autophagy pathway consists of four complexes: the unc51-like kinase (ULK) complex, which is necessary for autophagy induction; the class III phosphatidyl inositol 3-kinase (PI3K) complex containing Beclin 1 (Becn1, also known as Atg6), which is necessary for membrane nucleation; and two ubiquitin-like conjugation systems, which are important for autophagosome expansion and closure (9) (Figure 1).
Figure 1.
Stages of the autophagy pathway. (a) Induction of autophagy is mediated by the unc51-like kinase (ULK) complex, which is regulated by mammalian target of rapamycin (mTOR). (b) Nucleation is mediated by class III phosphatidyl inositol 3-kinase (PI3K). Nucleation produces an isolation membrane (phagophore). (c) The double membrane is extended by the ubiquitin-like conjugation systems of LC3 and Atg12. LC3 is conjugated to phosphatidylethanolamine (PE) and is incorporated into the autophagosome. (d) Mature autophagosomes fuse with lysosomes to produce autolysosomes.
Several signaling complexes regulate the onset of autophagosome formation. The best-studied regulator is mammalian target of rapamycin (mTOR), which negatively regulates autophagy during nutrient-rich conditions. mTOR consists of two distinct complexes, mTORC1 and mTORC2. mTORC1 is involved primarily in autophagy, whereas the role of mTORC2 in autophagy is not well defined (10). mTORC1 acts as a nutrient sensor by integrating signals from growth factors, including those in the insulin pathway, to indicate the availability of nutrients and energy. mTORC1 directly regulates the induction of autophagy by interacting with the ULK complex to suppress its kinase activity under nutrient-replete conditions (11–13). The ULK complex consists of Ulk1 and two additional proteins, FIP200 and Atg13, both of which are required for Ulk1 localization to the isolation membrane. Under nutrient-deficient conditions, the ULK complex is active and leads to recruitment of the PI3K complex to the isolation membrane (i.e., phagophore). Stimulation of PI3K results in increased production of phosphatidyl inositol 3-phosphate, which further positively regulates autophagosome formation (14). The PI3K complex consists of the class III PI3K (Vps34), Beclin 1, and Vps15. Although the default state under nutrient-rich conditions is autophagy inhibition, basal levels of autophagy have been detected under nutrient-replete conditions in multicellular organisms, suggesting additional mechanisms of autophagic regulation (15–17). For example, cytokines and immune signaling pathways regulate autophagy in mammals (18). Recently, a genome-wide siRNA screen identified 219 genes that inhibited basal autophagy independently of mTORC1. Surprisingly, most of these genes encoded cytokines, chemokines, and other cellular signaling molecules regulated by cytokines (19). An important area of investigation is to define the underlying principles of how this large and diverse set of signaling molecules regulate autophagy, especially in the intestine.
Several studies have shown that the endoplasmic reticulum (ER) is crucial for autophagosome formation and serves as a membrane source for autophagosomes. Electron tomographic analysis showed a direct extension between the ER and autophagosomal membranes (20, 21). In addition to the ER, other membranes, such as the nuclear, outer mitochondrial, and plasma membranes, may also be involved in autophagosome formation (22–24). Atg9, a membrane protein essential for autophagy, traffics between the trans-Golgi, endosomes, and autophagosome precursors, suggesting that the Golgi complex and endosome membranes may also serve as sources for the autophagosomal membrane (25, 26). Investigators recently showed that Rab11 and TCB1D14 (a Rab11GAP) are present on recycling endosomes and regulate autophagosome formation through interaction with Ulk1 (27), suggesting a role for recycling endosomes in the induction of autophagy. A major challenge in identifying the membrane source that initiates autophagosome formation in a given cell type is a lack of stable membrane markers for each potential membrane source.
Atg12 and LC3 are the two ubiquitin-like conjugation systems involved in the elongation step (reviewed in Reference 4). Atg12 is covalently conjugated to Atg5 by Atg7 and Atg10, which function as E1- and E2-like enzymes, respectively. The resulting Atg5-Atg12 conjugate noncovalently interacts with Atg16L1. This Atg16L1 complex localizes to the expanding autophagosome and dissociates during maturation of the autophagosomal membrane (4). Mammalian Atg16L1 differs from yeast Atg16 in that Atg16L1 has a multiple C-terminal WD repeat domain (28). The function of this WD repeat domain is not well understood; one possibility is that this domain may be involved in additional protein interactions for the regulation of specific subtypes of autophagy.
In the second ubiquitin-like conjugation system, LC3 is conjugated to the head group of phosphatidylethanolamine (PE). This conjugation event is mediated by Atg4B, which cleaves the precursor form of LC3 to expose the lipid conjugation site at the C-terminal glycine residue. Here Atg7 and Atg3 function as the E1- and E2-like enzymes, respectively (4). In mammals, other orthologs of LC3, GABARAP and GATE-16, are also conjugated to PE and localize on the autophagosome membrane (29).
In vitro experiments have shown that the Atg5-Atg12 conjugate interacts with Atg3 to facilitate PE conjugation to LC3. The Atg5-Atg12 conjugate behaves as an E3-like enzyme in the LC3 conjugation system (30). However, Atg16 is not essential for LC3 conjugation in yeast. The Atg16 complex may be involved in the recruitment of LC3 to the phagophore membrane. Artificial expression of the Atg16L1 complex on the plasma membrane results in LC3 lipidation at this site (31). Interestingly, the Golgi-resident protein Rab33B can interact with Atg16L1 to promote autophagy (32). Studies in yeast have shown that LC3 expression levels regulate the size of autophagosomes (33), possibly through FNBP1L, which is involved in the generation of membrane curvature (34). However, FNBP1L is dispensable for cellular autophagy but is required for xenophagy of intracellular Salmonella. Additionally, LC3 may also be involved in the completion of autophagosome formation. Incomplete autophagosomes accumulate in Atg3-deficient fibroblasts and in cells expressing a dominant negative form of Atg4B (35).
In the final stages of autophagy, the autophagosome maturation step, autophagosomes fuse with lysosomes to form autolysosomes for degradation. However, autophagosomes fuse with early and late endosomes as well as with multivesicular bodies (termed amphisomes) (36). Autolysosomes mature as more autophagosomes and lysosomes fuse. During the termination phase, lysosomes are tubulated and fragmented for renewal, a process regulated by mTOR (37).
Several pleiotropic functions of autophagy proteins apart from those of the classical autophagy pathway have been described. LC3 localizes to phagosomal membranes, double-membraned vesicles for positive-strand RNA virus replication, and even on non-membrane-enclosed protein aggregates (38–40). It has been difficult to distinguish between canonical autophagy and noncanonical autophagy because these noncanonical functions may encompass aspects of the canonical pathway. For example, Hwang et al. (41) recently showed that the Atg16L1 complex is required for interferon (INF)-γ-mediated antiviral defense and is independent of autophagy induction and lysosomal degradation. Additionally, the assays used to commonly detect autophagy can lead to ambiguous results. The widely used autophagy assay is biochemical detection of LC3 lipidation (conversion of LC3-I to LC3-II) and detection of LC3-II either on immunoblots or by the presence of punctate LC3-positive dots within cells as detected by microscopy. An increase in LC3-II can indicate either an enhancement in autophagosome formation or a block in autophagosome maturation and requires additional approaches to distinguish between these two possibilities. As mentioned above, LC3 puncta may represent the presence of LC3 on nonautophagosomal structures. Thus, although LC3-II formation is considered to be the hallmark feature of autophagosome formation, it is not a completely specific marker for classical autophagy, and other methods (e.g., electron microscopy) are required for classification of canonical versus noncanonical autophagy (42).
GENOME-WIDE ASSOCIATION STUDIES REVEAL A ROLE FOR AUTOPHAGY IN INFLAMMATORY BOWEL DISEASE
IBD is a chronic and debilitating condition of the gastrointestinal tract. CD and ulcerative colitis (UC) are the two major forms of IBD. Both forms of IBD are characterized by damage to the intestinal epithelial lining that forms a barrier between luminal contents and the mucosal immune system. Peak incidence for IBD is in early adult life, although individuals of any age can be affected. There is epidemiological evidence for a genetic basis for both major forms of IBD. Such evidence includes studies of concordance rates in monozygotic twins and in family members of those who develop the disease. In addition to genetics, environmental and microbial factors likely play a role in disease pathogenesis (43).
Recent years have seen dramatic progress in understanding the genetic basis of IBD due to comprehensive GWAS that have identified ~100 common genetic variants that are associated with increased disease susceptibility (8). These studies have been invaluable in identifying previously unsuspected biological pathways (including autophagy) that play a role in disease pathogenesis. One of the first genes implicated in CD susceptibility was NOD2, which has multiple mutations in its C terminus that are strongly associated with CD (44, 45). NOD2 is an intracellular pattern recognition receptor (PRR) and modulates Toll-like receptor (TLR) signaling and nuclear factor-κB (NF-κB) activity. Recent studies have shown that NOD2 acts as a sensor for intracellular bacteria by triggering autophagy through interaction with ATG16L1 (46, 47). Interestingly, the gene encoding ATG16L1 also has a single prominent CD susceptibility locus that encodes a point mutation (T300A, also known as Ala197Thr) (8). The Ala197Thr variant lies within the conserved WD repeat domain; however, the functional impact of this mutation is still unclear. As mentioned above, this domain is important for protein-protein interactions; therefore, this variant may interfere with its binding or with the recruitment of other proteins in the canonical and noncanonical autophagy pathways. For example, Travassos et al. (46) and Cooney et al. (47) have shown that cells with either a NOD2 or an ATG16L1 variant show impaired induction of autophagy upon stimulation with muramyl dipeptide, a NOD2 ligand. Additionally, these two gene variants show impaired antigen presentation in dendritic cells by loss of MHC class II localization to autophagic compartments and to the cell surface upon muramyl dipeptide simulation. This defect has functional consequences, as impaired antigen presentation leads to loss of CD4+ T cell responses following infection. These studies have attributed functional consequences to the NOD2 and ATG16L1 variants and have linked these disease-associated variants to autophagic bacterial control. These findings underscore the importance of innate immunity in IBD pathogenesis.
Further emphasizing the role of antibacterial autophagy in CD pathogenesis is another gene related to autophagy, that encoding immunity-related guanosine triphosphate (IRGM), which has multiple CD susceptibility loci. One single-nucleotide polymorphism is associated with a 20-kb deletion upstream of the IRGM gene. This deletion impacts IRGM expression levels and autophagic clearance of intracellular bacteria, including Mycobacterium bovis and Salmonella typhimurium (48–51). For IRGM-dependent clearance of M. bovis, IRGM localizes to the bacteria-containing compartment, which is later targeted to the lysosome for degradation. A second IRGM single-nucleotide polymorphism is associated with a synonymous variant (c.313C>T) that decreases IRGM expression through interaction with miR-196. This variant also shows deregulated antibacterial autophagy of adherent-invasive Escherichia coli (52).
Interestingly, NOD2, ATG16L1, and IRGM have been associated only with CD, not with UC. These GWAS have provided tremendous insight into cellular pathways contributing to pathogenic mechanisms for IBD, but only a small portion of the genetic variation has been accounted for. Identification and characterization of additional genes and pathways will require more sophisticated studies and are future goals of geneticists in this area. In addition, the precise mechanisms by which polymorphisms in autophagy genes contribute to CD pathogenesis need further elucidation. However, autophagy appears to be important to the mucosal immune system (Figure 2). The role of autophagy in both innate and adaptive immune defense and its applications in intestinal homeostasis are discussed in more detail in the remainder of this review.
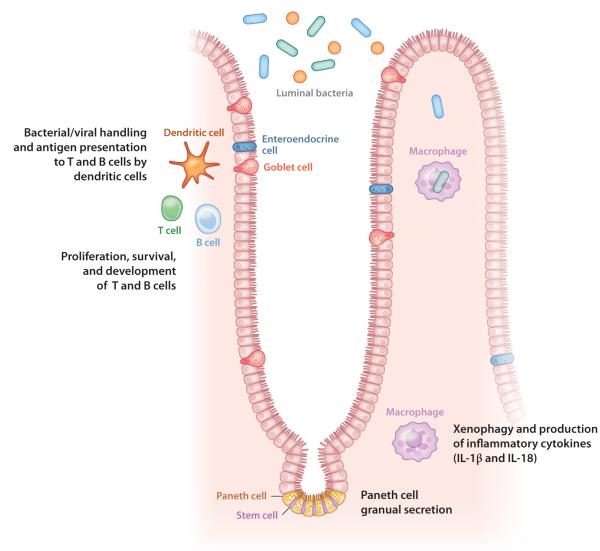
Figure 2.
Potential cellular targets of autophagy proteins in the intestine. Autophagy proteins are required for a variety of cellular functions that may be relevant to the intestine. These functions include lymphocyte development and survival, antigen presentation by dendritic cells, xenophagy and cytokine secretion by monocyte-derived cells, and antimicrobial peptide secretion by Paneth cells.
ROLE OF AUTOPHAGY IN INFLAMMATION
In 2008, an important study proposed a mechanism by which autophagy proteins could suppress intestinal inflammation. Saitoh et al. (53) found that Atg16L1 regulates endotoxin-mediated inflammasome activation in mice during injury. TLRs are a family of PRRs that mediate innate immune responses upon recognition of various microbial components (54). Treatment of macrophages with lipopolysaccharide (LPS), a TLR4 ligand, stimulated production of tumor necrosis factor (TNF)-α and IL-6 independently of Atg16L1 expression; however, IL-1β production was markedly enhanced in LPS-treated Atg16L1- and Atg7-deficient macrophages. Importantly, TLR4 stimulation of primary macrophages by LPS did not induce canonical autophagy, suggesting that additional roles of autophagy proteins are required for the control of IL-1β production in LPS-treated macrophages (53). Additionally, treatment of Atg16L1-deficient macrophages with noninvasive, commensal, gram-negative bacteria (Enterobacter aerogenes, Escherichia coli, and Klebsiella pneumoniae) also induced excess IL-1β production (53). Stimulation with ligands for TLR2 or TLR5 did not lead to increased IL-1β production, potentially indicating a specific role for TRIF (Tir domain–encoding adaptor-inducing IL-1β) signaling in the production of IL-1β and IL-18. In support of this hypothesis, macrophages doubly deficient for Atg16L1 and TRIF did not produce increased IL-1β in response to LPS stimulation, suggesting that Atg16L1-deficient macrophages showed enhanced IL-1β production in response to LPS through a TRIF-dependent mechanism (53).
Saitoh et al. (53) also found that levels of IL-1β mRNA and pro-IL-1β protein were unchanged in Atg16L1-deficient macrophages compared with controls, suggesting that autophagy proteins negatively regulate posttranslational processing of IL-1β. LPS stimulation resulted in the cleavage of caspase-1 through activation of inflammasomes, leading to increased IL-1β production. This finding suggested a functional link between these two pathways. The inflammasome is a multiprotein scaffold that mediates the cleavage and activation of caspase-1, which results in the activation and release of IL-1β and IL-18. Cytosolic receptors from the NOD-like receptor (NLR) family are critical components of the inflammasome and interact with the adaptor molecule ACS, which initiates the cleavage of caspase-1. In macrophages, increased activation of the NALP3 (NLRP3) inflammasome pathway elevates IL-1β and IL-18 production (55). In a follow-up study, LPS-treated, Atg5-deficient macrophages also showed inflammasome activation. Such activation led to atherosclerotic progression in mice that contained macrophages deficient for Atg5 (5).
The in vivo role of Atg16L1 in intestinal inflammation was evaluated by using a dextran sodium sulfate (DSS) model of chemically induced mucosal damage (53). Chimeric mice lacking Atg16L1 in hematopoietic cells showed decreased survival and increased weight loss upon DSS treatment compared with controls. The levels of proinflammatory cytokines IL-1β and IL-18 were elevated in sera of DSS-treated, Atg16L1-deficient chimeric mice and were reduced by the administration of anti-IL-1β and anti-IL-18 antibodies (53). These studies demonstrated that excessive production of cytokines in the absence of autophagy drives the development of colitis. These studies were recently corroborated in human CD patients. Plantinga et al. (56) reported increased production of IL-1β and IL-6 in response to stimulation with NOD2 ligands in monocytes derived from patients harboring the Atg16L1 T300A risk allele. In contrast to the findings in mice, increased IL-1β production in humans seems to be a result of increased transcription (56). These studies suggest that Atg16L1 may mediate the balance between NOD2-induced autophagy and cytokine production. Additionally, on the basis of these studies, IL-1β may emerge as an attractive therapeutic target for CD patients harboring the Atg16L1 risk allele.
The mechanism by which autophagy proteins regulate inflammasome activation is not completely understood. Generation of reactive oxygen species (ROS) plays an important role in caspase-1 activation induced by stimulatory molecules such as ATP, silica, uric acid crystals, and asbestos (57). Treatment with the ROS scavenger N-acetyl-l-cysteine suppressed IL-1β production from Atg16L1-deficient macrophages (53). Additionally, ROS levels in LPS-stimulated macrophages are higher in Atg16L1-deficient versus wild-type macrophages, suggesting that ROS accumulation in the absence of autophagy proteins plays a key role in driving inflammation. However, subsequent studies have shown that the NADPH oxidases are dispensable for inflammasome activation and that mitochondrial dysfunction drives inflammasome activation (58, 59). Additional studies have shown that autophagy proteins regulate Nalp3-dependent inflammation by preserving mitochondrial integrity and by inhibiting the release of mitochondrial DNA (60). Recently, researchers showed that activation of either the AIM2 or NLRP3 inflammasomes induces autophagy and targets active inflammasomes for degradation, indicating the existence of a negative feedback loop (61). These findings suggest that autophagy proteins negatively regulate inflammasome activation through suppression of ROS accumulation, inhibition of inflammasome components, and degradation of signals that activate inflammasomes.
Another important pathway regulated by autophagy genes is the type I IFN. The autophagy pathway is required for type I IFN production by plasmacytoid dendritic cells (DCs) after stimulation with single-stranded RNA viruses. Here, the autophagy pathway delivers cytosolic viral replication intermediates to endosomal TLR7, which is required for type I IFN production (62). Conversely, Atg5 deletion in macrophages increases type I IFN production in response to RNA viruses due to enhanced signaling through RIG-1-like receptors (RLRs), which also recognize single-stranded RNA viruses. This enhancement in RLR signaling in the absence of autophagy proteins is due mainly to the increased ROS production associated with dysfunctional mitochondria, as antioxidant treatment blocks RLR signaling (58). Additionally, INF-β production induced by double-stranded DNA is negatively regulated by Atg9a, but not by Atg7, in mouse embryonic fibroblasts (25). Collectively, these studies suggest that autophagy can both positively and negatively regulate type I IFN production by both autophagy-dependent and autophagy-independent mechanisms. Recently, Lei et al. (63) showed that the mitochondrial proteins NLRX1 and TUFM interact with Atg5-Atg12 and Atg16L1 to negatively regulate RLR-induced type I INF production and to activate autophagy, establishing a link between an NLR protein and virally induced autophagy through mitochondrial proteins.
ANTIMICROBIAL AUTOPHAGY
In addition to the role of the autophagy apparatus in macroautophagy, this apparatus can also be used to target and destroy invasive pathogens in a selective manner, a process referred to as xenophagy (64). This is a rapidly expanding area, and several lines of evidence have shown that autophagy can play both antibacterial and probacterial roles through various pathways, which are discussed below. The concepts are relevant to intestinal biology, as there are several well-recognized invasive pathogens in the intestine, including species of Salmonella and Shigella.
Intracellular bacterial pathogens can be engulfed by autophagosomes and delivered to lysosomes. This process typically leads to bacteria death in the acidic environment of lysosomes. Group A Streptococcus (GAS) is one well-characterized example of this model. After endosomal escape, cytoplasmic GAS becomes enveloped by autophagosomes and delivered to lysosomes for degradation (65). A similar mechanism is utilized for clearance of other invading pathogens such as S. typhimurium, Shigella flexneri, and Mycobacterium tuberculosis (18). Salmonella initially evades destruction by residing within phagosome-like structures termed Salmonella-containing vacuoles (SCVs). Only when damage mediated by the type III secretion apparatus occurs to the SCVs does the bacterium escape into the cytosol, where it is then targeted for autophagic degradation in the lysosome (66). Interestingly, pathogens have evolved mechanisms to evade degradation by either blocking autophagy or preventing delivery to lysosomes. Shigella evades targeting to autophagosomes by inhibiting its Atg5-binding protein, VirG. This inhibition is accomplished through production and secretion of IcsB through the Shigella type III secretion system (67). M. tuberculosis persists within phagosomes by evading fusion with lysosomes. However, activation of autophagy by rapamycin or IFN-γ results in phagosome maturation into phagolysosomes (68). Additionally, in vivo clearance of Toxoplasma gondii requires Atg5; however, the mechanism of degradation is independent of autophagosome formation. Here, Atg5 is required for the recruitment of IFN-γ-inducible p47 GTPase IIGP1 (Irga6) to the vacuole membrane that contains T. gondii to mediate clearance of this pathogen in vivo (69). Thus, autophagy proteins can mediate clearance of intracellular pathogens through both autophagosome-dependent and autophagosome-independent pathways.
Recent genetic studies in humans have further implicated the importance of autophagy proteins in the pathogenesis of tuberculosis (TB). IRGM was identified as a risk locus for TB susceptibility in a West African population. This −216T variant provides protection against disease caused by infection with the Euro-American lineage of M. tuberculosis but not against TB caused by M. africanum or M. bovis. Interestingly, the IRGM variants associated with IBD were not involved in TB susceptibility (70). Additionally, a genome-wide siRNA screen conducted to identify host factors involved in the replication of M. tuberculosis demonstrated a predominance of genes involved in the regulation of the autophagy pathway (71).
Some pathogens, however, are able to replicate in autophagosomes or lysosomes and have evolved mechanisms to activate autophagy or accelerate delivery into lysosomes. Francisella tularensis, for example, is resistant to degradation in lysosomes and is able to escape the phagosome to become subsequently engulfed by the autophagosome. Delivery of Francisella to the autophagosome results in increased microbial replication (72). Coxiella burnetii can also replicate in autophagosomes. Induction of autophagy by amino acid deprivation increased the number of C. burnetii vacuoles and bacterial load in Chinese hamster ovary cells (73). Yersinia pseudotuberculosis actually activates autophagy in macrophages, subverts autophagosomes for bacterial replication, and inhibits acidification of bacteria-containing autophagosomes (74). Brucella abortus is another pathogen that can subvert autophagy and survive in a Brucella-containing vacuole (BCV). The BCV is targeted to the ER, where it acquires autophagic features (aBCV). aBCVs are required for the completion of its life cycle and cell-to-cell spreading. aBCV formation requires autophagy-initiation proteins (Ulk1, Beclin 1, and Atg14L); however, it does not require autophagy-elongation proteins (Atg5, Atg16L1, Atg4B, Atg7, and LC3B) (75). These studies suggest that autophagosomes may serve as a protected intracellular niche for some bacterial pathogens, allowing such pathogens to evade degradation.
The mechanisms by which intracellular pathogens are targeted by the autophagy system are unknown. Interception at multiple points in the intracellular life cycle of pathogens likely helps mediate their recognition by autophagy proteins. The mechanisms used to target intracellular bacteria to autophagosomes are similar to those used in other types of selective autophagy. Adaptor proteins such as p62, Nbr1, and Ndp52 target various bacterial pathogens to nascent isolation membranes of autophagosomes (76). Travassos et al. (46) demonstrated that both Shigella infection and Listeria infection depend upon NOD1-induced recruitment of ATG16L1 to sites of bacterial entry. Using a variant of NOD2 associated with CD, these authors also showed that autophagocytic clearance of Shigella is significantly impaired (46). Investigators recently showed that the cytosolic lectin Galectin 8 recruits NDP52 to damaged SCVs, which may be involved in the activation of antibacterial autophagy (77). Additionally, another recent study suggested that infection with either Shigella or Salmonella triggers acute amino acid starvation in response to membrane damage, which results in the induction of autophagy (78).
In addition to pathogen recognition, autophagy proteins can directly mediate delivery of pathogens to lysosomes for degradation. The self-ligand and cell surface receptor SLAM is responsible for the recruitment of class III PI3K and Beclin 1 to gram-negative bacterial phagosomes, facilitating the fusion of phagosomes and lysosomes. Additionally, SLAM is involved in the activation of the antibacterial NADPH oxidase NOX2 on phagosomes (79). Furthermore, Huang et al. (80) showed that activation of TLR or Fcγ receptors during phagocytosis and bacterial autophagy recruits LC3 to the phagosome in a manner dependent on NOX2 generation of ROS. Collectively, these studies show that autophagy proteins facilitate bacterial degradation by mediating the formation and maturation of phagolysosomes.
As in cases of bacterial infections, autophagy inhibition can be either detrimental or protective in cases of viral infection. During herpes simplex virus infection, the viral protein ICP4.5 targets Beclin 1 to inhibit autophagosome initiation, leading to neurovirulence (81). The human immunodeficiency virus-1 (HIV-1) activates the mTOR pathway to inhibit autophagy. Inactivation of autophagy helps HIV-1 evade both innate and adaptive immune responses, including degradation in lysosomes, which results in increased transfer of infection to CD4+ T cells (82). These studies show that viruses have developed several strategies to prevent recognition or degradation by host autophagy.
In other cases, viruses can use components of the autophagy pathway to facilitate their survival and replication. In the case of HIV, autophagy assists in the processing of HIV envelope precursor protein Gag and extracellular viral release. The HIV protein Nef acts as an antiautophagic maturation factor and protects HIV from degradation by inhibiting later stages of autophagy through interaction with Beclin 1 (83). Picornaviruses such as poliovirus and coxsackievirus replicate in the gastrointestinal epithelium. Infection with picornaviruses results in an increased number of double-membraned cytoplasmic vesicles, whose surfaces are used as sites for viral RNA replication (39, 84). Poliovirus replicase proteins colocalize with LC3, and stimulation or inhibition of autophagy results in increased or decreased poliovirus yield, respectively (39). Replicase protein 2B induces conversion of LC3-II and recruits it to the replication membrane. Replicase proteins 3A and 2BC convert these membranes into double-membraned vesicles that contain LC3 and LAMP1 (39, 85, 86). Similarly, infection with rotavirus, which causes infantile viral gastroenteritis, also recruits LC3 to viral replication membranes (87).
The precise mechanisms by which autophagy proteins promote viral replication need to be better defined. Taken together, the studies suggest that autophagy functions to provide membrane sites for viral replication. Additionally, autophagy proteins, independently of their canonical role, can assist in viral replication. For example, coronaviruses use the unlipidated form of LC3 (LC3-I) to assist in the biogenesis of ER-derived, double-membraned viral replication sites (88). Taken together, these studies show that viruses utilize autophagy proteins both in a canonical and in a noncanonical fashion.
In contrast to these findings, the Atg16L1 complex and LC3, independently of induction of autophagy, inhibit the formation of a membranous murine norovirus (MNV) replication complex in response to IFN-γ in macrophages. MNV is a ubiquitous norovirus that is prevalent in mouse facilities around the world. MNV in humans is responsible for the majority of epidemic nonbacterial gastroenteritis cases (89). The MNV replication complex is distinct from that of autophagosomes in that it is not a concentric double-membraned structure and does not contain cytoplasmic contents or organelles. These studies emphasize that autophagy proteins, in addition to their canonical degradative role, have independent roles in multiple aspects of innate immune defense during infection.
REGULATION OF AUTOPHAGY BY IMMUNE SIGNALING
TLRs were historically the first class of PRRs to be connected with autophagy. In the intestine, TLRs are expressed on epithelial cell surfaces and endosomes. TLR signaling in the intestine is essential for homeostasis, as activation of TLRs leads to cellular proliferation, IgA induction, and secretion of antimicrobial peptides (90, 91). TLRs are activated by recognition of microbial-associated molecular patterns (MAMPs), thus allowing commensal microbiota to activate autophagy in intestinal cells that express these receptors.
TLRs can regulate autophagy induction (92, 93) and can recruit autophagy proteins to phagosomal membranes (38). A MAMP library screen for autophagy induction in RAW macrophages found recognition of single-stranded RNA by TLR7 to be a potent activator of autophagy (93). TLR7 recognizes viral replication intermediates and is localized on endosomes. Lee et al. (62) showed that autophagy is required for type I IFN–mediated control of vesicular stomatitis virus (VSV) in plasmacytoid DCs. Induction of autophagy by TLRs requires the MyD88 adaptor protein. TRIF and MyD88 indirectly interact with Beclin 1 to induce autophagy by sequestering Beclin 1 from Bcl-2 (also known as B cell CLL/lymphoma 2) (94). Another study showed that Beclin 1 may also be released from Bcl-2 during TLR signaling following phosphorylation of Bcl-2 by JNK-1 (95). High-mobility group box 1 (HMGB1), an extracellular damage-associated molecular pattern (DAMP) molecule, also regulates autophagy through binding of Beclin 1 and displacement of Bcl-2 (96). In Drosophila, recognition of VSV by Tol7 is required for activation of autophagy (97). Interestingly, activation of autophagy by TLR7 accelerates the elimination of Mycobacteria (BCG) from macrophages, suggesting that activation of autophagy through signaling of TLR7, a viral recognition receptor, can enhance bacterial elimination (93).
LPS induction of TLR4 also activates autophagy in a manner dependent on class III PI3K and p38 MAPK activation. As does TLR7, TLR4-mediated induction of autophagy increases the elimination of Mycobacteria from macrophages (92). Additionally, macrophage stimulation with TLR2 and TLR4 ligands results in the recruitment of Beclin 1 and LC3-II to the phagosome. The recruitment of LC3 to the phagosome depends on Atg5 and Atg7 and results in fusion of the phagosome with the lysosome. Interestingly, double-membrane vesicles are not associated with phagosomes, suggesting that TLR signaling results in the direct recruitment of LC3 to the phagosomal membrane. The recruitment of LC3 to phagosomes following activation of either TLRs or FCγRs depends on ROS generation by NOX2 (80). A recent report showed that the autophagy protein Rubicon, independently of its autophagic activity, may be involved in the recruitment of NADPH oxidase and in ROS production upon TLR stimulation in macrophages (98). These studies suggest that TLR signaling and TLR downstream effectors can enhance the elimination of microorganisms through the recruitment of autophagy proteins.
NLRs have also been implicated in the induction of autophagy. NLR signaling in autophagy induction was first implicated in Drosophila. Recognition of a peptidoglycan molecule by the intracellular PRR PGRP-LE resulted in the induction of autophagy, which was necessary to prevent intracellular growth of Listeria monocytogenes (99). Both Nod1 and Nod2 also induce autophagy in mammalian cells. In mice exposed to Nod1 and Nod2 ligands following thioglycolate priming, Nod-dependent induction of autophagy was seen in isolated macrophages. Rapamycin or LPS induction of autophagy was unaffected in Nod1- or Nod2-deficient mice, suggesting that this induction of autophagy was independent of TLR signaling and the canonical autophagy pathway. Both Nod1 and Nod2 coimmunoprecipitated with Atg16L1 and colocalized at the plasma membrane when overexpressed. This pattern was independent of RIPK-2, a mediator of Nod signaling. Both endogenous and overexpressed Atg16L1 and Nod2 localize at sites of bacterial entry following infection with Shigella (46). In a related study, NOD2 stimulation by muramyl dipeptide (MDP) also induced autophagy in DCs. In contrast to what was reported by Travassos et al. (46), another group found that induction of autophagy by NOD2 stimulation in DCs depends on RIPK-2 signaling (47). Recent work in an intestinal epithelial cell line has shown that RIPK-2 positively regulates Nod2-induced autophagy through activation of p38 MAPK and repression of a negative regulator of Nod2-induced autophagy (100). DCs also showed defective autophagy in response to MDP in the presence of a CD-associated variant of either NOD2 or ATG16L1. Expression of either variant or loss of function of Atg5, Atg7, or Atg16L1 resulted in loss of MHC class II localization within the autophagic compartment and failure of MHC class II to relocate to the cell surface (47). Collectively, these studies suggest that PRRs are important mediators of autophagy in response to bacterial stimuli. It will be interesting to see whether colocalization of PRRs and autophagy proteins in distinct cellular locations can serve as a platform for MAMP-mediated signaling. In contrast to Nod2, NLRP4 has inhibitory effects on autophagy through inhibition of both the initiation and maturation steps of the autophagy pathway (101). NLRP3, NLRP4, and other NLRs interact with Beclin 1. In addition, NLRP4 interacts with the class C vacuolar protein-sorting complex and inhibits the maturation of endosomes and autophagosomes (101). TLRs and NLRs have a common motif consisting of a leucine-rich repeat (LRR), which is shared in many proteins with uncharacterized function (i.e., a screen of the human proteins showed that 375 proteins contain an LRR domain) (102). Many of these molecules are of unknown function with regard to innate immunity and autophagy but warrant further investigation.
A multitude of additional immune signaling molecules may also regulate autophagy. For example, IFN-γ induces autophagy and antimycobacterial activity in macrophages. This induction of autophagy depends on the expression of immunity-related p47 GTPases (50, 68). NF-κB activation represses TNF-α-induced autophagy (103). The canonical IκB family of kinases (IKKs) activates autophagy in response to starvation, independently of NF-κB (104, 105). One of the mechanisms through which IKKs induce autophagy is by activation of JNK-1, which (as discussed above) releases Beclin 1 from Bcl-2 (104). Cathelicidin, a vitamin D3–derived antimicrobial peptide, is an important inducer of antimicrobial autophagy and has a key role in cellular defense against M. tuberculosis and HIV-1 (106, 107). Furthermore, vitamin D is required for IFN-γ-mediated antimicrobial activity against M. tuberculosis (108).
AUTOPHAGY AND ADAPTIVE IMMUNITY
Autophagy is required for the function and development of many distinct regulatory cell subsets in the intestine and has thus been implicated in the pathogenesis of IBD. Autophagy is involved in several adaptive immune functions, including antigen presentation by DCs, and in the development, differentiation, and homeostasis of T and B cells. The potential role of autophagy in IBD-associated adaptive immune responses is discussed below.
Antigen-presenting cells (APCs) link innate and adaptive immune responses. Autophagy plays a critical role in MHC class II–dependent antigen presentation by DCs. In the intestine, DCs can capture enteric bacteria that have breached the mucosal barrier. DCs are also capable of sampling luminal bacteria by extending dendrites into the lumen or by interacting with epithelial M cells. There are several different subsets of DCs in the intestine. CD103+ DCs maintain tolerance toward commensal bacteria, whereas other subsets mediate effector immune responses against pathogens (109). A unique macrophage-like DC subset in the intestine is able to produce large amounts of IL-23 (a TH17 cytokine implicated in CD pathogenesis) in response to bacterial products. These F4/80+ DCs are involved in intestinal granuloma formation in a mouse model of intestinal inflammation (110). Bacteria-derived ATP induces differential TH17 subsets through APC activation, which exacerbates a T cell–mediated model of colitis (111). As mentioned above, autophagy is required for the recognition of viral replication products in plasmacytoid DCs (62). Autophagy also enhances the efficacy of vaccination by increased antigen presentation in DCs (112). Additionally, autophagy is necessary in DCs for MHC class II presentation in vivo to protect against lethal herpes simplex virus infection (113). However, the role of autophagy in DC subsets implicated in intestinal inflammation remains to be explored.
In addition to MHC class II presentation, autophagy is also required for MHC class I presentation of intracellular antigens to CD8+ T cells (114). Investigators recently showed that cross-priming of CD8+ T cells failed to occur in a mouse model of chronic granulomatous disease (p47phox−/− mice) in response to fungal pathogens due to defective autophagy. Induction of autophagy by poly(I:C) restored defective cross-presentation in p47phox−/− mice (115).
Autophagosomes can engulf cytosolic components to deliver cellular and microbial products to MHC class II compartments (116). Autophagosomes labeled with GFP-LC3 directly fuse with MHC class II compartments in both epithelial cells and APCs. Additionally, fusion of antigens to LC3 results in delivery to MHC class II compartments via autophagy (117). Studies in lymphoblastic cells infected with the Epstein-Barr virus provided the first evidence that autophagy can deliver endogenous antigens for presentation on MHC class II to CD4+ T cells. These studies reported that long-lived nuclear antigen EBNA1 is presented to CD4+ T cells in an autophagy-dependent manner (118). A recent study showed that autophagy negatively regulates T cell activation by destabilizing the immunological synapse between T cells and DCs (119). Additional support for the critical role of autophagy in MHC class II–dependent antigen presentation to CD4+ T cells was provided by a study that showed that impaired autophagy in the thymic epithelium results in altered selection of certain MHC class II–restricted T cells and autoimmunity (120). In addition to the involvement of autophagy in mediating antigen presentation, autophagy also participates in the proliferation and survival of T cells (121, 122). Double-membraned autophagosomes are present in both human and murine T cells, and autophagy mediates the survival and proliferation of memory T cells through the clearance of mitochondrial mass (123, 124). Canonical autophagy (class III PI3K and Vps34) is required for the survival of naïve T cells. Vps34-deficient T cells show increased mitochondrial mass and the accumulation of ROS (125). Additionally, impaired autophagy has been implicated in ER stress and in impaired calcium homeostasis in T cells (122). Autophagy can also influence T cell function. For example, autophagy can negatively regulate NF-κB activation in effector T cells by selective degradation of Bcl10 (a mediator of NF-κB signaling) via p62 binding (126). Dysregulated homeostasis of memory T cells has also been implicated in the pathogenesis of colitis. As discussed above, Atg5 deficiency in the thymus results in an altered T cell repertoire and thus leads to colitis, suggesting that autophagy is required for appropriate selection of self-antigen-specific CD4+ T cells (120).
There is emerging evidence for the involvement of the IL-23/TH17 pathway in the pathogenesis of IBD (127). IL-23 receptor polymorphisms are associated with the development of IBD (128). A CD161-expressing TH17 subset is associated with inflammatory infiltrate of CD lesions (129). In addition, CD models demonstrated a pathogenic role for IL-17 (130, 131). Moreover, increased IL-23-responsive innate lymphoid cells were associated with IBD (132). However, the role of autophagy in these T cell subsets is not known.
B cells also play an important role in intestinal homeostasis and control of inflammation. B cells can produce cytokines under inflammatory conditions. Bregs are a B cell subset that produce IL-10 under inflammatory conditions and that can improve ongoing colitis (133, 134). Case reports from IBD patients have shown an exacerbation of colitis after B cell depletion therapy by using an anti-CD20 monoclonal antibody (rituximab) (135, 136). Additionally, the B1 B cell subset suppresses the development of colitis (137). Autophagy has been implicated in B lymphocyte development and in the maintenance of B1 B cells (138). Additionally, Beclin 1 is essential for the development of both T and B lymphocytes (139). Antigen presentation to CD4+ T cells by B cells also involves autophagy (140, 141).
Autophagy plays a role in many other immune processes. In neutrophils, autophagy is involved in the formation of neutrophil extracellular traps (NETs), which capture microbes through antimicrobial extracellular chromatin structures (142). The inhibition of autophagy blocks NET formation (143). Along with presentation of exogenous antigens, autophagy is also involved in the regulation of self-responses. Autophagy is important in clearing apoptotic cells (144–146). Uncleared apoptotic cells serve as a major source of self-antigens and can exacerbate chronic colitis (147).
INTESTINAL BARRIER FUNCTION
Epithelial cells line the inner surface of the intestine and create an important barrier to the luminal contents. Intestinal stem cells are located at base invaginations termed crypts of Lieberkühn. These stem cells rapidly and perpetually produce multiple, distinct, differentiated lineages: absorptive enterocytes, mucus-secreting goblet cells, hormone-secreting enteroendocrine cells, and antimicrobial-secreting Paneth cells (which are present in the small intestine and appear in the human colon during metaplasia) (148). Together, these cells form a dynamic barrier where the majority of epithelial lineages migrate upward out of crypts onto the mucosal surface. Paneth cells migrate to the crypt base, where they intermingle with stem cells. There are few established links between autophagy proteins and epithelial functions.
To date, the major link between autophagy and the intestinal epithelium is Paneth cells. This lineage expresses several microbial PRRs, including Nod2 and TLRs (149, 150), which can stimulate the release of Paneth cell granule contents (151–153). Paneth cells are highly specialized in the production, packaging, and secretion of antimicrobial peptides such as lysozyme, cryptdins, and RegIIIγ. These proteins can confer protection against pathogens and alter the composition of the commensal microbiota (154).
These secretory functions may have implications in IBD pathogenesis, as loss of multiple IBD susceptibility genes has been linked to Paneth cell dysfunction. Atg16L1 hypomorphic mice have defects in the packaging and secretion of antimicrobial proteins. Loss of Atg5 and Atg7 results in similar defects, suggesting a common role for these autophagy proteins in regulating Paneth cell function. CD patients homozygous for the T300A variant display similar morphological defects in Paneth cells (154a, 154b). Additionally, Paneth cells in mice deficient for the ER stress response gene Xbp1 appear to undergo premature apoptosis (155). Loss of Nod2 may also result in Paneth cell dysfunction, as Nod2-deficient mice show diminished defensin production (151, 156). Moreover, Paneth cells may sense nutritional state and impact stem cell function via mTORC1 (157). However, the role of autophagy in this function remains to be explored. Future studies geared toward understanding the impact of Paneth cell dysfunction in relation to autophagy protein deficiency will be crucial for understanding the pathogenesis of IBD. One idea is that Paneth cells function to shape the mucosa-associated microbiome. In addition, the role of autophagy proteins in stem cells and other lineages warrants further investigation.
CONCLUSION
A strength of the field is detailed analysis concerning the role of autophagy proteins in specified cell immune types. This is particularly true for the interaction of innate immune cells and microbes. Although much of this work is not performed in intestinal cells, this work is relevant to the intestine (Figure 2). The link between autophagy protein mutations and IBD suggests additional important avenues of investigation. These include study of the role of autophagy proteins in primary intestinal cells from mice and humans as well as evaluation of the role of specific mutations in these cells (158, 159). Such endeavors will be challenging, and phenotypes will likely be subtle and more complex than those in lineage knockouts that have been used successfully to elucidate potential functions of autophagy proteins. For example, using a mouse model that is globally hypomorphic for Atg16L1 expression, we found that the Paneth defect discussed above required an environmental trigger, in this case persistent infection of mice with MNV. This viral trigger was also required to induce the CD-like pathology in DSS-treated Atg16L1 hypomorphic mice. Importantly, there was no alteration in the levels of infection by any strain of MNV when wild-type mice and Atg16L1 hypomorphic mice were compared. In addition, there was no defect in infection by Salmonella in these mice (160). Thus, the mechanisms of xenophagy may not be as relevant in this model. In further support of this idea, uropathogenic E. coli (a microbe that has a reservoir in the intestine) readily infects wild-type but not Atg16L1 hypomorphic mice (161). Another major challenge is that mice with loss of function of Atg16L1 and Nod2 do not develop spontaneous intestinal inflammation, indicating that we do not understand the environmental factors required to trigger disease.
SUMMARY POINTS.
Autophagy is likely important for intestinal homeostasis, as evidenced by the multiple autophagy genes that contain mutations in patients with inflammatory bowel disease.
Several cell types in the intestine—including T and B cells, macrophages, dendritic cells, and Paneth cells—may be targeted by altered autophagy function.
Microbes can be cleared by autophagy (xenophagy). Several microbial species have developed evasion strategies to avoid being killed.
An unexpected role of autophagy proteins in the intestine is the control of secretion in Paneth cells.
Glossary
- Autophagy
literally defined as self-eating; the controlled catabolism of cellular components delivered to lysosomes by autophagosomes
- Inflammatory bowel disease (IBD)
a disease of spontaneous inflammation of the intestine; caused by genetics and the environment
- Autophagosome
a double-membraned structure that engulfs cytoplasmic components that in turn fuse with lysosomes
- Beclin 1
also known as Atg6; binds PI3K to trigger autophagy
- LC3
also known as Atg8; is transiently conjugated to phosphatidylethanolamine during membrane extension. This form is a marker of active autophagy
- Atg16L1
an enzyme in complex with Atg5 and Atg12 that extends the autophagosomal membrane; contains a gene mutation implicated in inflammatory bowel disease
- Xenophagy
selective autophagy of intracellular microbes
- Nod2/NOD2 (nucleotide-binding oligomerization domain–containing protein 2)
a pattern recognition receptor that binds muramyl dipeptide; contains multiple mutations implicated in inflammatory bowel disease
- IL-1β
the active form is a product of caspase-1 cleavage, is activated during inflammation, and is involved in a wide variety of activities
- Paneth cell
an intestinal epithelial lineage that is located at crypt bases and that secretes a wide array of antimicrobial proteins and peptides
Footnotes
DISCLOSURE STATEMENT The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
The Annual Review of Physiology is online at http://physiol.annualreviews.org
LITERATURE CITED
- 1.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–61. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razani B, Feng C, Coleman T, Emanuel R, Wen H, et al. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15:534–44. doi: 10.1016/j.cmet.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol. Rev. 2012;245:147–63. doi: 10.1111/j.1600-065X.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- 8.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Klionsky DJ. The regulation of autophagy—unanswered questions. J. Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 2009;29:157–71. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett. 2010;584:1313–18. doi: 10.1016/j.febslet.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–84. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 16.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–89. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 17.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 18.Virgin HW, Levine B. Autophagy genes in immunity. Nat. Immunol. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell. 2010;18:1041–52. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–85. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–37. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 22.English L, Chemali M, Desjardins M. Nuclear membrane-derived autophagy, a novel process that participates in the presentation of endogenous viral antigens during HSV-1 infection. Autophagy. 2009;5:1026–29. doi: 10.4161/auto.5.7.9163. [DOI] [PubMed] [Google Scholar]
- 23.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA. 2009;106:20842–46. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webber JL, Tooze SA. New insights into the function of Atg9. FEBS Lett. 2010;584:1319–26. doi: 10.1016/j.febslet.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates au-tophagosome formation via Rab11- and ULK1-positive recycling endosomes. J. Cell Biol. 2012;197:659–75. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 29.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 30.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 31.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell. 2008;19:2916–25. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–98. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huett A, Ng A, Cao Z, Kuballa P, Komatsu M, et al. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J. Immunol. 2009;182:4917–30. doi: 10.4049/jimmunol.0803050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 2008;19:4651–59. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seglen PO. Where endocytosis and autophagy meet: the amphisome. In: Fønhus M Sporstøl, Mousavi SA, Berg T., editors. Hepatic Endocytosis. Transworld Res. Netw; Trivandrum, India: 2008. pp. 61–100. [Google Scholar]
- 37.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–46. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–57. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 39.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–28. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 41.Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, et al. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 44.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–28. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 45.Hampe J, Frenzel H, Mirza MM, Croucher PJ, Cuthbert A, et al. Evidence for a NOD2-independent susceptibility locus for inflammatory bowel disease on chromosome 16p. Proc. Natl. Acad. Sci. USA. 2002;99:321–26. doi: 10.1073/pnas.261567999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 47.Cooney R, Baker J, Brain O, Danis B, Pichulik T, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 48.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-γ-inducible LRG-47. Science. 2003;302:654–59. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 50.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 51.Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) J. Immunol. 2007;179:6963–72. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 52.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat. Genet. 2011;43:242–45. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 53.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–68. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 54.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 55.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–86. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 56.Plantinga TS, Crisan TO, Oosting M, van de Veerdonk FL, de Jong DJ, et al. Crohn’s disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60:1229–35. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 57.Franchi L, Muñoz-Planillo R, Núñez G. Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 2012;13:325–32. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:2770–75. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Bruggen R, Koker MY, Jansen M, van Houdt M, Roos D, et al. Human NLRP3 inflammasome activation is Nox1-4 independent. Blood. 2010;115:5398–400. doi: 10.1182/blood-2009-10-250803. [DOI] [PubMed] [Google Scholar]
- 60.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 2012;13:255–63. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 63.Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–46. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 66.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281:11374–83. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 67.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 68.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 69.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Intemann CD, Thye T, Niemann S, Browne EN, Amanua Chinbuah M, et al. Autophagy gene variant IRGM −261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar D, Nath L, Kamal MA, Varshney A, Jain A, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–43. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 72.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA. 2006;103:14578–83. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutierrez MG, Vázquez CL, Munafó DB, Zoppino FC, Berón W, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981–93. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 74.Moreau K, Lacas-Gervais S, Fujita N, Sebbane F, Yoshimori T, et al. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol. 2010;12:1108–23. doi: 10.1111/j.1462-5822.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- 75.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010;12:836–41. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 77.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–18. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–75. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 79.Berger SB, Romero X, Ma C, Wang G, Faubion WA, et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 2010;11:920–27. doi: 10.1038/ni.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA. 2009;106:6226–31. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 82.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–69. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009;186:255–68. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong J, Zhang J, Si X, Gao G, Mao I, et al. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008;82:9143–53. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suhy DA, Giddings TH, Jr, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74:8953–65. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 2007;81:12543–53. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J. Virol. 2006;80:6061–71. doi: 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, et al. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–8. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, et al. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999;5:607–25. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Santaolalla R, Fukata M, Abreu MT. Innate immunity in the small intestine. Curr. Opin. Gastroenterol. 2011;27:125–31. doi: 10.1097/MOG.0b013e3283438dea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159–69. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 92.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–21. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 2008;283:33175–82. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, et al. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36:658–67. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang CS, Lee JS, Rodgers M, Min CK, Lee JY, et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012;11:264–76. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, et al. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 2008;9:908–16. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Homer CR, Kabi A, Marina-Garcia N, Sreekumar A, Nesvizhskii AI, et al. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J. Biol. Chem. 2012;287:25565–76. doi: 10.1074/jbc.M111.326835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jounai N, Kobiyama K, Shiina M, Ogata K, Ishii KJ, Takeshita F. NLRP4 negatively regulates autophagic processes through an association with Beclin1. J. Immunol. 2011;186:1646–55. doi: 10.4049/jimmunol.1001654. [DOI] [PubMed] [Google Scholar]
- 102.Ng AC, Eisenberg JM, Heath RJ, Huett A, Robinson CM, et al. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl. 1):4631–38. doi: 10.1073/pnas.1000093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, et al. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J. Biol. Chem. 2006;281:30373–82. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 104.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–31. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Comb WC, Cogswell P, Sitcheran R, Baldwin AS. IKK-dependent, NF-κB-independent control of autophagic gene expression. Oncogene. 2011;30:1727–32. doi: 10.1038/onc.2010.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009;6:231–43. doi: 10.1016/j.chom.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 107.Campbell GR, Spector SA. Hormonally active vitamin D3 (1α,25-dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection. J. Biol. Chem. 2011;286:18890–902. doi: 10.1074/jbc.M110.206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, et al. Vitamin D is required for IFN-γ-mediated antimicrobial activity of human macrophages. Sci. Transl. Med. 2011;3:104ra2. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat. Rev. Immunol. 2008;8:435–46. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mizoguchi A, Ogawa A, Takedatsu H, Sugimoto K, Shimomura Y, et al. Dependence of intestinal granuloma formation on unique myeloid DC-like cells. J. Clin. Investig. 2007;117:605–15. doi: 10.1172/JCI30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, et al. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 112.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 2009;15:267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 113.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–39. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.English L, Chemali M, Duron J, Rondeau C, Laplante A, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 2009;10:480–87. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.De Luca A, Iannitti RG, Bozza S, Beau R, Casagrande A, et al. CD4+ T cell vaccination overcomes defective cross-presentation of fungal antigens in a mouse model of chronic granulomatous disease. J. Clin. Investig. 2012;122:1816–31. doi: 10.1172/JCI60862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA. 2005;102:7922–27. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–96. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 119.Wildenberg ME, Vos AC, Wolfkamp SC, Duijvestein M, Verhaar AP, et al. Autophagy attenuates the adaptive immune response by destabilizing the immunologic synapse. Gastroenterology. 2012;142:1493–503. e6. doi: 10.1053/j.gastro.2012.02.034. [DOI] [PubMed] [Google Scholar]
- 120.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 121.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J. Immunol. 2011;186:5313–22. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 123.Stephenson LM, Miller BC, Ng A, Eisenberg J, Zhao Z, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–35. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J. Immunol. 2009;182:4046–55. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 125.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc. Natl. Acad. Sci. USA. 2012;109:8670–75. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36:947–58. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Abraham C, Cho JH. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu. Rev. Med. 2009;60:97–110. doi: 10.1146/annurev.med.60.051407.123757. [DOI] [PubMed] [Google Scholar]
- 128.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–63. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kleinschek MA, Boniface K, Sadekova S, Grein J, Murphy EE, et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J. Exp. Med. 2009;206:525–34. doi: 10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Investig. 2006;116:1310–16. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Z, Zheng M, Bindas J, Schwarzenberger P, Kolls JK. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm. Bowel Dis. 2006;12:382–88. doi: 10.1097/01.MIB.0000218764.06959.91. [DOI] [PubMed] [Google Scholar]
- 132.Geremia A, Arancibia-Cárcamo CV, Fleming MP, Rust N, Singh B, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 2011;208:1127–33. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fillatreau S, Gray D, Anderton SM. Not always the bad guys: B cells as regulators of autoimmune pathology. Nat. Rev. Immunol. 2008;8:391–97. doi: 10.1038/nri2315. [DOI] [PubMed] [Google Scholar]
- 134.Mizoguchi A, Bhan AK. A case for regulatory B cells. J. Immunol. 2006;176:705–10. doi: 10.4049/jimmunol.176.2.705. [DOI] [PubMed] [Google Scholar]
- 135.El Fassi D, Nielsen CH, Kjeldsen J, Clemmensen O, Hegedus L. Ulcerative colitis following B lymphocyte depletion with rituximab in a patient with Graves’ disease. Gut. 2008;57:714–15. doi: 10.1136/gut.2007.138305. [DOI] [PubMed] [Google Scholar]
- 136.Goetz M, Atreya R, Ghalibafian M, Galle PR, Neurath MF. Exacerbation of ulcerative colitis after rituximab salvage therapy. Inflamm. Bowel Dis. 2007;13:1365–68. doi: 10.1002/ibd.20215. [DOI] [PubMed] [Google Scholar]
- 137.Shimomura Y, Mizoguchi E, Sugimoto K, Kibe R, Benno Y, et al. Regulatory role of B-1 B cells in chronic colitis. Int. Immunol. 2008;20:729–37. doi: 10.1093/intimm/dxn031. [DOI] [PubMed] [Google Scholar]
- 138.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–14. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 139.Arsov I, Adebayo A, Kucerova-Levisohn M, Haye J, MacNeil M, et al. A role for autophagic protein Beclin 1 early in lymphocyte development. J. Immunol. 2011;186:2201–9. doi: 10.4049/jimmunol.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Watanabe K, Tsubata T. Autophagy connects antigen receptor signaling to costimulatory signaling in B lymphocytes. Autophagy. 2009;5:108–10. doi: 10.4161/auto.5.1.7278. [DOI] [PubMed] [Google Scholar]
- 141.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J. Exp. Med. 2011;208(13):2625–32. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–35. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 143.Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 145.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, et al. Microtubule-associated protein 1 light chain 3α (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA. 2011;108:17396–401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146.Li W, Zou W, Yang Y, Chai Y, Chen B, et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J. Cell Biol. 2012;197:27–35. doi: 10.1083/jcb.201111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mizoguchi A, Mizoguchi E, Smith RN, Preffer FI, Bhan AK. Suppressive role of B cells in chronic colitis of T cell receptor α mutant mice. J. Exp. Med. 1997;186:1749–56. doi: 10.1084/jem.186.10.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Symonds DA. Paneth cell metaplasia in diseases of the colon and rectum. Arch. Pathol. 1974;97:343–47. [PubMed] [Google Scholar]
- 149.Lala S, Ogura Y, Osborne C, Hor SY, Bromfield A, et al. Crohn’s disease and the NOD2 gene: a role for Paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 150.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000;1:113–18. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 151.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal α-defensin expression. Gut. 2004;53:1658–64. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rumio C, Besusso D, Palazzo M, Selleri S, Sfondrini L, et al. Degranulation of Paneth cells via Toll-like receptor 9. Am. J. Pathol. 2004;165:373–81. doi: 10.1016/S0002-9440(10)63304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–26. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 154a.Cadwell K, Patel KK, Komatsu M, Virgin HW, 4th, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–52. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154b.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, et al. A key role for autophagy and the autophagy gene Atg1611 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Simms LA, Doecke JD, Walsh MD, Huang N, Fowler EV, Radford-Smith GL. Reduced α-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut. 2008;57:903–10. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 157.Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–95. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–65. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 159.Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, Stappenbeck TS. Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338:108–13. doi: 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang C, Mendonsa GR, Symington JW, Zhang Q, Cadwell K, et al. Atg16L1 deficiency confers protection from uropathogenic Escherichia coli infection in vivo. Proc. Natl. Acad. Sci. USA. 2012;109:11008–13. doi: 10.1073/pnas.1203952109. [DOI] [PMC free article] [PubMed] [Google Scholar]